SUMMARY
Centrosome amplification is a common feature of many cancer cells, and it has been previously proposed that centrosome amplification can drive genetic instability and so tumorigenesis. To test this hypothesis, we generated Drosophila lines that have extra centrosomes in ~60% of their somatic cells. Many cells with extra centrosomes initially form multipolar spindles, but these spindles ultimately become bipolar. This requires a delay in mitosis that is mediated by the spindle assembly checkpoint (SAC). As a result of this delay, there is no dramatic increase in genetic instability in flies with extra centrosomes, and these flies maintain a stable diploid genome over many generations. The asymmetric division of the larval neural stem cells, however, is compromised in the presence of extra centrosomes, and larval brain cells with extra centrosomes can generate metastatic tumors when transplanted into the abdomens of wild-type hosts. Thus, centrosome amplification can initiate tumorigenesis in flies.
INTRODUCTION
Centrosomes are the main microtubule organizing centers in animal cells, and they comprise a pair of centrioles surrounded by an amorphous pericentriolar material (PCM) (Bornens, 2002; Kellogg et al., 1994). Centrosomes play an important part in organizing many cell processes, particularly during mitosis where they organize the poles of the mitotic spindle. The idea that centrosome amplification can contribute to tumorigenesis was first proposed by Theodor Boveri almost one hundred years ago (Boveri, 2008; Wunderlich, 2002). Boveri was aware that malignant cells often had an abnormal complement of chromosomes, and he had shown that the presence of extra centrosomes in sea urchin embryos invariably led to chromosome missegregation, as the chromosomes were randomly distributed among the spindle poles formed by the multiple centrosomes. This elegant hypothesis, however, was largely ignored as the discovery of oncogenes led to the idea that tumorigenesis is a multistep process involving the accumulation of several mutations or epigenetic changes that ultimately give rise to a cancer cell. Nevertheless, it remains a fact that genetic instability is a common feature of many different types of cancer.
Centrosome amplification is a common feature of many cancer cells (D’Assoro et al., 2002a, 2002b; Nigg, 2006; Pihan et al., 1998, 2001). Moreover, levels of centrosome amplification are often correlated with levels of genetic instability (Brinkley, 2001; Ghadimi et al., 2000; Lingle et al., 2002). Thus, it is now widely assumed that centrosome amplification inevitably leads to genetic instability, and that this can be a significant factor in the generation of fully transformed cancer cells. In support of this possibility, it has recently been shown that inducing genetic instability in mice can increase the rates of tumor formation in some, but not all, tissues (Weaver et al., 2007).
Centrosome amplification, however, does not necessarily lead to spindle multipolarity (Quintyne et al., 2005; Ring et al., 1982). In at least some cell types, extra centrosomes can “cluster” together during mitosis, and the cells often ultimately divide in a bipolar fashion. Indeed, it is thought that many cancer cells in culture have evolved mechanisms to cluster their centrosomes during mitosis so they avoid generating high (and potentially lethal) levels of aneuploidy during every round of cell division (Brinkley, 2001). Thus, the consequences of amplifying centrosomes within the context of a normal developing organism are far from clear.
In flies and humans, the protein kinase SAK/PLK4 plays a critical part in initiating centriole duplication, and the overexpression of this protein can drive centriole overduplication in cells (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Kleylein-Sohn et al., 2007). In this study, we have used stable Drosophila transgenic lines overexpressing SAK to drive centrosome amplification in ~60% of somatic cells. This has allowed us to assess the long-term consequences for an organism of having cells with too many centrosomes. Surprisingly, we find that cells with extra centrosomes invariably divide in a bipolar fashion in vivo, and the presence of extra centrosomes does not generate large-scale genetic instability. The asymmetric division of the neural stem cells (neuroblasts), however, is perturbed, and ~10% of these cells ultimately divide symmetrically. Most importantly, we show that the transplantation of brain cells with too many centrosomes can induce the formation of metastatic tumors in normal hosts.
RESULTS
Flies Overexpressing GFP-SAK Have Too Many Centrosomes in ~60% of Their Somatic Cells but Are Viable and Fertile
It has previously been shown that the overexpression of the centriole replication protein SAK/PLK4 leads to the formation of extra centrosomes in cells (Bettencourt-Dias et al., 2005; Habedanck et al., 2005; Kleylein-Sohn et al., 2007; Peel et al., 2007). To analyze the consequences of centrosome amplification within the context of a developing multicellular organism, we analyzed stable transformed Drosophila lines that expressed a GFP-SAK fusion protein under the control of the Ubiquitin promoter. This promoter is expressed at moderately high levels in all cells (Lee et al., 1988), and it leads to a dramatic overexpression of centriole duplication proteins, as these proteins are normally expressed at very low levels in cells (Peel et al., 2007). We generated several independent transformed lines, all of which showed similar degrees of centrosome amplification; we analyzed two of these lines in detail. Both lines behaved in an essentially indistinguishable manner in all the experiments reported here, so we simply refer to them as SAKOE lines, unless otherwise stated.
Quantification of centrosome number in SAKOE third-instar larval brain cells revealed that ~60% of these cells contained more than two centrosomes (Figures 1A–1C), and we obtained similar results in larval imaginal disc cells (data not shown). Note that in this and all subsequent experiments dots were only counted as centrosomes if they were stained by both centriolar and PCM or microtubule (MT) markers. Usually, 3 to 6 centrosomes were present in the cells with extra centrosomes, but cells with higher numbers were occasionally observed. To confirm that these structures were centrosomes, we performed an electron microscopy (EM) analysis of fixed whole-mount brains. In wild-type (WT) brains, two centrioles were identified at each spindle pole (n = 4) (Figure 1D). In contrast, half of the spindle poles we examined in SAKOE cells contained multiple centrioles (n = 9/18) (Figure 1E). Thus, the extra centrosomes we observe in the SAKOE cells contain morphologically recognizable centrioles.
Figure 1. Overexpression of SAK Induces Centrosome Amplification and Delays Development.
(A and B) Immunostaining of G2 wild-type (WT) (A) and SAKOE (B) neuroblasts with the centriole marker D-PLP (left, green in merged panel), the PCM marker γ-tubulin (middle, red in merged panel), and DNA (blue in merged panel). In the WT neuroblast the two centrosomes are asymmetric (see text for details) and only one contains high levels of γ-tubulin. In the SAKOE neuroblast eight centrosomes are present and these contain varying amounts of γ-tubulin.
(C) Quantification of centrosome number in WT (white bars) and SAKOE (red bars) neuroblasts. Note that we only scored D-PLP dots as centrosomes if they also contained some PCM.
