Abstract
Gene expression is controlled by regulatory elements that can be located far away along the chromosome or in some cases even on other chromosomes. Genes and regulatory elements physically associate with each other resulting in complex genome-wide networks of chromosomal interactions. Here we describe several well-characterized cases of long-range interactions involved in activation and repression of transcription. We speculate on how these interactions may affect gene expression and outline possible mechanisms that may facilitate encounters between distant elements. Finally, we propose that a genome-wide network analysis may provide new insights into the logic of long-range gene regulation.
Introduction
Gene expression is controlled through the combined action of multiple trans-acting factors that bind to regulatory elements. Detailed studies of several model loci, such as the beta-globin locus and the HoxD cluster [e.g. 1-4], have shown that regulatory elements that control them can be spread out over very large genomic regions, and can be located far from the gene(s) they control. An important question therefore is how sets of widely spaced elements can act coordinately to regulate a specific target gene.
Over the last couple of years new insights have been obtained into the mechanisms by which regulatory elements can act over large genomic distances [for reviews see e.g. 2, 5-9]. It is now becoming increasingly clear that a common mechanism by which distal elements regulate genes involves the formation of direct physical associations between these elements and their target gene(s) 10-17. Thus, large genomic distances along chromosomes that separate genes and their control elements are overcome by direct spatial clustering resulting in the formation of chromatin loops. Furthermore, genes can in some cases be regulated by elements located on different chromosomes by a process that involves physical associations between different chromosomes (“trans-interactions”) 18-21.
These novel and exciting observations have been obtained using powerful new molecular technologies, described below in more detail that can detect physical interactions between genomic elements. Examples of long-range interactions have been obtained in a variety of organisms, suggesting that gene regulation through direct physical association with regulatory elements and/or other genes is a common and conserved phenomenon. Based on these results we have proposed that the organization of a genome inside a living cell resembles a complex three-dimensional network of interacting genomic elements 9, 22. In some cases, e.g. in the case of enhancers, chromosomal interactions appear to be very specific and to require defined proteins to mediate associations between particular sets of genomic elements. In other cases, interactions appear less specific. Examples of the latter include apparently non-specific clustering of heterochromatic loci or association of groups of expressed genes in a limited number of transcriptionally active nuclear neighborhoods (e.g. in transcription factories, 23-28). Here we review our current understanding of the role of chromosomal interactions in gene activation and repression. We describe long-range control of several well-studied loci and review the role of trans-acting factors in mediating genomic interactions. We speculate on what drives the specificity of these interactions and on the biochemical mechanisms by which clustering of genes and elements can result in up- or down regulation of gene expression, which is still very poorly understood. Finally, we describe the challenges ahead, and propose that comprehensive study of genome-wide networks of chromosomal interactions may reveal new insights in gene regulation and chromosome biology.
Spacing of genes and regulatory elements suggests abundant long-range control
Even a cursory look at the human genome immediately reveals that coding exons are relatively sparse and that large “empty” genomic regions often separate genes. Current estimates indicate that only a few percent of the human genome encodes for protein 29-31. The function of the remaining part, especially intergenic regions, of the genome has been debated for a long time. Initially it was proposed that a large fraction of the non-protein coding portion of the genome is not functional. However, large-scale efforts to map regulatory elements in the human genome have revealed that the number of functional elements is significantly larger than the number of genes, and that the fraction of functional non-coding DNA is likely to be much larger than previously assumed. For instance, the ENCODE (Encyclopedia of DNA elements) consortium analyzed 1% of the human genome and reported many more experimentally defined putative regulatory elements than were expected based on the level of base-conservation 32. Although regulatory elements are certainly enriched in regions directly surrounding genes 33, these observations also confirm previous more anecdotal observations that regulatory elements can be located far from their target genes, and can also be found in so-called “gene deserts”, large genomic regions (over 500 Kb) that do not appear to contain any genes [e.g. 34].
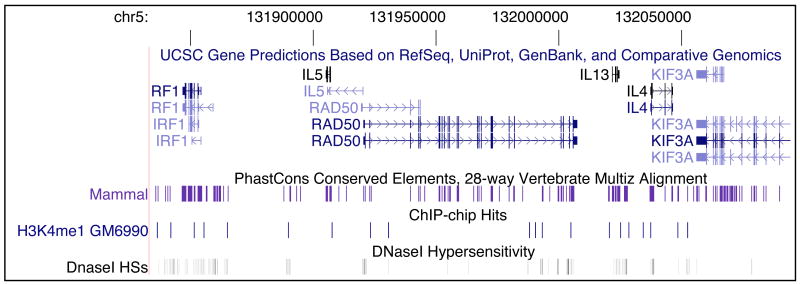
Based on these and other genome-wide studies it becomes apparent that the large open spaces that separate genes likely contain many regulatory elements. This is illustrated in Figure 1 that shows a 260 kb region of human chromosome 5 containing the TH2 locus. The figure presents a snapshot of the UCSC genome browser that displays the locations of genes, conserved elements, DNaseI hypersensitive sites (a common hallmark of regulatory elements) and locations of mono-methylated Lysine 4 of Histone H3 (a mark found at many enhancers 35). The presence of many putative regulatory elements located far from promoters suggests that a significant number of these elements must act over large genomic distances, sometimes up to hundreds of kilobases, to exert control over their target genes. Clearly, long-range phenomena must be widespread throughout complex genomes, and determination of functional relationships between genes and distant regulatory elements will be crucial for a full understanding of genome regulation.
Figure 1.
An overview of a 260 kb region on human chromosome 5 containing the TH2 locus encompassing the IL5, RAD50, IL4 and IL13 genes reveal that many putative elements are located far from any promoter. The image is a typical screenshot of the UCSC human genome browser (http://genome.cse.ucsc.edu/cgi-bin/hgGateway). Indicated are the positions of genes (UCSC gene predictions), conserved elements found in 28 vertebrates [determined using the Phastcons program described by Siepel et al. 102], DNaseI Hypersensitive sites [data from 32] and the locations of Histone H3 monomethylation at Lysine 4 [data from 32, 103]. All these features are marks for putative regulatory elements. The DNaseI hypersensitive sites located at the 3′ end of the RAD50 gene represent the Locus Control Region that regulates IL5, IL4 and IL13 expression (see the section of TH2 regulation in the main text).
Long-range control involves interactions between genomic elements
It has been suggested for many years that long-range control involves direct physical association of genomic elements resulting in formation of chromatin loops 1, 2, 7, 36, 37. However, due to technical limitations, detection of such loops had remained elusive. Development of the Chromosome Conformation Capture (3C) technology a few years ago allowed, for the first time, the detection of physical interactions between distant genomic elements 38. 3C has rapidly become an established research tool, and application of this technology has resulted in very convincing evidence for the widespread formation of long-range looping interactions between genes and regulatory elements.
