Abstract
A critical process that builds and maintains the eukaryotic cilium is intraflagellar transport (IFT). This process utilizes members of the kinesin-2 superfamily to transport cargo into the cilium (anterograde transport) and a dynein motor for the retrograde traffic. Using a novel RNAi knockdown method, we have analyzed the function of the homodimeric IFT kinesin-2, Kin5, in Tetrahymena ciliary transport. In RNAi transformants, Kin5 was severely downregulated and disappeared from the cilia, but cilia did not resorb, although tip structure was affected. After deciliation of the knockdown cell, cilia regrew and cells swam, which suggested that Kin5 is not responsible for the trafficking of axonemal precursors to build the cilium, but could be transporting molecules that act in ciliary signal transduction, such as guanine nucleotide exchange proteins (GEFs). Gef1 is a Tetrahymena ciliary protein, and current coimmunoprecipitation and immunofluorescence studies showed that it is absent in regrowing cilia of the knockdown cells lacking ciliary Kin5. We suggest that one important cargo of Kin5 is Gef1 and knockdown of Kin5 results in cell lethality.
Introduction
Recent studies suggest that Tetrahymena cilia may contain evolutionary precursors of important metazoan signaling pathways [1], [2], [3]. From studies on Chlamydomonas [4], it has become apparent that most motile protistan and non-motile sensory metazoan cilia, including human primary cilia, are built by intraflagellar transport (IFT) whose molecular constituents are largely conserved. However, the details of ciliary cargo transport, especially of transport of signaling and membrane components into the cilium, although critical for sensory cilia function, have not yet been fully elucidated. It has been shown that certain axonemal components are transported in the cilium by a heterotrimeric kinesin-2 [5], which in Tetrahymena is probably represented by the motor proteins, Kin1 and Kin2 [6] (protein notation taken from kinesin literature standards). In previous findings, we characterized Kin5 as a new, probably essential, IFT motor for intraciliary transport in Tetrahymena [7], which could be involved in the transport of certain membrane components as part of signaling systems. We cloned the 2508 bp coding region of Kin5 and identified it as a member of the kinesin-2 subfamily, whose closest phylogenetic neighbors were Osm3, a C. elegans homodimeric IFT motor protein that is responsible for building the distal segments of sensory cilia in that organism [8], [9], [10] and Kif17, originally identified as a mammalian neuronal protein involved in the transport of NMDA receptor [11] and later shown also to transport ciliary receptors [12]. Northern and western blots localized Kin5 to the cilium. With immunofluorescence, Kin5 was shown to be present in a punctate pattern along the cilium, colocalizing with orthologs of Chlamydomonas IFT complex proteins [13], including members of both IFT complex A (IFT 139/140), the complex probably involved in retrograde transport and complex B (IFT 57, 81, 88, 172) the anterograde transport complex necessary for building the axoneme [14]. Because permeabilization for immunolocalization was performed in the presence of AMP-PNP, it is likely that Kin5 is attached to both the doublet microtubules and the transport complexes.
We wanted to explore the function of Kin5 as a Tetrahymena IFT motor possibly involved in the intraciliary transport of membrane-associated signaling molecules toward the cilium tip. To do this, we proposed to alter the amount of Kin5 produced in the cell and observe how the phenotype is altered. Typically, Tetrahymena contains a somatic macronucleus and a germline micronucleus. Using biolistic transformation and a mutant construct based on the work of Cassidy-Hanley et al. [15], we were able to disrupt the macronuclear KIN5 locus. However, after phenotypic assortment, we failed to generate a somatic knockout, which indicated that this was probably an essential gene whose knockout would lead to lethality. We then decided to investigate whether RNAi production for gene knockdown of KIN5 was feasible in Tetrahymena.
In this study, we define a successful technology for KIN5 knockdown by an inducible short hairpin RNA (shRNA), with relevant conditions and controls. We characterize the KIN5 knockdown phenotype by cell survival studies to confirm that KIN5 is an essential gene, and we discuss the difficulties this presents in the knockdown experiments. We present data to indicate that Kin5 has novel intraciliary transport cargos, some probably related to the placement of membrane and signaling molecules within the cilium, rather than to the transport of components of the motile axoneme, and we discuss similarities and possible differences between the functions of Kin5 and Osm3 or Kif17.
Results
Construction of pK5KOAs.40/pInv2 and cell transformation
Our initial goal was to deplete Kin5 from the cells by integrating an inducible shRNA into the genome. T. thermophila expresses two redundant β-tubulin genes, BTU1 and BTU2. The pMTT1-BICH3 construct designed by Gorovsky's laboratory is able to target the IAG48[G1] sequence behind the powerful inducible metallothionein MTT1 promoter to the BTU1 coding region by biolistic transformation [16]. The IAG48[G1] sequence can be replaced by other sequences of interest that will be transcribed when a heavy-metal divalent cation such as Cd2+ is added. To introduce an inducible RNAi construct into the macronuclear genome, we replaced the IAG48[G1] sequence with a sequence which when transcribed would fold to produce a short double-stranded RNA. A short sequence (K5KOAs.40) based on 19 nucleotides from the KIN5 stalk coding region was chosen, which was unique to the KIN5 gene as verified by a blast search of the Tetrahymena genome (http://www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=t_thermophila). We then designed a construct containing the coding sequence, a loop of nine bases, the antisense sequence of the coding region and a termination codon (Figure 1A). We hypothesized that this construct, K5KOAs.40, would transcribe into a short double-stranded RNA that would act in an RNAi mechanism to suppress the expression of KIN5. A similar scheme was used to construct Inv2 which employed the same 19-nt sequence, but in the inverse orientation (Figure 1A), a type of scrambled sequence that was routinely used as a control for RNAi experiments. Using the HindIII and BamHI sites, the constructs were ligated into the modified MTT1 plasmid, here pK5KOAs.40 and pInv2, respectively and used for biolistic transformation as described by Cassidy-Hanley et al. [15].
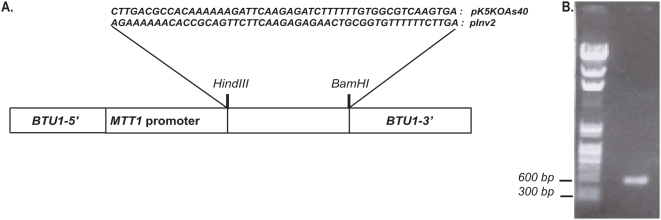
Figure 1. pK5KOAs40/pInv2 Vector Constructs.
A. Schematic diagram showing nucleotide sequences of K5KOAs.40 and Inv2 for shRNA inserted into the MTT1-containing vector. B. PCR verification of KO insert. Left lane: marker DNA; Right lane: Expected PCR product on genomic DNA based on MTTI.s and BTU1.a primers for the KO insert. An identical band was seen with the Inv2 insert for Inv2 cells.
