Abstract
Seven-transmembrane (G-protein coupled) receptors are key regulators of normal physiology and a large number of diseases, and this family of receptors is the target for almost half of all drugs. Cell culture models suggest that homodimerization and heterodimerization of 7-transmembrane receptors regulate processes including specificity of ligand binding and activation of downstream signaling pathways, making receptor dimerization a critical determinant of receptor biology and a promising new therapeutic target. To monitor receptor dimerization in cell-based assays and living animals, we developed a protein fragment complementation assay based on firefly luciferase to investigate dimerization of chemokine receptors CXCR4 and CXCR7, two 7-transmembrane receptors with central functions in normal development, cancer, and other diseases. Treatment with chemokine ligands and pharmacologic agents produced time- and dose-dependent changes in reporter signal. Chemokines regulated reporter bioluminescence for CXCR4 or CXCR7 homodimers without affecting signals from receptor heterodimers. In a tumor xenograft model of breast cancer, we used bioluminescence imaging to measure changes in receptor homodimerization in response to pharmacologic agents. This technology should be valuable for analyzing function and therapeutic modulation of receptor dimerization in intact cells and living mice.—Luker, K. E., Gupta, M., Luker, G. D. Imaging chemokine receptor dimerization with firefly luciferase complementation.
Keywords: bioluminescence, seven-transmembrane receptors
Recent studies suggest that multiple types of transmembrane receptors exist as homodimeric or heterodimeric complexes, rather than monomers. In particular, receptor dimerization is emerging as a key paradigm for seven-transmembrane (7-TM) receptors (also known as G-protein-coupled receptors), the family of receptors targeted most commonly by clinically approved pharmaceutical agents. Formation of receptor homodimers and heterodimers has been implicated in essentially all aspects of the biology of 7-TM receptors, including intracellular trafficking, ligand binding, pharmacologic inhibition, signal transduction, and internalization (1). However, homodimerization and heterodimerization of 7-TM receptors and the consequences of blocking these complexes remain poorly understand and controversial (2), and very little is known about dynamics of receptor interactions in a living animal.
Beyond the relevance for receptor biochemistry and signaling, complexes of 7-TM receptors also represent promising new therapeutic targets to modulate functions of these molecules in vivo. One relevant example is CXCR4, a 7-TM chemokine receptor with key functions in normal physiology and diseases including cancer, rheumatoid arthritis, and HIV (3). Blocking homodimerization of CXCR4 with a synthetic peptide interrupts downstream signaling pathways and chemotaxis, suggesting a novel approach to inhibit CXCR4 that likely can be applied to other 7-TM receptors (4). To translate this treatment strategy from cell culture assays to animal models and ultimately patients, it will be essential to develop noninvasive imaging assays to quantify changes in receptor dimerization in vivo.
Dimerization of 7-TM receptors has been analyzed with a variety of experimental techniques, including coimmunoprecipitation and resonance energy transfer methods based on fluorescence or bioluminescence (FRET and BRET, respectively). FRET and BRET techniques have the advantage of monitoring receptor complexes in intact cells, allowing effects of biological ligands and pharmacologic agents on receptor conformations to be quantified in real time. Although a limited number of studies have imaged BRET in living mice, autofluorescence may limit detection of green fluorescent protein (GFP) emission from BRET, and the technique has not been used to monitor receptor complexes in vivo (5). Therefore, no imaging reporter system presently exists that can detect dimerization of 7-TM receptors and responses of these complexes to physiological signals or pharmacologic intervention.
To image dynamics of receptor dimers in cell-based assays and living mice, we used a protein fragment complementation assay (PCA) based on firefly luciferase (6). This system was used to analyze homodimers and heterodimers of chemokine receptors CXCR4 and CXCR7. CXCR7 recently has been characterized as a receptor for chemokines CXCL11 and CXCL12, the latter being the cognate ligand for CXCR4 (7). Similar to CXCR4, CXCR7 promotes growth of several different types of cancer in mouse xenografts models, identifying CXCR7 as a potential new diagnostic and therapeutic target in breast, prostate, lung, and other cancers (8, 9). Using the firefly luciferase PCA, we were able to monitor magnitude and kinetics of conformational changes in CXCR4 and CXCR7 dimers in cell-based assays following treatment with physiological ligands or small molecules targeted to each receptor. The reporter system also allowed changes in receptor complexes to be detected and quantified in an orthotopic xenograft model of breast cancer. These results provide new insights into dynamics of CXCR4 and CXCR7 dimerization and establish an imaging reporter strategy to validate and optimize therapeutic agents targeted to receptor complexes.
MATERIALS AND METHODS
DNA constructs
Luciferase complementation plasmids in vector pEF for NLuc-416 and CLuc-398 were provided by Alnawaz Rehemtulla (University of Michigan, Ann Arbor, MI, USA). Mammalian expression and lentiviral vector constructs for human CXCR4 fused to NLuc-416 in plasmids pEF and FUGW, respectively, were described previously (10). To generate CXCR4-CLuc, we used polymerase chain reaction (PCR) to amplify CXCR4 and fuse it to CLuc 398 in pEF (10). Human CXCR7 was amplified by reverse transcriptase-PCR from human breast cancer cell line MCF-7 and inserted into the pEF, replacing CXCR4 in the NLuc and CLuc constructs. Human β2-adrenergic receptor (plasmid template provided by Roger Sunahara, University of Michigan, Ann Arbor, MI, USA) was amplified by PCR and inserted into pEF to produce β2-AR-NLuc-416. For all fusion proteins, the amino acid linker sequence was AAAQISYASRGGGSSGGG. All PCR products were verified by DNA sequencing. Sequences of PCR primers are provided in Supplemental Table 1.
Lentiviral vectors for these reporter constructs were prepared in vectors FUGW or FUPW (10, 11). CXCR4 and CXCR7 fusion proteins were digested with BglII and XbaI and inserted into the blunted PacI site of FUGW (NLuc constructs) or FUPW (CLuc constructs). Complementation reporters were expressed from a human EF-1α promoter.
Mouse CXCL12-α was amplified by PCR from the SDF-degrakine plasmid (gift of Lishan Su, University of North Carolina, Chapel Hill, NC, USA) and inserted into the BamHI and EcoRI sites of pUB6/V5-HisA (Invitrogen, Carlsbad, CA, USA). The ubiquitin promoter and CXCL12 were removed with BglII and EcoRI and inserted into the blunted PacI site of FUGW to create FUGW-CXCL12.
Cells
Human embryonic kidney 293 cells stably expressing large T antigen from SV40 virus (293T) were from Open Biosystems (Huntsville, AL, USA), and human breast cancer cell line MDA-MB-231 and mouse NIH 3T3 cells were purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). All cells were cultured in Dulbecco modified Eagle medium (DMEM; Invitrogen), 10% fetal bovine serum, 1% glutamine, and 0.1% penicillin/streptomycin/gentamicin. Cells were maintained in a 37°C incubator with 5% CO2.
To generate stable cell lines coexpressing various combinations of CXCR4-NLuc, CXCR4-CLuc, CXCR7-NLuc, and CXCR7-CLuc homodimers and heterodimers, we prepared recombinant lentiviruses expressing each of these proteins as described previously (11, 12). The 293T or 231 cells first were transduced with supernatants containing CXCR4-NLuc or CXCR7-CLuc in FUGW. Transduction efficiency was 100% as determined by fluorescence microscopy. These cells then were transduced with supernatants for CXCR4-CLuc or CXCR7-CLuc in FUPW. Again, transduction efficiency for CLuc constructs was 100%, as determined by fluorescence microscopy for mPlum. Batch populations of transduced cells were used for all experiments. 3T3 cells were transduced with FUGW-CXCL12 or FUGW as a control were generated in the same manner.
