Abstract
Although microRNAs have been investigated extensively in cancer research, little is known regarding their response to noxious agents in apparently healthy tissues. We analyzed the expression of 484 miRNAs in the lungs of rats exposed to environmental cigarette smoke (ECS) for 28 days. ECS down-regulated 126 miRNAs (26.0%) at least 2-fold and 24 miRNAs more than 3-fold. We previously demonstrated that 107 of 4858 genes (2.9%) and 50 of 518 proteins (9.7%) were up-regulated by ECS in the same tissue, which is consistent with the role of microRNAs as negative regulators of gene expression. The most remarkably down-regulated microRNAs belonged to the families of let-7, miR-10, miR-26, miR-30, miR-34, miR-99, miR-122, miR-123, miR-124, miR-125, miR-140, miR-145, miR-146, miR-191, miR-192, miR-219, miR-222, and miR-223, which regulate stress response, apoptosis, proliferation, angiogenesis, and expression of genes. In contrast, miR-294, an inhibitor of transcriptional repressor genes, was up-regulated by ECS. There was a strong parallelism in dysregulation of rodent microRNAs and their human homologues, which are often transcribed from genes localized in fragile sites deleted in lung cancer. Five ECS-down-regulated microRNAs are known to be affected by single nucleotide polymorphisms. Thus, changes in microRNA expression are an early event following exposure to cigarette smoke.—Izzotti, A., Calin, G. A., Arrigo, P., Steele, V. E., Croce, C. M., De Flora, S. Downregulation of microRNA expression in the lungs of rats exposed to cigarette smoke.
Keywords: mirnome, transcriptome, proteome, microarrays
MicroRNAs (miRNAs) are small noncoding RNAs of 19–24 nucleotides in length that are important in the regulation of crucial biological processes, such as cell growth (1), apoptosis (2), development (3), differentiation (4), and endocrine homeostasis (5). Recently, miRNA expression has been linked to cancer development (6). The first evidence came from the discovery that miR-15a and miR-16-1 are down-regulated or deleted in most patients with chronic lymphocytic leukemia (7). It has been proposed that alterations in miRNA genes play a critical role in the pathophysiology of many, perhaps, all human cancers (8). Several miRNAs have been associated with human lung cancer, (e.g., let-7g and miR-125) (9). MiRNA silencing is the evolutionary development of an archetypal defense system against invasion by foreign genes and viral infections (10). In general, high phylogenic conservation might result in high sensitivity toward genotoxic damage, as already demonstrated for mitochondria (11).
Many studies analyzed the global expression of miRNAs, referred to as mirnome, in cancer vs. noncancer tissues (8) and also in cardiovascular diseases (12). The use of miRNA microarrays makes it possible to perform profiling studies that evaluate differences in cancer-relevant miRNAs. MiRNA alterations bear biological relevance because they directly affect gene expression and protein synthesis (13).
Despite the prominent biological importance of miRNAs, little is known regarding their response to environmental carcinogens. In fact, only a few studies have explored miRNA responses to the cellular stress induced by chemical and physical agents. For instance, no significant alteration in the expression of miRNAs was detected in lymphoblastoid cells exposed in vitro to gamma radiation, whereas a global increase in miRNA expression was detected after exposure to sodium arsenite or induction of folate deficiency (14).
These premises prompted us to explore mirnome alterations in response to exposure to cigarette smoke (CS). There is overwhelming evidence that CS plays a major role in the epidemiology of lung cancer, cancer at other sites, and a variety of chronic degenerative diseases (15,16,17). Yet, it is difficult to reproduce the carcinogenicity of CS in animal models and to explore the mechanisms of CS as a complex mixture rather than to evaluate the effects of its individual carcinogenic components (18). Environmental CS (ECS), which is a form of indoor air pollution resulting from the mixture of sidestream CS and that portion of mainstream CS that is released by actively smoking individuals into ambient air, contains many free radicals and redox active compounds (19). We previously evaluated the effects of ECS in rodents at the level of genome (20), transcriptome (21,22,23), and proteome (22, 23).
The goal of the present study was to analyze miRNA alterations in the lung, which is the main target organ for ECS carcinogenicity (17). The results presented here provide evidence that exposure of rats to ECS mainly results in down-regulation of a number of miRNAs involved in the carcinogenesis process.
MATERIALS AND METHODS
Animals and treatments
Sixteen Sprague-Dawley rats (Harlan Italy, Correzzana, Milan, Italy), weighing 315–320 g, were either exposed to ECS for 4 wk or kept in filtered air for the same period of time (sham smoke). The housing and treatments of animals were in accordance with U.S. National Institutes of Health, Italian, and institutional guidelines.
