Abstract
Objective
Brain-derived neurotrophic factor (BDNF) plays an important role in the survival, differentiation, and outgrowth of select peripheral and central neurons throughout adulthood. There is growing evidence suggesting that BDNF is involved in the pathophysiology of mood disorders.
Methods
Ten SNPs across the BDNF gene were genotyped in a sample of 1,749 Caucasian Americans from 250 multiplex bipolar families. Family-based association analysis was employed with three hierarchical bipolar disorder models to test for association between SNPs in BDNF and the risk for bipolar disorder. In addition, an exploratory analysis was performed to test for an association of the SNPs in BDNF and the phenotypes of rapid cycling and episode frequency.
Results
Evidence of association (p<0.05) was found with several of the SNPs using multiple models of bipolar disorder; one of these SNPs also showed evidence of association (p<0.05) with rapid cycling.
Conclusion
These results provide further evidence that variation in BDNF affects the risk for bipolar disorder.
Keywords: brain-derived neurotrophic factor, bipolar disorder, association study, single nucleotide polymorphism
Introduction
While family, twin, and adoption studies have provided strong evidence for a significant genetic component in bipolar disorder (Craddock and Jones, 1999), it is clear that multiple genes must contribute to disease susceptibility (McGuffin and Katz, 1989; Gershon, 1990). Brain-derived neurotrophic factor (BDNF), which belongs to the neurotrophin family, plays an important role in the survival, differentiation, and outgrowth of select peripheral and central neurons throughout adulthood (Schinder, 2000; Huang and Reichardt, 2001). There is growing convergent evidence suggesting that BDNF is involved in the pathophysiology of mood disorders and in the mechanism of action of therapeutic agents (Sheline et al., 1996, 2003; Chen et al., 2001; Fukumoto et al., 2001; Duman, 2002; Karege et al., 2002; Nestler, 2002; Shirayama et al., 2002; Coyle et al., 2003; Shimizu et al., 2003; Chen et al., 2004; Ogden et al, 2004).
Several human studies have shown that serum BNDF levels were decreased in patients with major depressive disorder when compared with the control group (Karege et al., 2002;Shimizu et al., 2002). Increased levels of BDNF immunoreactivity were found in postmortem hippocampal tissue among antidepressant treated bipolar patients, compared with antidepressant untreated bipolar subjects (Chen, et al., 2001). Neuroimaging studies suggested that decreased BDNF levels might account for structural brain changes in bipolar patients (Sheline et al., 1996, 2003; Manji, et al., 2001).
Studies in animal models have suggested that BDNF expression might be a downstream target of antidepressant treatments and that BDNF might provide protection against stress-induced neuronal damage (Duman, 2002; Nestler et al., 2002; Coyle et al., 2003). In animal models of depression, an antidepressant-like effect was produced by the infusion of BDNF into the rat midbrain (Siuciak et al., 1997; Shirayama et al., 2002). The level of BDNF mRNA in the rat hippocampus was increased by electroconvulsive seizures, and the down-regulation of BDNF mRNA was blocked by the chronic administration of electroconvulsive seizures (Lindeforts, et al., 1995; Nibuya et al., 1995). Chronic administration of lithium or valproate increased BDNF expression in rat brain (Fukumoto et al., 2001). BDNF ablation affects the reactivity of the serotoninergic system to the stress hormone corticosterone (Hensler et al., 2007).
Two family studies of bipolar disorder have reported linkage to chromosome 11p13, where BDNF is located (Detera-Wadleigh et al., 1999; McInnes et al., 1996). Significant association of bipolar disorder with polymorphic markers genotyped in BDNF has been reported in two family based studies (Neves-Pereira et al., 2002; Sklar et al., 2002). Both studies genotyped the Val66Met polymorphism (rs6265) and reported over-transmission of the common Val66 allele to individuals with bipolar disorder. This finding was confirmed in a case-control study of unrelated subjects from the NIMH Genetics Initiative (Lohoff et al., 2005). Further evidence of association of the Val66Met polymorphism was reported in three small childhood-onset samples: two using a family-based design (Geller et al., 2004; Strauss, 2005) and one employing a case-control approach (Strauss et al., 2004). In addition to the Val66Met polymorphism, Strauss et al found significant evidence of association between bipolar disorder and four other BDNF SNPs (Strauss, 2005). A novel microsatellite polymorphism, which is located about 1.0 kb upstream of the translation initiation site of the BDNF gene, was reported to confer susceptibility to bipolar disorder (Okada et al., 2006). In contrast to these positive findings, a number of groups have failed to replicate the association of BDNF with the risk of bipolar disorder (Hong et al., 2003; Nakata et al., 2003; Kunugi et al., 2004; Oswald et al., 2004; Skibinska et al., 2004; Neves-Pereira et al., 2005; Green et al., 2006; Liu et al., 2007; Kanazawa et al., 2007). Several investigators also studied the association between BDNF and rapid-cycling: Muller et al found several markers across BNDF, including the Val66Met polymorphism, were significantly associated with both bipolar disorder and rapid-cycling (Muller et al., 2006); Green and colleagues did not find evidence of association between bipolar disorder and the Val66Met polymorphism, but their study of a subset of bipolar subjects with rapid cycling showed significant association with this missense polymorphism (Green et al., 2006).
Most association studies of bipolar disorder have limited their analyses of BDNF polymorphisms to a single marker: the Val66Met polymorphism (Geller et al., 2004; Green et al., 2006; Kunugi et al., 2004; Skibinska et al., 2004; Lohoff et al., 2005). The sample sizes for most studies were relatively small. To ensure a rigorous evaluation of the role of BDNF in bipolar disorder susceptibility, the present study has analyzed a large sample of American Caucasian multiplex bipolar disorder families. Ten single nucleotide polymorphisms (SNPs) that spanned BDNF, including Val66Met, were genotyped, and family-based association analysis was employed. Using this rigorous approach, we found that several BDNF SNPs were associated with the risk for bipolar disorder.
Materials and Methods
Sample
Multiplex families with bipolar disorder were recruited at 10 centers across the United States: Indiana University (with a satellite site at University of Louisville), Johns Hopkins University, the NIMH Intramural Research Program, Rush-Presbyterian Medical Center in Chicago, University of California at Irvine, University of California at San Diego, University of Chicago, University of Iowa, University of Pennsylvania, and Washington University in St. Louis. All participating sites’ institutional review boards approved this study. All subjects received a personal assessment with the Diagnostic Interview for Genetic Studies (DIGS) by a trained interviewer. Medical records were sought for any subject with a history of mental health treatment. First-degree relatives of each proband were interviewed as well with the DIGS and the FIGS (family interview for genetic studies). Pedigree extension through affecteds was carried out as much as possible given resources available to the sites. Each participant was diagnosed by a best estimate procedure involving two senior clinicians reviewing all available information and agreeing on all Axis I diagnoses. A third clinician tiebreaker was consulted when consensus was not reached (about 1–2% of cases). All affected subjects were at least 16 years of age. The ascertainment and assessment of the study participants were previously described (NIMH Genetics Initiative Bipolar Group, 1997).
