Abstract
The ubiquitin proteasome system (UPS) degrades abnormal proteins and most unneeded normal proteins, thereby playing a critical role in protein homeostasis in the cell. Proteasome inhibition is effective in treating certain forms of cancer, while UPS dysfunction is increasingly implicated in the pathogenesis of many severe and yet common diseases. It has been previously shown that doxorubicin (Dox) enhances the degradation of a UPS surrogate substrate in mouse hearts. To address the underlying mechanism, in the present study, we report that 1) Dox not only enhances the degradation of an exogenous UPS reporter (GFPu) but also antagonizes the proteasome inhibitor-induced accumulation of endogenous substrates (e.g., β-catenin and c-Jun) of the UPS in cultured NIH 3T3 cells and cardiomyocytes; 2) Dox facilitates the in vitro degradation of GFPu and c-Jun by the reconstituted UPS via the enhancement of proteasomal function; 3) Dox at a therapeutically relevant dose directly stimulates the peptidase activities of purified 20S proteasomes; and 4) Dox increases, whereas proteasome inhibition decreases, E3 ligase COOH-terminus of heat shock protein cognate 70 in 3T3 cells via a posttranscriptional mechanism. These new findings suggest that Dox activates the UPS by acting directly on both the ubiquitination apparatus and proteasome.
Keywords: cardiomyocytes, COOH-terminus of heat shock protein cognate 70-interacting protein, heat shock protein 70
targeted proteolysis by the ubiquitin (Ub)-proteasome system (UPS) includes two main steps: 1) the attachment of a series of Ub molecules to the target protein molecule via a process known as ubiquitination and 2) degradation of the ubiquitinated proteins by the proteasome (32, 36, 50, 53). Ubiquitination is a cascade of enzymatic reactions catalyzed collaboratively by the Ub-activating enzyme (E1), Ub-conjugating enzymes (E2s), Ub ligases (E3s), and occasionally a Ub chain elongation factor (E4) (23). A series of recent studies using genetic manipulations on specific E3s have demonstrated that altered ubiquitination of specific proteins yields a significant negative impact on cardiac function (11, 25, 51). The degradation of polyubiquitinated proteins is generally carried out by the 26S proteasome. UPS dysfunction has been implicated in a number of diseases, including cardiomyopathies, although its pathophysiological significance remains largely undefined (32, 50, 53). A major impediment is the lack of means to enhance proteasomal function.
Cardiotoxicity is a major hurdle limiting the use of doxorubicin (Dox), a potent anticancer agent of the anthracycline family (34). The mechanism by which Dox injures the heart remains to be fully elucidated (34). Oxidative stress from Dox metabolites may be an important cause, but antioxidant therapy could not completely abolish the cardiotoxicity, suggesting that additional mechanisms are involved (28). Perturbation of calcium handling and selective inhibition of cardiac muscle-specific gene expression, for instance, have also been implicated (34). More recently, several reports have indicated that Dox activates proteasome-mediated degradation of specific transcription factors (17, 38). Moreover, the proteasome has been believed to be used as a carrier for the translocation of Dox from the cytoplasm to the nucleus; thereby, its function could be altered by Dox (5, 19–21). Using a recently validated full-length protein reporter (GFPdgn) for the UPS, we (24) have demonstrated previously in intact mice that Dox enhances the degradation of a surrogate UPS substrate. However, the mechanism by which Dox activates the UPS remains to be elucidated.
In the present study, we sought to test whether Dox facilitates the degradation of endogenous bona fide substrates of the UPS and, more importantly, to determine how Dox acts on the UPS. Our findings suggest that Dox promotes the degradation of bona fide endogenous UPS substrates and activates the UPS by acting directly on both ubiquitination apparatuses and the proteasome.
MATERIALS AND METHODS
Materials.
Antibodies (A-150) against the COOH-terminal of heat shock protein cognate 70-interacting protein (CHIP), ubiquitin conjugation enzymes (F-340), the energy regenerating solution (B-10), and Ub (U-100) were purchased from Boston Biochem (Cambridge, MA), and mouse monoclonal anti-green fluorescent protein (GFP; clone2), rabbit polyclonal anti-c-Jun (N), and horseradish peroxidase-conjugated goat anti-mouse/rabbit secondary antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). 26S and 20S proteasomes, MG132, MG262, and epoxomycin (EPO) were purchased from BioMol (Plymouth Meeting, PA). Dox, cycloheximide (CHX), and rabbit polyclonal anti-Ub antibody were purchased from Sigma-Aldrich (St. Louis, MO). The ECL-plus enhanced chemiluminescence detection kit was from Amersham Bioscience (Piscataway, NJ). BCA reagents and the immunoprecipitation kit were from Pierce Biotechnology (Rock, IL). Recombinant human c-Jun was from Promega (Madison, WI), and rabbit anti-heat shock protein 70 (Hsp70) antibodies from Stressgen (Ann Arbor, MI).