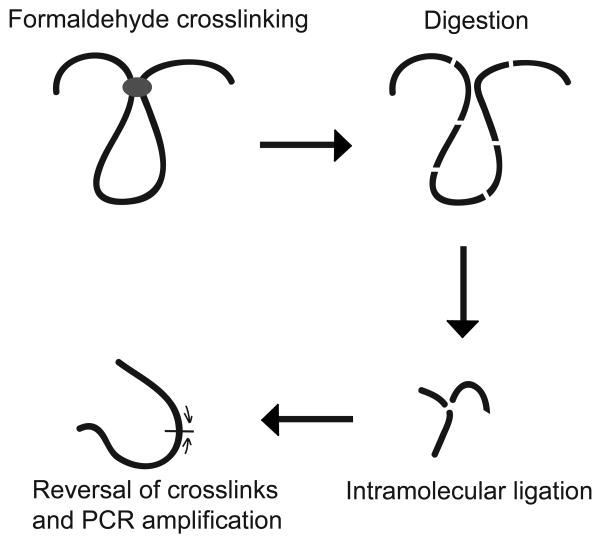
The 3C technology has been described and reviewed in detail elsewhere 5, 22, 38-43) and is schematically outlined in Figure 2. Briefly, the 3C technology relies on formaldehyde cross-linking of interacting chromatin segments inside cells followed by identification of pairs of interacting DNA elements using PCR. Formaldehyde cross-links two interacting chromatin segments through protein complexes that are bound to these segments. After cross-linking of intact cells chromatin is solubilized and DNA is gently fragmented using a restriction enzyme. The digested DNA is subsequently ligated under very dilute conditions to strongly favor intra-molecular ligation over intermolecular ligation. This results in preferential ligation of the ends of cross-linked restriction fragments to each other. Finally, the cross-links are reversed and DNA is purified.
Figure 2.
Representation of chromosome conformation capture (3C) technology. Cells are treated with formaldehyde to induce crosslinks between interacting chromatin fragments (center oval). Chromatin is then digested with a restriction enzyme and ligated at dilute DNA concentrations to induce intramolecular ligation. Crosslinks are reversed and ligation products are then quantified by PCR (arrows indicate PCR primers).
The 3C assay thus results in a very complex and genome-wide mixture of ligation products, which is referred to as a 3C library. Since formation of a ligation product is the direct consequence of the cross-linking of two chromatin segments, detection of a specific ligation product is evidence for direct physical association of the genomic elements in intact cells. The frequency with which a ligation product is formed is a relative measure for the frequency of interaction. It is important to note that 3C does not allow direct determination of absolute interaction frequencies (i.e. the number of cells that display the interactions at a given time point), but some estimates of true interactions frequencies can be obtained through modeling of 3C data using well-established polymer models 38, 44, and changes in patterns of 3C interactions can be used to deduce relative changes in chromatin folding and compaction [e.g. 44, 45].
The presence and relative abundance of a specific ligation product in the 3C library is typically determined using quantitative or semi-quantitative PCR using locus-specific primers. To do so one first predicts the sequence of the ligation product that is formed by the two genomic elements and then designs a pair of PCR primers in order to detect the presence of this specific ligation product. Design of 3C experiments requires careful planning and analysis of 3C data requires appropriate controls. These issues have been described in detail elsewhere 22, 43.
3C was originally used to determine the three-dimensional folding of yeast chromosome III, to map interactions between telomeres and to determine the kinetics of formation and loss of trans-interactions between homologous and non-homologous chromosomes respectively during meiosis 38. Subsequently, 3C has been used to detect and study long-range looping interactions involved in regulation of several gene clusters, such as the alpha- and beta-globin loci 10, 16, 17, that have served for a long time as models for long-range gene regulation. More recently a growing number of 3C studies have identified many more interactions between enhancers and their target genes [e.g. 14, 15, 46]. Therefore, it appears that looping is a very general and abundant mode of action for gene regulatory elements.
Below we describe some of these loci in more detail to illustrate the role of long-range looping interactions in gene regulation.
The alpha and beta-globin loci
The alpha and beta-globin loci have served as model systems for long-range gene regulation for many years 4, 37, 47. Many aspects of gene regulation by distal regulatory elements, including the role of long-range looping interactions, have been discovered through analysis of these two gene clusters. A comprehensive description of the regulation of these loci is beyond the scope of this review. Here we focus only on the recent discovery and analysis of looping interactions and their roles in globin expression.
The beta-globin locus contains a set of developmentally controlled genes (4 in mouse, and 5 in human) that encode variants of the beta-chain of hemoglobin. The beta-globin genes are regulated by a single element, the Locus Control Region (LCR) that is located approximately 25 kb upstream of the most proximal ε-globin gene (Figure 3). The LCR was originally identified as an element that confers position-independent and copy number-dependent expression to linked genes in transgenic mice 48. The LCR is a relatively large element (∼20 kb) that contains prominent DNase I hypersensitive sites 49. These sites define sub-regions of the LCR and represent regions of open chromatin that are bound by multi-protein complexes. For instance, several of the hypersensitive sites contain DNA binding sites that are recognized by transcription factors, such as GATA-1 and EKLF that play critical roles in activation of the beta-globin genes 50, 51.
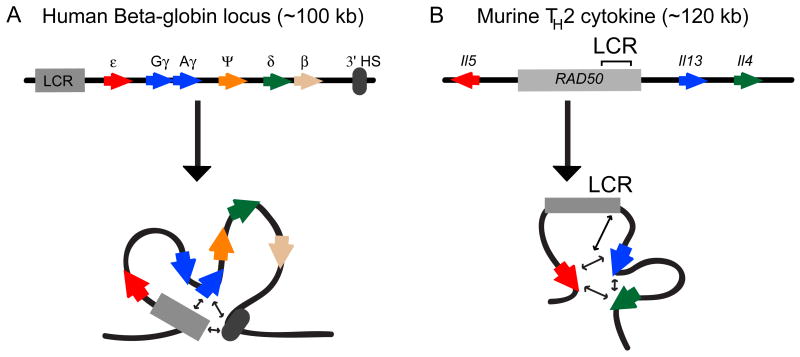
Figure 3.
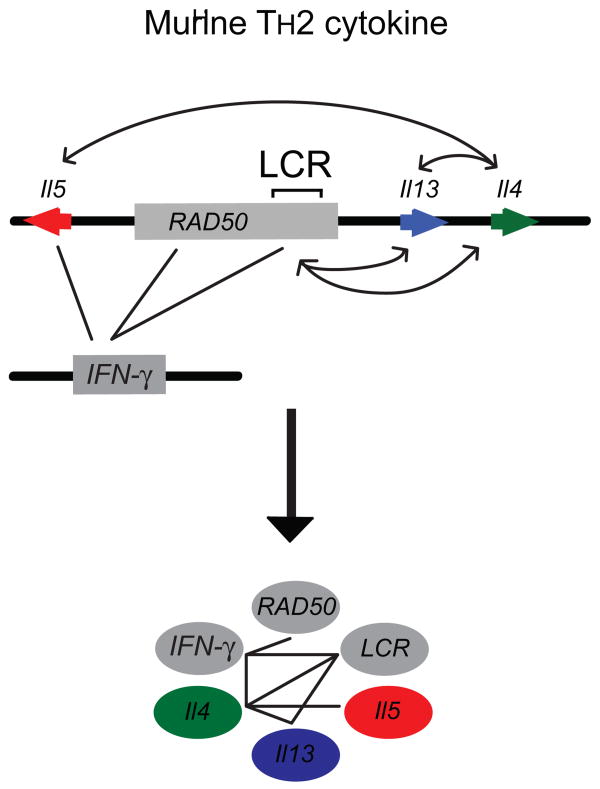
Long-range interactions involved in activation of transcription. A. Depicted is the linear organization of the human Beta-globin locus. The locus control region (LCR) and 3′ hypersensitive (3′HS1) site are depicted in gray. The ε, Gγ- and Aγ-, δ-, β-globin genes are indicated in red, blue, orange, green, and peach, respectively. The LCR and 3′HS1 are found to associate with the γ-globin genes upon its activation resulting in the depicted loop structure. B. Depicted is the linear organization of the T helper type 2 cytokine locus. Il5, RAD50 (whose 3′ region contains the locus control region), Il13, and Il4 are indicated in red, gray, blue, and green, respectively. The promoter regions of Il5, Il13, and Il4 are found to interact with each other as well as with the LCR region.