(D and E) EM micrographs of selected thin serial sections of WT (D) and SAKOE (E) neuroblast spindle poles. In the WT cell only two centrioles were detected at the spindle pole (red and yellow arrows). In this SAKOE cell three centrioles were detected at the spindle pole (red, yellow, and orange arrows).
(F) A graph showing the percentage of WT (white bars), SAKOE (red bars), DSas-4 (light gray), and DSas-4,SAKOE (dark gray) pupae that formed between the 5th and 10th day of development. More than 95% of SAKOE pupae eclosed as adults, which is similar to WT controls. Scale bar (A and B) = 10 µm; (D and E) = 0.5 µm.
We have previously shown that DSas-4 mutant flies (that lack centrioles and centrosomes) are morphologically normal and are only slightly delayed in development compared to WT flies (Basto et al., 2006). Surprisingly, we found that SAKOE flies were also morphologically normal (Figure S1 available online), but they exhibited a much longer delay in development compared to WT and DSas-4 mutants (Figure 1F). To test if this developmental delay was caused by the presence of extra centrosomes, we overexpressed GFP-SAK in a DSas-4 mutant background. These flies contained no detectable centrioles (data not shown) and, like DSas-4 mutants alone, they were only slightly delayed in development (Figure 1F). We conclude that it is the presence of extra centrosomes in SAKOE flies that delays their development.
Despite the delay in development, adult SAKOE flies were viable and fertile, although a significant fraction of eggs laid by SAKOE females (~60%) died early in development due to an accumulation of mitotic errors (Peel et al., 2007). Nevertheless, transgenic fly lines containing extra centrosomes in ~60% of their somatic cells can be maintained in the laboratory as viable and fertile stocks for many generations (at present we have maintained these stocks for nearly 2 years).
Mitosis in Cells with Extra Centrosomes
These findings indicate that the presence of extra centrosomes in the majority of somatic cells in an organism is compatible with normal development and long-term survival. This suggests that extra centrosomes do not dramatically interfere with cell division and cell-cycle progression. To better understand how cells divide in the presence of extra centrosomes we examined fixed SAKOE third-instar larval brain cells.
It has previously been shown that centrosomes behave asymmetrically in WT neuroblasts, with one centrosome associated with more PCM and nucleating more MTs than the other throughout interphase and during the early stages of mitosis (Rebollo et al., 2007; Rusan and Peifer, 2007). This asymmetry was noticeable during early mitosis in WT neuroblasts (Figure 1A and Figure 2A) but was often not apparent in SAKOE neuroblasts with extra centrosomes (Figure 1B and Figure 2B). In these early mitotic cells, all centrosomes were associated with PCM and MTs and the centrosomes were often of different sizes, but it was usually not possible to identify a single “dominant” centrosome, either in terms of PCM recruitment or MT nucleation (Figure 2B). Thus, centrosome asymmetry appears to be disrupted in neuroblasts with extra centrosomes.
Figure 2. Mitosis in Cells with Extra Centrosomes.
(A–F) Immunostaining of WT (A, C, and E) and SAKOE (B, D, and F) mitotic neuroblasts with D-PLP (left, green in merged panels), α-tubulin (2nd panel, red in 4th panel), Cnn (3rd panel, red in 5th panel), and DNA (blue in merged panels). In the WT prophase cell (A), the arrow highlights the dominant centrosome that contains more PCM and nucleates more microtubules (MTs). In the SAKOE prophase cell (B), seven centrosomes are present, but there is no single dominant centrosome. In the WT metaphase (C) and anaphase (E) cells, a centrosome is located at each pole of the spindle. In the SAKOE metaphase (D) and anaphase (F) cells, several centrosomes are clustered at the poles of each spindle (arrowheads) while others are not (arrows). The centrosomes that are not clustered at the spindle poles appear to contain less Cnn and are not associated with robust astral MTs (compare this to the situation in the prophase cell shown in B). Scale bar = 10 µm.
In WT brains, 98% of cells (n = 250 cells) formed a bipolar spindle by metaphase (Figure 2C, see also Figure S2). Surprisingly, 93% of cells with extra centrosomes, (n = 500 cells) had also formed a bipolar spindle by metaphase (Figure 2D and Figure S2). Usually, several of the centrosomes were clustered at the poles of the spindle, but we also often observed centrosomes that were not associated with either pole (Figure 2D, see also Figure S3). In metaphase and anaphase cells, the non-pole-associated centrosomes usually contained less PCM than the centrosomes located at the poles, and they were usually not associated with robust asters of MTs (arrows, Figures 2D–2F, see also arrows in Figure 3E), suggesting that they were partially inactivated. Importantly, we made similar observations on the clustering and partial inactivation of extra centrosomes in living SAKOE brain cells (Figure S3 and Movie S1–Movie S4).
Figure 3. Mad2 and Ncd Are Required for the Suppression of Spindle Multipolarity.
(A–C) Immunostaining of mad2,SAKOE mitotic neuroblasts with D-PLP (left, green in merged panels), α-tubulin (2nd panel, red in 5th panel), Cnn (3rd panel, red in 6th panel), and DNA (blue in merged panels). For comparison with WT controls see Figures 2A and 2E. Tripolar anaphases (A), anaphases with lagging chromatids (arrow in B), and polyploid cells (C) are often seen in mad2,-SAKOE brains but very rarely in SAKOE neuroblasts (see Table 1). Note that mad2,SAKOE neuroblasts can also contain much larger numbers of extra centrosomes than is ever seen in SAKOE cells (~30 in the cell shown here).
(D and E) Immunostaining of ncd,SAKOE mitotic neuroblasts with D-PLP (left panel, green in merged panels), α-tubulin (2nd panel, red in 4th panel), Cnn (3rd panel, red in 5th panel), and DNA (blue in merged panels). For comparison with WT control neuroblasts see Figures 2C and 2E. In ncd,SAKOE neuroblasts multipolar metaphases can be detected (D) but almost all anaphases are bipolar (E). Scale bar = 10 µm.
(F) A graph showing the percentage of WT (white bars), SAKOE (red bars), mad2 (light blue), mad2,-SAKOE (dark blue), ncd (bright green), and ncd,SAKOE (dark green) pupae that formed between the 5th and 10th days of development. Note that the WT and SAKOE data shown here are the same as that shown in Figure 1A, as these experiments were performed at the same time.