Live cell imaging
For transient transfections, 293T cells were transfected in 6-well plates with 3.5 μg of each reporter plasmid and 0.25 μg of plasmid for Gaussia luciferase (Promega Corp, Madison, WI, USA). In competition experiments, cells also were cotransfected with an equal amount of unfused CXCR4 or CXCR7 or FUGW vector control. One day after transfection, cells were plated into 96-well plates at a density of 1 × 104 cells/well using a Multidrop 384 dispensing system (Labsystems, Thermo Fisher Scientific, Waltham, MA, USA), and experiments were performed the subsequent day. Gaussia luciferase activity was measured in a parallel set of quadruplicate wells for each transfection condition and used to normalize firefly luciferase reporter signals. Stably transduced cell lines were plated in 96-well plates in the same manner and used for experiments 1 day later. Immediately before each assay, cells were switched to DMEM containing 0.2% bovine serum albumin (Probumin; BD Biosciences, San Jose, CA, USA). Quadruplicate samples were used for all experimental conditions. Cells were treated with various concentrations of CXCL11, CXCL12 (R&D Systems Inc, Minneapolis, MN, USA), AMD3100 (Sigma-Aldrich, St. Louis, MO, USA), TF14013 (gift of Nobutaka Fujii, Kyoto University, Kyoto, Japan), CCX733, or CCX754 (gifts of ChemoCentryx Inc, Mountain View, CA, USA), as described in figure legends. For selected experiments, cells were treated with 0.4 M sucrose in DMEM with 0.2% BSA or 50 μM chloroquine (reagents from Sigma). Incubation times for each agent are listed in figure legends.
Bioluminescence imaging of live cells was performed on a cryogenically cooled camera system (IVIS 100; Caliper Life Sciences, Mountain View, CA, USA) as described previously (6), using 0.5- to 5-min acquisition times, high sensitivity, and field of view B. After imaging, total protein per well was assayed by sulforhodamine B staining as described previously (13). Bioluminescence imaging data were normalized to total protein, and data were expressed as fold increase relative to untreated controls in each experiment.
Western blots
Membrane preparations from stably transduced cell lines were prepared as described previously (14). Lysates were probed with 1:500 dilutions of antibodies to firefly luciferase (Promega). Primary antibodies were detected with a 1:2000 dilution of secondary anti-goat antibody conjugated with horseradish peroxidase. Blots were developed with enhanced chemiluminescence (ECL) reagent (Amersham Biosciences Corp., Piscataway, NJ, USA).
ELISA
CXCL12 levels in supernatants of 3T3-CXCL12 and 3T3-GFP cells after 4 h of incubation were quantified by ELISA according to the manufacturer’s directions (Quantikine; R&D Systems).
In vivo bioluminescence imaging
All animal procedures were approved by the University of Michigan Committee for use and care of animals. Type 231 cells stably expressing CXCR4-NLuc and CXCR4-CLuc or CXCR7-NLuc and CXCR7-CLuc were injected bilaterally into the fourth inguinal mammary fat pads of 6- to 8-wk-old female nude mice (Taconic, Hudson, NY, USA) (2×106 breast cancer cells/fat pad) as described previously (12). Cells were coimplanted with 1 × 106 3T3 cells stably expressing CXCL12 and GFP or GFP alone. To regulate receptor dimerization, mice were injected intraperitoneally with 5 mg/kg AMD310 or subcutaneously with 100 mg/kg CCX754 or matched vehicle control. Imaging was performed at times before and after injection, as listed in figure legends.
Bioluminescence imaging was performed on an IVIS Spectrum system (Caliper) as described previously (15). Images were obtained for 5 min on high sensitivity and FOV C. Imaging data were quantified as photon flux using Living Image software (Caliper). Data for each tumor were normalized to photon flux prior to treatment to account for differences in tumor sizes in various mice.
Statistics
Data for photon flux or fold induction of bioluminescence were plotted as mean values with se. Pairs of data were analyzed by two-tailed paired t test to determine statistically significant differences (P<0.05) (GraphPad Prism; GraphPad, San Diego, CA, USA).
RESULTS
Firefly luciferase PCA for monitoring complexes of CXCR4 and CXCR7
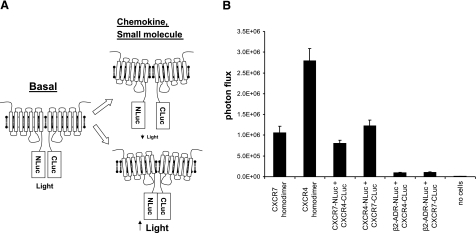
We previously have developed a PCA based on N- and C-terminal fragments of firefly luciferase (NLuc and CLuc, respectively) that have minimal background association and high reconstitution of bioluminescence when brought together by interacting proteins (6). To develop an imaging assay for receptor dimerization, we fused the intracellular C termini of CXCR4 or CXCR7 to either NLuc or CLuc, respectively. Changes in conformation of chemokine receptors in homodimeric or heterodimeric complexes will alter the efficiency of luciferase complementation, providing a quantitative measurement of receptor dynamics (Fig. 1A). Chemokine receptors remain functional when fused to luciferase fragments (10), so the reporter molecules will allow imaging of receptor complexes during physiological signaling.
Figure 1.
Firefly luciferase protein fragment complementation reporter for chemokine receptor dimers. A) Schematic diagram of PCA. NLuc and CLuc fragments fused to intracellular C termini of chemokine receptors associate to produce light under basal conditions. Chemokine ligands or small molecules alter association of luciferase fragments, depicted as conformational changes in receptor pairs, changing the bioluminescent signal for cell-based and in vivo imaging. B) Type 293T cells were transiently transfected with indicated complementation plasmids, cultured in medium with 10% serum, and assayed 48 h after transfection. Reporter bioluminescence was quantified in intact cells using a CCD-camera device and normalized to Gaussia luciferase activity as a transfection control (n=4/condition).
As an initial test of the assay system, we transiently transfected 293T cells with pairs of NLuc and CLuc fusion receptors so that cells coexpressed CXCR4 and CXCR7 homodimers or both orientations of CXCR4 and CXCR7 heterodimers. To determine specificity of receptor interactions, we cotransfected cells with CXCR4- or CXCR7-CLuc fusion proteins and β2-adrenergic receptor-NLuc. β2-Adrenergic receptor is a well-characterized 7-TM receptor with no known interactions with either CXCR4 or CXCR7. We also transfected cells with each fusion protein alone to control for bioluminescence from individual luciferase fragments.