CU522 cells contain a point mutation in the BTU1 gene, which renders them taxol-sensitive [16]; replacement of the BTU1 gene upon incorporation of the plasmid transforms them to taxol-resistant. After transformation of the CU522 cells with the pK5KOAs.40 or pInv2 constructs, the cells were kept at 20 µM paclitaxel, for phenotypic assortment, selecting for more and more copies of the K5KOAs.40 construct in the macronucleus as growth proceeded for a few months [17]. Using BTU1.a and MTT1.s primers in a PCR reaction, we verified that the K5KOAs.40 and Inv2 constructs were integrated into the correct loci (Fig 1B). After allowing for phenotypic assortment, single-cell isolates were grown up to stationary phase for further experimentation.
Induction of shRNA via the MTT1 promoter
The metallothionein gene, MTT1, responds to stress and to heavy metals. In particular, Cd2+ serves to induce transcription by the promoter greater than 200-fold over control conditions [18]. In order to test whether our construct would work to knockdown KIN5 by an RNAi mechanism and to define the time course of knockdown in Tetrahymena, we performed RT-PCR in a linear response range for qualitative analysis on samples following the addition of CdCl2, initially at 5.0 µg/ml concentration. We saw a much stronger response to Cd2+ when the clones were grown in 10 mM Tris-HCl, pH 7.5, as compared to 2× proteose peptone (2XPP). All further experiments were performed in Tris under starvation conditions. A non-ciliary control RNA (PGM1) was examined simultaneously with KIN5 RNA.
When cells were exposed to 5.0 µg/ml Cd2+ in the absence of transformation, both KIN5 RNA and PGM1 RNA remained qualitatively unchanged for 7 h and both messages persisted for at least 24 h (Figure 2A).
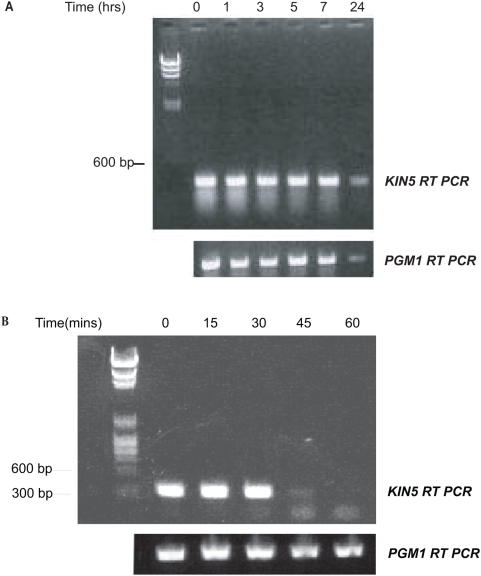
Figure 2. Stability of KIN5 and PGM1 messages.
A. CU522 cells grown in starvation conditions+5.0 µg/ml Cd2+ prior to transformation showing comparable relative stabilities of the KIN5 and PGM1 mRNA. B. Time course of degradation of KIN5 message after shRNA induction in K5KOAs.40 cells using 5.0 µg/ml Cd2+. RT-PCR products resolved on a 1% agarose gel. Left lane: DNA markers. The KIN5 message decreases at 45 min post-induction and is eliminated at 60 min. The PGM1 message remains constant.
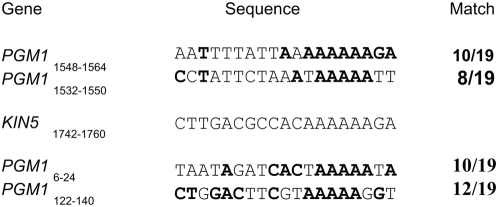
Figure 2B shows that after Cd2+ addition, the KIN5 message remained unchanged only in the samples taken 0, 15 and 30 min post-induction. By 45 min, the message greatly decreased and finally by 60 min, the message was no longer seen. There was no change in the PGM1 message in the KO cells for the entire time period, even though there is identity between the 19-nt KIN5 region and various stretches of the PGM1 gene in as many as 12 of the 19-nt positions (Figure 3). We conclude that the transcribed K5KOAs.40 construct specifically eliminated KIN5 RNA within 1 hour of induction, which suggests that the construct strategy was successful, probably working as hypothesized, via shRNA transcription and an RNAi mechanism.
Figure 3. Sequence Comparison of K5KOAs.40 vs. PGM1.
The indicated PGM1 coding regions show high degree of homology to the 19 nt (positions 1742–1760) chosen for KIN5 shRNA yet KIN5 shRNA does not affect PGM1.
Optimization of [Cd2+]
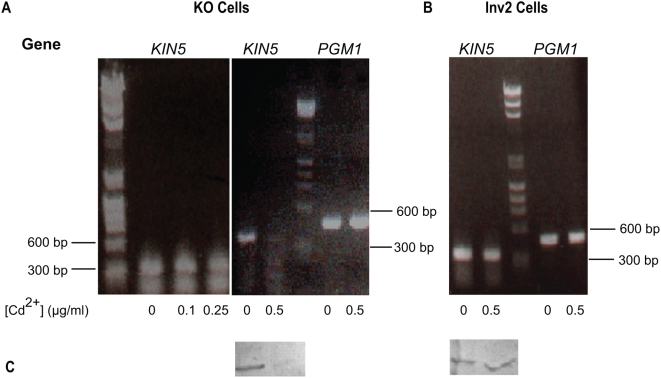
Viability and motility could also be affected by Cd2+, in concentrations that could be toxic under starvation conditions. In 5.0 µg/ml Cd2+, the KO cell population was not viable much beyond 1 h. In untransformed and Inv2 transformed cells in this Cd2+ concentration, viability also dropped sharply within a few hours, so that less than 10% of the population persisted for 24 h. Many cells ruptured, presumably because of failure of osmoregulation, which is a non-specific effect of heavy metal poisoning [19]. We set about to find the optimal [CdCl2] that would elicit the RNAi response without affecting the control cell viability or causing the cells to rupture. We chose concentrations of 0, 0.1, 0.25, and 0.5 µg/ml CdCl2 and performed RT-PCR at 0, 6, and 24 h on KO and Inv2 cell populations. At 0 and 6 h, the KIN5 message was present in similar amounts in all the samples. By 24 h, in KO cells, the KIN5 message was unchanged in 0, 0.1 or 0.25 µg/ml Cd2+, but essentially absent in 0.5 µg/ml CdCl2, while the PGM1 levels remained unaffected (Fig 4A). Both the KIN5 and PGM1 messages remained unaffected in Inv2 control cells from 0 to 24 h at 0.5 µg/ml Cd2+ (Figure 4B). The is confirmed by the loss of Kin5 protein in the KO cells upon cadmium induction whereas Kin5 levels remain unchanged in the Inv2 cells (Figure 4C)
Figure 4. Optimization of KIN5 shRNA.
