Abstract
The motor protein SecA drives the transport of polypeptides through the ubiquitous protein channel SecYEG. Changes in protein-nucleotide binding energy during the hydrolytic cycle of SecA must be harnessed to drive large conformational changes resulting in channel opening and vectorial substrate polypeptide transport. Here, we elucidate the ATP hydrolysis cycle of SecA from Escherichia coli by transient and steady-state methods. The basal ATPase activity of SecA is very slow with the release of ADP being some 600-fold slower than hydrolysis. Upon binding to SecYEG the release of ADP is stimulated but remains rate-limiting. ADP release is fastest in the fully coupled system when a substrate protein is being translocated; in this case hydrolysis and ADP release occur at approximately the same rate. The data imply that ADP dissociation from SecA is accompanied by a structural rearrangement that is strongly coupled to the protein interface and protein translocation through SecYEG.
Keywords: ATPase, SecYEG, steady-state kinetics, transient kinetics, translocon
ATP-dependent molecular motors couple hydrolytic power to mechanical movement in a controlled and directional fashion. Distortions in the structure of the active site driven by different chemical stages in the ATP hydrolysis cycle can be relayed across large distances, resulting in conformational changes in the ATPase and its interacting partners. Hence, ATP binding, formation of the initial ADP:Pi complex or release of either of the products can be coupled to different motions required to drive the protein machine. The reverse can also be accomplished in the capture of free energy e.g., by the F1F0-ATP synthase (1, 2).
The subject of this study, SecA, is an ATPase and the active component of the bacterial protein translocation machinery that also contains the protein channel SecYEG (3, 4). It peripherally associates with the membrane, receives unfolded secretory proteins and facilitates their passage across the inner membrane (5, 6). The structure of SecA reveals a single nucleotide binding site situated between 2 RecA-like folds, a scaffold, wing and preprotein cross-linking domain (7).
Addition to SecA of preprotein and SecYEG reconstituted into an acidic phospholipid membrane promotes protein translocation and an increase in the rate of ATP hydrolysis (4, 8). This corresponds to the energy-transducing activity and is a multistep process (9–11). How the structure of SecA or the channel (12, 13) relate to specific stages of the ATP coupled reaction is unclear.
Although there is still controversy in the field as to the nature and purpose of homo-oligomerisation in SecA, an increasing body of evidence suggests that SecA is dimeric in free solution, but dissociates into monomers as a consequence of the protein transport reaction (14–21). Additional rearrangements within SecA (14, 19, 21, 22) and SecYEG (21, 23, 24) have also been reported. These studies have been crowned by an X-ray structure of the complex (one copy of each) that reveals the nature and consequences of the interaction (25).
Knowledge of the nature and timing of individual steps of the hydrolytic cycle and associated changes in the conformation of motor, channel and substrate polypeptide holds the key to understanding the protein translocation reaction. Previous studies on the ATPase cycle of isolated SecA have found the basal hydrolytic turnover to be rather slow, with the release of the product ADP being significantly slower than the hydrolysis step (26–29). This low basal activity has been determined to be an effect of regulation of the ATPase by magnesium (14, 28). Cation binding to an allosteric site, distinct from that associated with catalysis increases the affinity of the enzyme for ATP while reducing its turnover (14); presumably a means to restrict futile hydrolysis.
In this study, we elucidate the ATPase reaction cycle in the context of the protein translocation reaction by analysis of the steady-state and pre-steady-state activity. Here, we address how the inhibited enzyme is stimulated in the presence of lipids, SecYEG and preprotein. This builds up to the description of the energy coupled ATPase activity that drives protein transport in Escherichia coli.
Results
SecYEG Proteoliposomes Stimulate the ATPase Activity of SecA, Reversing the Inhibitory Effects of Magnesium.
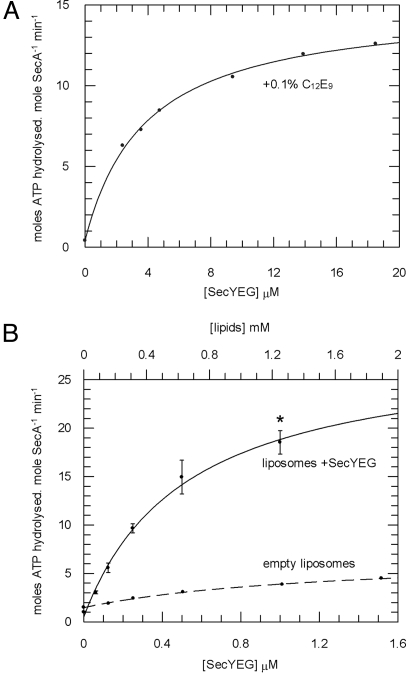
To determine the effect of interaction with SecYEG on the activity of SecA, a steady-state ATPase assay was performed in the presence of a range of concentrations of SecYEG solubilised in the detergent polyoxyethylene(9)dodecyl ether (C12E9) (Fig. 1A; the activity of SecA was insensitive to C12E9). The addition of detergent-solubilised SecYEG stimulated the ATPase activity of SecA and the titrations revealed a Kd,app for SecYEG of 3.9 μM (Table 1). Proteoliposomes containing SecYEG and E. coli polar lipids (PL) were tested for their ability to activate the ATPase activity of SecA in a similar way (Fig. 1B and Table 1). The Kd,app of SecA for SecYEG in proteoliposomes was 0.26 μM (assuming a 50% right-side out reconstitution). The ATPase rate in this case was stimulated such that the inhibitory effects of magnesium were now fully reversed (Table 2). Next, the steady-state parameters (KM and kcat) were defined under these conditions and were found to be the same as in the absence of allosterically-bound magnesium (Table 2) (14).