A whole-body exposure to ECS was achieved by using a smoking machine (model TE-10; Teague Enterprises, Davis, CA, USA). We used a mixture of sidestream (89%) and mainstream smoke (11%), mimicking an exposure to high-dose ECS. Kentucky 1R3 reference cigarettes, with a declared content of 22.8 mg total particulate matter and 1.46 mg nicotine each, were purchased from the Tobacco Research Institute (University of Kentucky, Lexington, KY, USA). We burned 5 cigarettes at one time, 6 h/day divided into 2 rounds of 3 h with a 3-h interval, 5 days/wk. This accounted for a whole-body exposure to the smoke generated by 600 cigarettes/wk.
After 4 wk, the rats were anesthetized deeply with the diethyl ether and killed by cervical dislocation. The lungs were collected, immersed in an RNA-stabilizing buffer, and frozen at −80°C.
RNA extraction, quantification, and evaluation of integrity
RNA samples pooled from the 2 treatment groups were homogenized in a buffer containing guanidinium-thiocyanate. After digestion with proteinase K (Boehringer, Mannheim, Germany), the material was sequentially treated with phenol and chloroform, digested with RNase-free DNase I (Sigma Chemical Co., St. Louis, MO, USA), and treated again with phenol and chloroform. Total RNA was pelleted on nitrocellulose filters by isopropanol and washed with 80% ethanol. The whole extraction procedure was performed by using an automated nucleic acid extractor (Genepure, 341; Applied Biosystems, Foster City, CA, USA).
RNA quantification was performed by NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Absorbance of the RNA samples was quantified at 260 and 280 nm, and the 260/280 ratio was calculated. The samples showed a 260/280 ratio > 1.9, which was assumed as an indicator of RNA purity.
Analysis of RNA structural integrity was performed by capillary electrophoresis by using the Bioanalyzer Agilent 2100 equipped with Agilent RNA 6000 nanochips (Agilent Technologies, Waldbronn, Germany). The presence of miRNA was confirmed by the detection of fragments in the 20–100 bp range.
MiRNA microarray analysis
MiRNA microarray profiling was conducted as described previously (24). Briefly, 5 μg of total RNA was used for hybridization on a custom miRNA microarray platform containing quadruplicates of 1117 miRNAs, 484 of which were from rodents, 627 from humans, and 6 from Arabidopsis thaliana, with annotated active sites. The whole list of miRNAs included in the microarray used is available at GEO database (registration number requested). These arrays contain gene-specific 40-mer oligonucleotide probes, spotted by contacting technologies and covalently attached to a polymeric matrix (24). The hybridized biotinylated transcripts were detected by streptavidin-Alexa647 conjugation, scanned on an Axon 4000B microarray scanner (Axon Instruments, Union City, CA, USA), and analyzed using Genepix Pro 6.0 (Axon Instruments).
MiRNA real time quantitative polymerase chain reaction (qPCR)
To validate microarray data, 3 of the miRNAs down-regulated by ECS, as shown by microarray analyses (let-7c, miR-34c, and miR-222), were analyzed by real time qPCR. These miRNAs were selected because of the importance of the regulated biological functions (Table 1). To this purpose, the miRNAs contained in 200 ng of total RNA were polyadenylated, and the first-strand cDNA was obtained by using the NCode™ miRNA First-Strand cDNA Synthesis Kit (Invitrogen Corporation, Carlsbad, CA, USA). Each cDNA sample (5 μl) was diluted 1:10 in water and then amplified in a PCR reaction mix containing SYBR green (25) by using a Universal qPCR Primer (Invitrogen Corporation) and a specific forward primer for each miRNA tested. The forward primer sequences were the entire mature miRNA sequences as reported in http://microrna.sanger.ac.uk/cgi-bin/sequences, i.e.: let-7c, TGAGGTAGTAGGTTGTATGGTT; miR-34c, AGGCAGTGTAGTTAGCTGATTGC; miR-222, AGCTACATCTGGCTACTGGGT.