Creation of GFPu-3T3 stable cell lines.
GFPu is a GFP modified with carboxyl fusion of degron CL1. It has proven to be a specific substrate for the UPS (2, 4). A plasmid harboring the GFPu expression cassette was donated by Dr. Ron Kopito of Stanford University (2). NIH 3T3 fibroblasts were purchased from the American Type Culture Collection and cultured in DMEM supplemented with 10% calf serum (HyClone, Logan, UT) and complete antibiotics. To generate stably transfected cell lines, 50–70% confluent cultures were transfected with the GFPu plasmid using FuGENE 6 transfection reagents (Roche Diagnostics, Indianapolis, IN). Two days after transfection, selection medium containing G418 was added. G418-resistant clones were isolated by 10 days after the addition of selection medium. Stably transfected clones were maintained in growth medium containing G418.
Neonatal rat ventricular myocytes (NRVMs) and adult mouse cardiomyocytes cultures were created as previously reported (4, 24). The protocol for the care and use of animals in this study was approved by University of South Dakota Institutional Animal Care and Use Committee.
Peptidase activity assays and in vitro Dox treatment of purified 20S proteasomes.
A total volume of 180-μl reaction mixture in Tris·HCl (pH 7.5) buffer containing purified 20S proteasomes (0.125 μg, BioMol) and various concentrations of Dox (0∼10 μM), H2O2 (30 or 150 μM), or proteasome inhibitors was incubated on ice for 30 min before 20 μl of the synthetic fluorogenic peptide Suc-LLVY-aminomethylcoumarin (AMC) (final concentration: 25 μM, Calbiochem) was added. The reaction mixture was then incubated at 37°C for 90 min. The reaction was stopped by the addition of 300 μl ethanol. A 2.0-ml aliquot of H2O was then added, and the fluorescence intensity of the samples was evaluated using a luminescence spectrometer (model LS 55, Perkin-Elmer). The excitation and emission wavelengths were 360 and 436 nm, respectively, for AMC products.
In vitro GFPu and c-Jun ubiquitination/degradation assays.
In vitro GFPu and c-Jun ubiquitination/degradation assays were performed using the purified 26S proteasome (BostonBiochem, Boston, MA) and the Ub-protein conjugation kit K-960 (BostonBiochem), which contains the full complement of purified Ub conjugation enzymes (E1, E2s, and E3s) from red blood cells, Ub, and the energy-regenerating system. Purified proteasomes and substrates were pretreated with Dox on ice for 30 min and then incubated with the ubiquitination reaction buffer for 60 min. The reaction was terminated by the addition of 3× Laemmli sample buffer, immediately followed by boiling for 5 min. The end products were then fractionated by 12% SDS-PAGE and detected by Western blot analysis.
Western blot analysis.
Protein extracts were fractionated by 10–12% SDS-PAGE and electrically transferred onto polyvinylidene difluoride membranes. Mouse or rabbit antibodies were used as the primary antibodies, and horseradish peroxidase-conjugated goat anti-mouse IgG or anti-rabbit antibodies were, respectively, used as secondary antibodies to detect the designated proteins. ECL-Plus detection reagents (Amersham Bioscience) and a VersaDoc imaging system (model 3000, Bio-Rad) were used to visualize and digitalize the Western blot images. The densitometry of Western blots was carried out using Quantity One software (Bio-Rad) as previously described (4). For the quantification of total ubiquitinated proteins (i.e., Fig. 3), all positive bands with a molecular weight greater than that of free Ub (7.6 kDa) were included.
Semiquantitative RT-PCR.
Isolation of total RNA and RT-PCR were performed as previously described (43). The primer sequences for CHIP and GAPDH were the same as those previously reported (22). The sequences of the primers specific for HSP70 were as follows: forward 5′-GGTGGTGCAGTCCGACATG-3′ and reverse 5′-TACGCCTCAGCGATCTCCTTC-3′. GAPDH was amplified as the internal control for potential variations in RNA sampling and the efficiency of reverse transcription. The numbers of PCR cycles were optimized for each transcript to avoid potential PCR saturation.