Surprisingly, the frequency of multi-polar and abnormal metaphase spindles was only slightly higher in SAKOE cells (2% and 5%, respectively, n = 500 cells) than in WT cells (0% and 2%, respectively, n = 250 cells) (Figure S2). And, by the time cells entered anaphase, the spindles were always bipolar in both WT and SAKOE brains (n = 200 cells and n = 400 cells, respectively), while the frequency of aneuploidy was only slightly higher in SAKOE brains (1.75%, n = 345 cells) compared to WT (0.7%, n = 150 cells) (Table 1). The mitotic index, however, was significantly higher in SAKOE brains (2.6% ± 0.5%, n = 7797 cells from 4 brains) compared to WT (1.9% ± 0.3%, n = 12,000 cells from 4 brains) (p < 0.05), indicating that mitosis takes longer than normal in cells that have extra centrosomes (Table 1). Thus, Drosophila somatic cells with extra centrosomes are delayed in mitosis but ultimately divide in a bipolar fashion.
Table 1.
Quantification of Mitotic Defects
| Mitotic Index |
% Polyploidy |
% Aneuploidy |
%Anaphase with Lagging Chromatids |
|
|---|---|---|---|---|
| WT | 1.9 ± 0.3 | ND | 0.7% | ND |
| SAKOE | 2.6 ± 0.5 | ND | 1.75% | ND |
| mad2 | 1.6 ± 0.5 | ND | 0.6% | 0.5% |
| mad2,SAKOE | 1.7 ± 0.5 | 12% | 10.0% | 12.0% |
| ncd | 2.2 ± 0.2 | ND | 1.0% | 1.0% |
| ncd,SAKOE | 5.2 ± 0.3 | 3.0% | 4.5% | 4.0% |
ND: not detected.
The Spindle Assembly Checkpoint Is Essential in Flies with Extra Centrosomes
These observations suggested that the spindle assembly check-point (SAC) might delay mitosis in cells with extra centrosomes. To test this hypothesis, we crossed the SAKOE lines to mad2 mutants. Although Mad2 is an essential component of the SAC, mad2 mutant flies are viable and fertile, indicating that the SAC is dispensable during unperturbed cell divisions in Drosophila (Buffin et al., 2007). The mad2,SAKOE flies were synthetically lethal and died as pupae, demonstrating that the SAC is absolutely essential for the viability of flies with extra centrosomes. The mad2,SAKOE larvae developed slightly faster than WT flies, at a rate comparable to mad2 mutants, and much faster than the SAKOE flies, demonstrating that it is the maintenance of an active SAC that causes the developmental delay in SAKOE flies (Figure 3F).
As expected, the mitotic index in mad2,SAKOE brains (1.7% ± 0.5%; n > 2000 cells) was lower than in WT or SAKOE brains (Table 1), confirming that the presence of extra centrosomes cannot delay mitosis in the absence of Mad2. In addition, we noticed a dramatic increase in the number of multipolar and defective spindles in mad2,SAKOE brain cells (Figure 3A–3C and Figure S2), and the levels of polyploidy, aneuploidy, and lagging chromosomes during anaphase were also dramatically increased (Table 1). Thus, the SAC is essential in cells with extra centrosomes to prevent spindle multipolarity and the subsequent generation of large-scale genetic instability, presumably because it allows extra time for bipolar spindle formation.
The Kinesin 14 Ncd Is Required for Efficient Centrosome Clustering
It has previously been shown that the minus-end-directed motor cytoplasmic Dynein plays an essential role in the clustering of supernumerary centrosomes (Quintyne et al., 2005). We tested whether another minus-end-directed motor, Ncd (HSET in vertebrates), could also be important for this process. Ncd is required for the efficient focusing of the spindle poles in fly cells (Goshima et al., 2005), but its activity is not absolutely essential in somatic cells and ncd mutants are viable (Endow and Komma, 1998; Skold et al., 2005). We expressed GFP-SAK in an ncd mutant background and found that the developmental rate of ncd,SAKOE flies was even slower than that of the SAKOE flies (Figure 3F), although the flies that hatched were all morphologically normal (data not shown). The ncd,SAKOE brains showed a dramatic increase in the rate of spindle multipolarity during prophase and metaphase (Figure 3D and Figure S2), but we still never observed any multipolar spindles during anaphase (Figure 3E, n = 230 cells). The levels of aneuploidy, polyploidy, and lagging chromosomes during anaphase were all slightly elevated in ncd,SAKOE brains compared to SAKOE or ncd mutant brains alone, while the mitotic index was much higher (Table 1). We conclude that cells with too many centrosomes have great difficulty in organizing a bipolar spindle in the absence of Ncd; nevertheless, these cells can still delay the exit from mitosis until bipolarity is eventually achieved.
Extra Centrosomes Lead to Abnormalities in the Asymmetric Divisions of Larval Neuroblasts
While centrosomes are not essential for somatic cell division (Basto et al., 2006; Hinchcliffe et al., 2001; Khodjakov et al., 2000; Uetake et al., 2007), the astral MTs generated by centrosomes seem to have a particularly important role during asymmetric divisions, when the spindles must align correctly with cortical polarity cues (Yu et al., 2006 and Siegrist and Doe, 2005). In DSas-4 mutant neuroblasts (that completely lack centrosomes), the anastral spindles have great difficulty in aligning properly with cortical cues and ~16% of neuroblasts ultimately divide symmetrically (Basto et al., 2006).
To test whether cells with extra centrosomes have defects in asymmetric cell division we initially examined the localization of the apical marker aPKC and the basal marker Miranda (Mira). In the majority of WT and SAKOE neuroblasts (91%, n = 80 and 79%, n = 150, respectively) the aPKC and Mira crescents were correctly localized on opposite sides of the neuroblasts (Figures S4A–S4C and S4E). We noticed, however, that in some SAKOE neuroblasts the cortical proteins were either misaligned or delocalized from the cortex (Figures S4D and S4E), while the metaphase plate was also sometimes not oriented correctly with respect to the polarity axis (Figure S4C).