A. Degradation of KIN5 message after shRNA induction in KO cells using 0–0.5 µg/ml Cd2+. Above: RT-PCR products resolved on a 1% agarose gel. At Cd2+ concentrations lower than 0.5 µg/ml, KIN5 mRNA is stable for 24 h. After 24 h in 0.5 µg/ml Cd2+, KIN5 mRNA is dramatically decreased, while PGM1 is unaffected. B. Effect of 0.5 µg/ml Cd2+ on KIN5 and PGM1 messages in Inv2 cells. KIN5 and PGM1 mRNA levels remain unaffected after 24 h. DNA markers shown: lines indicate 600 and 300 bp. C. Effect of 0.5 µg/ml Cd2+ on Kin5 protein levels in KO and Inv2 cells. Corresponding KO (left) and Inv2 (right) cell homogenates 12 h post-induction at either 0 or 0.5 µg/ml Cd2+ and blotted with K5T1 Ab to Kin5. While the Kin5 protein is severely knocked down in the KO cells upon shRNA induction, Kin5 levels remain unaffected in Inv2 cells under similar conditions.
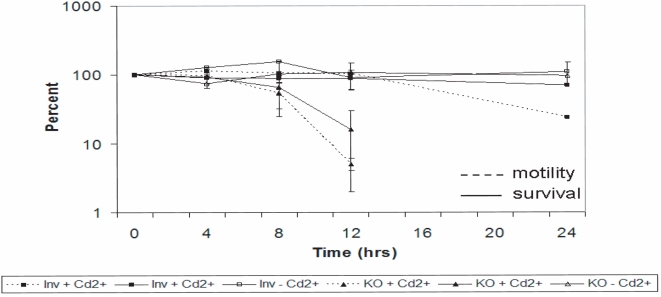
To show that Cd2+ was not toxic at 0.5 µg/ml, we measured cell survival with or without KIN5 knockdown, compared to untreated controls. In cells containing the Inv2 construct with or without Cd2+, survival at 24 h decreased only slightly with addition of cadmium (Figure 5). This suggests that Cd2+ toxicity at this concentration was minimal. Cells containing the KO construct without Cd2+ also survived as expected, but when Cd2+ was added so that Kin5 was knocked down, only about 10% of the original population survived. For the initial 12 h, the time course of loss of viability as measured by cell count paralleled the loss of KIN5 RNA and of survival measured by motility (Figure 5), implying that the reduced population under study remained viable. By 12 h, most of the KO cells were moving very slowly, while overall cell shape remained unchanged, nuclear morphology was normal and cilia were still beating. Taken together, these observations suggested that the KIN5 knockdown phenotype produced loss of normal coordinated movement and then lethality, which is consistent with our earlier finding that KIN5 is an essential gene. We concluded that most of the lethality seen at 12 h in the K5KOAs.40 population was not due to the presence of Cd2+, but rather to the loss of KIN5 mRNA and its consequences and that, at this time, there are still enough viable cells for study. In further experiments, we used 0.5 µg/ml Cd2+ and 12 h exposure to study the effects of KIN5 knockdown on Kin5 localization.
Figure 5. Effect of 0.5 µg/ml Cd2+ on K5KOAs.40 and Inv2 cell survival.
Initial population indicated as 100%. Solid lines: survival measured by cell count. Dashed lines: Survival measured by cell motility. Essentially, all surviving cells in the culture are motile.
Localization of Kin5
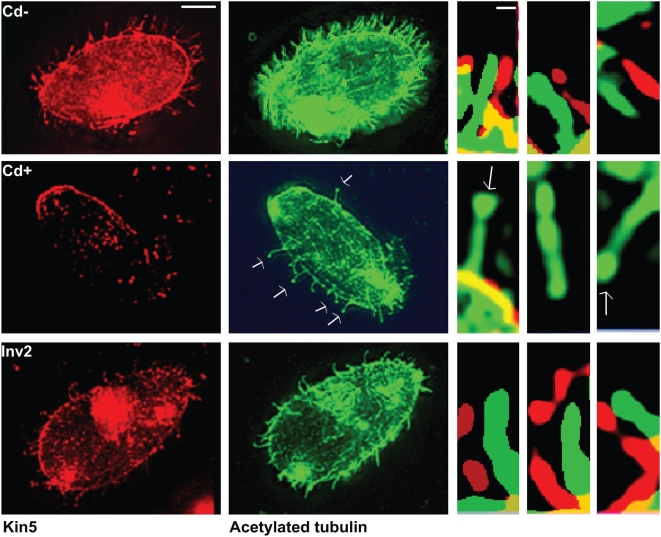
From previous studies [7], Kin5 localized to cilia in a punctate pattern, which is consistent with activity as an intraciliary transport motor. To confirm that loss of KIN5 RNA resulted in loss of Kin5 in cilia, we used a specific Kin5 Ab [7]. Immunofluorescence microscopy confirmed that after 12 h exposure to 0.5 µg/ml Cd2+, Kin5 was greatly reduced in K5KOAs.40 cells, while Kin5 in Inv2 cells was unaffected (Figure 4). The Kin5 signal completely disappeared from cilia upon exposure to CdCl2, although the cilia seemed otherwise unaffected. Some cilia, however, were found to have bulbous tips, as if tip elongation were blocked. In contrast, Kin5 remained in the cilia of Inv2 cells after 12 h in CdCl2 (Figure 6).
Figure 6. Kin5/tubulin immunofluorescence in cells without deciliation.
Top panel: K5KOAs.40 cells with 0 µg/ml Cd2+; middle panel: K5KOAs.40 cells with 0.5 µg/ml Cd2+ (12 h); bottom panel: Inv2 cells with 0.5 µg/ml Cd2+ (12 h). Right panels: Enlarged cilia showing the presence or absence of punctate pattern of Kin5 antibody (red) localization offset from the continuous tubulin localization (green). Scale bars: (left) 10 µm; (right) 1 µm. In K5KOAs.40 cells treated with Cd2+, Kin5 fluorescence is missing in the cell body and the cilia.