TABLE 1.
miRNAs that were down-regulated >3-fold and below the statistical significance threshold in the lung of rats exposed to ECS
| miRNA | Rodent miRNA
|
Human miRNA homologues
|
||||
|---|---|---|---|---|---|---|
| Sham/ECS ratio | P | Sham/ECS ratio | P | Location on fragile sitea | Regulated function | |
| let-7ab | 4.1 | 0.005 | 5.3 | 0.005 | 11q23-24 | Cell proliferation, oncogene (RAS) activation, angiogenesis |
| let-7b | 4.1 | 0.003 | 3.0 | 0.014 | — | Same as let-7a |
| let-7c | 4.5 | 0.021 | 5.9 | 0.051 | 21q11.1 | Same as let-7a |
| let-7f | 5.2 | 0.006 | 3.4 | 0.031 | 9q22.3 | Same as let-7a |
| miR-10a | 3.7 | 0.005 | NA | Angiogenesis | ||
| miR-26a | 3.1 | 0.012 | 2.6 | 0.021 | 3p23-21.31 | TGF expression |
| miR-30a | 3.2 | 0.021 | NA | — | Cell adhesion, protein repair, stress response (NF-κB activation), cell cycle, oncogene (EGF) activation | |
| miR-30c | 8.6 | 0.001 | 4.9 | 0.034 | — | Same as miR-30a |
| miR-34b | 3.0 | 0.003 | 4.3 | 0.022 | 11q23-24 | P53 effector (direct transcriptional targets for P53) |
| miR-34c | 3.1 | 0.005 | 3.3 | 0.023 | — | Same as miR-34b |
| miR-99b | 3.4 | 0.013 | 2.6 | 0.019 | 21q11.1 | Apoptosis |
| miR-122a | 4.0 | 0.004 | 1.3 | NS | — | Stress response |
| miR-123-prec | 3.1 | 0.006 | 4.2 | 0.017 | 9q33 | Angiogenesis |
| miR-124a-precb | 7.0 | 0.008 | NA | — | Stress response | |
| miR-125a-precb | 8.1 | 0.022 | 4.3 | 0.014 | 11q23-24 | Oncogene (ERBB2) activation |
| miR-125b | 3.3 | 0.003 | 5.4 | 0.050 | 21q11.1 | Stress response |
| miR-140sb | 4.4 | 0.027 | NA | — | P53 effector | |
| miR-145-prec | 4.6 | 0.001 | 4.3 | 0.050 | 5q32 | Protein repair, angiogenesis |
| miR-146-precb | 3.6 | 0.013 | 2.3 | 0.037 | — | Stress response (NF-κB activation) |
| miR-191-prec | 9.2 | 0.006 | 7.0 | 0.014 | — | Cell proliferation |
| miR-192 | 4.2 | 0.009 | 2.8 | 0.042 | — | Oncogene (RAS) activation |
| miR-219-prec | 5.3 | 0.004 | 1.4 | NS | — | Stress response, oncogene (ELK1, FOS) activation |
| miR-222-prec | 3.9 | 0.043 | 3.8 | 0.016 | — | Cell proliferation, angiogenesis |
| miR-223-prec | 4.7 | 0.021 | 6.6 | 0.017 | — | Protein repair, oncogene (RAS) activation |
The primers were purchased from TIB Molbiol (Berlin, Germany). The fluorescence intensity was acquired at the annealing step of each amplification cycle. The signal was normalized for the average background of each sample and calculated over the first 5 amplification cycles (Rotor-Gene 6.1.81 software; Corbett Life Sciences, Mortlake, Australia). Specificity of the amplicons was checked by performing the “melting curve” analysis.
Analysis of data
After local background subtraction, raw data were log transformed, normalized, and analyzed by GeneSpring® software version 7.2 (Silicon Genetics, Redwood City, CA, USA). Expression data were median centered by using the GeneSpring normalization option, which normalizes both per gene and per array. Comparisons between ECS and sham were done by evaluating the fold variation of the mean values of quadruplicate data generated for each miRNA. In addition, the statistical significance of the differences was evaluated by using the GeneSpring ANOVA tool and Bonferroni multiple testing correction. Correlations between rodent and human miRNAs were evaluated by simple regression analysis using the StatView software (Abacus Concept, Berkeley, CA, USA). Differences with P < 0.05 were taken as statistically significant.
RESULTS
Dysregulation of miRNAs in the lung of ECS-exposed rats
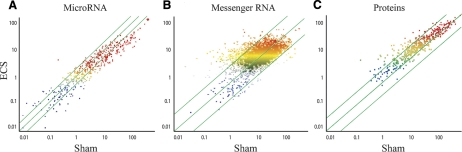
Figure 1A shows a scatter plot comparing the intensity of expression of 484 miRNAs in the lungs of sham-exposed and ECS-exposed rats. By excluding poorly expressed miRNAs, falling in the lowest quartile of expression intensity, exposure to ECS resulted in at least a 2-fold down-regulation of 126 miRNAs (26.0%) and in up-regulation of 7 miRNAs (1.4%). By selecting dysregulated miRNAs according to more restrictive criteria, (i.e., at least a 3-fold variation accompanied by a statistically significant difference between ECS and sham) 24 miRNAs were down-regulated and only 1 was up-regulated after exposure to ECS.
Figure 1.