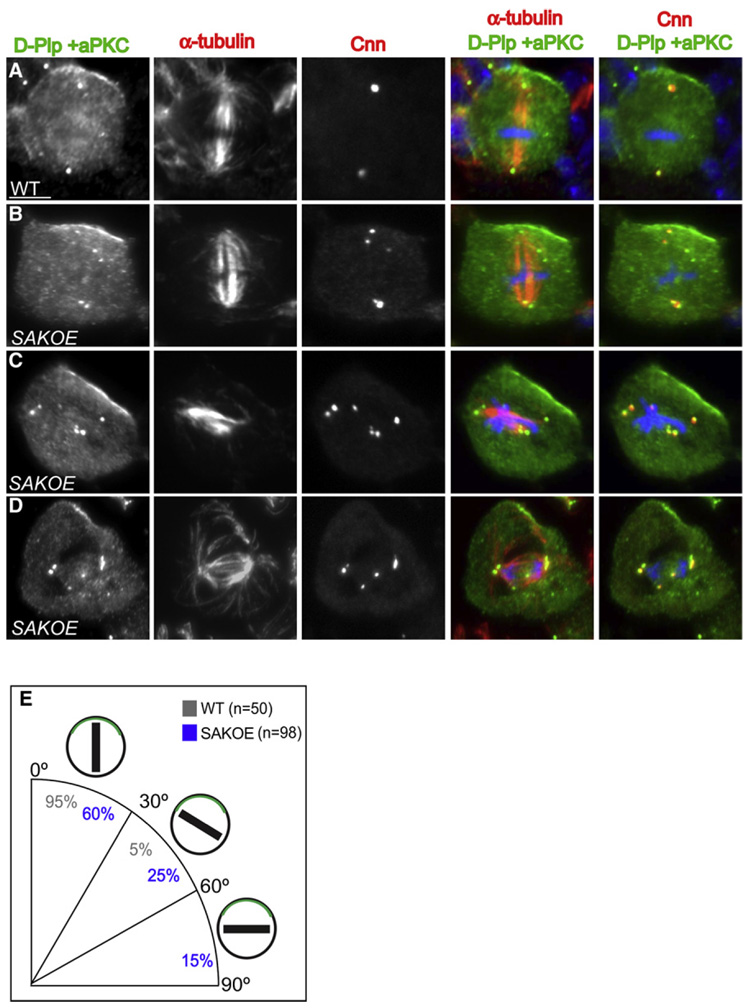
To characterize this alignment defect in more detail, we stained SAKOE third-instar larval neuroblasts with anti-α-tubulin antibodies and determined spindle orientation relative to the cortical aPKC crescent. In 95% (n = 50) of WT neuroblasts the spindle was aligned within 30° of the center of the aPKC apical crescent (Figures 4A and 4E), but this was true in only 60% (n = 98) of SAKOE neuroblasts that contained extra centrosomes (Figures 4B and 4E). Moreover, a significant fraction of spindles in SAKOE neuroblasts with extra centrosomes (15%, n = 98) were organized at right angles to the aPKC crescent (Figures 4C and 4E), which was not seen in WT cells. Importantly, these spindle alignment defects were not detected in SAKOE cells that contained only two centrosomes (Figures S5A and S5B). Taken together these data demonstrate that neuroblasts with extra centrosomes have relatively subtle problems in establishing and/or maintaining the localization of cortical cues, and more significant problems in aligning their spindles correctly with these cues.
Figure 4. Asymmetric Cell Division Is Perturbed in Neuroblasts with Extra Centrosomes.
(A–D) Immunostaining of WT (A) and SAKOE (B–D) neuroblasts with D-PLP and aPKC (left panel, green in merged panels), α-tubulin (2nd panel, red in 4th panel), Cnn (3rd panel, red in 5th panel), and DNA (blue in merged panels). In WT metaphase neuroblasts (A) the spindle is always aligned with the polarity axis (defined by the cortical aPKC crescent), as is the case in ~60% of SAKOE neuroblasts with extra centrosomes (B). In ~40% of SAKOE neuroblasts with extra centrosomes (C and D), however, the spindle fails to align properly with the aPKC crescent; spindle misalignment is also detected in some anaphase cells (D).
(E) Quantification of mitotic spindle alignment in WT and SAKOE neuroblasts that have extra centrosomes. In the schematic diagram of cells, the green crescent represent aPKC while the black bars represent the mitotic spindle. Scale bar = 10 µm.
To investigate why neuroblasts with extra centrosomes have trouble properly aligning their spindles, we examined the distribution of Mud, the Drosophila homolog of NuMA. This protein has been implicated in spindle positioning, and it is normally concentrated at centrosomes, with the apical centrosome usually containing more Mud than the basal centrosome (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006) (Figure S6A). In neuroblasts with extra centrosomes, however, the asymmetric centrosome staining of Mud was usually not evident, and, in many SAKOE neuroblasts (20%, n = 90), very little Mud could be detected at any of the centrosomes (Figures S6B and S6C). Thus, the centrosomal localization of Mud is perturbed in cells with extra centrosomes, potentially explaining, at least in part, why spindle orientation is perturbed.
In Drosophila neuroblasts redundant mechanisms cooperate to ensure that the process of asymmetric division is extremely robust, and although many mutants have initial defects in aligning their spindles with cortical cues, most mutant cells ultimately divide asymmetrically (Bowman et al., 2006 ; Izumi et al., 2006; Siegrist and Doe, 2005; Siller et al., 2006). To test whether SAKOE neuroblasts ultimately divided asymmetrically we followed the behavior of the MT-associated protein Jupiter::GFP (Karpova et al., 2006) in living neuroblasts. As described above, multipolar spindles were often seen at early stages of mitosis (Figures 5B and 5C), but these usually resolved themselves into a bipolar spindle, and the cells divided asymmetrically (~91%, n = 46, Figure 5B). In ~9% of cases, however, the cells with extra centrosomes divided symmetrically (Figure 5C), something we never observed in WT brains (Figure 5A, n = 30) (Basto et al., 2006).
Figure 5. Symmetric Divisions in SAKOE Neuroblasts.
(A–C) The dynamics of Jupiter::GFP (Jup::GFP) in living WT (A) and SAKOE (B and C) neuroblasts. WT neuroblasts always divide asymmetrically to produce two cells of different sizes. In neuroblasts with extra centrosomes both asymmetric (B) and symmetric divisions (C) are observed.
(D–F) Quantification of the number of neuroblasts in the central brain region. Immunostaining of WT (D) and SAKOE (E) brain lobes with Miranda (red) and Hoechst for labeling DNA (blue). (F) A graph showing the average number of neuroblasts in WT (white bars) and SAKOE (red bars) central brain lobes. Scale bar (A–C) = 5 µm. Error bars represent standard deviation (SD).
To test whether these asymmetric division defects could lead to an amplification of the neuroblast pool, we counted the number of central brain neuroblasts in WT and SAKOE brains (Figures 5D–5F). The number of neuroblasts inSAKOEbrain lobes (55 ± 10.4, n = 40 lobes from 20 animals) was slightly, but significantly (p < 0.01), increased when compared to WT brains (45 ± 9.4, n = 40 lobes, from 20 animals). Thus, the presence of extra centrosomes leads to an expansion in the number of stem cells in larval brains.
Centrosome Amplification Can Initiate Tumorigenesis
A long-standing question in cancer biology has been whether the presence of extra centrosomes within a tissue can drive tumorigenesis. SAKOE adult flies showed no obvious evidence of tumor growth (data not shown), but this is not surprising as there is little cell division in adult flies and very few mutations give rise to tumors in adults. To overcome this potential problem, transplantation assays have been developed where larval imaginal discs or brains are transplanted into the abdomen of WT adult hosts (Caussinus and Gonzalez, 2005; Gonzalez, 2007; Woodhouse et al., 1998). Transplanted WT tissue can survive in adult hosts for several weeks without overproliferating or forming tumors. In contrast, the transplantation of tissue from several mutants leads to tissue overproliferation and the formation of metastatic tumors within the WT host.