A whole genome screen was completed in 644 families consisting of 2,909 individuals (Edenberg et al., 1997; Detera-Wadleigh et al., 1997; Rice et al., 1997; Stine et al., 1997; Foroud et al., 2000; Dick et al., 2003; Willour et al., 2003; Zandi et al. 2003). Each family was carefully examined for its informativeness for family-based association study. A total of 250 multiplex families, consisting of 1,749 individuals (771 males, 978 females), were selected for genotyping based on two criteria: all family members were European American; and DNA was available from both parents for at least one affected individual in each family. This selection not only provided a more ethnically homogeneous sample, but also maximized its power for a family-based test of association.
Phenotype Definitions
A set of three hierarchical disease definitions were used in the statistical analysis for bipolar disorder. Model 1 was the narrowest disease definition and included as affected only those individuals meeting criteria for either bipolar I or schizoaffective bipolar disorders (657 affected). Model 2 included as affected individuals meeting criteria for model 1, plus individuals diagnosed with bipolar II disorder (785 affected). Model 3 was the broadest disease definition and included as affected all individuals meeting criteria for models 1 and 2, plus individuals with recurrent unipolar depression (950 affected). We considered two models for our definition of unaffected individuals. In the first, we used very liberal criteria: for each hierarchical disease model, an unaffected individual was defined as anyone who was not affected under that model (Model 1: 1,053 unaffected; Model 2: 925 unaffected; Model 3: 760 unaffected). In the second, more conservative definition, we did not consider anyone to be unaffected; rather, the individuals who did not meet criteria for being affected were classified as unknown.
Rapid cycling was defined as the occurrence of four or more distinct episodes of depression and mania or hypomania within twelve months. The rapid cycling variable met DSM-IV criteria. Episode frequency was defined as the total number of distinct episodes suffered by the bipolar patient divided by the total number of years since the first episode occurred.
SNP genotyping
BDNF is located on chr11p13 (Maisonpierre, et al., 1991) and is approximately 67 kb in size (NM_170731). SNPs across BDNF (Figure 1) were selected from public databases, such as dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and HapMap (http://www.hapmap.org/). Tag SNPs were chosen within the CEU population of the HapMap database (r2 = 0.8, MAF ≥ 0.05). The Val66Met polymorphism (rs6265) was among the genotyped SNPs. SNP genotyping was performed using a modified single-nucleotide extension reaction, with allele detection by mass spectrometry (Sequenom MassArray system, Sequenom, San Diego, CA). To determine allele frequencies and to test the assays, SNPs were genotyped in a set of 40 unrelated Caucasian individuals obtained from Coriell and examined for deviations from Hardy-Weinberg equilibrium. SNPs with minor allele frequencies greater than 0.10 were preferentially genotyped in the study sample; all SNPs were in Hardy-Weinberg equilibrium. All genotypic data on the subjects were checked for Mendelian inheritance errors using the program PEDCHECK (O’Connell and Weeks, 1998). Marker allele frequencies and heterozygosities were estimated using the USERM13 (Boehnke, 1991) option of the MENDEL linkage computer program. Deviation from Hardy-Weinberg equilibrium was assessed for each SNP using Haploview (Barret, et al., 2005). The genotyped SNPs in BDNF, along with their chromosomal positions and minor allele frequencies, are shown in Table 1.
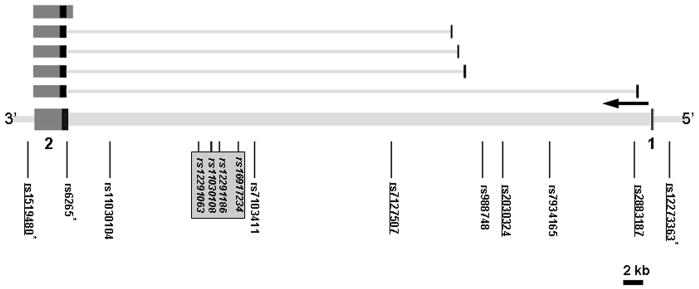
Figure 1. BDNF gene structure and SNP location.
The BDNF gene structure is drawn based on the transcript NM_170731 (encoding isoform b), with five alternative forms shown above: NM_170732 (isoform a), NM_170733 (isoform a), NM_001709 (isoform a), NM_170734 (isoform c), NM_170735 (isoform a), in order from bottom to top; other splicing forms have also been reported. SNP positions are shown below. Exons are numbered and indicated in black, the untranslated regions in dark gray and introns as light gray bars connecting the 2 exons. The direction of transcription of all isoforms is right to left, as indicated by the horizontal arrow. The SNPs which demonstrated significant association with bipolar disorder are underlined. The SNPs showing evolutionary conservation are indicated by asterisks. Four SNPs showing nominal significance (p < 0.05) in recent GWAS studies are in italics and boxed (Personal communication with Dr. Francis McMahon; Baum et al., 2007; The Wellcome Trust Case Control Consortium, 2007). The size of the gene is scaled by a 2 kb bar at lower right.
Table 1.
BDNF SNPs: chromosome position, minor allele frequency and association with bipolar disorder
| Average PDTc | |||||||
|---|---|---|---|---|---|---|---|
| SNP # | SNP | Positiona | Location† | MAFb | Model 1 | Model 2 | Model 3 |
| 1 | rs1519480 | 27,632,288 | downstream | 0.30 | 0.005 | 0.02 | 0.03 |
| 2 | rs6265 | 27,636,492 | exon 2Val66met | 0.21 | 0.40 | 0.26 | 0.06 |
| 3 | rs11030104 | 27,641,093 | intron 1 | 0.22 | 0.78 | 0.87 | 0.35 |
| 4 | rs7103411 | 27,656,701 | intron 1 | 0.23 | 0.64 | 0.69 | 0.27 |
| 5 | rs7127507 | 27,671,460 | intron 1 | 0.32 | 0.002 | 0.009 | 0.009 |
| 6 | rs988748 | 27,681,321 | intron 1 | 0.23 | 0.55 | 0.58 | 0.21 |
| 7 | rs2030324 | 27,683,491 | intron 1 | 0.47 | 0.02 | 0.04 | 0.16 |
| 8 | rs7934165 | 27,688,559 | intron 1 | 0.49 | 0.12 | 0.24 | 0.58 |
| 9 | rs2883187 | 27,697,668 | intron 1 | 0.47 | 0.02 | 0.04 | 0.14 |
| 10 | rs12273363 | 27,701,435 | upstream | 0.20 | 0.05 | 0.12 | 0.10 |
Positions along Chromosome 11 are based on dbSNP build 125/human genome build 34.1.