Scatter plots showing the relations between the expression of miRNAs (A; present study), messenger RNAs (B; ref. 22), and proteins (C; ref. 23) in sham-exposed rats and ECS-exposed rats. Color reflects the intensity of expression, from blue (lowest) to red (highest). Diagonal green lines indicate 2-fold variation intervals of the ECS/sham ratio.
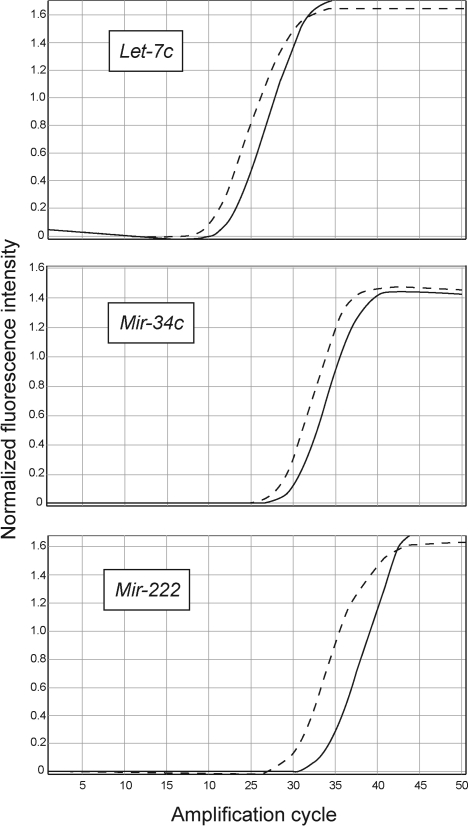
To validate the results of microarray analyses, 3 selected miRNAs (let-7c, miR-34c, and miR-222) were also analyzed by qPCR. Figure 2 shows the amplification cycles for these miRNAs in the lungs of ECS-exposed rats (full lines) and sham-exposed rats (dashed lines). In agreement with microarray data, all 3 miRNAs were down-regulated by ECS, and specifically, let-7c was down-regulated 4.6-fold (vs. 4.5 by microarray), miR-34c was down-regulated 3.6-fold (vs. 3.1), and miR-222, 8.9-fold (vs. 3.9).
Figure 2.
Quantitative real-time PCR curves evaluating miRNA expression in the lungs of sham-exposed rats (dashed lines) vs. ECS-exposed rats (solid lines).
Functions of ECS-dysregulated miRNAs
The biological functions of miRNAs were inferred from the available literature and by selecting genes having a context score > 0.31, as reported in Targetscan (http://www.targetscan.org), Miranda (http://www. microrna.org), and Sanger Institute (http://microrna. sanger.ac.uk) databases. MiR-294 was the only miRNA whose expression was significantly (P=0.05) and considerably (10.7-fold) up-regulated after exposure of rats to ECS. This miRNA modulates gene transcription and recognizes transcriptional repressor genes as the main target. As shown in Table 1, the 24 miRNAs down-regulated by ECS are involved in a variety of functions. The most relevant in the carcinogenesis process are stress response, cell proliferation, angiogenesis, expression of oncogenes, tumor suppressor genes, and genes encoding for tumor-related growth factors.
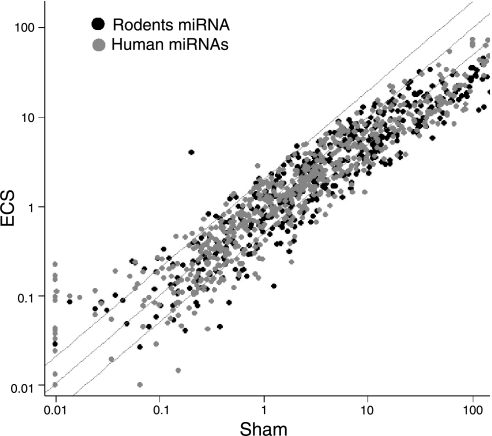
Interspecies comparison of rodent with human miRNAs
The scatterplot shown in Fig. 3 compares the effect of ECS on the expression of rodent miRNAs with their human homologues. Of the 24 rodent miRNAs down-regulated by ECS at least 3-fold and to a statistically significant extent, 20 had the human homologues spotted on the same array (Table 1). With 1 exception (miR-122a), all responded to ECS similarly to their rodent counterparts, with a significant correlation between the ECS/sham ratio variations in the 2 species (r=0.446, P<0.05).
Figure 3.
Scatter plot showing the overlap between the expression of rodent miRNAs (black symbols) and their human homologues (gray symbols) spotted on the same microarray.
It is noteworthy that 5 ECS-down-regulated human miRNA homologues, identified with asterisks in Table 1, are known to be transcribed by polymorphic genes (26, 27).