To test whether SAKOE larval brains could form tumors when transplanted into WT hosts we expressed α-Tubulin-GFP (Tub-GFP) in SAKOE flies so that we could follow the behavior of the transplanted tissue. We used brain tumor (brat) mutant brains as a positive control (Betschinger et al., 2006; Bello et al., 2006; Lee et al., 2006b). We did not detect any overproliferation when Tub-GFP, WT brains were transplanted into WT hosts (Figures 6A and 6E, n=90),but 36%of Tub-GFP, brat brains formed tumors (Figures 6B and 6E, n = 50) and several of the injected hosts went on to develop one or more GFP-positive metastases far from the original site of injection (n = 11/18).We found that 14% of Tub-GFP, SAKOE#2 brains (n = 104) and 20% of Tub-GFP, SAKOE#1 brains (n = 60) formed tumors when transplanted into WT hosts (Figures 6C and 6E), and several of these hosts went on to develop one or more GFP-positive metastases far from the original site of injection (Figure 6D, n = 5/15 for SAKOE#2, and n = 6/20 for SAKOE#1 brains). The overproliferation of either the Tub-GFP, brat, Tub-GFP, SAKOE#2, or Tub-GFP, SAKOE#1 tissues invariably led to the premature death of the WT host within 10 to 15 days of injection.
Figure 6. Transplantation of SAKOE Brain Tissue Induces Tumor Formation in WT Hosts.
(A–C) Pictures of WT hosts transplanted with either Tub-GFP,WT (A), Tub-GFP,brat (B), or Tub-GFP,SAKOE (C) brains at 14 days post-injection. (A) The GFP fluorescence of the WT brain is still detectable in the host, but the brain has not detectably overproliferated (arrow). The GFP fluorescence of the brat (B) and SAKOE transplanted brains (C) has expanded to fill the abdomen of the host.
(D) Picture of a GFP-positive metastasis in the eye of a WT host.
(E) A table showing the frequency of tumor and metastases formation in WT hosts transplanted with the different types of brain tissue.
DISCUSSION
Here we have examined the long-term consequences of having too many centrosomes in a complex multicellular organism. Surprisingly, we find that flies with extra centrosomes are viable, fertile, and can be maintained in the laboratory as a stable diploid stock for many generations. We show that cells with extra centrosomes almost invariably divide in a bipolar fashion in vivo, and the presence of extra centrosomes does not generate large-scale genomic instability. Nevertheless, tissues with extra centrosomes have the ability to overproliferate and form tumors when transplanted into WT hosts. We conclude that centrosome amplification is sufficient to promote tumorigenesis in flies.
Cell Division with Extra Centrosomes
Our observations reveal that, in vivo, the presence of extra centrosomes does not lead to large-scale genetic instability in somatic cells. As cells with extra centrosomes enter mitosis, most of the centrosomes are active and nucleate robust asters of MTs. As mitosis proceeds, however, many of the extra centrosomes become clustered together to form two dominant poles that assemble a bipolar mitotic spindle. This phenomenon of centrosome clustering has been described in several systems (Murphy, 2003; Quintyne et al., 2005; Ring et al., 1982). In addition, however, we find that many extra centrosomes do not become clustered at the spindle poles. Instead, these extra centrosomes appear to be gradually inactivated, and they organize less PCM and nucleate fewer MTs as mitosis proceeds. We do not understand the mechanism of this inactivation, but it could result from a competition for limiting supplies of PCM components. Perhaps the centrosomes that cluster together can communally organize more PCMand so nucleate more MTs than isolated centrosomes. This would then provide a negative feed-back loop as mitosis progresses so that, eventually, the isolated centrosomes are inactivated. Whatever its mechanism, the inactivation of isolated centrosomes ensures that they do not form extra spindle poles efficiently.
Cells with extra centrosomes are delayed in mitosis, and this delay is maintained by the SAC. It is possible that the presence of extra centrosomes somehow directly maintains the activity of the SAC, although previous reports suggest that this is not the case (Sluder et al., 1997). We suspect, therefore, that sister chromatids may be inefficiently aligned on multipolar spindles, and these improperly attached kinetochores ensure the maintenance of SAC activity until a bipolar spindle has formed.
The SAC is not normally essential for fly development, as mad2 mutant flies lack the SAC but are viable and fertile (Buffin et al., 2007). Flies with too many centrosomes, however, completely depend on the SAC as mad2,SAKOE flies exhibit high levels of spindle multipolarity and genetic instability and do not survive to adulthood. Thus, the SAC is essential to allow enough time for cells with extra centrosomes to organize bipolar spindles. Interestingly, while SAKOE flies are severely delayed in development, the mad2,SAKOE flies develop faster than WT flies, indicating that the SAC-dependent delay in mitosis also slows the development of SAKOE flies.
It has previously been shown that centrosome clustering in cells with extra centrosomes is dependent on the activity of dynein (Quintyne et al., 2005). Here we show that another minus-end-directed motor, Ncd (HSET in vertebrates), also plays a role in this process. Ncd is not essential for Drosophila development, and ncd mutants develop at normal rates with few mitotic defects (Endow and Komma, 1998; Skold et al., 2005; this work). We found that ncd,SAKOE flies were severely delayed in development, exhibited elevated levels of spindle multipolarity during early mitosis, had a dramatically increased mitotic index, but ultimately divided in a bipolar fashion. We conclude that Ncd enhances the efficiency of bipolar spindle formation in cells with extra centrosomes, but it is not absolutely essential, and the SAC ensures that these cells do not exit mitosis until they have formed a bipolar spindle.
Extra Centrosomes in Nonsomatic Tissues
The development of flies with extra centrosomes in their somatic tissues is delayed but otherwise appears to proceed normally. It is known, however, that the role of the centrosome differs between embryonic and somatic tissues in Drosophila. While somatic fly cells can tolerate the absence of centrosomes, these organelles are essential for early embryonic development in flies (Dix and Raff, 2007; Stevens et al., 2007; Varmark et al., 2007). Although SAKOE flies are viable and fertile, we note that ~60% of SAKOE embryos accumulate mitotic defects and die during early embryonic development (Peel et al., 2007). Moreover, the mechanisms that ensure bipolar spindle formation in the presence of extra centrosomes may be absent in male germ cells; the presence of extra centrioles or centriole fragments in these cells leads to the formation of multipolar spindles and to male sterility (Dix and Raff, 2007; Martinez-Campos et al., 2004). The overexpression of SAK does not lead to centrosome amplification in male germ cells (Peel et al., 2007), presumably explaining why SAKOE male flies are fertile. Interestingly, we could not detect any uncoordinated behavior in flies with extra centrosomes, suggesting that cilia assembly and function are unaffected by the presence of extra centrioles (Baker et al., 2004; Basto et al., 2006; Martinez-Campos et al., 2004).