Position within or near gene
Minor allele frequency in European Americans in this study
P-value of Average PDT statistic for association between the SNPs and bipolar disorder. Significantly associated SNPs are shown in bold. Analyses are reported for the narrow definition of unaffected in which everyone who is not affected is classified as unknown rather than unaffected.
Statistical analyses
Linkage disequilibrium (LD) was evaluated between the SNPs using the program Haploview (Barret, et al., 2005). The extent of LD was evaluated using Lewontin’s standardized disequilibrium coefficient (D′) and delta-squared (r2). The program Tagger (de Bakker et al., 2005) was used to examine how well the genotyped SNPs captured information from the ungenotyped SNPs. The Haploview program was also used to examine the haplotype block structures, where the blocks were defined using the criteria proposed by Gabriel (Gabriel, et al., 2002; Reich, et al., 2001).
Family-based association was performed for each of the genotyped SNPs using the three hierarchical bipolar disorder models. The pedigree disequilibrium test (PDT; Martin et al., 2000) implemented in the UNPHASED package (Dudbridge, 2003) was used to test SNPs for their association with bipolar disorder. The PDT utilizes data from all available trios in a family, as well as discordant sibships. Evidence for association is assessed based on the overtransmission of a particular allele to affected individuals, and the greater frequency of that particular allele in affected individuals than in their unaffected siblings. In the liberal definition of unaffected, the PDT statistic would include both concordant and discordant transmissions. When using the conservative definition of unaffected, the PDT statistic would only include transmissions to affected individuals. In all analyses, the “average-PDT” statistic, which averages the association statistic across all families, was used to test for association between the SNPs and bipolar disorder (Martin et al., 2000).
There were only 24 families in our sample with at least one member having the rapid cycling phenotype. As a result, there was no power to perform family-based association analyses. Therefore, a case-control approach was taken by selecting one rapid cycling case from each of the 24 families. A control was defined as a subject with bipolar disorder who did not have rapid cycling and one such subject was randomly selected from each of the remaining 226 families. The Pearson Chi-square test was performed to test for an association between the BDNF SNPs and the rapid cycling diagnosis.
Exploratory analyses were also performed on the phenotype of episode frequency. From each family, the bipolar subject with the greatest episode frequency was selected for analysis. Due to the skewness of the distribution, the distribution was dichotomized. Those patients in the top 1/3 of the distribution (83 subjects) were classified as having high episode frequency. Those patients in the bottom 1/3 of the distribution (83 patients) were classified as having low episode frequency. The Pearson Chi-square test was used to test for an association between episode frequency classification and BDNF SNP genotype.
Results
Ten SNPs were genotyped across BDNF. The pairwise LD among adjacent SNPs was high (D′ > 0.98) and all the SNPs fell into a single haplotype block (Figure 2). To determine whether these SNPs adequately assessed the variation within BDNF, the program Tagger (de Bakker et al., 2005) was used to examine how well the genotyped SNPs capture information from ungenotyped SNPs. Eight of the ten genotyped SNPs were in the HapMap database and could be assessed; these 8 SNPs captured 77.3% of all HapMap alleles (of MAF ≥ 0.05) at r2 > 0.8, and 88.6% alleles at r2 > 0.5; the mean r2 was 0.86. Two SNPs were not included in HapMap and therefore could not be assessed; therefore, these numbers underestimate the coverage of BDNF.
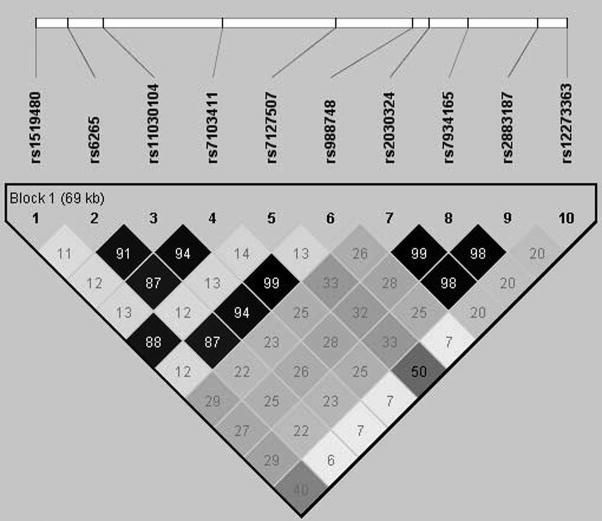
Figure 2.
Pairwise linkage disequilibrium (D′) between the SNPs genotyped in BDNF. Empty boxes indicate SNPs are in complete LD (D′=1), dark grey boxes are in high linkage disequilibrium.
Initial PDT analyses employing a liberal definition of unaffected detected some evidence of association (p<0.03) with two SNPs (rs1519480 and rs7127507) in BDNF across all three hierarchical bipolar disorder models. However, when PDT analyses were performed using the conservative definition of unaffected, much stronger evidence of association (p<0.005) with these two SNPs was observed using the narrowest disease model (Table1). Two additional SNPs (rs2030324 and rs2883187) provided evidence of association (p<0.05) when the two narrower models of disease were employed, and one more SNP (rs12273363) provided evidence only for the narrowest model.