Fig. 1.
SecYEG alleviates the inhibition of SecA by magnesium. The steady-state ATPase activity of SecA was measured under various conditions. The data were fitted to Eq. 1 (SI Equations) for the determination of Kd,app (Table 1). ATP turnover was measured in the presence of 0.1% C12E9 and increasing concentrations of SecYEG (A) and liposomes (B) made with (solid line) and without (dashed line) SecYEG. *, the conditions used to define the Michaelis–Menten parameters for ATP hydrolysis (Table 2).
Table 1.
Calculated kinetic parameters for wild type SecA in the context of various translocation partners
| Mg2+ | SecYEG (soluble in 0.1% C12E9) | SecYEG (proteoliposomes) | proOmpA | Kd,app[SecA·ligand], μM |
|---|---|---|---|---|
| + | Ligand titrated | − | − | 3.88 ± 0.31 |
| + | − | Ligand titrated | − | 0.52 ± 0.11* |
| + | − | +1 μM | Ligand titrated | 0.047 ± 0.017 |
The steady-state ATPase activity of purified wild type SecA was measured with increasing concentrations of translocation ligand, and fitted according to a 1-site ligand binding equation (Eqs. 1 or 3 in SI Equations) to determine values for Kd,app.
*, the Kd,app for SecA binding to SecYEG proteoliposomes is 0.26 μM assuming a 50% right-side out reconstitution.
Table 2.
Calculated kinetic parameters for wild type SecA in the context of various translocation partners
| Condition | KM[ATP], μM | kcat, min−1 | kcat, s−1 | kcleave, s−1 |
|---|---|---|---|---|
| +Mg2+ | 0.32 ± 0.03* | 0.56 ± 0.01* | 0.01 ± 1.7 × 10−4 | 6.14 ± 0.02† |
| −Mg2+ | 50.8 ± 3.2* | 22.4 ± 0.28* | 0.37 ± 4.6 × 10−3 | 30.4 ± 0.44 |
| +Mg2+; 1 μM SecYEG in proteoliposomes (* in Fig. 1B) | 51.1 ± 7.8 | 15.9 ± 0.62 | 0.27 ± 0.01 | 11.5 ± 0.07 |
| +Mg2+; 1 mM SecYEG in proteoliposomes; 0.75 μM proOmpA (* in Fig. 2B) | 46.1 ± 6.6 | 456.3 ± 17.21 | 7.6 ± 0.27 | 17.9 ± 0.15 |
The steady-state ATPase activity of purified wild type SecA was measured with increasing concentrations of ATP, and fitted according to Eq. 2 in SI Equations to determine values for KM[ATP] and kcat. Values for kcleave were derived from the pre-steady-state ATPase data shown in Figure 5 and are compared with the values for kcat in seconds. SE from the fitting procedure is shown.
*Data published in ref. 14.
†This value was determined to be 1.9 s−1 by quenched flow rapid mixing, the difference explained by the inherent inaccuracies of this method.
Steady-State Analysis of the ATPase Activity Associated with the Active Transport of proOmpA.