The LCR activates target genes that are located up to 80 kb away and it had been proposed for years that this regulation involves direct interactions between protein complexes bound to the LCR and complexes bound to the gene promoters 36, 37. 3C analysis of the murine beta-globin locus demonstrated that such looping interactions indeed occur 10. The LCR was found to be in direct physical contact with the expressed globin gene. These interactions were also observed using an entirely independent technology, RNA-TRAP 52. Furthermore, a subsequent 3C study found that the LCR – globin gene interaction is developmentally controlled 53. Specifically, in fetal stages the LCR interacts with the highly expressed gamma globin genes, whereas in adult stages the LCR associates with the beta globin gene. These observations confirmed, for the first time, the long-standing hypothesis that long-range control can involve direct physical communication between genes and distal regulatory elements.
Evidence for a prominent role of transcription factors in mediating these long-range interactions has come from studies that employed cell systems in which specific transcription factors were deleted. For instance fetal liver cells from EKLF knock-out mice display a significant reduction in beta-globin expression and a similarly diminished physical association between the LCR and the globin genes, as detected by 3C 54. Furthermore roles for the transcription factor GATA-1 and the co-factor FOG1 has been identified 13. In this study, mouse cells were generated that expressed GATA-1 fused to the Estrogen Receptor ligand-binding domain (GATA-1-ER). The fusion protein binds its cognate DNA binding sites in an estrogen or tamoxifen-dependent manner. This cell system allows controlled GATA-1 binding to the LCR and the globin promoters. 3C analysis showed that looping interactions between the LCR and the beta globin gene was dependent upon, and coincident with GATA-1 recruitment to the LCR and the globin genes. This study also showed that the GATA-1 - FOG1 interaction is critical in mediating the formation of the LCR-globin gene chromatin loop. Finally, recent work has revealed a role for the Ikaros protein in developmentally regulated switching of the interaction between the LCR with the gamma-globin gene to interaction with the adult beta-globin gene 55. Thus, multiple protein factors cooperate to mediate and control long-range interactions and beta-globin activation.
The alpha globin locus is comprised of alpha-globin genes that encode variants of the alpha chain of hemoglobin. Expression of the alpha genes is regulated by a set of remote regulatory elements, characterized by DNase I hypersensitive sites located 40-60 kb upstream of the alpha globin genes. These distal regulatory elements recruit a number of transcription factor complexes that play critical roles in activating the downstream alpha globin genes (e.g. GATA1, GATA-2 and EKLF) 17, 56. Two recent 3C studies have shown that these regulatory elements directly associate with the activated alpha globin genes through long-range looping 16, 17. The molecular basis of these long-range interactions has not been established yet, but it seems likely that large protein complexes containing multiple transcription factors play important roles, as is the case for the beta-globin locus.
The T helper type 2 cytokine locus
Looping interactions have also been observed in the T helper type 2 (TH2) cytokine locus 11. In this case the pattern of interactions is more complicated than those observed in the globin loci with multiple types of elements associating with each other in a cell type specific manner.
The 120 kb TH2 locus on mouse chromosome 11 contains three coordinately expressed cytokine genes, Il4, Il5 and Il13. These genes are under the general control of a LCR located ∼15 kb upstream of Il13 and 60 kb downstream of Il5 57, 58. The LCR is ∼ 25 kb in size and located within the 3′ end of the ubiquitously expressed RAD50 gene. 3C studies of the Th2 locus identified multiple looping interactions 11. First, it could be shown that the promoters of the three genes interact with each other, even in cells that are not derived from the T-cell lineage (e.g. fibroblasts). Second, 3C analysis of T-cells showed that the LCR directly associates with each of the promoters of the three cytokine genes (Figure 3). Interestingly, these looping interactions were observed in all cells from the T-cell lineage, but irrespective of the expression of the cytokine genes. In TH2 cells that express the cytokine genes, the interactions between the LCR and the gene promoters becomes significantly more prominent. Finally, two transcription factors, STAT6 and GATA-3, known to affect expression of the cytokine genes, were shown to play a role in sustaining interactions between the LCR and the cytokine promoters 11.
These results suggest that the three gene promoters interact ubiquitously even when not expressed. In cells from the T-cell lineage the LCR then associates with the cluster of gene promoters, generating a poised state prior to transcription. This poised state of associated promoters and enhancers would allow for rapid induction of transcription upon T-cell activation. Combined the results obtained for the TH2 locus provide a compelling example of the intricate network of interactions that can occur among genes and regulatory elements to drive cell type specific gene expression.
In addition to the specific transcription factors described above, other chromosome structural proteins may be involved in facilitating the formation of specialized higher order chromatin looping events at the TH2 locus. For instance, one interesting report identified the protein SATB (special AT-rich sequence binding protein 1) as a global organizer of the locus 59. SATB1 binds multiple sequences throughout the TH2 Locus and 3C and Chip-loop (which is used to analyze particular immunoprecipitated chromatin fragments for looping patterns) assays showed that upon TH2 activation the locus reorganizes into a specific chromosome configuration of several small loops that are anchored at their base by SATB1. A critical role for this conformation for transcription was supported by the observation that that SATB1 is also required for activation of genes within the locus. Therefore, SATB1 is responsible, at least in part, for organizing the locus into a transcriptionally active chromatin structure 59.
Trans interactions
In general genes are regulated by cis-acting elements, in which genes and elements are located on the same chromosome. However, trans-regulation, in which genes are regulated by elements located on different chromosomes can also occur. Trans-regulation has been studied extensively in Drosophila. In this organism homologous chromosomes are tightly synapsed. Paired homologs provide a structure in which an enhancer on one homolog can regulate a target gene located in cis, as well as the homologous allele located in trans, a process referred to as transvection 60.
It has been shown that transvection involves direct interactions between genes and enhancers. For example, the Abdominal-B gene in Drosophila is under the control of many enhancers, including IAB5, which is located 50 kb downstream of the transcription start site. By using confocal imaging of nascent transcripts, Ronshaugen and colleagues observed a direct association between the Abdominal-B promoter and the IAB5 enhancer. The image also suggests that this interaction more often occurs in trans 61.
Several recent studies have shown that trans-regulation also occurs in mammalian genomes, both between homologous and between non-homologous chromosomes. One interesting case of specific and functional interactions between homologous chromosomes involves the two X-chromosomes in female cells. In order to adjust the level of expression of X-linked genes to the level observed in male cells, one of the two X-chromosomes is mostly silenced 62, 63. For this process to function appropriately, cells have to count the number of X-chromosomes present and then inactivate only one of the two copies. X-chromosome inactivation is initiated at the X-chromosome inactivation center (Xic). Recently it was found that the two Xic's transiently associate at the time of development when X-inactivation is initiated 20, 21. Deletion mutants of the Xic indicated that this interaction was a critical step in the process of counting X-chromosomes and part of the pathway for ensuring only one of the two X-chromosome was inactivated. The precise mechanistic role of this association during the series of events that leads to inactivation of only one of the two X-chromosomes is far from clear. Some clues as to the molecular mechanism of Xic-trans interactions are now coming to light, suggesting roles for the insulator protein CTCF and transcription throughout the Xic region 64. It is likely that other factors also play roles as indicated by the discovery of a region of the Xic that is distinct from the CTCF binding sites that facilitates pairing of the X-chromosomes 65.