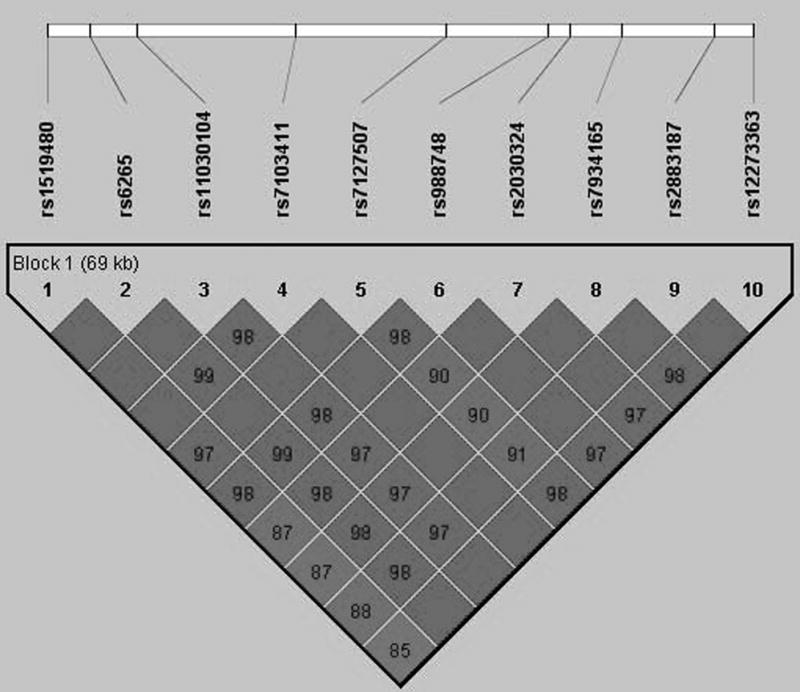
Several of the SNPs between rs1519480 and rs7127507 did not provide evidence of association using any of the three disease models. To better understand how a gene that appeared to be in a single block based on D′ estimates of LD could have such a pattern of association, we reviewed the pattern of LD using the r2 statistic. As shown in Figure 3, an unusual pattern of LD was identified. The two SNPs with the greatest evidence of association with bipolar disorder, rs1519480 and rs7127507, were in very high LD with each other (r2=0.88), but had little evidence of LD with the SNPs between them (r2<0.14). The intervening SNPs were in high LD with each other (r2>0.87) and with the SNP flanking rs7127507 (r2>0.87). The genotypes at rs1519480 and rs7127507 were 97% concordant. Data from HapMap were reviewed and these two SNPs are in nearly absolute LD in that dataset as well.
Figure 3.
Pairwise linkage disequilibrium (r2) between the SNPs genotyped in BDNF. Color scheme: white (r2=0), shades of grey (0<r2<1), black (r2=1).
SNP rs7127507 was also found to be associated with the phenotype of rapid cycling (p < 0.05; genotype counts in controls: CC:CT:TT=25:87:98; genotype counts in rapid cycling patients: CC:CT:TT=6:5:8). No other SNP provided evidence of association with rapid cycling. The previously reported Val66Met polymorphism (rs6265) was not significantly associated with either bipolar disorder or rapid cycling; there was a trend (p=0.06) toward an overtransmission of the common Val allele using the broadest bipolar disorder model. None of the SNPs were associated with episode frequency.
Discussion
This study presents a thorough evaluation of the evidence for association between bipolar disorder and the BDNF gene. We found several SNPs showing evidence of association with bipolar disorder. However, when we use the same phenotypic definition as Sklar and colleagues (Sklar et al., 2002), our results with SNP rs6265 using Model 1 are consistent with the previous report (p=0.40, T/U=654/644, where T=transmissions from heterozygous parent to affected offspring, U=non-transmissions). The study of Sklar et al, employed about 25% of the same subjects used in this study. We also analyzed the data by excluding the first 97 families ascertained trios from which were included in a previous report (Sklar et al., 2002). The same LD structure and a similar pattern of significant association results were obtained in this sample independent of the Sklar study. SNPs rs1519480 and rs7127507 were still significant across all three hierarchical bipolar disorder models (p<0.05); rs2030324 and rs2883187 still showed evidence of association (p<0.05) in the two narrower disease models.
It is not clear how best to define unaffected persons in multiplex pedigrees for bipolar disorder. Onset of bipolar disorder can occur throughout the life span; however, the median age of onset for the first major mood episode is about 20. Some authors report multiple peaks in age of onset data, with a subgroup of patients having their first episode in the 30’s or later (Leboyer et al., 2005; Lin et al., 2006). It is also clear that bipolar illness represents the extreme endpoint of a spectrum of mood instability, and that minor variants may not come to clinical attention (Angst, 1998; Akiskal and Pinto, 1999). In this context it is not unreasonable to define only affected individuals and regard undiagnosed persons as having an unknown affected status. The fact that BDNF produced more significant evidence for association in this model may suggest that our family database includes a number of persons with subclinical bipolar phenotypes carrying BDNF “risk” variants, who detracted from the association evidence when they were classed as unaffected.
It is interesting to see that SNP rs7127507 is significantly associated with both bipolar disorder and rapid cycling in our study. Muller and colleagues also reported that several markers on the gene BNDF had significant association with both bipolar disorder and rapid cycling, and suggested that their significant findings on bipolar disorder were mainly driven by the rapid cycling patients (Muller et al., 2006). Since we have a relatively small number of rapid cycling patients in our sample, and there is only one SNP associated with rapid cycling, we plan to collect more rapid cycling cases to confirm this observation.
Although we have performed the association tests for 10 SNPs, all these SNPs are in high LD, and fall in one haplotype block, as indicated in Figure 2; therefore, it would not be appropriate to correct for 10 tests, since these SNPs are highly correlated. However, since we employed three different nested phenotype definitions and two nested definitions of unaffected individuals (i.e., 6 models in total), an overly conservative Bonferroni correction was applied to our results, Both SNP rs1519480 and rs7127507 are still significantly associated with bipolar disorder after this correction (P<0.05). Since there is only one phenotype model for rapid cycling and these analyses were exploratory, we did not perform any statistical correction to that association result.
Many previous studies have focused on the missense polymorphism Val66Met (rs6265) (Geller et al., 2004; Kunugi et al., 2004; Neves-Pereira et al., 2002; Skibinska et al., 2004; Lohoff et al., 2005; Green et al., 2006; Liu et al., 2007; Kanazawa et al., 2007). Neves-Pereira et al. (2002) reported a significant association (p-value=0.00064) using 283 trios. Lohoff et al. (2005) performed a case-control (621 BPI cases vs. 998 controls) study and reported the Val allele was significantly increased in BPI patients when compared to the controls. Among their 621 BPI cases, 213 were used in this study, which accounts for 12% of this sample. Sklar et al (2002) found a marginally significant overtransmission of the Val allele to the bipolar proband (p-value=0.04) in their Hopkins sample (136 trios); however, this finding was not confirmed when they employed 189 trios from the NIMH bipolar initiative (p-value=0.09), which accounted for about 25% of our sample. They also did not find significant association in their UK sample (p-value=0.24) with a sample 145 trios. There are also a number of case-control studies which have failed to replicate the association of bipolar disorder with BDNF (Hong et al., 2003; Nakata et al., 2003; Kunugi et al., 2004; Oswald et al., 2004; Skibinska et al., 2004; Neves-Pereira et al., 2005; Green et al., 2006; Liu et al., 2007). These conflicting reports could be due to the use of differing study designs, variable ancestry of the samples, and the wide range of sample sizes which directly affect the power of the samples to detect association.