Statistics.
Means ± SD are presented where applicable. An unpaired Student's t-test was used for the determination of statistic probabilities. P values of <0.05 were considered significant.
RESULTS
Dox enhances GFPu degradation in both 3T3 cells and NRVMs.
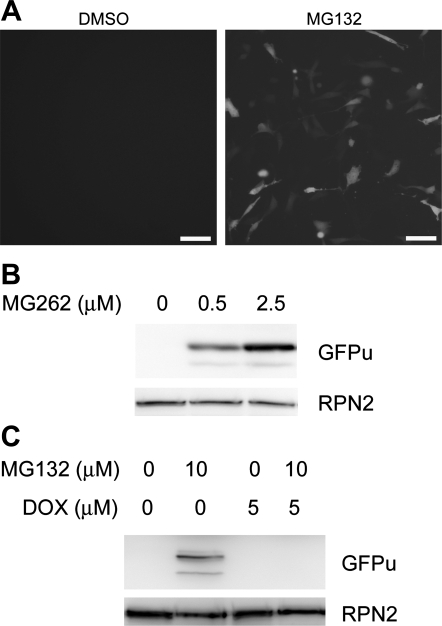
To efficiently monitor the dynamic changes in the proteolytic function of the UPS, we created multiple clonal stable 3T3 cell lines that constitutively expressed a surrogate substrate for the UPS, a modified GFP (GFPu) that was created by fusion of the yeast CL1 degron to the COOH-terminus of an enhanced GFP. The dependence of GFPu degradation on the UPS in NIH 3T3 cells was verified by the dose-dependent accumulation of GFPu protein by proteasome-specific inhibitors MG132 and MG262 (Fig. 1). This was in agreement with previous reports (2, 26) using cultured NRVMs and HEK-293 cells. Importantly, MG-132-induced GFPu protein accumulation was abolished by cotreatment with Dox (5 μM; Fig. 1C). However, RPN2 (a subunit of the 19S proteasome) did not appear to be significantly affected by the treatments and was used as a loading control (Fig. 1, B and C).
Fig. 1.
Doxorubicin (Dox) enhances GFPu degradation in 3T3 cells. A: epifluorescence micrographs of GFPu stably transfected 3T3 cells 6 h after treatment with a proteasome inhibitor (MG132; 10 μM) or vehicle control (DMSO). GFPu is a green fluorescent protein modified with carboxyl fusion of degron CL1. Scale bar = 10 μm. B: representative Western blot images showing dose-dependent increases in GFPu by MG262. C: Western blot images showing that MG132-induced GFPu accumulation was abolished by cotreatment with Dox. In both A and B, the same membranes were reprobed for RPN2 (a subunit of the 19S proteasome) to illustrate that the amount of total proteins loaded to the lanes absent of GFPu signals was not less than the lanes where GFPu was detected.
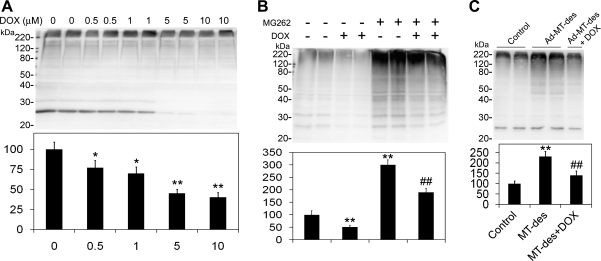
From the lysate of cultured NRVMs cotransfected with high titers of GFPu adenoviruses (Fig. 2A) and mutant desmin adenoviruses, GFPu was coimmunoprecipitated with anti-Ub antibodies, and higher-molecular-weight species (ubiquitinated) of GFPu were evident (Fig. 2, B and C). Dox treatment (1 μM) significantly reduced these GFPu proteins with the desmin protein levels remaining unaffected in these cells (Fig. 2, D and E). In cultured cardiomyocytes from adult GFPdgn transgenic mice (24), Dox-induced decreases in GFPdgn (a shorter version of GFPu) became more pronounced when protein synthesis was blocked by CHX (25 μM; Fig. 2, F and G), suggesting that the degradation of GFPdgn is enhanced by Dox. These in vitro experiments using cardiomyocytes and nonmyocytes verified our previous in vivo findings showing that Dox enhances UPS-mediated degradation of a surrogate substrate protein.