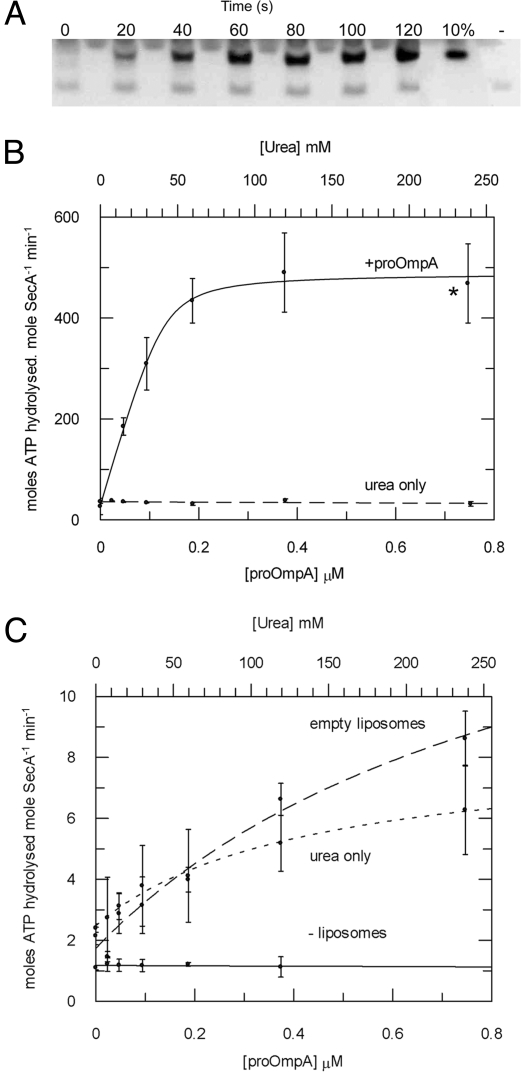
A very efficient translocation reaction was reconstituted from SecA, SecYEG proteoliposomes and the preprotein translocation substrate proOmpA (Fig. 2A). This correlated with a remarkable stimulation of the ATPase activity by ≈1,000-fold compared with the basal rate (Fig. 2B and Table 2). The urea required to maintain the substrate in a translocation-competent unfolded state was shown to have no effect in this respect (Fig. 2B). The effect of proOmpA on SecA ATPase activity in the presence of empty liposomes (lipid vesicles containing no SecYEG) showed only a marginal stimulation compared with the control titration with urea (Fig. 2C). A titration of proOmpA into an assay containing only SecA in the absence of any liposomes showed absolutely no effect of preprotein on ATPase activity, thus this transport-associated activation was entirely dependent on lipids and the presence of SecYEG (Fig. 2C). The activation was measured in the presence of different concentrations of proOmpA and the apparent affinity for the translocation sites was close to the SecA concentration. Therefore, the data were fitted according to a 1-site tight ligand binding equation (Eq. 3, SI Equations) to determine: Kd,app[pOA] = 47 ± 17 nM (Table 1). The Michaelis–Menten parameters were redefined for the saturated fully activated reaction [conditions shown by the asterisk in Fig. 2B (see also Table 2)]. This time, the large increase in kcat was not accompanied by a reduced affinity for ATP (increased KM).
Fig. 2.
Steady-state analysis of the translocation-coupled ATPase activity stimulated by membrane-bound SecYEG and proOmpA. (A) Aliquots of an ATPase assay described below were stopped at 20-s intervals and the amount of successfully translocated proOmpA was analyzed by protease protection and Western blot. These were compared with a measure of 10% of the total proOmpA in each reaction and a negative control carried out in the absence of ATP (−). (B) The ATPase activity of SecA (0.05 μM) was measured in the presence of 1 mM ATP in TKM buffer with 1 μM SecYEG proteoliposomes in TKM buffer, and increasing concentrations of proOmpA in urea (solid line); a control experiment applied the same concentrations of urea without proOmpA (dashed line). The datasets were fitted according to Eq. 3 (SI Equations) to reveal values for Kd,app (Table 1). *, the conditions used to define the Michaelis–Menten parameters for ATP hydrolysis (Table 2). (C) The ATPase activity of SecA in the presence of proOmpA was measured as in B in the absence of any lipids (solid line), in the presence of empty liposomes (dashed line), and with urea only, in the presence of empty liposomes (dotted line).
Rate-Limiting Step in the ATP Hydrolytic Cycle of SecA Is Release of ADP.
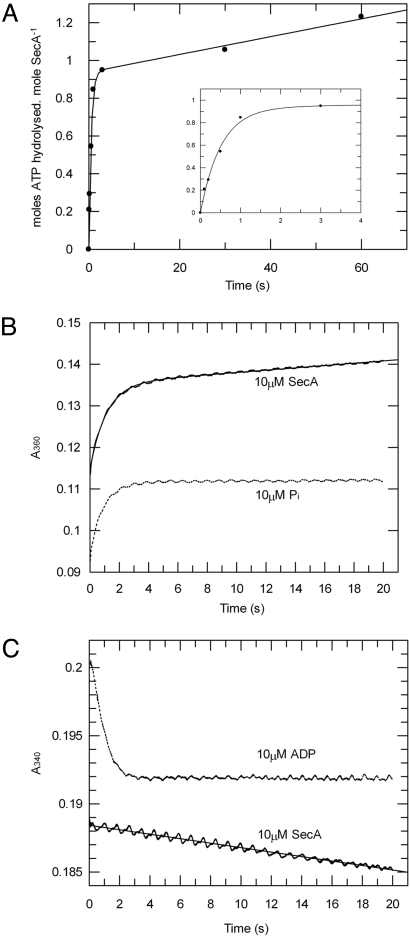
The rate of ATP cleavage in the active site of isolated SecA in the presence of magnesium was measured by quenched-flow rapid mixing, which revealed a burst of ATP hydrolysis before the establishment of the steady state (Fig. 3A). This demonstrates that the rate-limiting step is not binding or hydrolysis of ATP, but something downstream in the reaction, in agreement with previous work (27–29). The data were fitted to a single exponential plus the linear component of the steady-state activity (Eq. 4 in SI Equations). The amplitude of the exponential phase is 0.94 moles of ATP per mole of SecA monomer, determining a stoichiometry of the first turnover of ATP of one. This states that each SecA monomer binds and hydrolyses ATP independently at the same rate, i.e., there is no cooperativity between the 2 sites in the dimer.