Under baseline conditions, bioluminescence was detected readily from all combinations of CXCR4 and CXCR7 homodimers and heterodimers. Reporter signal was greatest from CXCR4 homodimers, with lesser amounts produced by CXCR7 homodimers and both NLuc and CLuc combinations of CXCR4 and CXCR7 heterodimers (Fig. 1B). The heterodimer pair of CXCR4-NLuc/CXCR7-CLuc produced significantly greater bioluminescence than the reverse orientation of CXCR4-CLuc/CXCR7-NLuc (P<0.05). Previous BRET studies reported that reciprocal changes in reporter proteins from one 7-TM receptor to another altered the magnitude of signal produced by heterodimers, indicating that BRET primarily detects changes in conformation between preformed receptor complexes (16). Similarly, differences in bioluminescence produced by reciprocal combinations of CXCR4 and CXCR7 fusion proteins suggest the firefly luciferase PCA also reports changes in conformation between receptors. Our data are consistent with prior research showing constitutive CXCR4 homodimers and CXCR4-CXCR7 heterodimers (4, 17,18,19), and the results indicate that CXCR7 also exists as homodimers. All combinations of CXCR4 and CXCR7 complexes had >10-fold more bioluminescence than heterodimeric complexes formed with β2-adrenergic receptor-NLuc, showing specificity of the PCA for detecting chemokine receptor complexes. Cells transfected with single NLuc or CLuc constructs had no detectable bioluminescence, verifying the requirement for both luciferase fragments to reconstitute enzyme activity (data not shown).
Ligand- and time-dependent changes in firefly luciferase PCA
Previous studies report conflicting data for ligand-dependent regulation of CXCR4 homodimers (16, 17, 19), and effects of chemokine ligands on CXCR7 homodimers have not been investigated. Chemokine CXCL11 binds to CXCR7, while CXCL12 binds to both CXCR4 and CXCR7 (7, 20). Having established feasibility of the assay technique, we generated stable populations of 293T cells for further biochemical analysis in cell-based assays. Cell surface expression of CXCR4 and CXCR7 reporter constructs was verified by flow cytometry (data not shown).
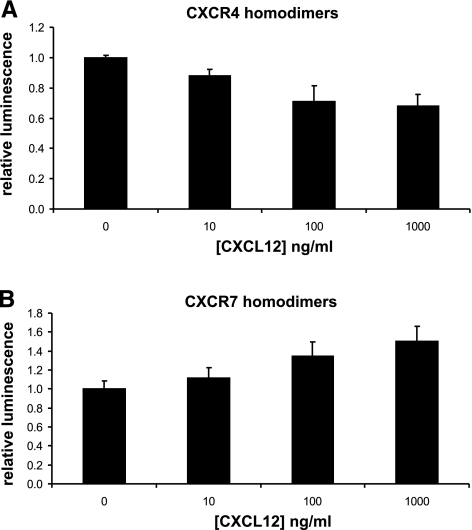
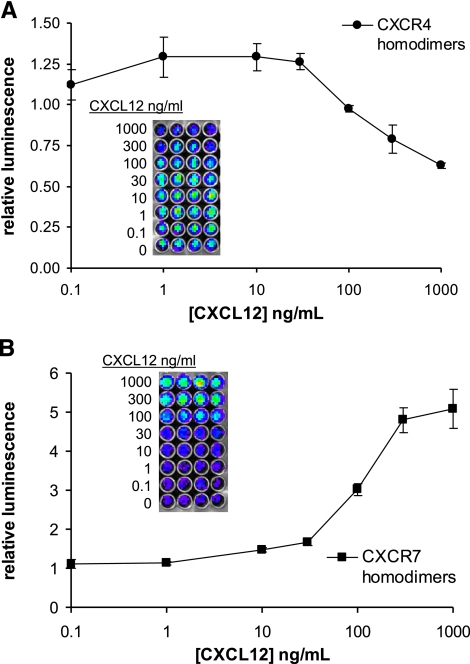
We treated stable 293T CXCR4 or CXCR7 homodimer reporter cells with increasing concentrations of CXCL12 and quantified bioluminescence immediately after adding chemokine. CXCL12 produced immediate dose-dependent effects on reporter bioluminescence for both CXCR4 and CXCR7 homodimers, although the direction of change differed for each chemokine receptor pair (Fig. 2). For CXCR4, incubation with CXCL12 decreased the magnitude of luciferase activity relative to untreated cells. Incubation with 10 ng/ml CXCL12 significantly reduced bioluminescence from CXCR4 homodimerization (P<0.05), and 1000 ng/ml decreased reporter activity by >30%. By comparison, CXCL12 increased bioluminescence from CXCR7 homodimers with significant effects at 100 ng/ml (P<0.05) and a peak increase of 50% at 1000 ng/ml chemokine. These data show immediate responses of the homodimerization PCA to chemokine ligands. Opposite responses of CXCR4 and CXCR7 homodimers to either decrease or increase bioluminescence, respectively, suggest that chemokines elicit distinct conformational changes in these receptors.
Figure 2.
Immediate effects of chemokine ligands on CXCR4 and CXCR7 homodimers. Type 293T cells expressing firefly luciferase PCA reporters for CXCR4 (A) or CXCR7 (B) homodimers were treated with increasing concentrations of CXCL12, and bioluminescence was quantified immediately after adding chemokine. Data were normalized to bioluminescence in control cells not incubated with CXCL12. Graphed values are means ± se (n=4) representative of 2 independent experiments.
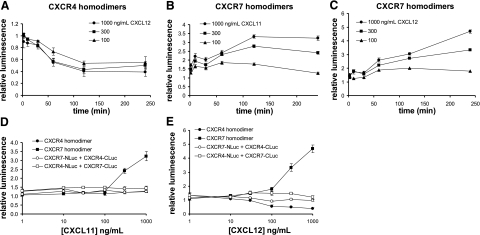
To further investigate responses of CXCR4 and CXCR7 homodimers to chemokines, we quantified changes in reporter bioluminescence following extended incubation with CXCL11 or CXCL12. We incubated stable 293T reporter cells with various concentrations of CXCL11 or CXCL12 for up to 4 h. For CXCR4, extended incubation with CXCL12 decreased complementation signals by up to 60% (Fig. 3A). The magnitude of this effect increased over time through 2 h and reached steady state levels for each concentration of chemokine, suggesting that prolonged exposure to ligand increases the extent of conformational changes in homodimeric complexes and/or affects more receptors. Treatment with CXCL11 did not alter bioluminescence from the CXCR4 homodimer reporter (Fig. 3D), showing a specific effect of CXCL12 on CXCR4 receptor complexes.
Figure 3.
Time and dose-dependent effects of chemokine ligands. A–C) Type 293T cells stably expressing firefly luciferase reporters for CXCR4 homodimers (A) or CXCR7 homodimers (B, C) were treated with increasing concentrations of indicated chemokine ligand through 4 h. Reporter activity in intact cells was quantified at various times, and data were normalized to bioluminescence at t = 0 in untreated cells. Data are means ± se. D, E) Type 293T cells expressing CXCR4 and CXCR7 homodimer and heterodimer reporters were treated for 4 h with increasing concentrations of CXCL11 (D) or CXCL12 (E) before quantifying luciferase activity in living cells (n=4/condition). Data were normalized to values at t = 0 for untreated cells; values are means ± se. Presented results are representative of 2 or more independent experiments.
Prolonged incubation with chemokines CXCL11 or CXCL12 also produced time- and dose-dependent changes in bioluminescence from CXCR7 homodimers (Fig. 3B, C). CXCL12 elicited relatively greater effects than CXCL11 on conformational changes and bioluminescence, with ≈5- and 3.5-fold induction over untreated control cells, respectively, at 1000 ng/ml. For CXCL11, changes in reporter bioluminescence from CXCR7 homodimerization peaked at 2 h, then remained steady or declined minimally, while incubation with 300 or 1000 ng/ml CXCL12 continued to increase luciferase activity through at least 4 h. These data demonstrate that changes in PCA signal (increase or decrease) are not determined solely by efficacy of chemokines but also are affected by relative differences in positions of luciferase fragments within the receptor dimer, similar to BRET or FRET (16).