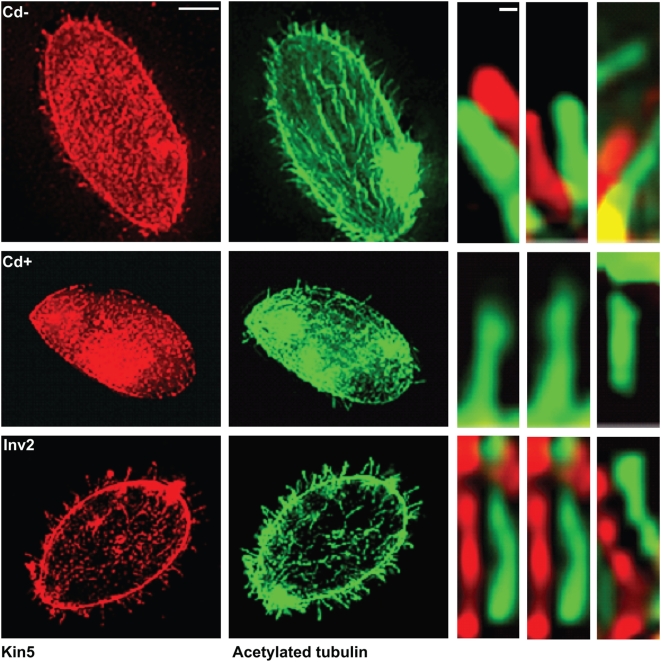
To address whether or not KIN5 knockdown due to RNAi affects axonemal assembly and function, we deciliated the cells just prior to CdCl2 treatment. After deciliation, the cells were initially non-motile. In the absence of CdCl2, the K5KOAs.40 cells were able to regenerate cilia and begin swimming after 2 h. The newly formed cilia contained Kin5. After CdCl2 treatment, the deciliated cells still grew cilia, but fewer and perhaps of shorter length. The cells became motile, but although some cytoplasmic Kin5 staining could be seen, ciliary Kin5 was completely absent. In contrast, the Inv2 cells continued to show Kin5 localization in the cilia after deciliation and CdCl2 treatment (Figure 7).
Figure 7. Kin5/tubulin immunofluorescence in cells after deciliation and ciliary regrowth for 2 h.
Top panel: K5KOAs.40 cells with 0 µg/ml Cd2+; middle panel: K5KOAs.40 cells with 0.5 µg/ml Cd2+ (12 h); bottom panel: Inv2 cells with 0.5 µg/ml Cd2+ (12 h). Right panels: Enlarged cilia showing the presence or absence of punctate pattern of Kin5 antibody (red) localization offset from the continuous tubulin localization (green). Scale bars: (left) 10 µm; (right) 1 µm. After deciliation, Cd2+ treated-K5KOAs.40 cells regrow cilia without Kin5.
Gef1 Transport by Kin5
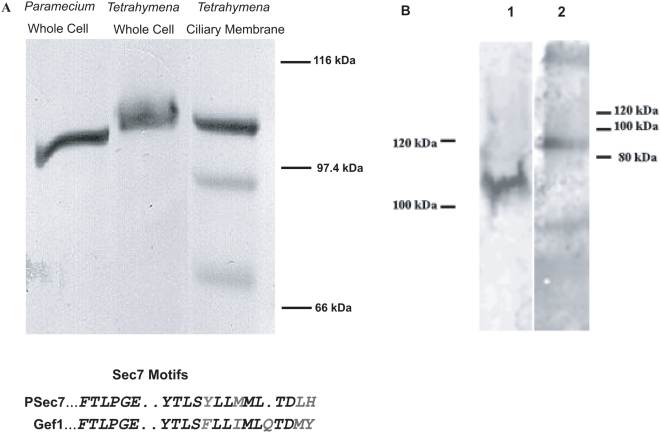
Kin5 has a close phylogenetic relationship to Kif17, a kinesin involved in the transport of NMDA-containing vesicles [11] and of cyclic nucleotide gated channels in mammalian ciliated cells [12] which suggested that Kin5 might also be involved in the transport of membrane proteins or signaling molecules. Gef1 is a Tetrahymena ciliary protein whose orthologue appears to be a guanine nucleotide exchange factor (GEF) of the Sec7 family, cloned in Paramecium. An antibody to the PH domain of the Paramecium protein (PSec7, [20]) recognized the corresponding protein fragment at ca. 100 kDa in Tetrahymena cilia, and therefore identified Gef1 (Figure 8). A pulldown using the Kin5 (K5T1) antibody also pulled down the Gef1 fragment, while in reverse, a pulldown of Gef1 using the PSec7 antibody also pulled down Kin5 (Figure 8), which suggests that Gef1 could be a cargo of Kin5. As a control, when the antibody was omitted, no bands were seen. If Gef1 was one such cargo of Kin5, transport of Gef1 should be disrupted in the K5KOAs.40 knockdown cells. We therefore examined Gef1 localization in KO and Inv2 cells before and after exposure to CdCl2. As anticipated, Gef1 identified by the PSec7 antibody localized along the cilium with Kin5 in the KO and Inv2 cells after 12 h if no Cd2+ was present and also in Inv2 cells when exposed to Cd2+ (Figure 9). Surprisingly, Gef1 was still present in the cilia of K5KOAs.40 cells after exposure to Cd2+, when Kin5 was no longer seen (Figure 9). This might occur if Kin5 transported Gef1 into the cilium and then released it along the ciliary membrane where Gef1 persisted even after ciliary Kin5 depletion. To test this hypothesis, we deciliated the K5KOAs.40 cells and examined the localization of Gef1 as cilia regrew in the absence vs. presence of Cd2+. In the absence of Cd2+, Gef1 and Kin5 both reappeared in the growing cilium, but in the presence of Cd2+, although the cilia regrew, neither Gef1 (Figure 10) nor Kin5 was present (Figure 7). We conclude that Gef1 is a likely Kin5 cargo that can be released from the transport apparatus to remain along the cilium.
Figure 8. Gef1 is a cargo of Kin5.
A. Immunoblot of Gef1. The Gef1 Ab identifies a ca. 100 kDa band (PSec7) from Paramecium, and an ortholog (Gef1) in Tetrahymena cell and ciliary membrane fractions. The Sec7 motif, which delineates a function in guanine nucleotide exchange, is found in both proteins (Grey letters indicate similarity). B. Co-immunoprecipitation of Kin5 and Gef1. 1: Kin5 immunoprecipitate probed with Gef1 Ab. 2: Gef1 immunoprecipitate probed with Kin5 Ab.
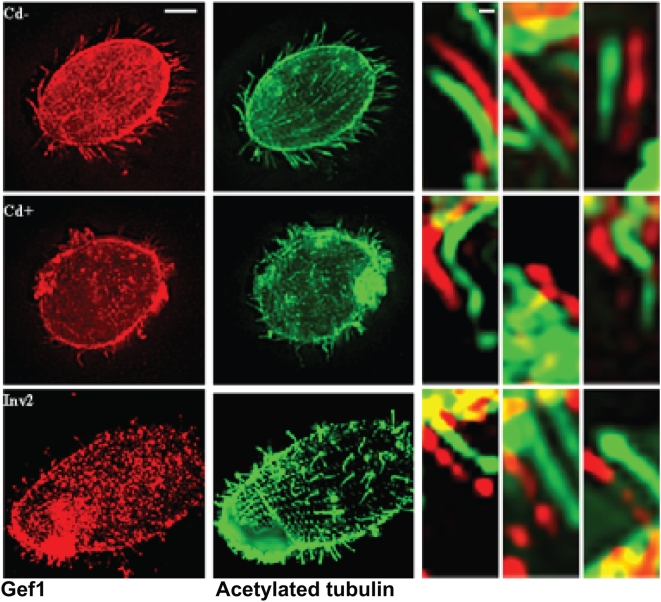
Figure 9. Gef1/tubulin immunofluorescence in cells without deciliation.