Fig. 2.
Dox enhances GFPu degradation in cultured ventricular myocytes. GFPu was forced to express in cultured neonatal rat ventricular myocytes (NRVMs) through adenoviral gene delivery. A: direct fluorescence micrographs of GFPu adenoviruses (Ad-GFPu)-infected NRVMs and control virus (Ad-β-Gal)-infected NRVMs. Scale bar = 10 μm. B: representative image of immunoblot (IB) analysis for GFPu in the immunoprecipitates (IP) of an anti-ubiquitin (Ub) antibody. C: Dox (10 μM) reduced GFPu protein levels in NRVMs. The full-length native GFPu band is indicated by an arrow (the same for D). D: GFPu and a mutant desmin (Des) were coexpressed in NRVMs. Compared with the treatment of the vehicle control (mock), Dox (10 μM for 24 h) significantly decreased the level of GFPu proteins [both the native form (indicated by the arrow) and the ubiquitinated forms, which have higher molecular weights] but not desmin protein. E: bar graph summarizing 4 independent repeats of the GFPu and desmin protein denisitometry data of the experiments shown in D. AU, arbitrary units. *P < 0.05 vs. mock. F: representative image of Western blot analysis for GFPdgn in the cycloheximide (CHX) chase experiment. Cardiomyocytes were isolated from adult mice expressing GFPdgn (a shorter form of GFPu) and cultured for 24 h before being treated with Dox or Dox + CHX. G: summary of the densitometry data from 2 repeats of the duplicated CHX chase experiments shown in F. *P < 0.05 vs. 0 h; #P < 0.05 and ##P < 0.01 vs. Dox.
Dox decreases endogenous ubiquitinated proteins and enhances the degradation of endogenous substrates of proteasomes.
Accumulation of ubiquitinated proteins is an important feature of proteasomal inhibition. To further analyze the effect of Dox on UPS proteolytic function, proteasome inhibition was induced by a proteasome inhibitor (MG262) in cultured NRVMs. Treatment of Dox at a dose of 10 μM or less significantly reduced the levels of Ub conjugates at the baseline condition (Fig. 3A) and markedly attenuated the accumulation of ubiquitinated proteins by proteasome inhibition caused by MG262 (Fig. 3B). We (26, 27) have previously reported that transgenic expression of a human desmin-related myopathy-linked mutant desmin in cardiomyocytes causes proteasome inhibition and accumulates ubiquitinated proteins. To further test whether Dox treatment can reduce mutant desmin expression-induced ubiquitinated protein accumulation, we overexpressed mutant desmin in cultured NRVMs via adenoviruses-mediated gene delivery. Consistent with our previous reports (26, 27), expression of mutant desmin caused significant increases in ubiquitinated proteins in the cells (Fig. 3C). Notably, the mutant desmin-induced accumulation of ubiquitinated proteins was also markedly attenuated by Dox treatment (2.5 μM; Fig. 3C).
Fig. 3.
Effects of Dox on the abundance of ubiquitinated proteins in NRVMs. Total protein extracts from cultured cells were fractionated by SDS-PAGE and analyzed for Ub conjugates using an anti-Ub antibody and Western blot analysis. For A–C, a representative Western blot image is shown at the top and a bar graph summarizing the densitometry data of Ub conjugates from 4 repeats is shown at the bottom. A: dose response of Dox on ubiquitinated proteins. B: NRVMs were treated with (+) or without (−) MG262 (5 μM) and/or Dox (2.5 μM) as indicated for 24 h in culture. *P < 0.05 and **P < 0.01 compared with 0 μM Dox; ##P < 0.01 compared with MG262 alone. C: effects of Dox on the increase of ubiquitinated proteins induced by mutant desmin (MT-des) expression. NRVMs were infected with adenoviruses expressing MT-des (Ad-MT-des) for 72 h before being treated with Dox (2.5 μM) for 24 h. *P < 0.05 compared with control; ##P < 0.01 compared with MT-des.