We also investigated regulation of CXCR4 and CXCR7 heterodimers by incubating 293T reporter cells with increasing concentrations of CXCL11 or CXCL12 for 4 h. Neither CXCL11 nor CXCL12 altered the magnitude of reporter bioluminescence from heterodimers, regardless of positions of NLuc or CLuc fusions to CXCR4 or CXCR7 (Fig. 3D, E). No changes in bioluminescence were evident at shorter incubation times (immediate through 2 h), excluding the possibility that effects of chemokines were transient (data not shown). Therefore, CXCR4 and CXCR7 form heterodimers that are not regulated by ligands for either receptor. These results are consistent with a previous study showing no effect of CXCL12 on coimmunoprecipitation of CXCR4 with CXCR7 (18).
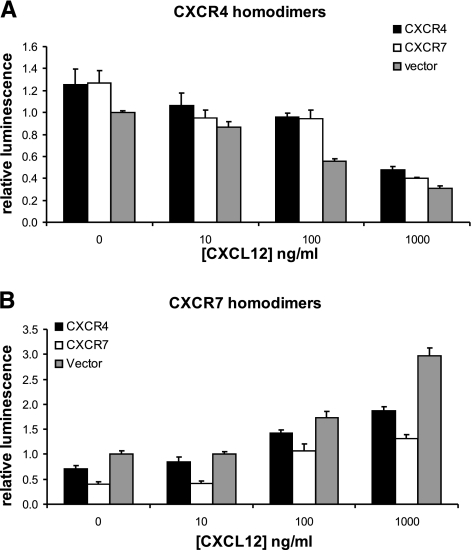
To further validate specificity of the assay, we performed competition experiments by cotransfecting 293T cells with equal amounts of CXCR4 or CXCR7 homodimer reporters and unfused CXCR4 or CXCR7. Forty-eight hours after transfection, cells were treated with 100 ng/ml CXCL12 for 2 h prior to assaying reporter bioluminescence. For the CXCR4 homodimer PCA, unfused CXCR4 or CXCR7 produced comparable (≈25%) increases in luciferase activity under baseline conditions as compared with vector control (P<0.05) (Fig. 4A). The increase in reporter signal caused by unfused competitors is consistent these receptor molecules altering basal conformation of CXCR4 homodimers, producing the opposite response to incubation with chemokine ligand. Treatment with CXCL12 reduced bioluminescence from CXCR4 homodimerization proportionately for cells without or with added receptors lacking a luciferase fragment. In cells transfected with the CXCR7 homodimerization PCA, unfused CXCR4 and CXCR7 reduced baseline bioluminescence by ≈30 and 60%, respectively (P<0.05 and P<0.01) (Fig. 4B). Incubation with CXCL12 produced comparable increases in luciferase activity from CXCR7 homodimerization while maintaining the same relative differences in bioluminescence observed under basal conditions. Collectively, these data confirm specificity of the PCA imaging assay and indicate that CXCR7 is a more effective competitor for CXCR4 in homodimeric complexes than CXCR4 is for CXCR7 homodimers.
Figure 4.
Competition with unfused receptors. Type 293T cells were transfected with equal amounts of plasmids for CXCR4 (A) or CXCR7 PCA homodimers (B) and unfused CXCR4 or CXCR7 or vector control. At 48 h after transfection, cells were treated for 2 h with increasing concentrations of CXCL12 before quantifying bioluminescence. Data were normalized to bioluminescence in cells transfected with vector control; values are means ± se (n=4). Results are representative of 2 independent experiments.
CXCR4 and CXCR7 dimerization reporters function in breast cancer cells
CXCR4 and CXCR7 promote growth of several different types of cancer cells, including breast, lung, and prostate, in mouse models of cancer (8, 9, 12, 21, 22). To demonstrate that the dimerization PCA functions in other cell types and establish a system for in vivo studies, we used human breast cancer cell line MDA-MB-231, a common model for primary and metastatic breast cancer. We generated stable cell lines expressing CXCR4 or CXCR7 homodimerization reporters or both combinations of CXCR4 and CXCR7 heterodimer reporters. These reporter cell lines show comparable responses to chemokines as 293T dimerization reporter cells (Fig. 5 and data not shown). These data show that the dimerization PCA has comparable responses in more than one cell type, indicating that this reporter technology can be used generally to analyze chemokine receptor complexes in a variety of different models.
Figure 5.
CXCR4 and CXCR7 homodimerization reporters in human breast cancer cells. MDA-MB-231 human breast cancer cells stably transduced with CXCR4 (A) or CXCR7 (B) homodimerization reporters were treated with increasing concentrations of CXCL12 for 4 h prior to assaying luciferase activity (n=4/condition, representative of 2 independent experiments). Data were normalized to luciferase activity in control cells not treated with CXCL12. Graphed values are means ± se. Insets show images of 96-well plates used for plotted data.
We used these cells to test reversibility of signal from the dimerization PCA following incubation with a chemokine ligand. We incubated 231 CXCR4 homodimerization cells with increasing concentrations of CXCL12 for 15 min, washed the cells to remove the ligand, and then measured bioluminescence after 30 min of incubation in chemokine-free medium (Fig. 6). Treatment with CXCL12 produced the expected dose-dependent decrease in luciferase activity from CXCR4 homodimers. Cells treated with either 10 or 100 ng/ml CXCL12 before removal of the chemokine recovered luciferase activity within 30 min to levels that slightly exceeded baseline bioluminescence, while signal reversal was more modest in cells treated with 1000 ng/ml CXCL12. Recovery of CXCR4 homodimerization signal to levels slightly greater than baseline is consistent with our prior work showing that treatment with low-to-modest levels of CXCL12 enhances subsequent signaling, potentially by recruiting new receptors to the cell surface (10, 23). These results establish reversibility of the dimerization PCA, particularly following stimulation with lower concentrations of chemokine ligand.
Figure 6.
Reversal of chemokine-dependent conformational changes in CXCR4 homodimers. Type 231 cells stably expressing CXCR4 homodimer reporters were treated for 15 min with various concentrations of CXCL12 before quantifying bioluminescence (no washout). Parallel wells of cells were treated identically with CXCL12, washed with PBS, and then incubated for 30 min without chemokine (30 min washout). Data were normalized to bioluminescence in control cells not treated with chemokine (NT) and graphed as means ± se (n=4/condition, representative of two independent experiments).
Regulation of conformational changes in CXCR4 and CXCR7 homodimers
In response to ligand binding, most 7-TM receptors internalize via clathrin-coated pits and then enter either recycling endosomes or acidified late endosomal and lysosomal compartments (24). While chemokines produce immediate changes in luciferase activity from CXCR4 and CXCR7 homodimers, our data show that the magnitude of change in bioluminescence increases over time. These results suggest that conformational changes in receptor complexes may be regulated by receptor trafficking after ligand binding.