Two of the SNPs providing evidence of association with bipolar disorder, rs1519480 and rs12273363, are located in highly conserved regions when compared with other vertebrate genomes in the 17-way Vertebrate MultiAlignment (UCSC Genome Browser, v. 141). This suggests that these two markers may play some important functional role. None of the SNPs genotyped in this study was on or near the alternative splicing sites that give rise to 5 known transcription isoforms of BDNF (www.ncbi.nlm.nih.gov). Therefore, the three significantly associated markers, rs7127507, rs2030324, and rs2883187, in the intronic region of the gene may play some other functional role(s) than alternative splicing. Such mechanisms are yet to be discovered. Interestingly, in two recently published genome-wide association studies (Baum et al., 2007; The Wellcome Trust Case Control Consortium, 2007), four additional SNPs in this intronic region (rs16917234, rs12291186, rs11030108, rs12291063) were found to be significantly associated with bipolar disorder in three independent datasets: NIMH, German and Wellcome Trust Case Control Consortium (Personal communication with Dr. Francis McMahon for the NIMH and German datasets). These four SNPs are shown in Figure 1. In contrast with the inconsistent reports on the Val66Met polymorphism, the cumulative evidence of association in the intron region suggests that mRNA processing, stability and BDNF expression levels play a role in the susceptibility for bipolar disorder.
The current study has several strengths. First, we employed a sample of 250 multiplex, well-characterized bipolar disorder families, with 402 trios in model 1, 471 trios in model 2 and 522 trios in model 3. Second, analyses were performed in a single ethnic group (European Americans). Third, we employed family-based tests of association on bipolar disorder, which minimize the effects of potential population stratification. Fourth, the most significant association analyses used only affected individuals. While this limits the informativeness of the PDT analysis by eliminating the contribution of discordant sib pairs, we have reduced the possible effects of erroneous subject classification. Fifth, 10 SNPs were genotyped across BDNF, with high LD between the SNPs. This ensured good coverage of the gene and maximized our power to detect association anywhere in BDNF.
In summary, we have found evidence of association between bipolar disorder and SNPs in BDNF, including SNPs near both ends of this gene. The evidence provided by us in this paper supports a role for BDNF as a pathophysiologic factor in mood disorders.
Acknowledgments
Sponsorships: This work was supported by MH59545.
Data and biomaterials were collected in four projects that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 1991–98, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, U01 MH46282, John Nurnberger, M.D., Ph.D., Marvin Miller, M.D., and Elizabeth Bowman, M.D.; Washington University, St. Louis, MO, U01 MH46280, Theodore Reich, M.D., Allison Goate, Ph.D., and John Rice, Ph.D.; Johns Hopkins University, Baltimore, MD U01 MH46274, J. Raymond DePaulo, Jr., M.D., Sylvia Simpson, M.D., MPH, and Colin Stine, Ph.D.; NIMH Intramural Research Program, Clinical Neurogenetics Branch, Bethesda, MD, Elliot Gershon, M.D., Diane Kazuba, B.A., and Elizabeth Maxwell, M.S.W.
Data and biomaterials were collected as part of ten projects that participated in the National Institute of Mental Health (NIMH) Bipolar Disorder Genetics Initiative. From 1999–03, the Principal Investigators and Co-Investigators were: Indiana University, Indianapolis, IN, R01 MH59545, John Nurnberger, M.D., Ph.D., Marvin J. Miller, M.D., Elizabeth S. Bowman, M.D., N. Leela Rau, M.D., P. Ryan Moe, M.D., Nalini Samavedy, M.D., Rif El-Mallakh, M.D. (at University of Louisville), Husseini Manji, M.D. (at Wayne State University), Debra A. Glitz, M.D. (at Wayne State University), Eric T. Meyer, M.S., Carrie Smiley, R.N., Tatiana Foroud, Ph.D., Leah Flury, M.S., Danielle M. Dick, Ph.D., Howard Edenberg, Ph.D.; Washington University, St. Louis, MO, R01 MH059534, John Rice, Ph.D, Theodore Reich, M.D., Allison Goate, Ph.D., Laura Bierut, M.D.; Johns Hopkins University, Baltimore, MD, R01 MH59533, Melvin McInnis M.D., J. Raymond DePaulo, Jr., M.D., Dean F. MacKinnon, M.D., Francis M. Mondimore, M.D., James B. Potash, M.D., Peter P. Zandi, Ph.D, Dimitrios Avramopoulos, and Jennifer Payne; University of Pennsylvania, PA, R01 MH59553, Wade Berrettini, M.D.,Ph.D.; University of California at Irvine, CA, R01 MH60068, William Byerley, M.D., and Mark Vawter, M.D.; University of Iowa, IA, R01 MH059548, William Coryell, M.D., and Raymond Crowe, M.D.; University of Chicago, IL, R01 MH59535, Elliot Gershon, M.D., Judith Badner, Ph.D., Francis McMahon M.D., Chunyu Liu Ph.D., Alan Sanders M.D., Maria Caserta, Steven Dinwiddie, M.D., Tu Nguyen, Donna Harakal; University of California at San Diego, CA, R01 MH59567, John Kelsoe, M.D., Rebecca McKinney, B.A.; Rush University, IL, R01 MH059556, William Scheftner, M.D., Howard M. Kravitz, D.O., M.P.H., Diana Marta, B.S., Annette Vaughn-Brown, MSN, RN, and Laurie Bederow, MA; NIMH Intramural Research Program, Bethesda, MD, 1Z01MH002810-01, Francis J. McMahon, M.D., Layla Kassem, PsyD, Sevilla Detera-Wadleigh, Ph.D, Lisa Austin, Ph.D, Dennis L. Murphy, M.D.
SNP genotyping by MALDI-TOF mass spectrometry used the facilities of the Center for Medical Genomics at Indiana University School of Medicine, which is supported in part by a grant from the Indiana Genomics Initiative (INGEN). INGEN is supported in part by the Lilly Endowment, Inc.