Centrosome Amplification and Tumor Formation
We find that brain cells with extra centrosomes can form tumors when injected into WT adult flies. The pathways that lead to tumor formation are complex, and the events that initiate this process remain controversial. Our work shows, however, that centrosome amplification is sufficient to initiate tumorigenesis in the fly.
It is not clear how centrosome amplification initiates tumor formation. Boveri originally hypothesized that extra centrosomes might promote tumorigenesis by promoting genetic instability. The nature of the link between aneuploidy and cancer, however, remains controversial. Recently, it has been shown that increased levels of aneuploidy in mouse models can promote tumor formation in certain tissues at later stages in life but suppress tumor formation upon exposure to certain carcinogens or upon the loss of particular tumor suppressor genes (Sotillo et al., 2007; Weaver et al., 2007). Thus, aneuploidy does not invariably lead to cancer formation. In SAKOE brains the rate of aneuploidy is low, although it is higher than that observed in WT brains (1.75% compared to 0.7%, respectively). It is possible that this modest increase in aneuploidy could allow cells with extra centrosomes to initiate tumor formation in flies.
Alternatively, previous studies in flies have shown that there is a correlation between defects in the asymmetric divisions of larval neural stem cells (neuroblasts) and the ability of injected mutant brain tissue to form tumors when transplanted into WT hosts (Caussinus and Gonzalez, 2005; Woodhouse et al., 1998). Defective asymmetric divisions can result in the expansion of the neuroblast population, which ultimately leads to overproliferation (Bello et al., 2006; Betschinger et al., 2006; Lee et al., 2006a, 2006b; Yu et al., 2006). We find that the asymmetric division of neuroblasts is perturbed in SAKOE brains, and this leads to an expansion of the neuroblast population—a defect that could allow SAKOE-injected brains to overproliferate and form tumors. Indeed, there is much interest in the idea that mutations in stem cells could be central to the generation of cancer (Al-Hajj and Clarke, 2004; Gonzalez, 2007). Importantly, although the increase in aneuploidy and the defects in asymmetric division are only seen in SAKOE cells that have extra centrosomes, we cannot rule out the possibility that SAK overexpression induces tumors via some other mechanism that is unrelated to centrosome amplification. Indeed, mice that are heterozygous for SAK have a variety of cell-cycle defects and have an increased incidence of spontaneous tumor formation (Swallow et al., 2005).
Our observations have important implications for understanding the potential link between centrosomes and cancer. In the literature it is often stated as fact that the presence of extra centrosomes in cells generates genetic instability. This assumption is based on the observation that extra centrosomes clearly lead to spindle multipolarity and genetic instability in some systems (Brinkley and Goepfert, 1998; Wunderlich, 2002) and the strong association between these two phenotypes in many cancer cells (D’Assoro et al., 2002a, 2002b; Fukasawa, 2005; Lingle et al., 1998, 2002; Lingle and Salisbury, 2000; Nigg, 2006; Pihan et al., 1998, 2001, 2003; Saunders, 2005). Our data demonstrate, however, that the presence of extra centrosomes does not inevitably lead to genetic instability in vivo, at least in a relatively simple organism like Drosophila. Instead, extra centrosomes are reasonably well tolerated in flies because several pathways cooperate to ensure that these cells ultimately divide in a bipolar fashion. Only when one or more of these pathways is compromised is large-scale genetic instability generated.
These findings highlight the possibility that the presence of extra centrosomes could prove to be an “Achilles heel” for many different cancers. Fly cells with too many centrosomes are viable, but they are much more reliant on certain pathways (such as the SAC) or proteins (such as Ncd) for their survival than normal cells. It seems plausible that inhibiting these pathways in cancer patients could effectively kill the cancer cells, while leaving normal cells relatively unharmed.
EXPERIMENTAL PROCEDURES
Generation of Transgenic Lines
P-element-mediated transformation vectors were generated by amplifying the complete SAK coding region from a full-length cDNA with att sites at either end for Gateway cloning (Invitrogen). PCR products were inserted into the Gateway pDONR Zeo vector and sequenced. This vector was recombined with the pUbq-GFPNT Gateway vector (R.B., unpublished data) to place full-length SAK with GFP at its N terminus under the control of the Ubiquitin (Ubq) promoter.
Fly Stocks
We used w67 and wf stocks as controls in our experiments. The majority of experiments described here were performed with two transgenic lines: Ubq-GFP-SAK#1 and Ubq-GFP-SAK#2. We recombined Ubq-GFP-SAK#2 with the DSas-4S2214 (Basto et al., 2006), mad2P (Buffin et al., 2007), and ncd1 (Endow and Komma, 1998) mutations by standard recombination methods. For live-cell imaging we crossed the following markers into the appropriate genetic backgrounds: Ubq-RFP-α-Tubulin (a gift from Saskia Suijkerbuijk and Jeroen Dobbelaere), Ubq-GFP-DSas-4 (Peel et al., 2007), and a MT-associated protein tagged to GFP, Jupiter:GFP (Karpova et al., 2006). Measurements of growth and survival rates were performed as previously described (Basto et al., 2006).
Antibodies
The following antibodies were used in this study: rabbit anti-DSas-4 (Basto et al., 2006), rabbit anti-D-PLP (Martinez-Campos et al., 2004), rabbit anti-Cnn (Lucas and Raff, 2007) (all at 1–2 µg/ml final concentration), guinea pig anti-Cnn (1:500, E. Lucas and J.W.R., unpublished data), rabbit anti-Mud (1:250) (Izumi et al., 2006), mouse anti-Miranda (1:20) (Ikeshima-Kataoka et al., 1997), rabbit anti-aPKC (1:500, SC-216, Santa Cruz Biotechnology, inc.), mouse anti-g-tubulin (1:1000; GTU88, Sigma), mouse anti-α-tubulin (1:1000: DM1a, Sigma), rabbit anti-phospho-Histone3 (1:2000, Upstate Biotechnology). All fluorescent secondary antibodies were obtained from Molecular Probes (Invitrogen).