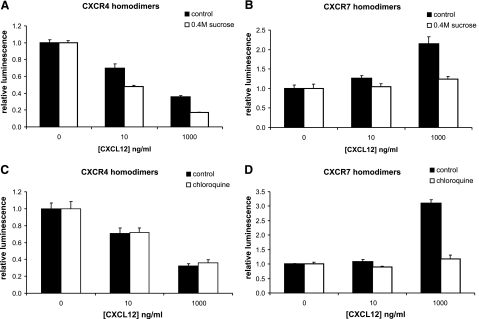
To investigate effects of selected steps in receptor trafficking on CXCR4 and CXCR7 homodimerization, we first performed experiments using hypertonic sucrose, which blocks clathrin-coated endocytosis (25). Stable MDA-MB-231 cell lines expressing either CXCR4 or CXCR7 homodimerization reporters were pretreated with 0.4 M sucrose for 30 min and then incubated with increasing concentrations of CXCL12 for 2 h. For CXCR4, treatment with hypertonic sucrose significantly enhanced the effect of CXCL12 to reduce bioluminescence (P<0.05) (Fig. 7A). Incubation with 1000 ng/ml CXCL12 reduced luciferase activity by ≈60% relative to untreated cells, while addition of sucrose to the incubation medium decreased the reporter signal by ≈80%. By comparison, hypertonic sucrose blocked the increase in bioluminescence produced by prolonged incubation of CXCR7 homodimers with CXCL12 (P<0.05 at 1000 ng/ml) (Fig. 7B). These data imply that conformational changes in CXCR4 homodimers are reduced by clathrin-dependent endocytosis, while conformational changes in CXCR7 homodimers are promoted by this pathway.
Figure 7.
Effects of endocytosis and acidified intracellular compartments on conformational changes. Type 231 CXCR4 (A, C) or CXCR7 (B, D) homodimer reporter cells were pretreated with medium containing 0.4 M sucrose or medium alone (A, B) for 30 min prior to 2 h of incubation with CXCL12 added to the pretreatment medium. Cells in panels C and D were incubated with CXCL12 in medium containing 50 μM chloroquine or vehicle control for 2 h before quantifying bioluminescence. Data were normalized to luciferase activity in cells not treated with CXCL12; graphed values are means ± se (n=4/condition, representative of two independent experiments).
We also analyzed functions of late endosomes and lysosomes on receptor complexes, using chloroquine or ammonium chloride to inhibit acidification of these compartments. Type 231 reporter cells were treated with low or high concentrations of CXCL12 for 2 h in the presence or absence of these pharmacologic agents. Neither chloroquine nor ammonium chloride altered the reduction in CXCR4 homodimerization reporter activity in response to CXCL12 (Fig. 7C). However, acidified intracellular compartments were necessary for CXCL12 to elicit conformational changes in CXCR7 homodimers during extended periods of incubation (Fig. 7D). Treatment with chloroquine or ammonium chloride almost completely abrogated effects of CXCL12 to increase luciferase activity from CXCR7 homodimers (P<0.05). Together with results using hypertonic sucrose, these data show that internalization and trafficking through acidified intracellular compartments promote conformational changes in CXCR7, but not CXCR4, homodimers.
Effects of specific inhibitors on receptor complexes
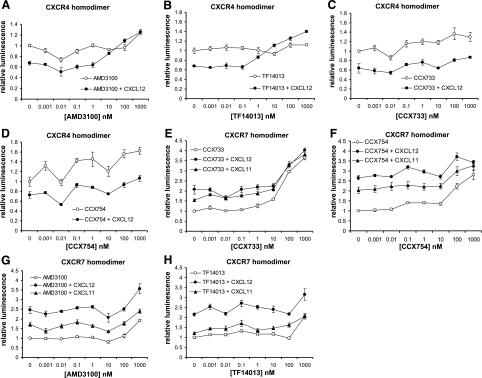
To further investigate the firefly luciferase PCA for monitoring conformational changes in receptor complexes, we used compounds that selectively block binding of CXCL12 to CXCR4 (AMD3100, TF14013) or CXCL11 and CXCL12 binding to CXCR7 (CCX733, CCX754) (7, 26, 27). We incubated 293T reporter cells with increasing concentrations of inhibitor alone for 30 min and then treated cells with a modest concentration of chemokine (100 ng/ml) for 2 h before quantifying bioluminescence. For CXCR4 homodimers, CXCL12 alone produced the expected decrease in reporter signal, and this effect was blocked by both CXCR4 inhibitors (Fig. 8A, B). Our EC50 values of ≈10 and 30 nM for TF14013 and AMD3100, respectively, are comparable to those reported from a BRET study of CXCR4 homodimerization (16). Treatment with 10 nM AMD3100 modestly activated the CXCR4 reporter without and with CXCL12. Potentially, this effect may be due to partial agonist effects of AMD3100 in some experimental systems, although such effects typically are seen at higher concentrations of compound (28). While BRET has shown opposite effects of CXCL12 and inverse agonists structurally related to TF14013 on CXCR4 homodimerization (16), TF14013 alone did not alter bioluminescence for CXCR4 complementation. However, the highest concentration of TF14013 combined with CXCL12 increased reporter signal above that for CXCR4 homodimers alone (P<0.05), suggesting allosteric regulation of ligand-regulated conformational changes in CXCR4 homodimerization (29). CXCL12-dependent reductions in CXCR4 homodimerization signal were not affected significantly by either CXCR7 inhibitor, although higher concentrations of CCX754 and to a lesser extent CCX733 produced modest increases in bioluminescence (Fig. 8C, D).
Figure 8.
Pharmacologic modulation of chemokine receptor dimerization. Stably transduced CXCR4 homodimer (A–D) or CXCR7 homodimer (E–H) reporter cells were incubated with vehicle control or increasing concentrations of CXCR4 inhibitor (AMD3100, TF14013) or CXCR7 inhibitor (CCX733, CCX754) for 2 h; 100 ng/ml CXCL12 (A–H), CXCL11 (E–H), or vehicle control then was added to cells for an additional 2 h in continued presence of inhibitor. Bioluminescence was quantified in living cells (n=4/condition). Data were normalized to control cells untreated with either inhibitor or chemokine and expressed as fold increase relative to control. Symbols represent means ± se. Data are representative of two independent experiments.
None of the tested inhibitors blocked CXCL11- or CXCL12-dependent increases in bioluminescence from CXCR7 homodimerization reporters. However, treatment with CXCR7 inhibitors alone enhanced peak reporter activity for CXCR7 homodimerization by ≈2.5- and 3.5-fold for CCX754 and CCX733, respectively, and significant increases (P<0.05) first were detected at 100 and 10 nM, respectively (Fig. 8E, F). Therefore, while these agents block binding of CXCL11 and CXCL12 to CXCR7, the compounds function like chemokine ligands with respect to CXCR7 homodimerization. We also observed that treatment with 1 μM of either CXCR4 inhibitor increased bioluminescence for CXCR7 homodimerization, indicating a loss of receptor selectivity at concentrations substantially above the EC50 (Fig. 8G, H). None of the tested compounds altered reporter bioluminescence for CXCR4-CXCR7 heterodimers (data not shown). Overall, these data establish that pharmacologic regulation of receptor complexes can be quantified in living cells with the firefly luciferase PCA.
Imaging receptor complexes in vivo
We used an orthotopic xenograft model of breast cancer to investigate the PCA for imaging complexes of CXCR4 and CXCR7 in vivo. We implanted mice with 231 cells stably expressing CXCR4 and CXCR7 homodimer reporters or CXCR4-CXCR7 heterodimers. For experiments with CXCR4 homodimers and heterodimers of CXCR4 and CXCR7, we also coimplanted murine fibroblasts stably expressing CXCL12 to reproduce secretion of this chemokine by stromal cells in the microenvironment of human breast cancer and stimulate conformational changes in CXCR4 complexes (30) (Supplemental Fig. 1).