Spilianakis and co-workers who studied the TH2 locus described one of the first cases of trans-regulation in mouse that involve non-homologous chromosomes 18. Using 3C they discovered that regulatory elements of the TH2 locus (described above) on chromosome 11 directly associate with the interferon gamma gene on chromosome 10. Interestingly, these interactions were cell type specific and appeared to play a role in coordinating expression of the two interacting loci. Importantly, a functional role for this trans-interaction was confirmed by demonstrating that deletion of a regulatory element in the TH2 locus (RHS7, a DNAseI hypersensitive site) on chromosome 11 abolished the inter-chromosomal interaction and affected expression of the Interferon gamma gene on chromosome 10 upon differentiation into TH2 cells.
Lomvardas and co-workers described another example of trans-regulation 19. These authors studied the mechanism by which each olfactory neuron expresses only one of many olfactory receptor (OR) genes present in the mouse genome. Previous work had identified the H enhancer that regulates a cluster of OR genes located 75 kb downstream of the enhancer 66. By employing a 3C-based methodology the H-enhancer was found to interact not only with OR genes located in cis, but also with OR genes located in trans 19. Interestingly, FISH studies confirmed that in a given neuron the enhancer interacts with only one target gene, either in cis or in trans, and this corresponded to the one OR gene that was expressed in that neuron 19.
Long-range interactions involving only one enhancer may provide an elegant explanation for the puzzling observation that only one OR gene is expressed per cell. However, deletion of the H-enhancer had surprisingly little effect on OR gene regulation in trans, suggesting that the mechanisms driving single OR gene expression are more complex than initially proposed 67, indicating that the role of the H-enhancer is either redundant or part of a more complicated mechanism to ensure single OR gene expression.
The examples described above involve very specific trans-interactions between genes and other genomic elements. Another class of trans-interactions involves non-specific association of expressed genes at sites of ongoing transcription (transcription factories). Each cell only contains a limited number of transcription factories, and at any factory multiple genes are engaged in transcription at any given point in time 23, 28. Direct, but apparently non-specific associations between active genes could be detected using a 3C-based approach 25. These associations likely occur as a result of the association of genes with the same transcription factory, which brings these genes in close spatial proximity. This class of trans-interactions may reflect a level of self-organization of the nucleus in which the expression status of a gene determines, at least in part, the nuclear neighborhood a gene resides in and thus which segments of the genome it has an opportunity to associate with 68.
Not all associations at transcription factories may be non-specific. Recently the groups of Fraser and Cook reported that some genes prefer to associate with the same transcription factory, and thus can be found to interact rather specifically 28, 69. These factories may be enriched in transcription factors that regulate a specific subset of genes.
How do enhancers find their target genes?
The observation that genes can cluster together and that regulatory elements directly associate with their target genes explains how elements can act over large genomic distances. However, 3C-based studies do not provide direct insights into the process(es) by which specific genomic elements can efficiently encounter one another in the context of complex nuclear organization.
The original looping model presumes that genes and elements are relatively free to move in three dimensions and can encounter each other by direct collisions 36, 37. The composition of protein complexes bound to both the regulatory element and the promoter would then determine affinity and specificity of the looping interactions. A limitation of the looping model is the fact that it does not include any direct explanation for how two genomic elements can “find” one another other than by chance. Given the complexity of the nucleus, and the number of genes and elements that are present, it seems that an active process of searching for interacting partners would greatly facilitate long-range gene control.
The “tracking model” is appealing because it does include an active searching mechanism. The model proposes that complexes bound to a regulatory element find their target gene through an active process that involves “tracking“ of the protein complex bound to the element along the chromatin fiber, dragging the regulatory element along. When an appropriate promoter is encountered a more stable complex is formed between regulatory element and the target gene.
An additional attractive aspect of the tracking model is the fact that it also provides a possible mechanism of action of insulator elements. Insulator elements are elements that can interfere with long-range gene regulation by preventing an enhancer from activating a target gene 70, 71. Insulators only block enhancer function when located in between the enhancer and the target promoter. This intriguing position-dependent phenomenon is not understood in detail. The tracking model would predict that insulators act as physical barriers that cannot be passed by the tracking enhancer complex. On the other hand, the fact that in some cases regulatory elements on one chromosome can regulate genes located on a different chromosome is not straightforwardly explained by the tracking model and would be more conveniently explained by a looping model.
Sub-nuclear structures may also play a role in bringing genes and other genomic elements together. For instance, in yeast tRNA genes associate transiently with the nucleolus, and therefore come in close spatial proximity to each other 72. Other examples include interactions of genes with transcription factories, which will passively and often non-specifically facilitate physical associations between expressed genes 25.
Recent observations have revealed active and rapid direct movement of genomic loci upon gene activation. These movements were dependent on nuclear actin 73, 74. These results rekindle a long-standing and contentious issue in the field regarding the role of nuclear actin and myosin. Active movement of loci could aid in bringing distant genes and regulatory elements together, e.g. at sub-nuclear structures such as transcription factories. Future studies will require detailed analyses of chromatin movements using sophisticated imaging methods combined with molecular manipulation of the actin and myosin machineries to link relocalization of loci to gene regulation at the level of single cells.
The mechanisms that facilitate long-range interactions and the role of nuclear organization in general are only just beginning to be understood and will likely be the focus of many studies in the next couple of years.
How could enhancer-gene interactions regulate transcription?
Little is known about the precise molecular mechanisms by which interactions between genomic loci modulate the process of transcription. Most models propose that enhancer-gene interactions somehow facilitate recruitment and/or stabilization of components of the transcription machinery. Recent work from the Higgs laboratory suggests that long-range interactions in the alpha-globin locus results in efficient transfer of RNA polymerase and other basal transcription factors from the distal elements to the alpha globin gene promoter 17. Roles for looping interactions in later steps of transcription have also been identified. For instance, studies of the role of the LCR in beta-globin gene expression have indicated that the LCR is important for enhancing transcription elongation 75, perhaps through recruitment of elongation factors. Whether these phenomena are specific for LCRs controlling highly expressed gene clusters, or are more widespread is currently not known.
Enhancers may also act by modulating the sub-nuclear position of their target gene and/or by stimulating their association with transcription factories. One example is again provided by analysis the beta-globin locus. Work by Ragoczy et al, has shown that there are changes in localization of the locus upon erythroid differentiation 76, 77. During maturation, the locus is localized away from the nuclear periphery and pericentric heterochromatin (PCH). This relocalization also coincides with a decrease in Pol II foci (i.e. transcription factories). These foci are no longer found throughout the nucleus, but are now found more towards interior regions of the chromosome with some reaching out to the periphery. Interestingly, the relocalization of the locus away from the periphery and its association with actively transcribing transcription factories is dependent upon the LCR. This points to the possibility that the LCR, and perhaps enhancers in general stimulate expression of target genes, at least in part, by facilitating their colocalization with active transcription factories 77. Similar models for enhancer action have been proposed by Cook and co-workers 78.