Fig. 3.
Transient kinetic analysis of SecA ATPase activity shows that ADP release is rate-limiting in the hydrolytic cycle. The pre-steady-state ATPase activity of SecA was measured in the presence of 1 mM ATP and 2 mM MgCl2. (A) The data from quenched-flow rapid mixing were fitted to Eq. 4 (SI Equations), to reveal kcleave as 1.9 s−1. (Inset) Expanded plot of the first 4 s. (B) Release of phosphate during ATP hydrolysis by 10 μM SecA was measured by stopped-flow rapid mixing using the EnzChek kit and compared with a control produced by mixing equimolar phosphate (10 μM) with the assay components (dotted line). The data for the ATPase (solid line) were fitted to Eq. 4 (SI Equations), determining kcat to be 0.013 s−1. (C) Release of ADP during ATP hydrolysis by SecA was measured by stopped-flow rapid mixing, using an enzyme-linked assay for ADP. As a control, the trace produced by mixing 10 μM ADP with the assay components is shown (dotted line). The data for the ATPase reaction (solid line) were fitted to a straight line to deduce the value for kcat as 0.013 s−1.
The release of phosphate from SecA during the pre-steady-state phase of the reaction was monitored by an enzyme-linked assay for free phosphate (Fig. 3B). This revealed a fast burst of phosphate release (limited by the rate of the linking enzymes), followed by a steady-state rate of 0.013 s−1. Therefore, phosphate release is not rate-limiting.
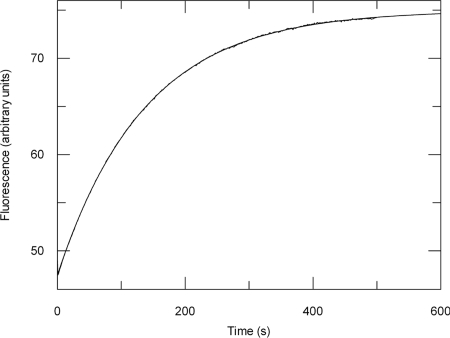
Measurement of ADP release after the initiation of the ATPase reaction showed that there was no exponential burst of free ADP as only a steady-state rate of 0.013 s−1 could be detected (Fig. 3C). To confirm that the rate constant for ADP release is equal to the steady-state rate, koff[ADP] was measured directly by the displacement of bound ADP by the fluorescent ADP analogue MANT-ADP (Fig. 4). As expected, koff[ADP] (0.0073 ± 1.1 × 10−5 s−1) was very similar to the kcat (0.01 ± 1.7 × 10−4 s−1) (Table 2). Hence, in accordance with previous findings (26–29), ADP release is the predominant rate-limiting step in the ATP hydrolytic cycle of SecA and the steady-state complex is SecA:ADP.
Fig. 4.
The rate constant for ADP release by SecA (koff[ADP]) is equal to the steady-state ATPase rate (kcat). koff[ADP] was measured by displacement of ADP bound to SecA by MANT-ADP, resulting in an increase in FRET between the intrinsic tryptophans in SecA and the MANT fluorophore. SecA (0.5 μM) preincubated with 2.5 μM ADP was mixed with 12.5 μM MANT-ADP in a stopped-flow device and the fluorescence change was measured using excitation light at 296 nm and a 399-nm emission cut-off filter. The data were fitted to a single exponential equation (Eq. 4), giving a koff[ADP] of 0.0073 ± 1.1 × 10−5 s−1.
Stimulation of ATPase Activity Is only Mildly Contributed by the Cleavage Rate.
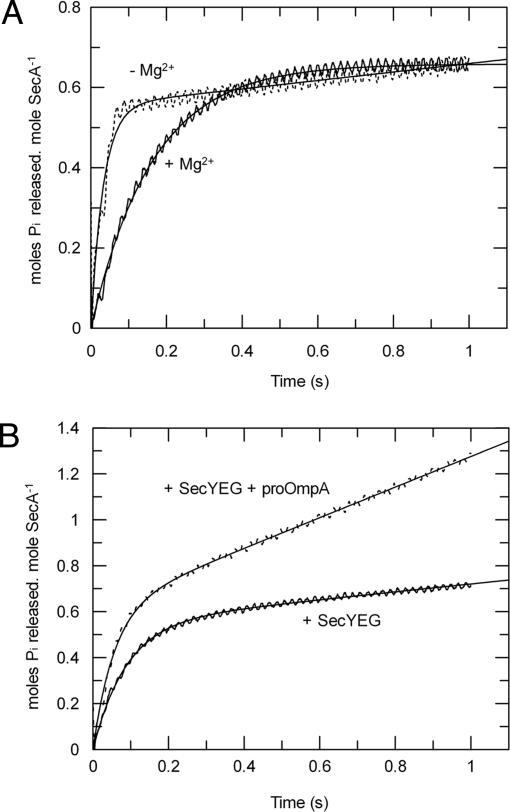
To examine the effects of inhibitory magnesium and the translocation components on the rate constant for ATP cleavage (kcleave), a faster and much more sensitive method to monitor the pre-steady-state ATPase activity was required. The fact that phosphate release is not rate-limiting meant that it could be exploited as a measure of the first turnover of the reaction by use of a fluorescently labeled phosphate-binding protein (Phosphate Sensor) (Fig. 5 A and B). A burst of phosphate release was observed in both low (≈1 μM) and inhibiting (2 mM) concentrations of magnesium (Fig. 5A). Therefore, the rate-limiting event, in both cases, is downstream of the phosphate-release step. The data were fitted (Eq. 4 in SI Equations) to reveal a 5-fold increase of kcleave when SecA is more active in the absence of magnesium, only a modest change compared with a 40-fold increase in kcat (Table 2).