References
- Akiskal HS, Pinto O. The evolving bipolar spectrum. Prototypes I, II, III, and IV. Psychiatr Clin North Am. 1999;22:517–34. doi: 10.1016/s0193-953x(05)70093-9. [DOI] [PubMed] [Google Scholar]
- Angst J. The emerging epidemiology of hypomania and bipolar II disorder. J Affect Disord. 1998;50:143–51. doi: 10.1016/s0165-0327(98)00142-6. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Baum AE, Akula N, Cabanero M, Cardona I, Corona W, Klemens B, Schulze TG, Cichon S, Rietschel M, Nöthen MM, Georgi A, Schumacher J, Schwarz M, Abou Jamra R, Höfels S, Propping P, Satagopan J, Detera-Wadleigh SD, Hardy J, McMahon FJ. A genome-wide association study implicates diacylglycerol kinase eta (DGKH) and several other genes in the etiology of bipolar disorder. Mol Psychiatry. 2007 May 8; doi: 10.1038/sj.mp.4002012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991;48:22–25. [PMC free article] [PubMed] [Google Scholar]
- Chen B, Dowlatshahi D, MacQueen GM, Wang JF, Young LT. Increased hippocampal BDNF immunoreactivity in subjects treated with antidepressant medication. Biol Psychiatry. 2001;50:260–265. doi: 10.1016/s0006-3223(01)01083-6. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24(18):4401–11. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyle JT, Duman RS. Finding the intracellular signaling pathways affected by mood disorder treatments. Neuron. 2003;38:157–60. doi: 10.1016/s0896-6273(03)00195-8. [DOI] [PubMed] [Google Scholar]
- Craddock N, Jones I. Genetics of bipolar disorder. J Med Genet. 1999;36:585–594. doi: 10.1136/jmg.36.8.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bakker PIW, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nature Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Yoshikawa T, Sanders AR, Goldin LR, Turner G, Rollins DY, Moses T, Guroff JJ, Kazuba D, Maxwell ME, Edenberg HJ, Foroud T, Lahiri D, Nurnberger JI, Jr, Stine OC, McMahon F, Meyers DA, MacKinnon D, Simpson S, McInnis M, DePaulo JR, Rice J, Goate A, Gershon ES. Initial genome scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 4, 7, 9, 18, 19, 20, and 21q. Am J Med Genet. 1997;74:254–62. doi: 10.1002/(sici)1096-8628(19970531)74:3<254::aid-ajmg4>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Detera-Wadleigh SD, Badner JA, Berrettini WH, Yoshikawa T, Goldin LR, Turner G. A high-density genome scan detects evidence for a bipolar-disorder susceptibility locus on 13q32 and other potential loci on 1q32 and 18p11.2. Proc Natl Acad Sci. 1999;96:5604–5609. doi: 10.1073/pnas.96.10.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick DM, Foroud T, Flury L, Bowman ES, Miller MJ, Rau NL, Moe PR, Samavedy N, El-Mallakh R, Manji H, Glitz DA, Meyer ET, Smiley C, Hahn R, Widmark C, McKinney R, Sutton L, Ballas C, Grice D, Berrettini W, Byerley W, Coryell W, DePaulo R, MacKinnon DF, Gershon ES, Kelsoe JR, McMahon FJ, McInnis M, Murphy DL, Reich T, Scheftner W, Nurnberger JI., Jr Genomewide linkage analyses of bipolar disorder: a new sample of 250 pedigrees from the National Institute of Mental Health Genetics Initiative. Am J Hum Genet. 2003;73:107–14. doi: 10.1086/376562. Erratum in: Am J Hum Genet 73:979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- Duman RS. Genetics of childhood disorders: XXXIX. Stem cell research, Part 3: regulation of neurogenesis by stress and antidepressant treatment. J Am Acad Child Adolesc Psychiatry. 2002;41:745–748. doi: 10.1097/00004583-200206000-00016. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Foroud T, Conneally PM, Sorbel JJ, Carr K, Crose C, Willig C, Zhao J, Miller M, Bowman E, Mayeda A, Rau NL, Smiley C, Rice JP, Goate A, Reich T, Stine OC, McMahon F, DePaulo JR, Meyers D, Detera-Wadleigh SD, Goldin LR, Gershon ES, Blehar MC, Nurnberger JI., Jr Initial genomic scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 3, 5, 15, 16, 17, and 22. Am J Med Genet. 1997;74:238–46. [PubMed] [Google Scholar]
- Foroud T, Castelluccio PF, Koller DL, Edenberg HJ, Miller M, Bowman E, Rau NL, Smiley C, Rice JP, Goate A, Armstrong C, Bierut LJ, Reich T, Detera-Wadleigh SD, Goldin LR, Badner JA, Guroff JJ, Gershon ES, McMahon FJ, Simpson S, MacKinnon D, McInnis M, Stine OC, DePaulo JR, Blehar MC, Nurnberger JI., Jr Suggestive evidence of a locus on chromosome 10p using the NIMH genetics initiative bipolar affective disorder pedigrees. Am J Med Genet. 2000;96:18–23. [PubMed] [Google Scholar]
- Fukumoto T, Morinobu S, Okamoto Y, Kagaya A, Yamawaki S. Chronic lithium treatment increases the expression of brain-derived neurotrophic factor in the rat brain. Psychopharmacology. 2001;158:100–106. doi: 10.1007/s002130100871. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Geller B, Badner JA, Tillman R, Christian SL, Bolhofner K, Cook EH., Jr Linkage disequilibrium of the brain-derived neurotrophic factor Val66Met polymorphism in children with a prepubertal and early adolescent bipolar disorder phenotype. Am J Psychiatry. 2004;161:1698–1700. doi: 10.1176/appi.ajp.161.9.1698. [DOI] [PubMed] [Google Scholar]
- Gershon ES. Genetics. In: Gooswin FK, Jamison KR, editors. Manic-depressive illness. New York: Oxford University Press; 1990. pp. 373–401. [Google Scholar]
- Green EK, Raybould R, Macgregor S, Hyde S, Young AH, O’Donovan MC, Owen MJ, Kirov G, Jones L, Jones I, Craddock N. Genetic variation of brain-derived neurotrophic factor (BDNF) in bipolar disorder: case-control study of over 3000 individuals from the UK. Br J Psychiatry. 2006;188:21–5. doi: 10.1192/bjp.bp.105.009969. [DOI] [PubMed] [Google Scholar]