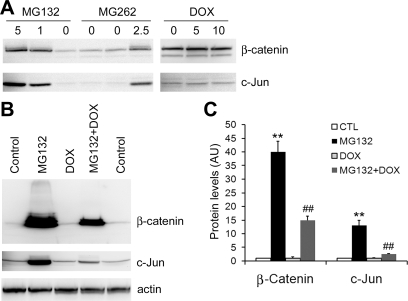
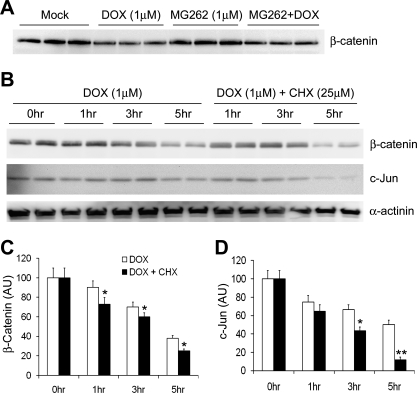
Posttranslational modifications of a protein molecule play an important role in the degradation of the protein by the UPS. To verify the prodegradation effect of Dox on ubiquitinated proteins, two endogenous typical proteasome substrates and important transcription factors, c-Jun and β-catenin, were further examined. β-Catenin participates in a large cytoplasmic protein complex containing the serine/threonine protein kinase glycogen synthase kinase (GSK)-3β, the tumor suppressor gene product adenomatous polyposis coli (APC), and axin/conductin (13). In the absence of Wnt, GSK-3β is constitutively active and promotes the degradation of β-catenin by NH2-terminal phosphorylation and subsequent ubiquitination and proteasomal targeting (13). The activity of c-Jun depends on its cellular concentration, and the latter is regulated at both synthesis and proteasome-dependent degradation (52). In addition, posttranslational modifications, such as phosphorylation and dephosphorylation in response to many stimuli, modulate both the activity and stability of the transcription factor. In NIH 3T3 cells, both c-Jun and β-catenin protein levels were low, and Dox did not affect their protein levels at the baseline condition (Fig. 4A). When the cells were exposed to proteasome inhibitor, these two substrates dramatically increased. Dox (2.5 μM) significantly decreased the elevation of c-Jun and β-catenin proteins induced by MG132 (Fig. 4, B and C). Cardiomyocytes contain more abundant β-catenin (13, 49). In NRVMs, both the baseline and proteasome-inhibitor-induced β-catenin protein expression were decreased by Dox treatment (Fig. 5A). Interestingly, the steady-state protein level of β-catenin did not seem to be significantly increased at 16 h after MG262 treatment in cultured NRVMs (Fig. 5A). This unexpected result was likely caused by a simultaneous decrease in β-catenin protein synthesis during a relatively long term of proteasome inhibition resulting from MG262 (1 μM). To minimize potential complications from changes in protein synthesis, we then carried out shorter-term experiments using adult mouse cardiomyocytes, which have a lower rate of protein synthesis under the in vitro culture condition (Fig. 5B). In these experiments (Fig. 5, B–D), we found that 1 μM Dox induced time-dependent decreases in the protein levels of both β-catenin and c-Jun within 5 h, and the effects became more pronounced when protein synthesis was blocked by pretreatment with CHX (25 mM). These data indicate that the decreases of these endogenous UPS substrates by Dox are at least in part attributable to enhanced degradation. In cardiomyocytes, the enhanced degradation of c-Jun and β-catenin is a potential mechanism underlying the cardiac toxicity of Dox.
Fig. 4.
Dox enhances the degradation of endogenous proteasome substrates c-Jun and β-catenin in 3T3 cells. A and B are images of Western blot. A: c-Jun and β-catenin protein levels in 3T3 cells treated with Dox or proteasome inhibitors MG132 and MG262. 3T3 cells were treated with different doses (in μM) of MG132, MG262, or Dox for 6 h. Cells were extracted with SDS lysis buffer. B and C: 3T3 cells were treated with MG132 (5 μM) and/or Dox (2.5 μM) for 12 h. The levels of β-catenin and c-Jun in total protein extracts were measured. Actin was probed as a loading control. A set of representative images is shown in B, and the quantitative data of 3 repeats are shown in C. **P < 0.01, MG132 group compared with control (CTL) group; ##P < 0.01, MG132 + Dox group compared with the MG132 group.
Fig. 5.