Top panel: K5KOAs.40 cells with 0 µg/ml Cd2+; middle panel: K5KOAs.40 cells with 0.5 µg/ml Cd2+ (12 h); bottom panel: Inv2 cells with 0.5 µg/ml Cd2+ (12 h). Right panels: Enlarged cilia showing the presence or absence of punctate pattern of Gef1 antibody (red) localization offset from the continuous tubulin localization (green). Scale bars: (left) 10 µm; (right) 1 µm. In KO cells treated with Cd2+, unlike Kin5, Gef1 remains in the cilia.
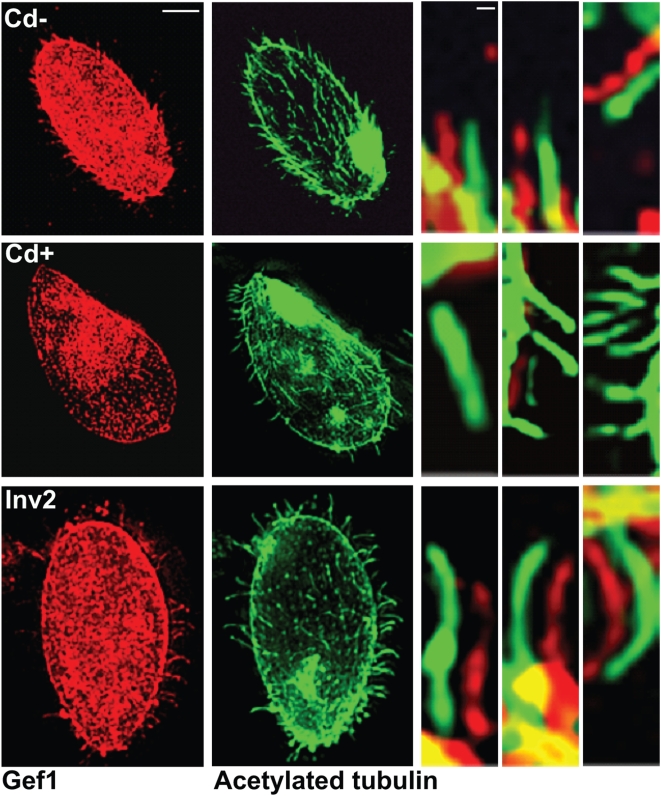
Figure 10. Gef1/tubulin immunofluorescence in cells after deciliation and ciliary regrowth for 2 h.
Top panel: K5KOAs.40 cells with 0 µg/ml Cd2+; middle panel: K5KOAs.40 cells with 0.5 µg/ml Cd2+ (12 h); bottom panel: Inv2 cells with 0.5 µg/ml Cd2+ (12 h). Right panels: Enlarged cilia showing the presence or absence of punctate pattern of Gef1 antibody (red) localization offset from the continuous tubulin localization (green). Scale bars: (left) 10 µm; (right) 1 µm. In control cells, Gef1, like Kin5, returns to the cilia. In KO cells treated with Cd2+, Gef1, like Kin5, does not return to the cilium.
Discussion
RNAi in Tetrahymena
For this study, we developed a method of knockdown of a specific macronuclear transcript in Tetrahymena by constructing an inducible shRNA. We relied on previous methodology [21]. We chose a 19 nt sequence specific to the gene of interest, KIN5, and linked the sense and antisense strands by a short hairpin which would be excised during processing [22]. We placed this behind the metallothionein promoter and used biolistic transformation to insert the construct into the genome. After selection for phenotypic assortment and addition of an inducer (Cd2+), we could demonstrate specific knockdown of the mRNA of interest, while a control transcript was unaffected. The inverse construct from the 19-nt sequence had no effect. This methodology should be applicable to shRNA production for knockdown of other genes of interest in Tetrahymena. A similar conclusion was reached by Howard-Till and Yao [23].
In our experiments, we transformed and selected cells grown to stationary phase in growth medium (proteose peptone), but for shRNA induction, following Nilsson [19], we moved the cells into a simplified buffer that does not support division and yielded cells that were more sensitive to Cd2+. Presumably, under growth conditions, more MTT1 protein will be synthesized to transport Cd2+ out of the cytoplasm. We found that exposure to 5.0 µg/ml Cd2+ produced a knockdown, but was highly toxic, leading to cell death within a few hours, even in control cells. Cells transformed with an RNAi control construct (Inv2) at an order of magnitude lower concentration of Cd2+ (0.5 µg/ml) were unaffected for at least 12 h, suggesting that there was a little Cd2+ toxicity at this time. In contrast, only about 10% of the cells transformed with the KO construct survived, whether survival was measured by number of motile cells or by cell counts, which is consistent with the production of shRNA for KIN5 knockdown by 12 h exposure to 0.5 µg/ml Cd2+. We attribute lethality mainly to the near absence of Kin5 in the cell, rather than to Cd2+ toxicity under these conditions. Lower concentrations of Cd2+ did not induce shRNA for up to 24 h. There is a window of Cd2+ concentration and time of exposure that maximizes shRNA production and minimizes Cd2+ toxicity. For essential proteins, like Kin5, it is important to demonstrate survival in control populations with the Cd2+ conditions chosen. Therefore, we used 0.5 µg/ml Cd2+and 12 h exposure as standard conditions, but flexibility should be possible.
KIN5 knockdown phenotype and Kin5 function in intraciliary transport
The anterograde transport of materials within many cilia and flagella is powered by at least two different motor complexes; a heterotrimeric kinesin-2 and a homodimeric kinesin-2. In Tetrahymena, Kin1 and Kin2 are motors of the heterotrimeric kinesin-2 whose individual knockout has little effect, but whose combined knockout leads to a resorption of cilia [6]. This suggests that, like Chlamydomonas Fla10 and Fla10h [5], the principal cargos of the Kin1/Kin2 motor complex are probably axonemal precursors, such as radial spoke proteins, necessary for the assembly and maintenance of the 9+2 cilium.
Kin5, the presumed homodimeric kinesin-2 characterized previously [7], is similar to Kin1/Kin2 in that the knockdown of this molecule results in its disappearance from cilia and eventual lethality. But, unlike the kin1/kin2 knockout phenotype, the KIN5 knockdown evidently does not cause ciliary resorption. Although forward progression is affected, cilia generally remain motile, until just before cell death. The disappearance implies that Kin5-based transport turns over recurrently to bring new motors, and presumably their cargos, into the cilium. Furthermore, the cargo transported by Kin5 cannot be critical for the maintenance of ciliary structure, except for perhaps at the cilium tip, or for motility, as demonstrated when cells are deciliated after knockdown. When RNAi for KIN5 is induced, even though KIN5 message remains, the cell regrowing the cilia end up with little or no Kin5. These cilia are motile. In Tetrahymena, Kin5 must at least in part be involved in a pathway of intraciliary transport where the cargo must presumably not include the axonemal precursors necessary for the building of the cilium.