- Hensler JG, Advani T, Monteggia LM. Regulation of Serotonin-1A Receptor Function in Inducible Brain-Derived Neurotrophic Factor Knockout Mice After Administration of Corticosterone. Biol Psychiatry. 2007 doi: 10.1016/j.biopsych.2006.10.015. in Press. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Huo SJ, Yen FC, Tung CL, Pan GM, Tsai SJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and mood disorders, age of onset and suicidal behavior. Neuropsychobiology. 2003;48:186–189. doi: 10.1159/000074636. [DOI] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF. Neurotrophins: roles in neuronal development and function. Annu Rev Neurosci. 2001;24:677–736. doi: 10.1146/annurev.neuro.24.1.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanazawa T, Glatt SJ, Kia-Keating B, Yoneda H, Tsuang MT. Meta-analysis reveals no association of the Val66Met polymorphism of brain-derived neurotrophic factor with either schizophrenia or bipolar disorder. Psychiatr Genet. 2007;17:165–170. doi: 10.1097/YPG.0b013e32801da2e2. [DOI] [PubMed] [Google Scholar]
- Karege F, Bondolfi G, Gervasoni N, Schwald M, Aubry JM, Bertschy G. Decreased serum brain-derived neurotrophic factor levels in major depressed patients. Psychiatry Res. 2002;109:143–148. doi: 10.1016/s0165-1781(02)00005-7. [DOI] [PubMed] [Google Scholar]
- Kunugi H, Iijima Y, Tatsumi M, Yoshida M, Hashimoto R, Kato T. No association between the Val66Met polymorphism of the brain-derived neurotrophic factor gene and bipolar disorder in a Japanese population: a multicenter study. Biol Psychiatry. 2004;56:376–378. doi: 10.1016/j.biopsych.2004.06.017. [DOI] [PubMed] [Google Scholar]
- Leboyer M, Henry C, Paillere-Martinot ML, Bellivier F. Age at onset in bipolar affective disorders: a review. Bipolar Disord. 2005;7:111–8. doi: 10.1111/j.1399-5618.2005.00181.x. [DOI] [PubMed] [Google Scholar]
- Lin PI, McInnis MG, Potash JB, Willour V, MacKinnon DF, DePaulo JR, Zandi PP. Clinical correlates and familial aggregation of age at onset in bipolar disorder. Am J Psychiatry. 2006;163:240–6. doi: 10.1176/appi.ajp.163.2.240. [DOI] [PubMed] [Google Scholar]
- Lindefors N, Brodin E, Metsis M. Spatiotemporal selective effects on brain-derived neurotrophic factor and trkB messenger RNA in rat hippocampus by electroconvulsive shock. Neuroscience. 1995;65:661–670. doi: 10.1016/0306-4522(94)00550-o. [DOI] [PubMed] [Google Scholar]
- Liu M, Ling SH, Li WB, Wang CY, Chen DF, Wang G. An association study between GRIN1, BDNF genes and bipolar disorder. Hereditas. 2007;29:41–6. doi: 10.1360/yc-007-0041. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Sander T, Ferraro TN, Dahl JP, Gallinat J, Berrettini WH. Confirmation of association between the Val66Met polymorphism in the brain-derived neurotrophic factor (BDNF) gene and bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet. 2005;139:51–3. doi: 10.1002/ajmg.b.30215. [DOI] [PubMed] [Google Scholar]
- Manji HK, Duman RS. Impairments of neuroplasticity and cellular resilience in severe mood disorders: implications for the development of novel therapeutics. Psychopharmacol Bull. 2001;35(2):5–49. [PubMed] [Google Scholar]
- Maisonpierre PC, Le Beau MM, Espinosa R, Ip NY, Belluscio L, de la Monte SM, Squinto S, Furth ME, Yancopoulos GD. Human and rat brain-derived neurotrophic factor and neurotrophin-3: gene structures, distributions, and chromosomal localizations. Genomics. 1991;10:558–568. doi: 10.1016/0888-7543(91)90436-i. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees: The Pedigree Disequilibrium Test. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin P, Katz R. The genetics of depression and manic-depressive disorder. Br J Psychiatry. 1989;155:294–304. doi: 10.1192/bjp.155.3.294. [DOI] [PubMed] [Google Scholar]
- McInnes LA, Escamilla MA, Service SK, Reus VI, Leon P, Silva S. A complete genome screen for genes predisposing to severe bipolar disorder in two Costa Rican pedigrees. Proc Natl Acad Sci. 1996;93:13060–13065. doi: 10.1073/pnas.93.23.13060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller DJ, de Luca V, Sicard T, King N, Strauss J, Kennedy JL. Brain-derived neurotrophic factor (BDNF) gene and rapid-cycling bipolar disorder: family-based association study. Br J Psychiatry. 2006;189:317–23. doi: 10.1192/bjp.bp.105.010587. [DOI] [PubMed] [Google Scholar]
- Nakata K, Ujike H, Sakai A, Uchida N, Nomura A, Imamura T. Association study of the brain-derived neurotrophic factor (BDNF) gene with bipolar disorder. Neurosci Lett. 2003;337:17–20. doi: 10.1016/s0304-3940(02)01292-2. [DOI] [PubMed] [Google Scholar]
- Nestler EJ, Gould E, Manji H, Buncan M, Duman RS, Greshenfeld HK, Hen R, Koester S, Lederhendler I, Meaney M, Robbins T, Winsky L, Zalcman S. Preclinical models: status of basic research in depression. Biol Psychiatry. 2002;52:503–528. doi: 10.1016/s0006-3223(02)01405-1. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Cheung JK, Pasdar A, Zhang F, Breen G, Yates P. BDNF gene is a risk factor for schizophrenia in a Scottish population. Mol Psychiatry. 2005;10:208–212. doi: 10.1038/sj.mp.4001575. [DOI] [PubMed] [Google Scholar]
- Neves-Pereira M, Mundo E, Muglia P, King N, Macciardi F, Kennedy JL. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: evidence from a family-based association study. Am J Hum Genet. 2002;71:651–655. doi: 10.1086/342288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibuya M, Morinobu S, Duman RS. Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci. 1995;15:7539–7547. doi: 10.1523/JNEUROSCI.15-11-07539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niculescu AB. Genomic studies of mood disorders -- the brain as a muscle? Genome Biol. 2005;6:215. doi: 10.1186/gb-2005-6-4-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIMH Genetics Initiative Bipolar Group. Genomic survey of bipolar illness in the NIMH genetics initiative pedigrees: a preliminary report. Am J Med Genet. 1997;74:227–237. doi: 10.1002/(sici)1096-8628(19970531)74:3<227::aid-ajmg1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Nurnberger JI, Jr, Blehar MC, Kaufman CA, York-Cooler C, Simpson SG, Friedman JH, Severe JB, Malaspina D, Reich T, Miller M, Bowman E, DePaulo R, Cloninger R, Robinson G, Mildin S, Gershon E, Maxwell E, Guroff J, Kirch D, Wynne D, Berg K, Tsuang M, Faraone S, Pepple J, Ritz AL. Diagnostic interview for genetic studies. Arch Gen Psychiat. 1994;51:849–849. doi: 10.1001/archpsyc.1994.03950110009002. [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden CA, Rich ME, Schork NJ, Paulus MP, Geyer MA, Lohr JB, Kuczenski R, Niculescu AB. Candidate genes, pathways and mechanisms for bipolar (manic-depressive) and related disorders: an expanded convergent functional genomics approach. Mol Psychiatry. 2004;9:1007–29. doi: 10.1038/sj.mp.4001547. [DOI] [PubMed] [Google Scholar]