As reported in Table 1, the ECS-down-regulated miRNAs are often transcribed from genes located in genomic fragile sites that are frequently deleted in human lung cancer.
DISCUSSION
The results obtained provide evidence that exposure of rats to ECS causes extensive alterations in miRNA expression in the lung. In the interpretation of these data, it should be taken into account that the examined lung samples contained a mixed population of cells involved in a variety of biological functions and characterized by various differentiation stages, ranging from stem cells to terminally differentiated cells. Evidence is increasing that cancer stem cells, a subpopulation of differentiated cells with self-renewal properties, play a major role in carcinogenesis (28).
All but one of the observed major alterations in miRNA expression involved down-regulation. Interestingly, previous studies using the same animals (22, 23) had shown that ECS up-regulates gene transcription (Fig. 1B) and protein expression (Fig. 1C). These opposite trends are in line with the well-established role of miRNAs as negative regulators of gene expression and protein synthesis. For instance, the miR-145-prec, which was down-regulated 4.6-fold by ECS, recognizes as a target the ubiquitin ligase E gene, whose expression was up-regulated 2.4-fold, but the homologue protein was up-regulated 4.8-fold. The miR-219-prec, which was down-regulated 5.3-fold by ECS, recognizes as a target the stress response Erg gene, whose expression was up-regulated 2.7-fold, but the homologue protein was up-regulated 2.9-fold. MiR-34c, which was down-regulated 3.1-fold by ECS, recognizes as a target the proapoptotic gene Bcl2-associated death agonist α, whose expression was modestly up-regulated (1.2-fold), but the homologue protein was up-regulated 2.3-fold. The global analysis of the microarray databases relative to mirnome (484 miRNAs), transcriptome (4858 genes), and proteome (518 proteins) will be the subject of further bioinformatic studies.
In cancer research, this mechanism had mainly been investigated by analyzing neoplastic cells (6,7,8,9). For the first time, we show here that miRNA alterations also occur as an early response in the target tissue of animals exposed in vivo to environmental carcinogens before cancer onset. From a quantitative point of view, miRNAs proved to be the most sensitive biomarker of exposure to ECS-induced alterations among the postgenomic end points evaluated. In fact, in the present study, ECS down-regulated >2-fold 126 of 484 miRNAs (26.0%) in the lung. In the same tissue, ECS up-regulated >2-fold 107 of 4858 genes (2.9%) and 50 of 518 proteins (9.7%) (22, 23). Therefore, miRNA analysis compares favorably with both transcriptome analysis and proteome analysis as a new biomonitoring tool in molecular medicine.
Moreover, the findings of the present study shed light on fundamental pathogenic mechanisms involved in the damage produced by CS in its main target organ. These mechanisms can be inferred by taking into account the functions of the ECS-dysregulated miRNAs. MiR-294, the only miRNA that was dramatically up-regulated after ECS exposure, regulates gene transcription and recognizes transcriptional repressors genes (e.g., zinc finger protein 697; AT-rich interactive domain 4A) as main targets. Accordingly, the increase of miR-294 expression causes silencing of transcriptional repressors, thereby resulting in a global increase of gene transcription as a response to ECS. In contrast, ECS-down-regulated miRNAs are involved in a variety of mechanisms playing a role in the carcinogenesis process and in other alterations of physiological functions caused by CS in the respiratory tract. Some of the detected alterations likely depend on ECS-induced DNA changes. Taking into account that promotion is the main mechanism of cigarette smoking in lung carcinogenesis (17), other miRNA alterations may reflect the occurrence of epigenetic events.
Several miRNAs involved in the activation of the NF-κB pathway (i.e., miR-30a, miR-146-prec, miR-132, and miR-155) were included in the microarray used. All of them were down-regulated by ECS, either more than 3-fold and below the statistically significant threshold (miR-30a, miR-146-prec) or to a lower extent (miR-132 1.7-fold, not significant, and miR-155 2.0-fold; P<0.05). These results are in line with proteome data generated by testing the lungs of the same animals, demonstrating that exposure to ECS increases the release of the NF-κB inhibitors α, β, and γ subunits, a mechanism resulting in NF-κB activation and nuclear translocation (23). NF-κB is known to play an important role in inflammation, which is typically associated with exposure to ECS. The observed general trend of miRNA down-regulation in the lung is also likely to contribute to inflammatory processes. In fact, a global down-regulation of miRNA expression was observed in effector T cells compared with naive cells (29). It is also noteworthy that miR-122 down-regulation is known to induce the expression of heme oxygenase-1 (30), a potent antioxidant activity that is dramatically up-regulated in the liver of mice transplacentally exposed to ECS (31) and, to a lesser extent, in the lungs of mice exposed to ECS and light (32).