Immunofluorescence Analysis of Brains
Brains were dissected and fixed as described previously (Martinez-Campos et al., 2004). Preparations were examined using either a Zeiss Axioskop II microscope with a CoolSnapHQ camera (Photometrics) with Metamorph software (Molecular Devices Corp.), or a Zeiss LSM 510 Meta scanning confocal system mounted an a Zeiss Axiophot II microscope, or a Perkin Elmer ERS Spinning Disc confocal system using ERS software mounted on a Zeiss Axiovert 200M microscope. All images were processed with Adobe Photoshop software: all images were adjusted using the same procedures that were applied to the whole image. Spindles were classified as multipolar when more than two centrosomes (revealed by costaining of D-Plp and Cnn) organized α-tubulin foci. Mitotic spindles were classified as abnormal when it was not possible to identify clearly spindle poles and/or when the morphology of the spindle was very disorganized.
The levels of aneuploidy and polyploidy were calculated in third-instar larval brain squashes as described previously (Basto et al., 2006). The mitotic index was calculated by fixing and squashing four brains of the appropriate genotype and then staining them with anti-phospho-histone H3 antibodies and Hoechst. Pictures of 5–10 fields of cells (typically containing 200–400 total cells per field) were obtained from each brain, and the ratio of mitotic (phospho-histone H3 positive) to nonmitotic cells was calculated for each field. Each field analyzed was counted as an “event” and thus used to calculate the standard deviation and the significance of the difference between the different datasets (p value) using the two-tailed t test function in Excel.
Analysis of spindle position relative to the apical aPKC crescent position was performed by staining fixed brains with anti-aPKC, anti-DPLP, anti-α-tubulin, and anti-Cnn antibodies. For SAKOE neuroblast analysis, we only scored mitotic cells with more than two centrosomes and where an aPKC crescent could be clearly distinguished. The angle between the spindle axis and the aPKC crescents was determined using the measurement tool in Metamorph. The data were analyzed for statistical significance using the two-tailed t test function in Excel.
Previous studies with anti-Mud antibodies descibed Mud localization at centrosomes and also at the apical cortex (Bowman et al., 2006; Izumi et al., 2006; Siller et al., 2006). In our hands, we could not detect Mud at the apical cortex, presumably because we used different fixation conditions designed to allow us to quantify centrosome number in the SAKOE cells.
Neuroblast numbers were obtained by counting the number of Miranda-positive central brain neuroblasts in 40 WT and 40 SAKOE brain lobes. From the 40 WT brain lobes we found a minimum and maximum number of 30 and 69 neuroblasts, respectively, with an average of 45 (SD ± 9.4). From the 40 SAKOE brain lobes we found a minimum and maximum number of 41 and 78 neuroblasts, respectively, with an average of 55 (SD ± 10.4). The significance of the difference between the two datasets was assessed using the two-tailed t test function in Excel.
Supplementary Material
Supplemental Data include Experimental Procedures, six figures, and four movies and can be found with this article online at http://www.cell.com/cgi/content/full/133/6/1032/DC1/.
ACKNOWLEDGMENTS
We thank Paloma Dominguez and Cayetano Gonzalez for teaching us how to perform abdominal injections and Allan Shearn for useful advice. Roger Karess kindly provided the mad2 mutant stock prior to publication. We thank Susana Godinho, Kwon Mijung, and David Pellman for sharing results before publication and for valuable discussions. We thank F. Matsuzaki for kindly providing antibodies for Mud and Miranda. We would like to thank H. Doerflinger, V. Mirouse, C. Dix, D. Rivers, E. Lucas, and J. Pines for helpful discussions and comments on the manuscript. This work was supported by Research Fellowships from Cancer Research UK (J.W.R. and K.B.), the Wellcome Trust PhD student-ships (N.P. and A.F.), and a Royal Society Dorothy Hodgkin award (R.B.).
REFERENCES
- Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- Baker JD, Adhikarakunnathu S, Kernan MJ. Mechanosensory-defective, male-sterile unc mutants identify a novel basal body protein required for ciliogenesis in Drosophila. Development. 2004;131:3411–3422. doi: 10.1242/dev.01229. [DOI] [PubMed] [Google Scholar]
- Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- Bello B, Reichert H, Hirth F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. doi: 10.1242/dev.02429. [DOI] [PubMed] [Google Scholar]
- Betschinger J, Mechtler K, Knoblich JA. Asymmetric segregation of the tumor suppressor brat regulates self-renewal in Drosophila neural stem cells. Cell. 2006;124:1241–1253. doi: 10.1016/j.cell.2006.01.038. [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- Boveri T. Concerning the origin of malignant tumours by Theodor Boveri. Translated and annotated by Henry Harris. J. Cell Sci. 2008;121(Suppl 1):1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- Bowman SK, Neumuller RA, Novatchkova M, Du Q, Knoblich JA. The Drosophila NuMA homolog Mud regulates spindle orientation in asymmetric cell division. Dev. Cell. 2006;10:731–742. doi: 10.1016/j.devcel.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Brinkley BR. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 2001;11:18–21. doi: 10.1016/s0962-8924(00)01872-9. [DOI] [PubMed] [Google Scholar]
- Brinkley BR, Goepfert TM. Supernumerary centrosomes and cancer: Boveri’s hypothesis resurrected. Cell Motil. Cytoskeleton. 1998;41:281–288. doi: 10.1002/(SICI)1097-0169(1998)41:4<281::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Buffin E, Emre D, Karess RE. Flies without a spindle check-point. Nat. Cell Biol. 2007;9:565–572. doi: 10.1038/ncb1570. [DOI] [PubMed] [Google Scholar]
- Caussinus E, Gonzalez C. Induction of tumor growth by altered stem-cell asymmetric division in Drosophila melanogaster. Nat. Genet. 2005;37:1125–1129. doi: 10.1038/ng1632. [DOI] [PubMed] [Google Scholar]
- D’Assoro AB, Barrett SL, Folk C, Negron VC, Boeneman K, Busby R, Whitehead C, Stivala F, Lingle WL, Salisbury JL. Amplified centrosomes in breast cancer: a potential indicator of tumor aggressiveness. Breast Cancer Res. Treat. 2002a;75:25–34. doi: 10.1023/a:1016550619925. [DOI] [PubMed] [Google Scholar]