- Okada T, Hashimoto R, Numakawa T, Iijima Y, Kosuga A, Tatsumi M, Kamijima K, Kato T, Kunugi H. A complex polymorphic region in the brain-derived neurotrophic factor (BDNF) gene confers susceptibility to bipolar disorder and affects transcriptional activity. Mol Psychiatry. 2006;11:695–703. doi: 10.1038/sj.mp.4001822. [DOI] [PubMed] [Google Scholar]
- Oswald P, Del-Favero J, Massat I, Souery D, Claes S, van Broeckhoven E. Non-replication of the brain-derived neurotrophic factor (BDNF) association in bipolar affective disorder: a Belgian patient-control study. Am J Med Genet. 2004;129B:34–35. doi: 10.1002/ajmg.b.30056. [DOI] [PubMed] [Google Scholar]
- Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50:211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, Lander ES. Linkage disequilibrium in the human genome. Nature. 2001;411(6834):199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- Rice JP, Goate A, Williams JT, Bierut L, Dorr D, Wu W, Shears S, Gopalakrishnan G, Edenberg HJ, Foroud T, Nurnberger J, Jr, Gershon ES, Detera-Wadleigh SD, Goldin LR, Guroff JJ, McMahon FJ, Simpson S, MacKinnon D, McInnis M, Stine OC, DePaulo JR, Blehar MC, Reich T. Initial genome scan of the NIMH genetics initiative bipolar pedigrees: chromosomes 1, 6, 8, 10, and 12. Am J Med Genet. 1997;74:247–53. doi: 10.1002/(sici)1096-8628(19970531)74:3<247::aid-ajmg3>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Schinder AF, Poo M. The neurotrophin hypothesis for synaptic plasticity. Trends Neurosci. 2000;23(12):639–45. doi: 10.1016/s0166-2236(00)01672-6. [DOI] [PubMed] [Google Scholar]
- Sheline YI, Gado MH, Kraemer HC. Untreated depression and hippocampal volume loss. Am J Psychiatry. 2003;160:1516–1518. doi: 10.1176/appi.ajp.160.8.1516. [DOI] [PubMed] [Google Scholar]
- Sheline YI, Wang PW, Gado MH, Csernansky JG, Vannier MW. Hippocampal atrophy in recurrent major depression. Proc Natl Acad Sci. 1996;93:3908–3913. doi: 10.1073/pnas.93.9.3908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu E, Hashimoto K, Okamura N, Koike K, Komatsu N, Kumakiri C, Nakazato M, Watanabe H, Shinoda N, Okada S, Iyo M. Alterations of serum levels of brain-derived neurotrophic factor (BDNF) in depressed patients without or with antidepressants. Biol Psychiatry. 2003;54:70–75. doi: 10.1016/s0006-3223(03)00181-1. [DOI] [PubMed] [Google Scholar]
- Shirayama Y, Chen AC, Nakagawa S, Russell DS, Duman RS. Brain-derived neurotrophic factor produces antidepressant effects in behavioral models of depression. J Neurosci. 2002;22:3251–3261. doi: 10.1523/JNEUROSCI.22-08-03251.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siuciak JA, Lewis DR, Wiegand SJ, Lindsay RM. Antidepressant-like effect of brain-derived neurotrophic factor (BDNF) Pharmacol Biochem Behav. 1997;56:131–137. doi: 10.1016/S0091-3057(96)00169-4. [DOI] [PubMed] [Google Scholar]
- Skibinska M, Hauser J, Czerski PM, Leszczynska-Rodziewicz A, Kosmowska M, Kapelski P. Association analysis of brain-derived neurotrophic factor (BDNF) gene Val66Met polymorphism in schizophrenia and bipolar affective disorder. World J Biol Psychiatry. 2004;5:215–220. doi: 10.1080/15622970410029936. [DOI] [PubMed] [Google Scholar]
- Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G. Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus. Mol Psychiatry. 2002;7:579–593. doi: 10.1038/sj.mp.4001058. [DOI] [PubMed] [Google Scholar]
- Stine OC, McMahon FJ, Chen L, Xu J, Meyers DA, MacKinnon DF, Simpson S, McInnis MG, Rice JP, Goate A, Reich T, Edenberg HJ, Foroud T, Nurnberger JI, Jr, Detera-Wadleigh SD, Goldin LR, Guroff J, Gershon ES, Blehar MC, DePaulo JR. Initial genome screen for bipolar disorder in the NIMH genetics initiative pedigrees: chromosomes 2, 11, 13, 14, and X. Am J Med Genet. 1997;74:263–9. [PubMed] [Google Scholar]
- Strauss J, Barr CL, George CJ, Devlin B, Vetro A, Kiss E, Baji I, King N, Shaikh S, Lanktree M, Kovacs M, Kennedy JL. Brain-derived neurotrophic factor variants are associated with childhood-onset mood disorder: confirmation in a Hungarian sample. Mol Psychiatry. 2005;10:861–7. doi: 10.1038/sj.mp.4001685. [DOI] [PubMed] [Google Scholar]
- Strauss J, Barr CL, George CJ, King N, Shaikh S, Devlin B. Association study of brain-derived neurotrophic factor in adults with a history of childhood-onset mood disorder. Am J Med Genet (Neuropsychiatr Genet) 2004;131B:16–19. doi: 10.1002/ajmg.b.30041. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007 Jun 7;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willour VL, Zandi PP, Huo Y, Diggs TL, Chellis JL, MacKinnon DF, Simpson SG, McMahon FJ, Potash JB, Gershon ES, Reich T, Foroud T, Nurnberger JI, Jr, DePaulo JR, Jr, McInnis MG. Genome scan of the fifty-six bipolar pedigrees from the NIMH genetics initiative replication sample: chromosomes 4, 7, 9, 18, 19, 20, and 21. Am J Med Genet B Neuropsychiatr Genet. 2003;121:21–7. doi: 10.1002/ajmg.b.20051. [DOI] [PubMed] [Google Scholar]
- Zandi PP, Willour VL, Huo Y, Chellis J, Potash JB, MacKinnon DF, Simpson SG, McMahon FJ, Gershon E, Reich T, Foroud T, Nurnberger J, Jr, DePaulo JR, Jr, McInnis MG National Institute of Mental Health Genetics Initiative Bipolar Group. Genome scan of a second wave of NIMH genetics initiative bipolar pedigrees: chromosomes 2, 11, 13, 14, and X. Am J Med Genet B Neuropsychiatr Genet. 2003;119:69–76. doi: 10.1002/ajmg.b.10063. [DOI] [PubMed] [Google Scholar]