The ECS-down-regulated miRNAs play pivotal functions in cell biology. Let-7s are a family of highly conserved archetypical miRNAs, poorly expressed or deleted in human lung tumors, recognizing the RAS oncogene as a target for their silencing activity (33). Under physiological conditions, let-7 is expressed at the highest levels in the lung compared with other tissues, and its inhibition results in an increased division of A549 lung cancer cells in vitro (34). Many genes involved in cell proliferation are silenced by let-7, including cyclins B1, E2, F, G2, and CDC2, 25, and 34 (34). Lung cancer patients with low let-7 expression survived a shorter time than those with high expression (35). Interestingly, the exogenous delivery of let-7 prevents the formation of lung tumors from premalignant lesions and shrinks tumors with activating RAS mutations (36). Our results indicate that let-7 down-regulation is an early event in the carcinogenesis process. It is likely, however, that let-7 down-regulation may still be reversible during early steps, being irreversible only in cancer-committed cells. Studies evaluating the dose-response effects of CS on miRNA expression and the reversibility of miRNA alterations on discontinuation of exposure to CS are now in progress.
Other ECS-down-regulated miRNAs are associated with the activation of tumor suppressor genes. For instance, miR-34 is directly transactivated by P53 and its expression induces cell cycle arrest (37). The finding that this miRNA is an early target for ECS is consistent with our previous studies demonstrating the important role of P53 in smoke-induced postgenomic alterations (21) and lung cancer development (38). Accordingly, the ECS-induced down-regulation of miR-34 may be interpreted as a contribution to cell proliferation that, in case of long-term exposure, can prevail on defensive mechanisms related to the activation of apoptotic pathways. The miR-34 gene, commonly deleted in human cancers, recognizes target genes that regulate cell cycle progression, apoptosis, and DNA repair (39).
The ECS-induced down-regulation of several miRNAs appears to be related to an induction of cell proliferation. This conclusion supports proteome data obtained by antibody microarray (23), which revealed a significant increase in several proteins involved in cell replication, including various cyclins, cyclin-dependent kinases, cell division cycle proteinases, and proliferating cell nuclear antigen in the lung of ECS-exposed rats. Some of the genes encoding for these proteins are direct targets for ECS-down-regulated miRNAs. Such is the case for cyclin F protein, which was significantly increased by ECS (23) and whose gene is a direct target for let-7 (34).
Our study demonstrates that ECS induces a down-regulation in many miRNAs involved in silencing of angiogenic activities. This is the case for miR-123 and miR-222, which are established regulators of the angiogenic phenotype (40). Several target genes for miR-34 are also known to regulate angiogenesis (37, 39). Down-regulation by ECS of let-7, miR-10a, and miR-145 promotes angiogenesis (Table 1). On the whole, these results provide evidence that ECS stimulates vessel growth in the lung of the exposed animals, a conclusion supported also by our previous findings that ECS increases the amounts of endothelin 1 receptor protein in rat lung (23) and the expression of genes encoding for angiogenin, angiopoietin, vascular endothelial growth factor (VEGF)-A, and VEGF-B in mouse lung (21). Moreover, exposure of mice to mainstream CS produced foci of cavernose proliferation of pulmonary blood vessels (41).
MiR-125a-prec and miR-125b were 2 of the most strikingly ECS-down-regulated human miRNA homologues. Interestingly, miR-125b is recognized as the homologue of C. elegans lin-4, the first miRNA discovered (42). MiR-125 plays an important role in lung carcinogenesis, its genetic targets including the ERBB2 proto-oncogene that encodes for the EGF receptor, which is highly expressed in carcinomas (43). MiR-125 genes are located in 11q23-q24 and 21q11.1, regions that are frequently deleted in lung cancer (9).
It is noteworthy that the miR-125a gene is affected by a G/U single nucleotide polymorphism at nucleotide 8 (27). The miR-125a U polymorphism blocks the processing of pri-miRNA to miRNA precursor (27), which probably renders this miRNA highly susceptible to those genotoxic agents, such as ECS, that induce miRNA down-regulation.
It has been reported that a relatively low level of variation occurs in miRNA genes, which is counteracted by an appreciable level of variation at their target genes (26). Nevertheless, 5 human homologues of ECS-down-regulated miRNAs are characterized by functionally relevant genetic polymorphisms. This finding suggests that the effects of ECS in the lung involve the interaction with not only the individual asset of genes encoding for metabolic and DNA repair activities but also those genes involved in miRNA functions. Accordingly, miRNA gene polymorphisms should be investigated in future studies to establish their contribution to the interindividual variability of susceptibility toward ECS.