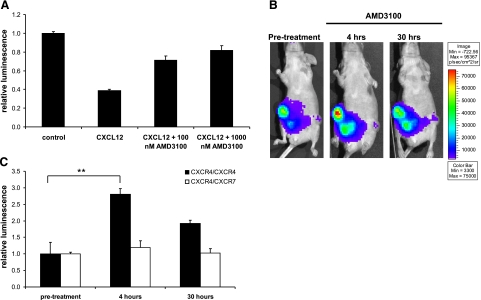
In cultured cells, extended incubation with CXCL12 reduced bioluminescence from the CXCR4 homodimer reporter, and the effect of CXCL12 on receptor complexes was inhibited by AMD3100. We hypothesized that CXCL12-producing fibroblasts in the tumor microenvironment would elicit a similar reduction in CXCR4 homodimer signal that could be increased by treating mice with AMD3100. As evidence supporting this model, we first tested the hypothesis in cell-based assays. We treated 231 CXCR4 homodimer PCA cells with CXCL12 and then added AMD3100 to medium containing this chemokine. Cells incubated with CXCL12 alone showed the expected reduction in bioluminescence from the CXCR4 homodimerization reporter (Fig. 9A). By comparison, treatment for 4 h with AMD3100 produced dose-dependent increases in luciferase activity (P<0.01), showing that AMD3100 can reverse conformational changes elicited by CXCL12.
Figure 9.
In vivo imaging of CXCR4 receptor dimers. A) Type 231 cells stably transduced with CXCR4 dimerization reporters were incubated in cell culture with 1 μg/ml CXCL12 for 15 min. AMD3100 or vehicle control then was added to CXCL12-containing medium for 4 h before quantifying bioluminescence in living cells (n=4/condition). Data are presented as mean ± se bioluminescence relative to values measured in cells not treated with CXCL12 or AMD3100 (representative of 2 independent experiments). B, C) Type 231 CXCR4 homodimer or CXCR4-CXCR7 heterodimer cells were coimplanted with CXCL12-producing fibroblasts into mammary fat pads of nude mice. B) Representative bioluminescence images of the same mouse prior to treatment with 7.5 mg/kg AMD3100 i.p. and then 4 and 30 h later. CXCR4 homodimer tumors are on the right side of the mouse; CXCR4-CXCR7 heterodimers are on the left. Bioluminescence is shown as a pseudocolor scale, with red the highest and blue the lowest levels of photons. C) Quantified data from in vivo imaging experiments for CXCR4 and CXCR4-CXCR7 tumors (n=4/tumor type). **P < 0.01.
To extend this model to animal studies, we obtained baseline bioluminescence images of mice implanted with tumor xenografts of CXCR4 homodimer or CXCR4-CXCR7 heterodimer reporter cells. Mice then were treated with a single dose of AMD3100 and imaged to quantify effects on dimerization. Four hours after administering AMD3100, bioluminescence from CXCR4 homodimers increased by ≈2.5-fold above pretreatment levels (P<0.01) (Fig. 9B, C). Bioluminescence from CXCR4 homodimer tumors subsequently decreased by 30 h. By comparison, AMD3100 had no effect on bioluminescence from CXCR4-CXCR7 heterodimers at either time point, showing specificity for CXCR4 homodimers. These data demonstrate that the CXCR4 homodimerization PCA can quantify in vivo inhibition of CXCL12-mediated conformational changes in receptor complexes.
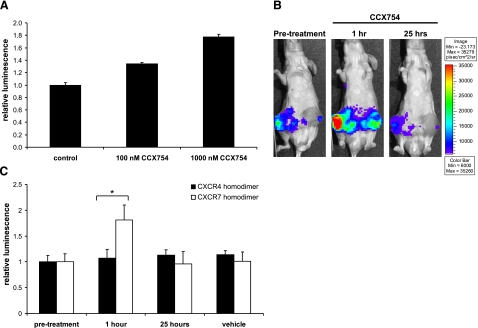
To investigate pharmacologic regulation of CXCR7 complexes in vivo, we used the CXCR7-specific compound CCX754. In cell-based assays, treatment with this compound significantly increased CXCR7 homodimerization in 231 reporter cells (Fig. 10A). We implanted a separate cohort of mouse 231 CXCR7 or CXCR4 homodimer reporter cells. Because cell culture studies revealed that CCX754 increased luciferase complementation, we coimplanted these tumors with fibroblasts transduced only with GFP. We imaged tumor-bearing mice before and after treatment with the CXCR7 modulator CCX754. Bioluminescence from CXCR7 homodimerization increased significantly by 1 h after treatment and returned to baseline levels the following day, demonstrating in vivo regulation of CXCR7 complexes (Fig. 10B, C). Treatment with CCX754 did not alter bioluminescence from CXCR4 homodimers, and neither CXCR4 nor CXCR7 reporter signals were affected by treatment with vehicle control. Collectively, these data establish the firefly luciferase PCA for monitoring specific pharmacologic regulation of chemokine receptor complexes in living animals.
Figure 10.
In vivo modulation of CXCR7 homodimers. A) CXCR7 homodimerization cells were treated in cell culture with CCX754 or vehicle control for 1 h prior to quantifying luciferase activity. Data were normalized to bioluminescence in control cells; values are means ± se (n=4, representative of two separate experiments). B, C) Female nude mice were implanted with mammary fat pad xenografts of 231 CXCR7 or CXCR4 homodimer cells and fibroblasts expressing GFP. B) Representative bioluminescence images of the same mouse bearing bilateral CXCR7 homodimer tumors prior to subcutaneous injection of 100 mg/kg CCX754 and then 1 and 25 h later. Pseudocolor scale shows highest and lowest levels of bioluminescence as red and blue, respectively. Note that this scale encompasses a smaller range of photon flux values than that shown in Fig. 9. C) Quantified bioluminescence imaging data from mice bearing CXCR7 or CXCR4 homodimer tumors (n=4 each). Data are normalized to bioluminescence in the same tumor prior to treatment to account for differences in tumor sizes. Graphed values are means ± se. *P < 0.05.
DISCUSSION
Previous studies have used resonance energy transfer techniques (FRET, BRET) and coimmunoprecipitation to analyze 7-TM receptor complexes in living cells. These studies suggest that receptor dimers are the functional units of 7-TM receptors with heterodimeric complexes modifying ligand binding, allosteric regulation of receptors, and activation of downstream signaling pathways. These cell culture studies indicate that dynamics of receptor homodimerization and heterodimerization are critical for normal and pathological functions of 7-TM receptors and point to receptor complexes as promising new targets for therapy. To bridge the gap between cell culture studies, in vivo physiology, and treatment, it is essential to develop quantitative assays for receptor dimerization in living animals.
We developed a firefly luciferase PCA to image dimeric complexes of chemokine receptors in intact cells and living mice. Using this system, we demonstrated constitutive association of CXCR4 and CXCR7 as homodimers and heterodimers. These data validate previous studies showing constitutive homodimerization of CXCR4 and heterodimerization of CXCR4 and CXCR7, and we demonstrate for the first time that CXCR7 also exists as preformed dimers (4, 17,18,19, 31). Chemokine ligands altered conformations of CXCR4 and CXCR7 homodimers, respectively, but neither CXCL11 nor CXCL12 affected reporter signals from heterodimeric complexes. CXCR4-CXCR7 heterodimers have been reported to alter CXCR4 signaling in cultured cells (18, 32), suggesting that fundamental differences in interactions between CXCR4 homodimers and CXCR4-CXCR7 heterodimers control outputs of downstream effector molecules. The firefly luciferase PCA provides a new method to link dynamics of receptor complexes to effects on signaling pathways in cell culture and animal models.