Clearly, much more studies are needed to unravel the mechanisms by which interactions between genes and elements facilitate transcriptional control.
Long-range interactions and gene repression
Long-range interactions are not only involved in gene activation, but also in gene silencing and heterochromatin formation. One such example is observed in Drosophila. By inserting a large region of heterochromatin into the brown (bw) gene, a null mutation, brownDominant allele (bwD), is created. Dernburg et al. were able to observe by FISH that the bwD allele now had a tendency to interact with centromeric heterochromatin (Figure 4). In addition, in heterozygotes the wild-type allele associates with the bwD allele and also becomes associated with centromeric heterochromatin, which results in silencing of both alleles. This suggests that the silencing of the bw gene is due to its long-range association with centromeric heterochromatin 79.
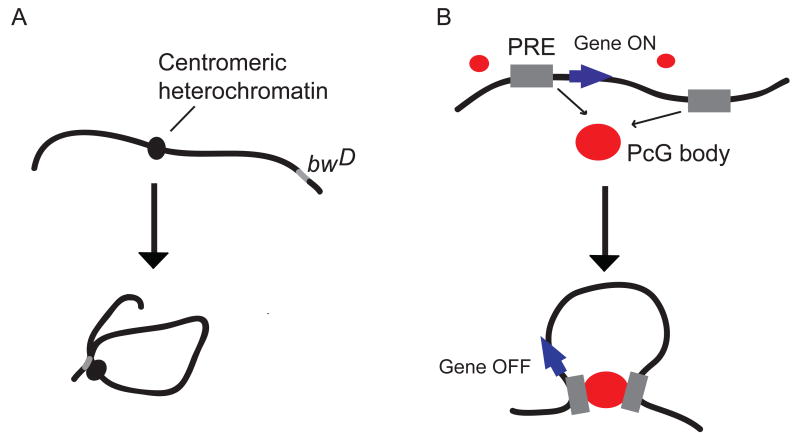
Figure 4.
Long-range interactions involved in repression of transcription. A. Depicted is the linear organization of the locus containing the Drosophila mutation, brownDominant (bwD) gene (shown in gray). The bwD is found associated with the centromeric heterochromatin (depicted as a solid oval). B. Depicted is the linear organization of a hypothetical locus containing two polycomb response elements (PRE) (shown as gray boxes) near a gene (green arrow). The gene is repressed when the PREs localize at PcG bodies (depicted as red circles) resulting in a chromatin loop.
Although still very poorly understood, we highlight other examples in which silencing and heterochromatin formation involves long-range interactions between genomic elements and genes.
Telomere clustering
Yeast telomeres are heterochromatic regions that are silenced by a group of proteins termed the silent information regulator (SIR) proteins 80. After initial recruitment via their interaction with Rap1 at telomeric ends, SIR proteins spread in cis resulting in formation of patches of heterochromatin that can be several kilobases in size 81, 82. Genes that are located near telomeres can become silenced in a SIR-dependent manner, a phenomenon that is referred to as telomere position effect 83.
When telomeres are visualized by immunostaining using antibodies against SIR proteins, a small set of clusters is observed 84. The number of SIR foci is much smaller than the number of telomeres, indicating that each focus contains multiple telomeres. Consistently, direct interactions between telomeres could be detected using 3C 38, 85. Although the functional benefit as to why telomeres cluster is still unknown one hypothesis is that clustering of these features results in a high local concentration of silencing factors and creates nuclear neighborhoods that are enriched in and could help recruit heterochromatin proteins that are present in limiting concentrations 86.
The mechanisms underlying long-range telomeric interactions are not understood in detail but a role for nuclear membrane association has been proposed. Two pathways anchor telomeres to the nuclear envelope. One pathway involves the Ku70/Ku80 dimer 87 while the other involves the protein Esc1p 88, 89. When both pathways are impaired the telomeres are completely delocalized from the periphery and clustering of telomeres is affected as is telomeric silencing 88-90.
Although these results suggest that membrane anchoring is essential for heterochromatin formation and long-range telomeric interactions, several lines of experiments indicate that this is not necessarily the case. First, membrane anchoring is not sufficient for cluster formation because in sir mutants telomeres remain associated with the periphery but no longer form clusters 89. Secondly, attachment to the nuclear envelope is not essential for heterochromatin formation and gene silencing at the silent mating type loci 90, 91. Third, in ku70Δ mutants telomere clusters can be re-established under conditions when SIR proteins are overproduced 91. Combined, these observations suggest that it is SIR-dependent heterochromatin itself that facilitates long-range interactions between heterochromatic loci. The apparent role of membrane anchoring could be indirect, in that proteins involved in tethering (Ku70, Esc1) also facilitate recruitment of SIR proteins. Such a self-associating property of SIR-bound chromatin domains would be consistent with other non-deterministic models mentioned above that propose that active and inactive chromatin domains in higher eukaryotes self-organize in distinct nuclear compartments 68.
Polycomb complex dependent silencing
Polycomb group (PcG) proteins are highly conserved factors that are involved in silencing of various developmental genes 92, 93. In Drosophila these proteins form complexes and bind to regulatory elements called PREs (Polycomb response elements) to silence nearby genes. One well-studied PRE is the Fab-7 element. This element is involved in gene regulation of the Abdominal-B gene. Interestingly, when multiple copies of Fab-7 are present throughout the genome, they have a tendency to interact with each other at so-called PcG bodies, subnuclear bodies enriched in PcG proteins (Figure 4) and these long-range interactions enhance the level of gene silencing of Abdominal B 94. The mechanism by which PREs affect gene activity and how their interactions enhance gene silencing is not understood. Interestingly, PREs are being transcribed to generate small non-coding RNAs and it was found recently that interactions between PREs are dependent upon the RNAi machinery 95. This is intriguing as small RNAs are also involved in formation of classical centromeric heterochromatin in Schizosaccharomyces pombe 96. These exciting observations suggest that RNA may play general roles in gene silencing and heterochromatin formation and that they may do so through mediating long-range associations.
The above experiments identified interactions between homologous PRE-elements. Lanzuolo and colleagues addressed the question of whether PRE interactions also occur between non-homologous PREs 97. By using FISH and 3C they studied the bithorax complex (BX-C). The BX-C locus contains the Ubx, AdbA and AbdB genes, as well as a set of different PREs including the bxd and Fab-7 elements that repress Ubx and AbdB respectively. FISH analysis showed that bxd and Fab-7, that are separated by 130 kb, are frequently co-localized in cells in which these elements repress their target genes. In contrast, in tissues where AbdB is expressed and Ubx is silenced the two PREs are not co-localized. They also found that when in a repressed state, the PREs colocalize with PcG bodies in more than 80% of the cases.
Furthermore, 3C studies of the organization of the entire 340 kb region encompassing BX-C showed that the abdA promoter interacted with all PREs and boundary elements within this region, as well as with other gene promoters and the 3′ ends of all homeotic genes in the region. Thus, multiple long-range interactions were detected between PREs and genes, between PREs and between genes themselves. These long-range interactions were proposed to play a role in stably maintaining the repressed state.
Does clustering of heterochromatic loci play a role in gene silencing?