In conclusion, the results obtained shed light on new pathogenic mechanisms exerted in the lung by CS. Furthermore, evidence is provided that mirnome alterations induced by ECS may be used as a new intermediate end point in lung cancer prevention studies.
Acknowledgments
This study was supported by the U.S. National Cancer Institute (contract N01-CN 53301).
References
- Cheng A M, Byrom M W, Shelton J, Ford L P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Guo M, Hay B A. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Karp X, Ambros V. Developmental biology. Encountering microRNAs in cell fate signaling. Science. 2005;310:1288–1289. doi: 10.1126/science.1121566. [DOI] [PubMed] [Google Scholar]
- Chen C Z, Li L, Lodish H F, Bartel D P. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- Poy M N, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald P E, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, Stoffel M. A pancreatic-islet specific miRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Garzon R, Fabbri M, Cimmino A, Calin G A, Croce C M. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Calin G A, Dumitru C D, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, Rassenti L, Kipps T, Negrini M, Bullrich F, Croce C M. Frequent deletions and down-regulation of micro-RNA genes miR-15 and miR-16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G A, Croce C M. MicroRNA–cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- Calin G A, Sevignani C, Dumitru C D, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce C M. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancer. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen I M, Cheng G, Wieland S, Volinia S, Croce C M, Chisari F V, David M. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature. 2007;449:919–922. doi: 10.1038/nature06205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balansky R, Izzotti A, Scatolini L, D'Agostini F, De Flora S. Induction by carcinogens and chemoprevention by n-acetylcysteine of adducts to mitochondrial DNA in rat organs. Cancer Res. 1996;56:1642–1647. [PubMed] [Google Scholar]
- Ji R, Cheng Y, Yue J, Yang J, Liu X, Chen H, Dean D B, Zhang C. MicroRNA expression signature and antisense-mediated depletion reveal an essential role of microRNA in vascular neointimal lesion formation. Circ Res. 2007;100:1579–1588. doi: 10.1161/CIRCRESAHA.106.141986. [DOI] [PubMed] [Google Scholar]
- Zong Q, Schummer M, Hood L, Morris D R. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc Natl Acad Sci U S A. 1999;96:10632–10636. doi: 10.1073/pnas.96.19.10632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit C J, Eddy K, Kelsey K T. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- Doll R, Peto R. The causes of cancer: quantitative estimates of avoidable risks of cancer in the United States today. J Natl Cancer Inst. 1981;66:1191–1308. [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services Washington, DC, USA: U.S. Government Printing Office; Reducing the health consequences of smoking, 25 years of progress. Office on Smoking and Health, DHSS Publication No. CDC-89–8411. 1989:43–54. [Google Scholar]
- International Agency for Research on Cancer Tobacco smoke and involuntary smoking. Lyon, France: International Agency for Research on Cancer; 2004 [Google Scholar]
- Hecht S S. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- Church D F, Pryor W A. Free-radical chemistry of cigarette smoke and its toxicological implications. Environ Health Perspect. 1985;64:111–126. doi: 10.1289/ehp.8564111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Bagnasco M, D'Agostini F, Cartiglia C, Lubet R A, Kelloff G J, De Flora S. Formation and persistence of nucleotide alterations in rats exposed whole-body to environmental cigarette smoke. Carcinogenesis. 1999;20:1499–1505. doi: 10.1093/carcin/20.8.1499. [DOI] [PubMed] [Google Scholar]
- Izzotti A, Cartiglia C, Longobardi M, Bagnasco M, Merello A, You M, Lubet R A, De Flora S. Gene expression in the lung of p53 mutant mice exposed to cigarette smoke. Cancer Res. 2004;64:8566–8572. doi: 10.1158/0008-5472.CAN-04-1420. [DOI] [PubMed] [Google Scholar]
- Izzotti A, Bagnasco M, Cartiglia C, Longobardi M, Camoirano A, Tampa E, Lubet R A, De Flora S. Modulation of multigene expression and proteome profiles by chemopreventive agents. Mutat Res. 2005;591:212–223. doi: 10.1016/j.mrfmmm.2005.03.032. [DOI] [PubMed] [Google Scholar]