Our results also establish that this imaging reporter can be used to quantify pharmacologic modulation of chemokine receptor dimerization in cells and mice. We monitored effects of CXCR4 inhibitors to block binding of CXCL12 to the receptor and indirectly regulate CXCR4 homodimerization. We also determined that two preclinical inhibitors of CXCR7 directly produce changes in homodimerization of CXCR7, in addition to previously validated inhibition of chemokine binding to this receptor (7). Our data suggest effects of these compounds to block tumor growth in animal models may be mediated in part by altering conformations of CXCR7 complexes, potentially controlling ligand binding or coupling to intracellular effectors.
Previous studies have used resonance energy transfer techniques (FRET or BRET) to investigate conformational changes in CXCR4 homodimers or complexes of CXCR4 with other 7-TM receptors in cell-based assays (4, 16, 17, 33). Results obtained with both techniques indicate that CXCR4 exists as preformed homo- and heterodimeric complexes, including a heterodimeric complex with CXCR7 (18). FRET techniques typically show modest changes in signal (≈30%) from conformational changes in CXCR4 following treatment with CXCL12. BRET assays of CXCR4 complexes typically show larger changes in signal in response to CXCL12 (≈2- to 3-fold), likely due to enzymatic amplification of energy transfer from Renilla luciferase to the acceptor fluorescent protein. FRET and BRET studies of CXCR4 homodimers typically have quantified changes in signal intensity after 5–10 min of incubation with CXCL12 or pharmacologic agent and have not analyzed effects of extended treatment with chemokine. Interestingly, a BRET study by Babcock et al. (19) analyzed signal changes following 1 h of incubation with CXCL12, but these authors found only minimal effects of ligand on CXCR4 complexes.
The firefly luciferase PCA described in this manuscript produces changes in reporter signals that are comparable to those reported for early changes in CXCR4 complexes measured with FRET or BRET techniques. Our data for immediate responses of CXCR4 and CXCR7 homodimeric complexes were obtained using 1–2 min of image acquisition, which is shorter than typically described for FRET and BRET experiments. The firefly luciferase PCA shows that conformational changes in CXCR4 and CXCR7 homodimeric complexes are amplified over time. For CXCR7 homodimers, progressive changes in bioluminescence require clathrin-dependent endocytosis and acidified intracellular compartments. These data suggest that conformational changes in CXCR7 homodimers continue to occur as this receptor traffics to lysosomes, an organelle where a large fraction of intracellular CXCR7 resides (34). It is unclear why Babcock et al. (19) found only minimal changes in CXCR4 complexes after 1 h, while we found that chemokine-dependent changes in CXCR4 homodimers increase over time. We note that immediate conformational changes in CXCR4 homodimers produced by our current system had greater variation than with prolonged incubation times. We currently are testing alternative linkers to improve the system as we use firefly luciferase PCA to further investigate molecular regulation of CXCR4 and CXCR7 complexes and effects of intracellular trafficking on conformational changes in dimers. Notably, this PCA allows us to move readily between cell-based and animal models as we pursue these studies.
CXCR4 is a key regulator of many different disease processes, including cancer, HIV, atherosclerosis, and rheumatoid arthritis, and CXCR7 also has been implicated in tumor progression and metastasis. Because of the central role of these receptors in disease, development of pharmacologic agents to inhibit or activate these receptors is an active area of investigation for many pharmaceutical companies and academic laboratories. The propensity of chemokine and other 7-TM receptors to heterodimerize adds previously under-appreciated complexity to targeting these molecules therapeutically. Moreover, pharmacologic agents directly targeting receptor dimers, rather than ligand-receptor binding, provide a promising new approach to regulate 7-TM receptors (35). Taken together, our studies show the utility of the PCA reporter for identifying new therapeutic agents targeted to chemokine receptor complexes and validating efficacy of these agents in vivo. Moreover, we expect this general imaging strategy can be implemented to quantify dimerization of other 7-TM receptors and different families of transmembrane receptors.
Supplementary Material
Acknowledgments
This work was supported in part by grant BCTR0503550 from the Susan G. Komen Foundation and U.S. National Institutes of Health (NIH) grants P50CA093990 and R24CA083099 for the University of Michigan Small Animal Imaging Resource. Experiments were performed and analyzed by all authors. K.E.L. and G.D.L. wrote the manuscript. The authors declare no conflicts of interest.
References
- Terrillon S, Bouvier M. Roles of G-protein-coupled receptor dimerization. EMBO Rep. 2004;5:30–34. doi: 10.1038/sj.embor.7400052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James J, Oliveira M, Carmo A, Iaboni D, Davis S. A rigorous experimental framework for detecting protein oligomerization using bioluminescence resonance energy transfer. Nat Methods. 2006;3:1001–1006. doi: 10.1038/nmeth978. [DOI] [PubMed] [Google Scholar]
- Luker K, Luker G. Functions of CXCL12 and CXCR4 in breast cancer. Cancer Lett. 2006;238:30–41. doi: 10.1016/j.canlet.2005.06.021. [DOI] [PubMed] [Google Scholar]
- Wang J, He L, Combs C, Roderiquez G, Norcross M. Dimerization of CXCR4 in living malignant cells: control of cell migration by a synthetic peptide that reduces homologous CXCR4 interactions. Mol Cancer Ther. 2006;5:2592–2599. doi: 10.1158/1535-7163.MCT-05-0261. [DOI] [PubMed] [Google Scholar]
- De A, Gambhir S. Noninvasive imaging of protein-protein interactions from live cells and living subjects using bioluminescence resonance energy transfer. FASEB J. 2005;19:2017–2019. doi: 10.1096/fj.05-4628fje. [DOI] [PubMed] [Google Scholar]
- Luker K, Smith M, Luker G, Gammon S, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A. 2004;101:12288–12293. doi: 10.1073/pnas.0404041101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns J, Summers B, Wang Y, Melikian A, Berahovich R, Miao Z, Penfold M, Sunshine M, Littman D, Kuo C, Wei K, McMaster B, Wright K, Howard M, Schall T. A novel chemokine receptor for SDF-1 and I-TAC involved in cell survival, cell adhesion, and tumor development. J Exp Med. 2006;203:2201–2213. doi: 10.1084/jem.20052144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanon M, McClanahan T, Murphy E, Yuan W, Wagner S, Barrera J, Mohar A, Verasteugui E, Zlotnik A. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Miao Z, Luker K, Summers B, Berahovich R, Bhojani M, Rehemtulla A, Kleer C, Essner J, Nasevicius A, Luker G, Howard M, Schall T. CXCR7 (RDC1) promotes breast and lung tumor growth in vivo and is expressed on tumor-associated vasculature. Proc Natl Acad Sci U S A. 2007;104:15735–15740. doi: 10.1073/pnas.0610444104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luker K, Gupta M, Luker G. Imaging CXCR4 signaling with firefly luciferase complementation. Anal Chem. 2008;80:5565–5573. doi: 10.1021/ac8005457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois C, Hong E, Pease S, Brown E, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–872. doi: 10.1126/science.1067081. [DOI] [PubMed] [Google Scholar]