It has been proposed that formation of heterochromatic clusters contributes to the initiation and/or maintenance of the heterochromatic state. Clustering of heterochromatin would result in formation of nuclear neighborhoods with high local concentration of heterochromatin factors 27, 86. When a gene is located in a heterochromatic neighborhood it would be more efficiently silenced. The observation that association of the wild type bw locus with centromeric heterochromatin is correlated with silencing of the gene is consistent with this idea. However, direct evidence that clustering contributes to heterochromatin formation and gene silencing has been difficult to obtain. The reason for this is the fact that factors that mediate clustering are often also required for local heterochromatin formation in cis. Perhaps heterochromatin formation is completely dependent on clustering so that any condition that affects clustering also affects local formation of heterochromatin and gene silencing. Alternatively, clustering may be an intrinsic property of heterochromatin and therefore any mutation or condition that affects heterochromatin formation will also disrupt heterochromatic interactions. One possible exception is the case described by Grimaud et al. in which RNAi components were found to be important for long-range interactions between PREs and effective silencing but not for local recruitment of PcG proteins 95.
Future perspective: a systems approach to study networks of genomic interactions
Over the last couple of years enormous progress has been made towards understanding long-range control of genes. Long-range interactions between genes and their regulatory elements have now been firmly established. Other types of interactions, e.g. clustering of heterochromatic regions and of groups of active genes, have also been detected. These observations point to the existence of extensive networks of long-range interactions that underlie long-range control of genes and shape the spatial organization of chromosomes.
These findings provide important answers to long-standing questions regarding both long-range control of genes and aspects of nuclear organization. However, these new insights also raise many new questions. How do enhancers find their target genes in the context of the crowded nucleus? How do these interactions result in changes in gene expression? What determines the specificity of long-range interactions? What protein complexes are involved? What is the role of the apparently less specific associations between heterochromatic regions or between expressed genes? To what extent do these interactions simply reflect self-organizing aspects of nuclear organization in which regions with similar epigenetic states associate with each other? In order to begin to answer these questions new experimental approaches are needed. In particular, better experimental model systems are needed that allow experimental manipulation of looping interactions so that the biochemical mechanisms by which long-range interactions mediate gene regulation can be directly probed. Three-dimensional imaging of living cells will be crucial to link movement of loci and their associations to gene control.
In addition to these more traditional approaches, we propose that a systems or network analysis approach will add valuable insights into the logic of long-range gene regulation. We believe the genome can be represented as a physical network of interacting elements 9, 22 and that mapping and analysis of these networks will provide novel insights into the logic of long-range gene regulation. For instance, comprehensive analysis of chromosomal interactions may reveal whether regulatory elements tend to regulate one or more genes, whether regulatory hubs are present (i.e. elements controlling large sets of genes, or genes receiving regulatory input from large sets of elements), whether there are differences in these connections for different classes of genes, and how these interactions change during development (Figure 5). One way to represent the intricate web of three-dimensional interactions that occur in cells is in the form of the widely used 2-dimensional network diagrams of nodes and edges. Such network models have proven to be very helpful to visualize, share and analyze these complex data sets.
Figure 5.
Network analysis of chromosomal interactions in the TH2 locus. Top panel: depicted is a linear organization of TH2 locus. Arrows indicate long-range interactions between genes and elements as detected in naïve T-cells 11, 18. Lower panel: long-range interactions can be conveniently presented as a network of interacting nodes.
Global analysis of the logic of long-range gene regulation by mapping the architecture of the network of chromosomal interactions may well be feasible with the advent of several newly developed high-throughput 3C-based technologies. These new methods allow comprehensive determination of the composition of 3C libraries, which is not possible by PCR. The 4C technologies (3C-on-Chip or Circular 3C) can be used to identify all genomic regions that interact with a given gene or element of interest 19, 25, 98, 99. 4C employs inverse PCR to amplify all DNA fragments that have become ligated to a genomic element of interest. The 5C technology (3C-Carbon Copy) is different form 4C in that it employs highly multiplexed ligation mediated amplification to detect up to millions of unique ligation products present in a 3C library. In contrast to 4C, 5C is not anchored on a single genomic element of interest, but can be used to determine all interactions between two large sets of genomic elements, e.g. between a set of enhancers and a set of putative target genes 100.
Initial datasets obtained by 4C and 5C already illustrate the power of these approaches. For instance the group of Ohlsson, who developed the “Circular-3C” variant, found that imprinted loci display a tendency to interact with each other 99. Another example is provided by application of the “3C-on Chip”-variant to analysis of the beta-globin locus 25. Using 4C, interactions between the restriction fragment containing hypersensitive site 2 within the LCR and the rest of the genome were determined. The majority of interactions with the locus were with sequences on the same chromosome regardless of whether the locus was active or inactive. Interestingly, it was found that the locus associated with 66 clusters on chromosome 7 when the locus is active and only 45 in brain when the locus is inactive. When analyzed further, it was found that in its active state the locus interacts with actively transcribed genes, such Uros, Eraf, and Kcnq1. In contrast, when the beta-globin locus was inactive, it interacted preferentially with other non-transcribed loci. Finally, 5C has been applied to further study the human beta-globin locus. 5C analysis not not only confirmed interactions that had previously been detected by 3C between the LCR and the gamma-globin gene and the 3′ HS1 but has also discovered a new chromatin looping interaction, between the LCR and a beta-globin pseudogene, a region that has been implicated in developmental control of the locus 100.
All these technologies promise to yield important new insights into the spatial organization of entire genomes. Although 3C-based technologies are powerful, they only provide static and population-averaged insights into the spatial conformations of chromosomes. Finally, it would be very beneficial to also develop alternative and complementary strategies, particularly using single-cell and time-resolved imaging, to explore genome organization. For example, recent work performed in the Fraser laboratory combines RNA and DNA FISH with 3C. They find that actively transcribed genes are found colocalized together in regions enriched for RNAP II (transcription factories). Further, this data was confirmed by 3C. (Osborne, 2004). Another recently developed imaging platform, three-dimensional structured illumination microscopy (3D-SIM), promises to greatly contribute to studies of genome organization at the single cell level 101. Ideally, information obtained from all these experimental approaches will be integrated to obtain a coherent view of the structural and functional organization of chromosomes. Clearly, these are exciting times and many new phenomena underlying genome regulation and organization may be discovered in the very near future.
Acknowledgments
Research in the Dekker lab is supported by grants from NIH (HG003143), the Keck Foundation and the Cystic Fibrosis Foundation.
Biographies
About the authors
Adriana Miele received a bachelor of science degree from Stonehill College in 2002 majoring in biology with a minor in chemistry. Currently, she is a PhD student of the Interdisciplinary Graduate Program at the University of Massachusetts Medical School in Worcester, MA. Her current thesis work is focused on analyzing and understanding the mechanism of long-range chromatin interactions in Saccharomyces cerevisiae.
Job Dekker is an Associate Professor in the Program of Gene Function and Expression and the Department of Biochemistry and Molecular Pharmacology at the University of Massachusetts Medical School. His group studies the spatial organization of genomes in relation to gene regulation. He obtained his bachelor of science degree in Biology in 1993 and his PhD in physiological Chemistry in 1997 from Utrecht University, The Netherlands. From 1998 till 2003 he was a post-doctoral fellow in the laboratory of Dr. Nancy Kleckner at Harvard University, during which time he developed the widely used Chromosome Conformation Capture technology.
Footnotes
Summary of the review article: Over the last few years important new insights into the process of long-range gene regulation have been obtained. Gene regulatory elements are found to engage in direct physical interactions with distant target genes and with loci on other chromosomes to modulate transcription. An overview of recently discovered long-range chromosomal interactions is presented, and a network approach is proposed to unravel gene-element relationships.