- D’Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002b;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- Dix CI, Raff JW. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr. Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow SA, Komma DJ. Assembly and dynamics of an anastral: astral spindle: the meiosis II spindle of Drosophila oocytes. J. Cell Sci. 1998;111:2487–2495. doi: 10.1242/jcs.111.17.2487. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Ghadimi BM, Sackett DL, Difilippantonio MJ, Schrock E, Neumann T, Jauho A, Auer G, Ried T. Centrosome amplification and instability occurs exclusively in aneuploid, but not in diploid colorectal cancer cell lines, and correlates with numerical chromosomal aberrations. Genes Chromosomes Cancer. 2000;27:183–190. [PMC free article] [PubMed] [Google Scholar]
- Gonzalez C. Spindle orientation, asymmetric division and tumour suppression in Drosophila stem cells. Nat. Rev. Genet. 2007;8:462–472. doi: 10.1038/nrg2103. [DOI] [PubMed] [Google Scholar]
- Goshima G, Nedelec F, Vale RD. Mechanisms for focusing mitotic spindle poles by minus end-directed motor proteins. J. Cell Biol. 2005;171:229–240. doi: 10.1083/jcb.200505107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H, Skeath JB, Nabeshima Y, Doe CQ, Matsuzaki F. Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature. 1997;390:625–629. doi: 10.1038/37641. [DOI] [PubMed] [Google Scholar]
- Izumi Y, Ohta N, Hisata K, Raabe T, Matsuzaki F. Drosophila Pins-binding protein Mud regulates spindle-polarity coupling and centrosome organization. Nat. Cell Biol. 2006;8:586–593. doi: 10.1038/ncb1409. [DOI] [PubMed] [Google Scholar]
- Karpova N, Bobinnec Y, Fouix S, Huitorel P, Debec A. Jupiter, a new Drosophila protein associated with microtubules. Cell Motil. Cytoskeleton. 2006;63:301–312. doi: 10.1002/cm.20124. [DOI] [PubMed] [Google Scholar]
- Kellogg DR, Moritz M, Alberts BM. The centrosome and cellular organization. Annu. Rev. Biochem. 1994;63:639–674. doi: 10.1146/annurev.bi.63.070194.003231. [DOI] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr. Biol. 2000;10:59–67. doi: 10.1016/s0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation. Genes Dev. 2006a;20:3464–3474. doi: 10.1101/gad.1489406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Wilkinson BD, Siegrist SE, Wharton RP, Doe CQ. Brat is a Miranda cargo protein that promotes neuronal differentiation and inhibits neuroblast self-renewal. Dev. Cell. 2006b;10:441–449. doi: 10.1016/j.devcel.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lee HS, Simon JA, Lis JT. Structure and expression of ubiquitin genes of Drosophila melanogaster. Mol. Cell. Biol. 1988;8:4727–4735. doi: 10.1128/mcb.8.11.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Salisbury JL. The role of the centrosome in the development of malignant tumors. Curr. Top. Dev. Biol. 2000;49:313–329. doi: 10.1016/s0070-2153(99)49015-5. [DOI] [PubMed] [Google Scholar]
- Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA. 1998;95:2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl. Acad. Sci. USA. 2002;99:1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas EP, Raff JW. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J. Cell Biol. 2007;178:725–732. doi: 10.1083/jcb.200704081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Campos M, Basto R, Baker J, Kernan M, Raff JW. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 2004;165:673–683. doi: 10.1083/jcb.200402130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TD. Drosophila skpA, a component of SCF ubiquitin ligases, regulates centrosome duplication independently of cyclin E accumulation. J. Cell Sci. 2003;116:2321–2332. doi: 10.1242/jcs.00463. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan GA, Purohit A, Wallace J, Knecht H, Woda B, Quesenberry P, Doxsey SJ. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 1998;58:3974–3985. [PubMed] [Google Scholar]
- Pihan GA, Purohit A, Wallace J, Malhotra R, Liotta L, Doxsey SJ. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 2001;61:2212–2219. [PubMed] [Google Scholar]
- Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63:1398–1404. [PubMed] [Google Scholar]
- Quintyne NJ, Reing JE, Hoffelder DR, Gollin SM, Saunders WS. Spindle multipolarity is prevented by centrosomal clustering. Science. 2005;307:127–129. doi: 10.1126/science.1104905. [DOI] [PubMed] [Google Scholar]
- Rebollo E, Sampaio P, Januschke J, Llamazares S, Varmark H, Gonzalez C. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- Ring D, Hubble R, Kirschner M. Mitosis in a cell with multiple centrioles. J. Cell Biol. 1982;94:549–556. doi: 10.1083/jcb.94.3.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W. Centrosomal amplification and spindle multipolarity in cancer cells. Semin. Cancer Biol. 2005;15:25–32. doi: 10.1016/j.semcancer.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Siegrist SE, Doe CQ. Microtubule-induced Pins/Galphai cortical polarity in Drosophila neuroblasts. Cell. 2005;123:1323–1335. doi: 10.1016/j.cell.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Siller KH, Cabernard C, Doe CQ. The NuMA-related Mud protein binds Pins and regulates spindle orientation in Drosophila neuroblasts. Nat. Cell Biol. 2006;8:594–600. doi: 10.1038/ncb1412. [DOI] [PubMed] [Google Scholar]
- Skold HN, Komma DJ, Endow SA. Assembly pathway of the anastral Drosophila oocyte meiosis I spindle. J. Cell Sci. 2005;118:1745–1755. doi: 10.1242/jcs.02304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G, Thompson EA, Miller FJ, Hayes J, Rieder CL. The checkpoint control for anaphase onset does not monitor excess numbers of spindle poles or bipolar spindle symmetry. J. Cell Sci. 1997;110:421–429. doi: 10.1242/jcs.110.4.421. [DOI] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11:9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. From stem cell to embryo without centrioles. Curr. Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swallow CJ, Ko MA, Siddiqui NU, Hudson JW, Dennis JW. Sak/Plk4 and mitotic fidelity. Oncogene. 2005;24:306–312. doi: 10.1038/sj.onc.1208275. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodja-kov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila. Curr. Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 2007;11:25–36. doi: 10.1016/j.ccr.2006.12.003. [DOI] [PubMed] [Google Scholar]
- Woodhouse E, Hersperger E, Shearn A. Growth, metastasis, and invasiveness of Drosophila tumors caused by mutations in specific tumor suppressor genes. Dev. Genes Evol. 1998;207:542–550. doi: 10.1007/s004270050145. [DOI] [PubMed] [Google Scholar]
- Wunderlich V. JMM—past and present. Chromosomes and cancer: Theodor Boveri’s predictions 100 years later. J. Mol. Med. 2002;80:545–548. doi: 10.1007/s00109-002-0374-y. [DOI] [PubMed] [Google Scholar]
- Yu F, Kuo CT, Jan YN. Drosophila neuroblast asymmetric cell division: recent advances and implications for stem cell biology. Neuron. 2006;51:13–20. doi: 10.1016/j.neuron.2006.06.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Data include Experimental Procedures, six figures, and four movies and can be found with this article online at http://www.cell.com/cgi/content/full/133/6/1032/DC1/.