- Izzotti A, Bagnasco M, Cartiglia C, Longobardi M, Balansky R M, Merello A, Lubet R A, De Flora S. Chemoprevention of genome, transcriptome, and proteome alterations induced by cigarette smoke in rat lung. Eur J Cancer. 2005;41:1864–1874. doi: 10.1016/j.ejca.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Liu C G, Calin G A, Meloon B, Gamliel N, Sevignani C, Ferracin M, Dumitru C D, Shimizu M, Zupo S, Dono M, Alder H, Bulrich F, Negrini M, Croce C M. An oligonucleotide microchip for genome-wide microRNA profiling in human and mouse tissues. Proc Natl Acad Sci U S A. 2004;101:9740–9744. doi: 10.1073/pnas.0403293101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Piana A, Cartigilia C, Longobardi M, De Flora S. Interplay between Helicobacter pylori and host gene polymorphisms in inducing oxidative DNA damage in the gastric mucosa. Carcinogenesis. 2007;28:892–898. doi: 10.1093/carcin/bgl208. [DOI] [PubMed] [Google Scholar]
- Saunders M A, Liang H, Li W-H. Human polymorphism at microRNAs and microRNA target sites. Proc Natl Acad Sci U S A. 2007;104:3300–3305. doi: 10.1073/pnas.0611347104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan R, Pak C, Jin P. Single nucleotide polymorphism associated with mature mirR-125a alters the processing of pri-miRNA. Human Mol Gen. 2007;16:1124–1131. doi: 10.1093/hmg/ddm062. [DOI] [PubMed] [Google Scholar]
- Sagar J, Chaib B, Sales K, Winslet M, Seifalian A. Role of stem cells in cancer therapy and cancer stem cells: a review [Electronic version] Cancer Cell Int. 2007;7:9. doi: 10.1186/1475-2867-7-9. doi:10.1186/1475-2867-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Neilson J R, Kumar P, Manocha M, Shankar P, Sharp P A, Manjunath N. MiRNA profiling of naïve, effector and memory CD8 T cells. PloS ONE. 2007;2:e1020. doi: 10.1371/journal.pone.0001020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y, Zheng J, Lambrecht R W, Bonkovsky H L. Reciprocal effects of micro-RNA-122 on expression of heme oxygenase-1 and hepatitis C virus genes in human hepatocytes. Gastroenterology. 2007;133:1166–1174. doi: 10.1053/j.gastro.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izzotti A, Balansky R M, Cartiglia C, Camoirano A, Longobardi M, De Flora S. Genomic and transcriptional alterations in mouse fetus liver after transplacental exposure to cigarette smoke. FASEB J. 2003;17:1127–1129. doi: 10.1096/fj.02-0967fje. [DOI] [PubMed] [Google Scholar]
- Izzotti A, Cartiglia C, Longobardi M, Balansky R M, D'Agostini F, Lubet R A, De Flora S. Alterations of gene expression in skin and lung of mice exposed to light and cigarette smoke. FASEB J. 2004;18:1559–1561. doi: 10.1096/fj.04-1877fje. [DOI] [PubMed] [Google Scholar]
- Johnson S M, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert K L, Brown D, Slack F J. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Johnson C D, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, Wilson M, Wang X, Shelton J, Shingara J, Chin L, Brown D, Slack F J. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, Stephens R M, Okamoto A, Yokota J, Tanaka T, Calin G A, Liu C G, Croce C M, Harris C C. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack F J. Oncomirs–microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- He L, He X, Lim L P, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson A L, Linsley P S, Chen C, Lowe S W, Cleary M A, Hannon G J. A microRNA component of the p53 tumor suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Flora S, Balansky R M, D'Agostini F, Izzotti A, Camoirano A, Bennicelli C, Zhang Z, Wang Y, Lubet R A, You M. Molecular alterations and lung tumors in p53 mutant mice exposed to cigarette smoke. Cancer Res. 2003;63:793–800. [PubMed] [Google Scholar]
- Chang T C, Wentzel E A, Kent O A, Ramachandran K, Mullendore M, Lee K H, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein C J, Arking D E, Beer M A, Maitra A, Mendell J T. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Zeiher A M, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- Balansky R, Ganchev G, Iltcheva M, Steele V E, D'Agostini F, De Flora S. Potent carcinogenicity of cigarette smoke in mice exposed early in life. Carcinogenesis. 2007;28:2236–2243. doi: 10.1093/carcin/bgm122. [DOI] [PubMed] [Google Scholar]
- Lee R C, Feinbaum R L, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Fujimoto N, Wislez M, Zhang J, Iwanaga K, Dackor J, Hanna A E, Kalyankrishna S, Cody D D, Price R E, Sato M, Shey J W, Minna J D, Peyton M, Tang X, Massarelli E, Herbst R, Threadgill D W, Wistuba I I, Kurie J M. High expression of ErbB family members and their ligands in lung adenocarcinomas that are sensitive to inhibition of epidermal growth factor receptor. Cancer Res. 2005;65:11478–11485. doi: 10.1158/0008-5472.CAN-05-1977. [DOI] [PubMed] [Google Scholar]