In C. elegans, unlike Tetrahymena, the orthologous homodimeric kinesin-2, Osm3, and the heterotrimeric kinesin are redundant in building the cilia-based sensillum body [10], while Osm3 alone is the IFT transporter necessary to build the distal ends [9]. Presumably, membrane receptors are enhanced at the distal ends of invertebrate sensilla. The mammalian homologue of Kin5, Kif17, is endogenously expressed in the ciliary layer of olfactory neurons and in MDCK cell cilia. Coimmunoprecipitation suggests that Kif17 complexes with olfactory cyclic nucleotide-gated channel (CNG) subunits in olfactory epithelium, implying that Kif17 is the motor for CNG intraciliary transport. CNG channels transfected into MDCK cells are transported into their primary cilia [12]. Similar to Tetrahymena, but unlike C. elegans, transfection with dominant negative Kif3a results in complete loss of cilia, whereas transfection with dominant negative Kif17 does not produce ciliary resorption. CNG channels are clustered at the distal ends of olfactory cilia [24], suggesting that like Osm3, Kif17 is particularly important in transporting receptors to the distal ends of the cilia. In this regard, knockdown of KIN5 may produce some ciliary shortening because Kin5 is necessary to build the mature cilia tip. However, this remains to be quantitated and the kinetics of Kin5 synthesis and turnover within the cilium remain to be defined.
Gef1 is a Kin5 cargo
One viable hypothesis is that, like Kif17, and perhaps Osm3, Kin5 transports specific membrane proteins such as guanylyl cyclase [1] into the Tetrahymena cilium. Gef1 could be a cargo of Kin5 that facilitated concentration of targeted membrane receptors. Based on immunolocalization and sequence homology, Gef1 is a PH domain-containing GEF that is itself probably a ciliary membrane peripheral protein. Gef1 and Kin5 coimmunoprecipitate and in untransformed, uninduced or Inv2 cells induced with Cd2+, Gef1 and Kin5 localize together along the cilia. When cells are deciliated in the absence of KIN5 shRNA induction, Gef1 and Kin5 reappear in a similar punctate pattern as new cilia grow. However, when cilia are examined after induction of KIN5 shRNA so that Kin5 is no longer seen, Gef1 is still found along the ciliary membrane. When these cells are deciliated, cilia regrow without Gef1. These results suggest that Gef1 is released from the Kin5 transport apparatus to become resident in the ciliary membrane independent of further transport. Gef1 remains to be fully characterized, but a GEF is also found resident in the connecting cilium of mammalian photoreceptors as well as in ciliated epithelial tissues [25], [26] and in primary cilia [27]. GEFs interact with G-proteins involved in protein import into specialized cellular compartments, for example in nuclear import [28] Similarly, these results might imply that a specific GEF, Gef1 for Tetrahymena, would be localized along the ciliary membrane, where it might operate to facilitate release of other cargo, particularly membrane channels and receptors, imported into the cilium.
Materials and Methods
Plasmid Construction & Biolistic Transformation
20 µg of pMTT1-BICH3 (obtained from Dr. Martin Gorovsky, Univ. of Rochester, NY) was digested with HindIII and BamHI and resolved on a 1% low-melt agarose (Fisher Scientific, Pittsburgh, PA). The ∼6.0 KB vector was gel isolated away from the ∼1.5 KB of IAG48[G1] sequence using the Qiaquick Gel Isolation Kit (Qiagen, Valencia, CA) in a final elution volume of 50 µl. 1 µg of pK5KOAs.40s sense (GATCCCTTGACGCCACAAAAAAGATTCAAGAGATCTTTTTGTGGCGTCAAGTGAA) and 1 µg of pK5KOAs.40a anti-sense (GGAACTGCGGTGTTTTTTCTAAGTTCTCTAGAAAAACACCGCAGTTCACTTTCGA) oligonucleotides were mixed together in a 50 µl volume, heated at 95°C for 2 min, and incubated at 37°C for 2.5 h. A similar reaction was done for the inverse control using pInv2s sense (GATCCAGAAAAAACACCGCAGTTCTTCAAGAGAGAACTGCGGTGTTTTTTCTTGAA) and pInv2a anti-sense (GTCTTTTTTGTGGCGTCAAGAAGTTCTCTCTTGACGCCACAAAAAAGAACTTTCGA) oligonucleotides. 5 µl of the oligonucleotide mix were ligated with 5 µl of pMTT1-BICH3 (HindIII/BamHI digested and gel isolated to remove IAG48[G1]). CaCl2 competent E coli (DH5α) were transformed with the ligation mix. After plasmid minipreps (Qiagen), restriction digests and PCR were used to verify correct insertion.
20 µg of each vector was digested with KpnI/SacII (New England Biolabs) and phenol∶chloroform purified. 4 µl of the digest were mixed with 40 µl of prepared gold particles for biolistic transformation of T. thermophila CU522. After the shootings using the basic biolistic transformation protocol [29] and the Model PDS-1000/He Biolistic Particle Delivery System (Biorad, Hercules, CA), the cells were resuspended in 50 ml 2× proteose peptone (PP) plus 1× anti-mycotic mix (Gibco, Grand Island, NY). After incubation at room temperature for 2 h, paclitaxel (LC Laboratories, Woburn, MA) was added to a concentration of 20 µM. After 3 days at 25°C, a small aliquot was cultured into a 5 ml tube with 2XPP+20 µM paclitaxel. Every 3–4 days, ∼20 µl of cells were recultured into 5 ml 2XPP media containing 20 µM paclitaxel in order to complete phenotypic assortment. After a period of a few months, they were cultured into 2XPP media without any paclitaxel, maintained for at least two weeks and recultured by similar methods to allow for complete phenotypic assortment. They were then inoculated into 2XPP media containing 20 µM paclitaxel and grown overnight. Single-cells were isolated in drops and then transferred to 2 ml 2XPP with 1× anti-mycotic solution and grown to produce small clonal colonies for 3 days to select for cells that had completed phenotypic assortment with respect to the K5KO construct. Integration of the appropriate constructs was verified by PCR using the following primers: BTU1.a: TGGTTTAGCTGACCGATTCAG; MTT1.s: GCTGCTCAAAACATAGCTCATTC. The reaction conditions were performed on the Geneamp PCR System 2400 (Perkin Elmer, Boston, MA) using 1 µl of genomic DNA as follows: 95°C – 2 min; 95°C – 45 sec, 57°C – 1 min, 72°C – 1 min for 35 cycles; 72°C – 10 min; 4°C hold. 5 µl of the reaction products were resolved on a 1% agarose gel. Cells incorporating the RNAi construct were designated K5KOAs.40 (KO cells); cells incorporating the inverse construct were Inv2 cells.