Fig. 5.
Control of the ATPase activity has only a minor contribution from the rate of ATP cleavage. The pre-steady-state ATPase activity of SecA was measured by stopped-flow rapid mixing using Phosphate Sensor. ATP hydrolysis by SecA was measured in isolation with 1 mM ATP in the presence (solid line) or absence (dotted line) of 2 mM MgCl2 (A) and in the presence of 1 mM ATP, 2 mM MgCl2, and 1 μM SecYEG reconstituted into proteoliposomes, with (solid line) or without (dotted line) the addition of 0.75 μM translocation substrate proOmpA (B). The data were fitted to Eq. 4 (SI Equations); the resulting values of ATP cleavage rate (kcleave) are shown in Table 2.
The assay was repeated with 2 mM magnesium and SecYEG proteoliposomes, in the presence (for active translocation) or absence of 0.75 μM proOmpA (Fig. 5B); a burst of phosphate release was observed in both cases (Table 2). The addition of SecYEG proteoliposomes was accompanied by a small increase in the kcleave by a factor of 2, toward the value obtained for the non-inhibited enzyme. Reconstitution of the full translocation reaction by addition of proOmpA had only a slight further effect on kcleave (3-fold increase with respect to basal levels); compared with a 1,000-fold activation of the steady-state activity. Therefore, when the cycle is fully coupled, ADP release becomes almost as fast as the cleavage of ATP [Table 2; compare kcat and kcleave for the basal and stimulated activity (upper and lower rows)]. In these conditions the concentration of the SecA:ATP complex will approach that of SecA:ADP, signifying a profound change in the steady-state complex.
Discussion
SecA is a helicase family ATPase whose activity is affected by interaction with other components of the bacterial protein translocation system: SecYEG, lipids and preprotein (8, 30). In this study, we aimed to clarify and build upon a number of other reports on the ATPase activity of SecA, to form a complete understanding of the kinetics of the nucleotide reaction chemistry. This can then form the basis for an understanding of the mechanism of the translocation reaction.
The predominant form of the steady-state complex during basal activity is SecA:ADP, consistent with the work of others (28, 29). This remains the case when the system is released from magnesium inhibition upon interaction with the membrane bound channels. In other words, a change in turnover rate is almost entirely controlled by promoting the release of ADP. This is in accordance with previous studies, showing nucleotide exchange to be controlled by translocation ligands, which in turn alleviate ATPase suppression (27, 30, 31). Finally, to reconstitute the entire translocation reaction, unfolded preprotein was added to the SecA:SecYEG complex in total E. coli proteoliposomes. This produced the fastest catalytic cycle in which the preprotein binds with a high apparent affinity (Kd,app = 47 nM) and promotes a highly active species of SecA (1,000-fold more active than the basal activity). The apparent affinity for this interaction is comparable with one obtained for translocation of proOmpA into IMVs containing over-expressed SecYEG (KM[proOmpA] = 180 nM) (32).
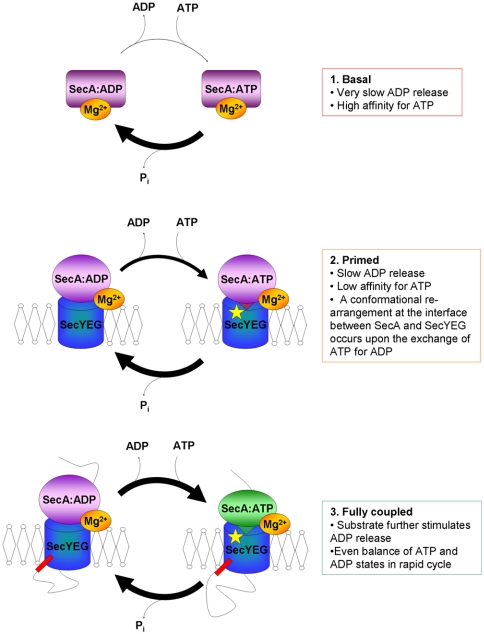
The KM[ATP] of the SecA-YEG complex is unaffected by the translocation of proOmpA (Table 2), therefore the primary effect of preprotein is to increase the off-rate for ADP. In this instance, kcat approaches kcleave, so that the steady-state complex shifts from being largely dominated by SecA:ADP to one populated by rapidly interconverting SecA:ATPand SecA:ADP (Fig. 6). This fundamental shift in the steady-state behavior from a dormant to an active form of SecA must require large scale changes of structure. Although these experiments do not report on the architecture, there is a possibility that they may involve momomerisation; reported to occur in conditions that promote protein translocation (17, 18, 21).