References
- 1.Kleinjan DA, van Heyningen V. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.West AG, Fraser P. Hum Mol Genet. 2005;14:R101–111. doi: 10.1093/hmg/ddi104. [DOI] [PubMed] [Google Scholar]
- 3.Spitz F, Gonzalez F, Duboule D. Cell. 2003;113:405–417. doi: 10.1016/s0092-8674(03)00310-6. [DOI] [PubMed] [Google Scholar]
- 4.Stamatoyannopoulos G. Exp Hematol. 2005;33:256–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dekker J. Trends Biochem Sci. 2003;28:277–280. doi: 10.1016/S0968-0004(03)00089-6. [DOI] [PubMed] [Google Scholar]
- 6.de Laat W, Grosveld F. Chromosome Res. 2003;11:447–459. doi: 10.1023/a:1024922626726. [DOI] [PubMed] [Google Scholar]
- 7.Chambeyron S, Bickmore WA. Curr Opin Cell Biol. 2004;16:256–262. doi: 10.1016/j.ceb.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Sexton T, Schober H, Fraser P, Gasser SM. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 9.Dekker J. Science. 2008;319:1793–1794. doi: 10.1126/science.1152850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Mol Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 11.Spilianakis CG, Flavell RA. Nat Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Garrard WT. Mol Cell Biol. 2005;25:3220–3231. doi: 10.1128/MCB.25.8.3220-3231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Mol Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 14.Jing H, Vakoc CR, Ying L, Mandat S, Wang H, Zheng X, Blobel GA. Mol Cell. 2008;29:232–242. doi: 10.1016/j.molcel.2007.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grass JA, Jing H, Kim SI, Martowicz ML, Pal S, Blobel GA, Bresnick EH. Mol Cell Biol. 2006;26:7056–7067. doi: 10.1128/MCB.01033-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou GL, Xin L, Song W, Di LJ, Liu G, Wu XS, Liu DP, Liang CC. Mol Cell Biol. 2006;26:5096–5105. doi: 10.1128/MCB.02454-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spilianakis CG, Lalioti MD, Town T, Lee GR, Flavell RA. Nature. 2005;435:637–645. doi: 10.1038/nature03574. [DOI] [PubMed] [Google Scholar]
- 19.Lomvardas S, Barnea G, Pisapia DJ, Mendelsohn M, Kirkland J, Axel R. Cell. 2006;126:403–413. doi: 10.1016/j.cell.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 20.Xu N, Tsai CL, Lee JT. Science. 2006;311:1149–1152. doi: 10.1126/science.1122984. [DOI] [PubMed] [Google Scholar]
- 21.Bacher CP, Guggiari M, Brors B, Augui S, Clerc P, Avner P, Eils R, Heard E. Nat Cell Biol. 2006;8:293–299. doi: 10.1038/ncb1365. [DOI] [PubMed] [Google Scholar]
- 22.Dekker J. Nat Methods. 2006;3:17–21. doi: 10.1038/nmeth823. [DOI] [PubMed] [Google Scholar]
- 23.Iborra FJ, Pombo A, Jackson DA, Cook PR. J Cell Sci. 1996;109:1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 24.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 25.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nat Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 26.Faro-Trindade I, Cook PR. Biochem Soc Trans. 2006;34:1133–1137. doi: 10.1042/BST0341133. [DOI] [PubMed] [Google Scholar]
- 27.Sexton T, Schober H, Fraser P, Gasser SM. Nat Struct Mol Biol. 2007;14:1049–1055. doi: 10.1038/nsmb1324. [DOI] [PubMed] [Google Scholar]
- 28.Osborne CS, Chakalova L, Mitchell JA, Horton A, Wood AL, Bolland DJ, Corcoran AE, Fraser P. PLoS Biol. 2007;5:e192. doi: 10.1371/journal.pbio.0050192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 30.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck B, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, Szustakowki J, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 31.Baltimore D. Nature. 2001;409:814–816. doi: 10.1038/35057267. [DOI] [PubMed] [Google Scholar]
- 32.ENCODE-consortium. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang ZD, Paccanaro A, Fu Y, Weissman S, Weng Z, Chang J, Snyder M, Gerstein MB. Genome Res. 2007;17:787–797. doi: 10.1101/gr.5573107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nobrega MA, Ovcharenko I, Afzal V, Rubin EM. Science. 2003;302:413. doi: 10.1126/science.1088328. [DOI] [PubMed] [Google Scholar]
- 35.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 36.Bulger M, Groudine M. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 37.Engel JD, Tanimoto K. Cell. 2000;100:499–502. doi: 10.1016/s0092-8674(00)80686-8. [DOI] [PubMed] [Google Scholar]
- 38.Dekker J, Rippe K, Dekker M, Kleckner N. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 39.Miele A, Gheldof N, Tabuchi TM, Dostie J, Dekker J. In: Current Protocols in Molecular Biology. Editon edn. Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. vol. John Wiley & Sons; Hoboken, N.J.: 2006. pp. 21.11.21–21.11-20. [Google Scholar]
- 40.Splinter E, Grosveld F, de Laat W. Methods Enzymol. 2004;375:493–507. doi: 10.1016/s0076-6879(03)75030-7. [DOI] [PubMed] [Google Scholar]
- 41.Dostie J, Dekker J. Nat Protoc. 2007;2:988–1002. doi: 10.1038/nprot.2007.116. [DOI] [PubMed] [Google Scholar]
- 42.Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 43.Simonis M, Kooren J, de Laat W. Nat Methods. 2007;4:895–901. doi: 10.1038/nmeth1114. [DOI] [PubMed] [Google Scholar]
- 44.Dekker J. Genome Biol. 2007;8:R116. doi: 10.1186/gb-2007-8-6-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gheldof N, Tabuchi TM, Dekker J. Proc Natl Acad Sci U S A. 2006;103:12463–12468. doi: 10.1073/pnas.0605343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang Q, Carroll JS, Brown M. Mol Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- 47.Higgs DR, Garrick D, Anguita E, De Gobbi M, Hughes J, Muers M, Vernimmen D, Lower K, Law M, Argentaro A, Deville MA, Gibbons R. Ann N Y Acad Sci. 2005;1054 doi: 10.1196/annals.1345.012. [DOI] [PubMed] [Google Scholar]
- 48.Grosveld F, van Assendelft GB, Greaves DR, Kollias G. Cell. 1987;51:975–985. doi: 10.1016/0092-8674(87)90584-8. [DOI] [PubMed] [Google Scholar]
- 49.Forrester WC, Takegawa S, Papayannopoulou T, Stamatoyannopoulos G, Groudine M. Nucleic Acids Res. 1987;15:10159–10177. doi: 10.1093/nar/15.24.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatoyannopoulos JA, Goodwin A, Joyce T, Lowrey CH. EMBO J. 1995;14:106–116. doi: 10.1002/j.1460-2075.1995.tb06980.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tewari R, Gillemans N, Wijgerde M, Nuez B, von Lindern M, Grosveld F, Philipsen S. EMBO J. 1998;17:2334–1341. doi: 10.1093/emboj/17.8.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carter D, Chakalova L, Osborne CS, Dai YF, Fraser P. Nat Genet. 2002;32:623–626. doi: 10.1038/ng1051. [DOI] [PubMed] [Google Scholar]
- 53.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. Nat Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 54.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keys JR, Tallack MR, Zhan Y, Papathanasiou P, Goodnow CC, Gaensler K, Crossley M, Dekker J, Perkins AC. Br J Haematol. 2008;141:398–406. doi: 10.1111/j.1365-2141.2008.07065.x. [DOI] [PubMed] [Google Scholar]