Cell Growth and Induction
KO and Inv2 cells were grown in 15 ml 2XPP for two days, spun down, resuspended in 5 ml starvation media (10 mM Tris, pH 7.5), incubated overnight without shaking, and diluted to 2.0×104 cells/ ml. CdCl2 was added to a final concentration, either in a range from 0 to 0.5 µg/ml or 5.0 µg /ml. Aliquots of motile cells were studied. For ciliary regrowth experiments, deciliation was performed by shear. Cells were routinely examined microscopically for the presence of motility and cilia, and then placed in new 2XPP. Return of motility and beating cilia were monitored for up to 2 h.
Cell Motility & Survival
Cell survival was measured by counting the cells in the motile population by hemocytometer. One to three readings were taken at each time point and a percentage survival calculated by comparison to t = 0. To ensure that the populations were composed of viable cells, motility was measured as the number of cell crossings at a pre-determined line in a one minute-time period. One to four readings were taken at t = 0 and every four hrs up to 24 hrs and calculated as a percentage corresponding to t = 0.
RNA Isolation & RT PCR
Total RNA was isolated from equal number of cells at various time points using an RNeasy mini prep kit (Qiagen). Afterwards, 1 µl was used as template for the One-Step RT PCR kit (Qiagen) using either KIN5 primers (tkin5.40s CCAGCAGCATAAGCTATGG; tkin5.40a ATGAAGACTGTTGCCGCCACC) or PGM1 primers (PGM3s AAAAGGTTAGTGGTTGTTAAGG; PGM3a CTTGTGTAAATCATACTTTATTT) in a total reaction volume of 25 µl. The reaction conditions were performed on the Geneamp PCR System 2400 (Perkin Elmer) as follows: 50°C – 30 min, 95°C – 15 min; 95°C – 45 sec, 57°C – 1 min, 72°C – 1 min for 35 cycles; 72°C – 10 min; 4°C hold. 5 µl of the reactions were resolved on a 1% agarose gel. Within the linear range of product production as described by the kit, the amount of product produced is at least qualitatively proportional to the amount of message, which was confirmed by gel analysis of repeated multiple replications of the PCR experiments.
Immunofluorescence
T. thermophila K5KOAs.40 and Inv2cells were incubated at 0.5 µg/ml CdCl2, spun down at different time points and resuspended in PHEM buffer (50 mM PIPES 50 mM HEPES 1 mM EGTA 2 mM MgS04) and an equal volume of Buffer A (PHEM buffer +4% paraformaldehyde +1% Triton X-100) containing 2 mM AMP-PNP (Sigma-Aldrich). After incubating for 5 min at room temperature, 1/20 volume of 10% Triton X-100 was added for 30 min. The cells were spun down (1100 g for 3 min) and washed 2× with TBST (10 mM Tris-HCl, pH 8.0 150 mM NaCl 0.05% Tween-20). For immunofluorescence microscopy, cells were incubated in the primary antibody for Kin5 [7], acetylated tubulin (Sigma-Aldrich), or Gef1 [20] for 15 min, washed two times in Buffer B (10 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.05% Tween-20, 3% bovine serum albumin, 5 mM CaCl2), incubated with Cy3 or Cy5 secondary antibody (Jackson Lab, West Grove, PA) at 1∶100 dilution for 15 min and washed once with Buffer B. Controls omitted primary antibody incubation. Gef1 is a ciliary protein defined by a rabbit polyclonal antibody (Gef1 Ab) based on a peptide sequence (IQLMGRFDLDEEKDT) from the PH domain of a cloned Paramecium ciliary protein, PSec7 [20] probably a guanine nucleotide exchange factor (GEF1). A Scanalytics EPR deconvolution system (Scanalytics Inc., Fairfax, VA) [30] was employed for 3D reconstruction. Matched images were subjected to pseudocoloring for comparison of localizations. For critical analysis, the gain on the red channel was set to saturate the cell cytoplasm and individual cilia were selected. Colocalization in red and green images of the same cilium was compared by offsetting the images slightly [7].
Western Blotting for Kin5 and Gef1
For western blotting, cilia fractions were prepared by shear or by dibucaine deciliation. After pelleting by centrifugation, membrane/matrix and axoneme fractions were prepared by demembranation in Triton ×100. Ciliary membranes were precipitated from the Triton supernate with 10%TCA and pelleted at 16000 g for 10 min. Normally, 15–20 µl samples derived from diluted packed cells calculated as about 104 Tetrahymena cells or cell equivalents (from an original sample containing 106 cells/ml) were used. Samples were run on a 7.5% SDS-PAGE gel and transferred to a nitrocellulose membrane (250 mA, 4 h) and blocked with blotto (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% TWEEN 20, 2 mM sodium azide, 2% non-fat milk) for one hour. The membrane was incubated overnight in blotto+affinity-purified K5T1 antibody (1∶100) or Gef1 Ab (1∶500). The membrane was washed three times in TBST (10 mM Tris-HCl, pH 8.0, 150 mM NaCl, 0.05% TWEEN 20, 2 mM sodium azide) for 5 min each and incubated with anti-rabbit secondary antibody (1∶4000 in blotto) conjugated with alkaline phosphatase (Sigma-Aldrich). After washing in TBST for 5 min each, the membrane was exposed to NBT/BCIP (KPL, Gaithersburg, MD).
Coimmunoprecipitation
To ensure clean results with protein immunoprecipitation, a mucocyst-less strain of T. thermophila (SB255) was employed. 15 ml of T. thermophila SB255 at stationary phase were spun down (1100 g for 3 min) and resuspended in 1.0 ml lysis buffer (0.15 M NaCl, 1% Triton-X-100, 50 mM Tris). After incubation on ice for 30 min, the tube was spun (13200 RPM for 1 min) in a table-top centrifuge to remove cell debris. 100 µl of K5T1 antibody or Gef1 antibody was added and incubated at 4°C with gentle shaking overnight. 100 µl of Protein-A coated sepharose beads (10% v/v) were added and incubated at 4°C for one h. The tube was centrifuged (13200 RPM for 1 min) to pellet the beads which were washed twice with 350 µl lysis buffer and resuspended in 50 µl lysis buffer. After brief vortexing, 8 µl was used for Western blot with the opposite antibody (K5T1 Ab used for the Gef1 immunoprecipitate and the Gef1 Ab for the K5T1 immunoprecipitate).