Fig. 6.
Schematic overview depicting the changes in steady-state complex of SecA during the translocation reaction. Both SecA and SecYEG are shown as single objects (which may correspond to dimers). Soluble SecA with Mg2+ bound to an allosteric binding site has low catalytic activity and the dominant steady-state complex is SecA:ADP. When SecA associates with the membrane, SecYEG can alleviate this inhibition, yet ADP release is still relatively slow. The presence of a preprotein substrate renders the system fully active: The apparent affinity for ATP remains the same but ADP release is stimulated, resulting in approximately equal amounts of SecA:ADP and SecA:ATP that are rapidly interconverting through the steady-state cycle. The binding of ATP by SecA (green) causes a change in the conformation at the SecA-YEG interface resulting in insertion of a loop of SecA into the channel in SecY (K268, yellow star) (21, 25). Hydrolysis of ATP brings about retraction of the loop of SecA from the SecYEG channel entrance.
A previous study showed that SecA bound by non-hydrolysable ATP analogues (either AMPPNP or ATPγS), but not by ADP, causes a conformational change at the entrance to the channel at residue K268 of SecY (21). In this context, it was interesting to learn that the structure of SecA-YEG in the ATP bound state reveals a helical loop of SecA that penetrates the channel at this very site (25). We now show that these 2 different conformations (ADP bound and ATP bound) represent the steady-state complex of SecA in basal and stimulated situations. These observations support the notion that the SecA-YEG interface toggles between 2 different conformations, one with the SecA loop inserted close to K268 (ATP bound) and one with SecA retracted (ADP bound). Together the translocation components and preprotein act as nucleotide exchange factors to effectively increase the rate of this conformational switching. We infer that the ADP-dissociation step is strongly connected to a key structural rearrangement of SecA and SecYEG, particularly at their interface and the entrance to the protein channel. The implication is that ADP release is coupled to a pivotal step in the translocation cycle.
Materials and Methods
Chemicals and Biochemicals.
All lipids were purchased from Avanti, C12E9 and DeoxyBigCHAP were from Anatrace and Biobeads from Bio-Rad; the EnzChek kit, Phosphate Sensor, MANT-ADP and precast gels were supplied by Molecular Probes (Invitrogen), Kinase-Glo reagent was from Promega, Chelating Sepharose Fast Flow from GE Healthcare Life Sciences, and all other reagents acquired from Sigma.
Over-Expression and Purification of Protein Translocation Reaction Components.
Wild type E. coli SecA and SecYEG were over-expressed and purified as before (14, 21). Protein quantification was determined using extinction coefficients based on amino acid analysis (Alta Biosceinces) of 99,000 M−1·cm−1 (SecA) and 139,000 M−1·cm−1 (SecYEG) (21). proOmpA Δ176–296 Strep was derived from plasmid pTrc99A and was over-expressed and purified as described in ref. 33. Protein quantification was calculated using the extinction coefficient, estimated according to the sequence, of 49,860 M−1·cm−1.
Reconstitution of SecYEG into Proteoliposomes and in Vitro Translocation.
Five milligrams of SecYEG was mixed with 4.5 mls of E. coli PL (20 mg/mL in 6% DeoxyBigCHAP) and reconstituted into proteoliposomes using Biobeads to remove the detergent. Translocation reactions were carried out at 25 °C in TKM buffer (50 mM TEA pH 7.5, 50 mM KCl, 2 mM MgCl2) with saturating SecYEG proteoliposomes (1 μM SecYEG), 0.05 μM SecA, 0.7 μM proOmpA and 1 mM ATP. Successfully translocated proOmpA substrate was protected from proteinase-K digestion and detected by Western blot analysis using an antibody raised against proOmpA.
Steady-State ATPase Assays.
Steady-state ATPase assays were carried out and monitored at 25 °C, using the EnzChek kit and a Lambda 25 spectrophotometer (Perkin–Elmer). The SecA concentration used was either 0.3 μM or 0.05 μM (monomer concentration), and assays were all carried out in TKM buffer. In each reaction, translocation reaction components were added up to the concentrations indicated in the text, before the initiation of the reaction by addition of 1 mM ATP. All of the data were fitted using GraFit (Erithacus), as described in SI Equations.
Quenched-Flow ATPase Assays.