Dox enhances the degradation of endogenous proteasome substrates c-Jun and β-catenin in cultured cardiomyocytes. A: representative image of Western blot for β-catenin protein levels in NRVMs treated with Dox (1 μM) and/or MG262 (1 μM) for 16 h. Note that Dox decreased β-catenin protein levels both at baseline and in the presence of a proteasome inhibitor. B–D: CHX chase experiments for β-catenin and c-Jun proteins. Cultured adult mouse cardiomyocytes were treated with Dox (1 μM) for the indicated times in the absence or presence of CHX (25 mM) pretreatment (1 h before Dox treatment). A representative cohort of Western blot images is shown in B, and quantitative data from 4 repeats are shown in C (β-catenin) and D (c-Jun), respectively. α-Actinin (a relatively stable cytoskeletal protein) was also probed with the same membranes. To minimize the impact from potential loading variations, both β-catenin and c-Jun signals were normalized to the same lane α-actinin signal. The average value of the 0-h time point (i.e., Dox untreated) was arbitrarily set as 100 AU and was used to quantify the other time points. *P < 0.05 and **P < 0.01, Dox + CHX group compared with CHX group at the same time point.
Dox facilitates the in vitro degradation of GFPu and c-Jun by the UPS through the enhancement of proteasome function.
All the evidence from the cell culture experiments described so far is consistent with the notion that Dox has a protein degradation-enhancing effect. However, the influence of protein synthesis is inevitably a compounding factor and could not be completely ruled out in those experiments. To address this issue, we performed in vitro ubiquitination and degradation of GFPu and c-Jun using a reconstituted UPS. For the GFPu degradation reaction, heat-inactivated protein extracts from cardiomyocytes infected with a high dose of Ad-GFPu were used as GFPu substrates. For c-Jun degradation assays, recombinant full-length c-Jun (Promega) was used. Western blot analyses of the substrate proteins (GFPu and c-Jun), as well as ubiquitinated proteins, at the end of the reactions showed that Dox (1 μM) significantly reduced the level of the native form of GFPu and the ubiquitinated form of GFPu (Fig. 6A, lanes f vs. e) and c-Jun (lane f of Fig. 6C, lanes f vs. e) and decreased the overall amount of Ub conjugates in the fully reconstituted ubiquitination and proteasome degradation system (Fig. 6, B and D, lanes f vs. e). In the assays shown in Fig. 6, B and D, lanes c and d, where the substrate protein GFPu (Fig. 6B) or c-Jun (Fig. 6D) was incubated with a fully reconstituted ubiquitination system in the absence of functional proteasomes, robust ubiquitination occurred because high-molecular-weight Ub conjugates were clearly detected; however, no statistically significant differences in the amount of Ub conjugates were discerned between the group in lane d (with Dox) and the group in lane c (without Dox), indicating that Dox does not have a general enhancing effect on the in vitro activity of the ubiquitination system. Also, notably, in the assay using the inactivated cell lysates containing GFPu as the substrate (Fig. 6B), the Ub conjugates were significantly reduced by Dox (lanes b vs. a) when the 26S proteasome was present but the ubiquitination system was absent (lanes a and b). This suggests that Dox can enhance 26S proteasomes-mediated degradation of preexisted ubiquitinated proteins independently of de novo ubiquitination. However, in the same experimental settings, when purified c-Jun was used as the substrate (Fig. 6D), Dox did not significantly alter the abundance of Ub conjugates (lanes b vs. a). This is likely because there were not proteasome-degradable preexisted ubiquitinated proteins in the purified c-Jun protein preparation. Taken together, these in vitro experiments prove that Dox at a relatively low dose can increase proteasome proteolytic function but not the activity of a general ubiquitination system.
Fig. 6.
Dox facilitates the degradation of GFPu and c-Jun in vitro. In A–D, a representative Western blot image is shown at the top and a bar graph is shown at the bottom to depict the densitometry data from 4 repeats. A and B: in vitro ubiquitination and degradation of GFPu in a cell-free system. Cardiomyocytes infected with a high dose of Ad-GFPu were homogenized, inactivated by heating, and used as GFPu substrates. The supernatant protein (15 μg) was incubated for 1 h in a degradation system with the indicated combinations of purified 26S proteasome (0.6 μg), a full complement of ubiquitination enzymes (E1–E3), and Dox (1 μM). GFPu (A) and ubiquitinated proteins (B) were analyzed using Western blots. The average density of the groups in lanes a, c, and e was set as 100 AU and used for calculating the relative densities of the groups in lanes b, d, and f, respectively. For statistics, lanes b vs. a, lanes d vs. c, and lanes f vs. e were compared (*P < 0.05). C and D: in vitro ubiquitination and degradation of c-Jun in a cell-free system. Recombinant full-length c-Jun (0.5 footprint units) was incubated in the same ubiquitination/degradation system as described in A and B. c-Jun (C) and Ub conjugates (D) were immunoblotted and quantified in the same way as described for A and B.