- 56.Anguita E, Hughes J, Heyworth C, Blobel GA, Wood WG, Higgs DR. EMBO J. 2004;23:2841–2852. doi: 10.1038/sj.emboj.7600274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee GR, Fields PE, Griffin TJ, Flavell RA. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 58.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 59.Cai S, Lee CC, Kohwi-Shigematsu T. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 60.Duncan IW. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 61.Ronshaugen M, Levine M. Dev Cell. 2004;7:925–932. doi: 10.1016/j.devcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 62.Avner P, Heard E. Nat rev Genet. 2001;2:59–67. doi: 10.1038/35047580. [DOI] [PubMed] [Google Scholar]
- 63.Chow JC, Yen Z, Ziesche SM, Brown CJ. Annu Rev Genomics Hum Genet. 2005;6 doi: 10.1146/annurev.genom.6.080604.162350. [DOI] [PubMed] [Google Scholar]
- 64.Xu N, Donohoe ME, Silva SS, Lee JT. Nat Genet. 2007;39:1390–1396. doi: 10.1038/ng.2007.5. [DOI] [PubMed] [Google Scholar]
- 65.Augui S, Filion GJ, Huart S, Nora E, Guggiari M, Maresca M, Stewart AF, Heard E. Science. 2007;318:1632–1636. doi: 10.1126/science.1149420. [DOI] [PubMed] [Google Scholar]
- 66.Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- 67.Fuss SH, Omura M, Mombaerts P. Cell. 2007;130:373–384. doi: 10.1016/j.cell.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 68.Misteli T. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Xu M, Cook PR. J Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gerasimova TI, Corces VG. Annu Rev Genet. 2001;35:135–208. doi: 10.1146/annurev.genet.35.102401.090349. [DOI] [PubMed] [Google Scholar]
- 71.Wallace JA, Felsenfeld G. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thompson M, Haeusler RA, Good PD, Engelke DR. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Curr Biol. 2006;16:825–831. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 74.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. J Cell Biol. 2007;179:1095–1103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sawado T, Halow J, Bender MA, Groudine M. Genes Dev. 2003;17:1009–1018. doi: 10.1101/gad.1072303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST. Chromosome Res. 2003;11:513–525. doi: 10.1023/a:1024939130361. [DOI] [PubMed] [Google Scholar]
- 77.Ragoczy T, Bender MA, Telling A, Byron R, Groudine M. Genes Dev. 2006;20:1447–1457. doi: 10.1101/gad.1419506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marenduzzo D, Faro-Trindade I, Cook PR. Trends Genet. 2007;23:126–133. doi: 10.1016/j.tig.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 79.Dernburg AF, Broman KW, Fung JC, Marshall WF, Philips J, Agard DA, Sedat JW. Cell. 1996;85:745–759. doi: 10.1016/s0092-8674(00)81240-4. [DOI] [PubMed] [Google Scholar]
- 80.Huang Y. Nucleic Acids Res. 2002;30:1465–1482. doi: 10.1093/nar/30.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moretti P, Freeman K, Coodly L, Shore D. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 82.Marcand S, Buck SW, Moretti P, Gilson E, Shore D. Genes Dev. 1996;10:1297–1309. doi: 10.1101/gad.10.11.1297. [DOI] [PubMed] [Google Scholar]
- 83.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 84.Gotta M, Laroche T, Formenton A, Maillet L, Scherthan H, Gasser SM. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bystricky K, Laroche T, van Houwe G, Blaszczyk M, Gasser SM. J Cell Biol. 2005;168:375–387. doi: 10.1083/jcb.200409091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maillet L, Boscheron C, Gotta M, Marcand S, Gilson E, Gasser SM. Genes Dev. 1996;10:1796–1811. doi: 10.1101/gad.10.14.1796. [DOI] [PubMed] [Google Scholar]
- 87.Laroche T, Martin SG, Gotta M, Gorham HC, Pryde FE, Louis EJ, Gasser SM. Curr Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- 88.Andrulis ED, Zappulla DC, Ansari A, Perrod S, Laiosa CV, Gartenberg MR, Sternglanz R. Mol Cell Biol. 2002;22:8292–8301. doi: 10.1128/MCB.22.23.8292-8301.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM. EMBO J. 2004;23:1301–1312. doi: 10.1038/sj.emboj.7600144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gartenberg MR, Neumann FR, Laroche T, Blaszczyk M, Gasser SM. Cell. 2004;119:955–967. doi: 10.1016/j.cell.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 91.Mondoux MA, Scaife JG, Zakian VA. Genetics. 2007;177:2019–2029. doi: 10.1534/genetics.107.079848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ringrose L, Paro R. Development. 2007;134:223–232. doi: 10.1242/dev.02723. [DOI] [PubMed] [Google Scholar]
- 93.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Cell. 2007;128:735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 94.Bantignies F, Grimaud C, Lavrov S, Gabut M, Cavalli G. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Grimaud C, Bantignies F, Pal-Bhadra M, Ghana P, Bhadra U, Cavalli G. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 96.Zofall M, Grewal SI. Cold Spring Harb Symp Quant Biol. 2006;71:487–196. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]
- 97.Lanzuolo C, Roure V, Dekker J, Bantignies F, Orlando V. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 98.Würtele H, Chartrand P. Chromosome Res. 2006;14:477–495. doi: 10.1007/s10577-006-1075-0. [DOI] [PubMed] [Google Scholar]
- 99.Zhao Z, Tavoosidana G, Sjölinder M, Göndör A, Mariano P, Wang S, Kanduri C, Lezcano M, Sandhu KS, Singh U, Pant V, Tiwari V, Kurukuti S, Ohlsson R. Nat Genet. 2006;38:1341–1347. doi: 10.1038/ng1891. [DOI] [PubMed] [Google Scholar]
- 100.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio EH, Krumm A, Lamb J, Nusbaum C, Green RD, Dekker J. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schermelleh L, Carlton PM, Haase S, Shao L, Winoto L, Kner P, Burke B, Cardoso MC, Agard DA, Gustafsson MG, Leonhardt H, Sedat JW. Science. 2008;320:1332–1336. doi: 10.1126/science.1156947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Siepel A, Bejerano G, Pedersen JS, Hinrichs AS, Hou M, Rosenbloom K, Clawson H, Spieth J, Hillier LW, Richards S, Weinstock GM, Wilson RK, Gibbs RA, Kent WJ, Miller W, Haussler D. Genome Res. 2005;15:1034–1050. doi: 10.1101/gr.3715005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Koch CM, Andrews RM, Flicek P, Dillon SC, Karaöz U, Clelland GK, Wilcox S, Beare DM, Fowler JC, Couttet P, James KD, Lefebvre GC, Bruce AW, Dovey OM, Ellis PD, Dhami P, Langford CF, Weng Z, Birney E, Carter NP, Vetrie D, Dunham I. Genome Res. 2007;17:691–707. doi: 10.1101/gr.5704207. [DOI] [PMC free article] [PubMed] [Google Scholar]