Quenched-flow measurements of pre-steady-state ATPase activity were carried out at 25 °C using a Hi-Tech Scientific RQF-63 apparatus. The reaction was initiated by mixing 50 μM SecA with 0.5 mM ATP in TKM buffer with the addition of 10% glycerol and then quenched at time-points with 20% trichloroacetic acid (TCA). Precipitated protein was removed by centrifugation at 13,000 × g for 5 min and the reaction neutralised by addition of Tris·HCl pH10 to 400 mM. After addition of BSA to 0.1 mg/mL, the amount of ATP remaining in the solution was quantified with Kinase-Glo reagent, measuring the luminescence with a SpectraMax M2 plate reader (Molecular Devices). Protein quantification was performed as described above and ATP concentrations were confirmed using the extinction coefficient at 259 nm of 15,400 M−1·cm−1.
Stopped-Flow Transient ATPase Assays.
All stopped-flow experiments were carried out at 25 °C using an Applied Photophysics SX-17NV apparatus measuring either absorbance or fluorescence. The buffer used was either TK (50 mM TEA pH 7.5, 50 mM KCl) or TKM buffer. Enzyme-linked assays measuring free phosphate utilised the EnzChek kit, measuring absorbance at 360 nm. Assays for release of ADP used the pyruvate kinase/lactate dehydrogenase enzyme-linked assay, with final concentrations 35 units/mL pyruvate kinase; 50 units/mL lactate dehydrogenase; 2 mM phospho(enol)pyruvate; 0.2 mM NADH. The quantity of ADP liberated was calculated from the change in absorbance at 340 nm, using the extinction coefficient of NADH (6,220 cm−1·M−1). Assays for free phosphate using Phosphate Sensor (MDCC-labeled phosphate-binding protein) were monitored by fluorescence using an emission filter with a cut-off at 455 nm, and an excitation wavelength of 430 nm. Phosphate sensor was present at 5 μM and a “phosphate mop” consisting of 200 μM 7-methyl guanosine and 0.01 units/mL purine nucleoside phosphorylase was included in all reactions. All transient kinetic data were fitted using GraFit (Erithacus) as described in SI Equations.
Measurement of ADP Release by SecA.
The rate of ADP release from SecA was measured by following displacement of bound ADP with an excess of 2′-(or-3′)-O-(N-methylanthraniloyl)-ADP (MANT-ADP). Binding of the MANT-ADP resulted in fluorescence resonance energy transfer (FRET) between the native tryptophans in SecA and the MANT moiety. MANT-ADP binds to SecA at least as tightly as ADP and the rate of displacement of ADP with MANT-ADP was independent of the MANT-ADP concentration used. SecA (0.5 μM) bound to ADP (2.5 μM) was mixed with MANT-ADP (12.5 μM) in TKM buffer (2 mM MgCl2). The reaction was followed by stopped flow fluorimetry at 25 °C, exciting the tryptophans with light at 296 nm and measuring the MANT fluorescence using a 399 nm emission cut-off filter.
Supplementary Material
Acknowledgments.
We thank Dr. Stuart Bellamy for assistance with quenched-flow experiments and Sir John Walker FRS and Dr. Mark Dillingham for stimulating discussions and the latter for critical reading of the manuscript. Some of the data were collected at the EMBO Practical Course in Transient Kinetics 2008. We thank Prof Mike Geeves, Dr. John Eccleston, and Chris Toseland for their assistance. This work was funded by Wellcome Trust Project Grant 084452, Biotechnology and Biological Sciences Research Council Project Grant BB/C503538/1, and a Ph.D. Studentship (to V.A.M.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809592106/DCSupplemental.
References
- 1.Abrahams JP, Leslie AGW, Lutter R, Walker JE. Structure at 2.8 angstrom resolution of F-1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 2.Boyer PD. The ATP synthase—a splendid molecular machine. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 3.Lill R, et al. SecA protein hydrolyzes ATP and is an essential component of the protein translocation ATPase of Escherichia coli. EMBO J. 1989;8:961–966. doi: 10.1002/j.1460-2075.1989.tb03458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brundage L, Hendrick JP, Schiebel E, Driessen AJ, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 5.Cunningham K, et al. SecA protein, a peripheral protein of the Escherichia coli plasma membrane, is essential for the functional binding and translocation of proOmpA. EMBO J. 1989;8:955–959. doi: 10.1002/j.1460-2075.1989.tb03457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arkowitz RA, Joly JC, Wickner W. Translocation can drive the unfolding of a preprotein domain. EMBO J. 1993;12:243–253. doi: 10.1002/j.1460-2075.1993.tb05650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hunt JF, et al. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science. 2002;297:2018–2026. doi: 10.1126/science.1074424. [DOI] [PubMed] [Google Scholar]