Interestingly, the enhancement by Dox on the proteasome in vitro was lost when Dox was used at a higher dose (10 μM or higher; data not shown).
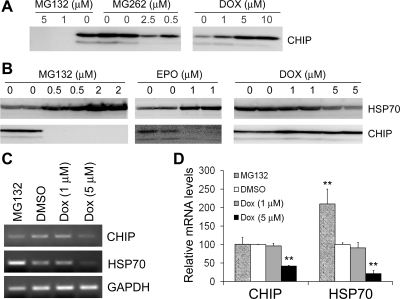
Dox and proteasome inhibitors regulate CHIP in opposing directions.
CHIP is a U box-containing Ub E3 ligase as well as a cochaperone of HSP70 (18). The abilities of CHIP to interact with molecular chaperones and function as a Ub ligase make CHIP the most attractive candidate of E3 ligase for ubiquitinating misfolded proteins, thereby likely playing a central role in protein quality control. CHIP may target non-native proteins for degradation by the proteasome (31). HSP70 itself is a typical CHIP target protein (39). Since Dox metabolites are known to increase ROS, which increase the production of misfolded proteins, we sought to examine the effect of Dox on the protein levels of CHIP and HSP70. We also included a most commonly used proteasome inhibitor (MG132) in our initial test. This is because our previous experiments have shown that Dox and proteasome inhibition have opposite effects on proteasome proteolytic function. To our surprise, MG132 unexpectedly and strikingly diminished CHIP protein levels in the cell even at relatively low doses (Fig. 7, A and B). To confirm that the findings were not unique to MG132 but rather common to proteasome inhibition, we subsequently tested a closely related proteasome inhibitor (MG262) and an inhibitor (EPO) of a different family. Very interestingly, three different proteasome inhibitors (MG132, MG262, and EPO) all dramatically decreased the CHIP protein level, whereas Dox markedly increased the CHIP protein level in 3T3 cells (Fig. 7, A and B). The changes in CHIP protein levels were accompanied by reciprocal changes in Hsp70 protein levels (Fig. 7B), consistent with the notion that Hsp70 is a target of CHIP (39).
Fig. 7.
Proteasome inhibitors decrease while Dox increases the COOH-terminal of heat shock protein cognate 70 (CHIP). A: representative Western blot images of CHIP in 3T3 cells treated with MG132, MG262, or Dox for 12 h at the indicated doses. B: Western blot analyses of CHIP and heat shock protein 70 (HSP70) in 3T3 cells treated with MG132, epoxomicin (EPO), or Dox at the indicated doses for 6 h. Experiments were repeated twice, and representative results are shown. C and D: semiquantitative RT-PCR analyses of CHIP and HSP70 transcript levels in 3T3 cells treated with MG132 (0.5 μM), Dox at the indicated doses, or DMSO (vehicle control) for 12 h. Representative gel images of the indicated PCR products are shown in C. GAPDH was included as the internal control against which CHIP and HSP70 were normalized. The average signal of DMSO-treated cells was set as 100 AU and used as the standard to calculate the relative transcript levels of other treatment groups. The results of 3 repeats are shown in D. **P < 0.01 vs. the DMSO group.
In addition, we examined the steady-state transcript levels of HSP70 and CHIP to determine whether the opposing effects of proteasome inhibition and Dox treatment on HSP70 and CHIP protein levels occurred at the transcription level or posttranscription level. The MG132 (0.5 μM) treatment significantly increased HSP70 transcript levels but failed to change CHIP in the 3T3 cells, suggesting that both increased synthesis and decreased degradation may contribute to the upregulation of HSP70 protein by proteasome inhibition. However, neither HSP70 nor CHIP transcript levels were significantly altered by Dox at 1 μM, whereas both were markedly decreased by 5 μM Dox at 12 h after treatment (Fig. 7, C and D), indicating that the Dox-induced increase in CHIP protein expression is transcription independent.
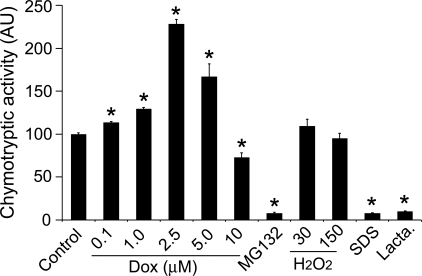
Dox directly regulates the peptidase activity of 20S proteasomes.