Acknowledgments
We would like to thank Martin Gorovsky and his laboratory for the pMTT1-BICH3 construct and Jacek Gaertig and Charles Guerra for initial help with the biolistic transformation procedure. This work was largely completed when Aashir Awan was a student in the Sue Golding Graduate Division and it forms part of his thesis.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: Salary and supply support for A.Awan and A. Bell was provided by University funds while they were graduate students. Both have completed their Ph.D.s and this paper forms part of that work, for which P.Satir has no extramural funding at present. Funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1.Linder JU, Engel P, Reimer A, Krüger T, Plattner H, et al. Guanylyl cyclases with the topology of mammalian adenylyl cyclases and an N-terminal P-type ATPase-like domain in Paramecium, Tetrahymena and Plasmodium. Embo J. 1999;18:4222–4232. doi: 10.1093/emboj/18.15.4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christensen ST, Guerra CF, Awan A, Wheatley DN, Satir P. Insulin receptor-like proteins in Tetrahymena thermophila ciliary membranes. Curr Biol. 2003;13:R50–R52. doi: 10.1016/s0960-9822(02)01425-2. [DOI] [PubMed] [Google Scholar]
- 3.Veland IR, Awan A, Pedersen LB, Yoder BK, Christensen ST. Cilia and Signaling Pathways in Mammalian Development, Health and Disease J. Nephrology. 2009:In Press. doi: 10.1159/000208212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbaum JL, Witman GB. Intraflagellar transport. [Review] Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 5.Piperno G, Mead K, Henderson S. Inner dynein arms but not outer dynein arms require the activity of kinesin homologue protein KHP1 (FLA10) to reach the distal part of flagella in Chlamydomonas. J Cell Biol. 1996;133:371–379. doi: 10.1083/jcb.133.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JM, Marsala C, Kosoy R, Gaertig J. Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol Biol Cell. 1999;10:3081–3096. doi: 10.1091/mbc.10.10.3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Awan A, Bernstein M, Hamasaki T, Satir P. Cloning and characterization of Kin5, a novel Tetrahymena ciliary kinesin II. Cell Motil Cytoskeleton. 2004;58:1–9. doi: 10.1002/cm.10170. [DOI] [PubMed] [Google Scholar]
- 8.Scholey JM. Intraflagellar transport. Annu Rev Cell Dev Biol. 2003;19:423–443. doi: 10.1146/annurev.cellbio.19.111401.091318. [DOI] [PubMed] [Google Scholar]
- 9.Snow JJ, Ou G, Gunnarson AL, Walker MR, Zhou HM, et al. Two anterograde intraflagellar transport motors cooperate to build sensory cilia on C. elegans neurons. Nat Cell Biol. 2004;6:1109–1113. doi: 10.1038/ncb1186. [DOI] [PubMed] [Google Scholar]
- 10.Pan X, Ou G, Civelekoglu-Scholey G, Blacque OE, Endres NF, et al. Mechanism of transport of IFT particles in C. elegans cilia by concerted action of kinesin–II and Osm-3 motors. J Cell Biol. 2006;174:1035–1045. doi: 10.1083/jcb.200606003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Setou M, Nakagawa T, Seog DH, Hirokawa N. Kinesin superfamily motor protein KIF17 and mLin-10 in NMDA receptor-containing vesicle transport. Science. 2000;288:1796–1802. doi: 10.1126/science.288.5472.1796. [DOI] [PubMed] [Google Scholar]
- 12.Jenkins PM, Hurd TW, Zhang L, McEwen DP, Brown RL, et al. Ciliary targeting of olfactory CNG channels requires the CNGB1b subunit and the kinesin-2 motor protein, KIF17. Curr Biol. 2006;16:1211–1216. doi: 10.1016/j.cub.2006.04.034. [DOI] [PubMed] [Google Scholar]
- 13.Cole DG. The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic. 2003;4:435–442. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen LB, Veland IR, Schrøder JM, Christensen ST. Assembly of primary cilia. Developmental Dynamics. 2008;237:1993–2006. doi: 10.1002/dvdy.21521. [DOI] [PubMed] [Google Scholar]
- 15.Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, et al. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaertig J, Thatcher TH, Gu L, Gorovsky MA. Electroporation-mediated replacement of a positively and negatively selectable beta-tubulin gene in Tetrahymena thermophila. Proc Natl Acad Sci USA. 1994;91:4549–4553. doi: 10.1073/pnas.91.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orias E, Flacks M. Macronuclear genetics of Tetrahymena. I. Random distribution of macronuclear genecopies in T. pyriformis, syngen 1. Genetics. 1975;79:187–206. doi: 10.1093/genetics/79.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dondero F, Cavaletto M, Ghezzi AR, La Terza A, Banni M, Viarengo A. Biochemical characterization and quantitative gene expression analysis of the multi-stress inducible metallothionein from Tetrahymena thermophila. Protist. 2004;155:157–168. doi: 10.1078/143446104774199565. [DOI] [PubMed] [Google Scholar]
- 19.Nilsson JR. Tetrahymena in Cytotoxicology: with special reference to effects of heavy metals and selected drugs. Europ J Protistol. 1989;25:2–25. doi: 10.1016/S0932-4739(89)80074-4. [DOI] [PubMed] [Google Scholar]
- 20.Nair S, Guerra C, Satir P. A Sec7-related protein in Paramecium. FASEB J. 1999;13:1249–1257. doi: 10.1096/fasebj.13.10.1249. [DOI] [PubMed] [Google Scholar]
- 21.Shang Y, Song X, Bowen J, Corstanje R, Gao Y, et al. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc Natl Acad Sci U S A. 2002;99:3734–3739. doi: 10.1073/pnas.052016199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paddison PJ, Caudy AA, Sachidanandam R, Hannon GJ. Short hairpin activated gene silencing in mammalian cells. Methods Mol Biol. 2004;265:85–100. doi: 10.1385/1-59259-775-0:085. [DOI] [PubMed] [Google Scholar]
- 23.Howard-Till RA, Yao MC. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol Cell Biol. 2006;26:8731–8742. doi: 10.1128/MCB.01430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flannery RJ, French DA, Kleene SJ. Clustering of cyclic-nucleotide-gated channels in olfactory cilia. Biophys J. 2006;91:179–188. doi: 10.1529/biophysj.105.079046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong DH, Pawlyk B, Sokolov M, Strissel KJ, Yang J, et al. RPGR isoforms in photoreceptor connecting cilia and the transitional zone of motile cilia. Invest Ophthalmol Vis Sci. 2003;44:2413–2421. doi: 10.1167/iovs.02-1206. [DOI] [PubMed] [Google Scholar]
- 26.Panizzi JR, Jessen JR, Drummond IA, Solnica-Krezel L. New functions for a vertebrate Rho guanine nucleotide exchange factor in ciliated epithelia. Development. 2007;134:921–931. doi: 10.1242/dev.02776. [DOI] [PubMed] [Google Scholar]
- 27.Nachury MV, Loktev AV, Zhang Q, Westlake CJ, Peränen J, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 28.Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Curr Biol. 1998;8:1403–1406. doi: 10.1016/s0960-9822(98)00023-2. [DOI] [PubMed] [Google Scholar]
- 29.Hai B, Gaertig J, Gorovsky MA. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 2000;62:513–531. doi: 10.1016/s0091-679x(08)61554-x. [DOI] [PubMed] [Google Scholar]
- 30.Femino AM, Fay FS, Fogarty K, Singer RH. Visualization of single RNA transcripts in situ. Science. 1998;280:585–590. doi: 10.1126/science.280.5363.585. [DOI] [PubMed] [Google Scholar]