- 8.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature domains of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 9.van der Wolk JP, de Wit JG, Driessen AJ. The catalytic cycle of the Escherichia coli SecA ATPase comprises two distinct preprotein translocation events. EMBO J. 1997;16:7297–7304. doi: 10.1093/emboj/16.24.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Economou A, Wickner W. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell. 1994;78:835–843. doi: 10.1016/s0092-8674(94)90582-7. [DOI] [PubMed] [Google Scholar]
- 11.Economou A, Pogliano J, Beckwith J, Oliver D, Wickner W. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell. 1995;83:1171–1181. doi: 10.1016/0092-8674(95)90143-4. [DOI] [PubMed] [Google Scholar]
- 12.Breyton C, Haase W, Rapoport TA, Kühlbrandt W, Collinson I. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature. 2002;418:662–665. doi: 10.1038/nature00827. [DOI] [PubMed] [Google Scholar]
- 13.van den Berg L, et al. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 14.Gold VA, Robson A, Clarke AR, Collinson I. Allosteric regulation of SecA: Magnesium-mediated control of conformation and activity. J Biol Chem. 2007;282:17424–17432. doi: 10.1074/jbc.M702066200. [DOI] [PubMed] [Google Scholar]
- 15.Woodbury RL, Hardy SJ, Randall LL. Complex behavior in solution of homodimeric SecA. Protein Sci. 2002;11:875–882. doi: 10.1110/ps.4090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alami M, Dalal K, Lelj-Garolla B, Sligar SG, Duong F. Nanodiscs unravel the interaction between the SecYEG channel and its cytosolic partner SecA. EMBO J. 2007;26:1995–2004. doi: 10.1038/sj.emboj.7601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Duong F. Binding, activation and dissociation of the dimeric SecA ATPase at the dimeric SecYEG translocase. EMBO J. 2003;22:4375–4384. doi: 10.1093/emboj/cdg418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Or E, Navon A, Rapoport TA. Dissociation of the dimeric SecA ATPase during protein translocation across the bacterial membrane. EMBO J. 2002;21:4470–4479. doi: 10.1093/emboj/cdf471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Osborne AR, Clemons WM, Jr, Rapoport TA. A large conformational change of the translocation ATPase SecA. Proc Natl Acad Sci USA. 2004;101:10937–10942. doi: 10.1073/pnas.0401742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tziatzios C, et al. The bacterial protein-translocation complex: SecYEG dimers associate with one or two SecA molecules. J Mol Biol. 2004;340:513–524. doi: 10.1016/j.jmb.2004.04.076. [DOI] [PubMed] [Google Scholar]
- 21.Robson A, Booth AE, Gold VA, Clarke AR, Collinson I. A Large Conformational Change Couples the ATP Binding Site of SecA to the SecY Protein Channel. J Mol Biol. 2007;374:965–976. doi: 10.1016/j.jmb.2007.09.086. [DOI] [PubMed] [Google Scholar]
- 22.Gelis I, et al. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam PC, Maillard AP, Chan KK, Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bostina M, Mohsin B, Kuhlbrandt W, Collinson I. Atomic model of the E. coli membrane-bound protein translocation complex SecYEG. J Mol Biol. 2005;352:1035–1043. doi: 10.1016/j.jmb.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Zimmer J, Nam Y, Rapoport TA. Structure of a complex of the ATPase SecA and the protein-translocation channel. Nature. 2008;455:936–943. doi: 10.1038/nature07335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shiozuka K, Tani K, Mizushima S, Tokuda H. The proton motive force lowers the level of ATP required for the in vitro translocation of a secretory protein in Escherichia coli. J Biol Chem. 1990;265:18843–18847. [PubMed] [Google Scholar]
- 27.Sianidis G, et al. Cross-talk between catalytic and regulatory elements in a DEAD motor domain is essential for SecA function. EMBO J. 2001;20:961–970. doi: 10.1093/emboj/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fak JJ, et al. Nucleotide exchange from the high-affinity ATP-binding site in SecA is the rate-limiting step in the ATPase cycle of the soluble enzyme and occurs through a specialized conformational state. Biochemistry. 2004;43:7307–7327. doi: 10.1021/bi0357208. [DOI] [PubMed] [Google Scholar]
- 29.Zito CR, Antony E, Hunt JF, Oliver DB, Hingorani MM. Role of a conserved glutamate residue in the Escherichia coli SecA ATPase mechanism. J Biol Chem. 2005;280:14611–14619. doi: 10.1074/jbc.M414224200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karamanou S, et al. Preprotein-controlled catalysis in the helicase motor of SecA. EMBO J. 2007;26:2904–2914. doi: 10.1038/sj.emboj.7601721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Natale P, Swaving J, van der Does C, de Keyzer J, Driessen AJ. Binding of SecA to the SecYEG complex accelerates the rate of nucleotide exchange on SecA. J Biol Chem. 2004;279:13769–13777. doi: 10.1074/jbc.M312892200. [DOI] [PubMed] [Google Scholar]
- 32.de Keyzer J, Van der Does C, Driessen AJ. Kinetic analysis of the translocation of fluorescent precursor proteins into Escherichia coli membrane vesicles. J Biol Chem. 2002;277:46059–46065. doi: 10.1074/jbc.M208449200. [DOI] [PubMed] [Google Scholar]
- 33.Osborne AR, Rapoport TA. Protein Translocation Is Mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.