- Smith M, Luker K, Garbow J, Prior J, Jackson E, Piwnica-Worms D, Luker G. CXCR4 regulates growth of both primary and metastatic breast cancer. Cancer Res. 2004;64:8604–8612. doi: 10.1158/0008-5472.CAN-04-1844. [DOI] [PubMed] [Google Scholar]
- Luker K, Pica C, Schreiber R, Piwnica-Worms D. Overexpression of IRF9 confers resistance to antimicrotubule agents in breast cancer cells. Cancer Res. 2001;61:6540–6547. [PubMed] [Google Scholar]
- Luker G, Rao V, Crankshaw C, Dahlheimer J, Piwnica-Worms D. Characterization of phosphine complexes of technetium(III) as transport substrates of the multidrug resistance P-glycoprotein and functional markers of P-glycoprotein at the blood-brain barrier. Biochemistry. 1997;36:14218–14227. doi: 10.1021/bi971931z. [DOI] [PubMed] [Google Scholar]
- Luker G, Bardill J, Prior J, Pica C, Piwnica-Worms D, Leib D. Noninvasive bioluminescence imaging of herpes simplex virus type 1 infection and therapy in living mice. J Virol. 2002;76:12149–12161. doi: 10.1128/JVI.76.23.12149-12161.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percherancier Y, Berchiche Y, Slight I, Volkmer-Engert R, Tamamura H, Fujii N, Bouvier M, Heveker N. Bioluminescence resonance energy transfer reveals ligand-induced conformational changes in CXCR4 homo- and heterodimers. J Biol Chem. 2005;280:9895–9903. doi: 10.1074/jbc.M411151200. [DOI] [PubMed] [Google Scholar]
- Toth P, Ren D, Miller R. Regulation of CXCR4 receptor dimerization by the chemokine SDF-1alpha and the HIV-1 coat protein gp120: a fluorescence resonance energy transfer (FRET) study. J Pharmacol Exp Ther. 2004;310:8–17. doi: 10.1124/jpet.103.064956. [DOI] [PubMed] [Google Scholar]
- Sierro F, Biben C, Martinez-Munoz L, Mellado M, Rashohoff R, Li M, Woehl B, Leung H, Groom J, Batten M, Harvey R, Martinez-A C, Mackay C, Mackay F. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock G, Farzan M, Sodroski J. Ligand-independent dimerization of CXCR4, a principal HIV-1 coreceptor. J Biol Chem. 2003;278:3378–3385. doi: 10.1074/jbc.M210140200. [DOI] [PubMed] [Google Scholar]
- Balabanian K, Lagane B, Infantino S, Chow K, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M, Bachelerie F. The chemokine SDF-1/CXCL12 binds to and signals through the orphan receptor RDC1 in T lymphocytes. J Biol Chem. 2005;280:35760–35766. doi: 10.1074/jbc.M508234200. [DOI] [PubMed] [Google Scholar]
- Phillips R, Burdick M, Lutz M, Belperio J, Keane M, Strieter R. The stromal derived factor-1/CXCL12-CXC chemokine receptor 4 biological axis in non-small cell lung cancer metastases. Am J Respir Crit Care Med. 2003;167:1676–1686. doi: 10.1164/rccm.200301-071OC. [DOI] [PubMed] [Google Scholar]
- Wang J, Shiozawa Y, Wang J, Wang Y, Jung Y, Pienta K, Mehra R, Loberg R, Taichman R. The role of CXCR7/RDC1 as a chemokine receptor for CXCL12/SDF-1 in prostate cancer. J Biol Chem. 2008;283:4283–4294. doi: 10.1074/jbc.M707465200. [DOI] [PubMed] [Google Scholar]
- Ding Z, Issekutz T, Downey G, Waddell T. L-selectin stimulation enhances functional expression of surfaces CXCR4 in lymphocytes: implications for cellular activation during adhesion and migration. Blood. 2003;101:4245–4252. doi: 10.1182/blood-2002-06-1782. [DOI] [PubMed] [Google Scholar]
- Drake M, Shenoy S, Lefkowitz R. Trafficking of G protein-coupled receptors. Circ Res. 2006;99:570–582. doi: 10.1161/01.RES.0000242563.47507.ce. [DOI] [PubMed] [Google Scholar]
- Heuser J, Anderson R. Hypertonic media inhibit receptor-mediated endocytosis by blocking clathrin-coated pit formation. J Cell Biol. 1989;108:389–400. doi: 10.1083/jcb.108.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. The bicyclam AMD3100 story. Nat Rev Drug Discov. 2003;2:581–587. doi: 10.1038/nrd1134. [DOI] [PubMed] [Google Scholar]
- Tamamura H, Hiramatsu K, Mizumoto M, Ueda S, Kusano S, Terakubo S, Akamatsu M, Yamamoto N, Trent J, Wang Z, Peiper S, Nakashima H, Otaka A, Fujii N. Enhancement of the T140-based pharmacophores leads to the development of more potent and bio-stable CXCR4 antagonists. Org Biomol Chem. 2003;1:3663–3669. doi: 10.1039/b306613b. [DOI] [PubMed] [Google Scholar]
- Zhang W, Navenot J, Haribabu B, Tamamura H, Hiramatu K, Omagari A, Pei G, Manfredi J, Fujii N, Broach J, Peiper S. A point mutation that confers constitutive activity to CXCR4 reveals that T140 is an inverse agonist and that AMD3100 and ALX40-4C are weak partial agonists. J Biol Chem. 2002;277:24515–24521. doi: 10.1074/jbc.M200889200. [DOI] [PubMed] [Google Scholar]
- Springael J, Urizar E, Costagliola S, Vassart G, Parmentier M. Allosteric properties of G protein-coupled receptor oligomers. Pharmacol Ther. 2007;115:410–418. doi: 10.1016/j.pharmthera.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Orimo A, Gupta P, Sgroi D, Arenzana-Seisdedos F, Delaunay T, Naeem R, Carey V, Richardson A, Weinberg R. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Lagane B, Chow K, Balabanian K, Levoye A, Harriague J, Planchenault T, Baleux F, Gunera-Saad N, Arenzana-Seisdedos F, Bachelerie F. CXCR4 dimerization and β-arrestin-mediated signaling account for the enhanced chemotaxis to CXCL12 in WHIM syndrome. Blood. 2008;112:34–44. doi: 10.1182/blood-2007-07-102103. [DOI] [PubMed] [Google Scholar]
- Hartmann T, Grabovsky V, Pasvolsky R, Shulman Z, Buss E, Spiegel A, Nagler A, Lapidot T, Thelen M, Alon R. A crosstalk between intracellular CXCR7 and CXCR4 involved in rapid CXCL12-triggered integrin activation but not in chemokine-triggered motility of human T lymphocytes and CD34+ cells. J Leukoc Biol. 2008;84:1130–1140. doi: 10.1189/jlb.0208088. [DOI] [PubMed] [Google Scholar]
- Sohy D, Parmentier M, Springael J. Allosteric transinhibition by specific antagonists in CCR2/CXCR4 heterodimers. J Biol Chem. 2007;282:30062–30069. doi: 10.1074/jbc.M705302200. [DOI] [PubMed] [Google Scholar]
- Boldajipour B, Mahabeleshwar S, Kardash E, Reichman-Fried M, Blaser H, Minina S, Wilson D, Xu Q, Raz E. Control of chemokine-guided cell migration by ligand sequestration. Cell. 2008;132:463–473. doi: 10.1016/j.cell.2007.12.034. [DOI] [PubMed] [Google Scholar]
- Gupta A, Decaillot F, Devi L. Targeting opioid receptor heterodimers: strategies for screening and drug development. AAPS J. 2006;8:E153–E159. doi: 10.1208/aapsj080118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.