The above experiments show that Dox can enhance the degradation of both endogenous and exogenous protein substrates of the 26S proteasome, suggesting that Dox is a potent activator of the UPS, likely by acting on the initiation of ubiquitination and by enhancing proteasome proteolytic function. To determine whether Dox also exerts a direct effect on proteasome intrinsic peptidase activities, we further tested the effect of Dox on the peptidase activity of purified 20S proteasomes in a test tube. In UPS-mediated proteolysis, peptide cleavage occurs inside the 20S proteasome (53). Three measurable peptidase activities (chymotrypsin-like, caspase-like, and trypsin-like activities) have been described in 20S proteasomes, and they are, respectively, performed by β5-, β2-, and β1-subunits. Among them, chymotrypsin-like activity appears to be the most important one and is commonly evaluated using a synthetic fluorogenic peptide substrate. Here, we found that the chymotrypsin-like activity of purified 20S proteasomes (BioMol) was significantly enhanced by in vitro direct treatment of Dox (from 0.1 to 2.5 μM) in a dose-dependent manner (Fig. 8). Dox also showed a proteasome-enhancing effect at 5 μM, although the effect appeared to be less robust than at 2.5 μM. When the Dox dose was increased to 10 μM, treated 20S proteasomes started to show significantly decreased peptidase activity (Fig. 8). These results demonstrate a direct activation effect of Dox on proteasome proteolytic function at concentrations ranging between 0.1 and 5.0 μM. The direct effects of Dox on 20S proteasomes are unlikely related to its oxidative property because H2O2 treatments (30 or 150 μM) did not produce significant effects on proteasome peptidase activity in the companying experiments, although SDS (0.1%) and bona fide proteasome inhibitors MG132 (10 μM) and clasto-lactacystin β-lactone (1 μM) were able to inhibit the activity very effectively (Fig. 8).
Fig. 8.
Dox directly regulates the peptidase activity of purified 20S proteasomes. Purified 20S proteasomes were pretreated with Dox at the indicated doses, MG-132 (10 μM), H2O2 (30 and 150 μM), SDS (0.1%), or clasto-lactacystin β-lactone (Lacta; 1 μM) on ice for 30 min before being assayed for chymotryptic activities. The mean value of DMSO-treated cells (control) was set as 100 AU and used to normalize the other treatment groups. Results of 4 repeats are shown. *P < 0.05 or 0.01 vs. controls.
DISCUSSION
Although Dox had been shown to enhance the degradation of a surrogate protein substrate of the UPS in mouse hearts (24), it was unclear whether the effect resulted from the direct action of Dox on the UPS or from Dox-induced modifications on the substrate, especially in light of the fact that Dox and its metabolites are known to increase ROS and ROS may increase the production of oxidized proteins. Being substrates, oxidized proteins may indirectly activate UPS-mediated proteolysis. After verifying that Dox also enhances the degradation of bona fide endogenous UPS substrates in cultured cells, the present study demonstrated, for the first time, that Dox at a dose range relevant to human therapeutic doses can directly increase UPS-mediated protein degradation in a cell-free system and even directly enhance peptide cleavage activities of the 20S proteasome in the test tube. Furthermore, we discovered that through a posttranscriptional mechanism, Dox upregulates, whereas proteasome inhibition downregulates, the protein levels of CHIP, a critical E3 ligase involved in protein quality control (30, 31, 39). These new discoveries suggest that Dox activates the UPS at least in part through acting directly on both ubiquitination apparatuses and the proteasome.
Dox at the clinically relevant dose range activates directly the proteasome.
Dox has been shown to both inhibit and activate UPS proteolytic activities in experimental settings (10, 24, 45). The present study reveals that a critical factor for this dichotomy is likely the dosage of Dox. By incubating purified 20S proteasomes with Dox in test tubes, we were able to demonstrate that Dox directly activated 20S proteasome peptidases at doses ranging between 0.1 and 5 μM but inhibited the proteasome at a higher dose (10 μM; Fig. 8). Consistently, we also observed using in vitro protein degradation assays that Dox at 1 μM enhanced the degradation of bona fide full-length substrate proteins by the fully reconstituted UPS (Fig. 6) but that Dox failed to do so at 20 μM (data not shown). From 5 to 10 μM, the dose increases by only a factor of 2, but the clinical relevance differs sharply. In clinical cancer chemotherapy, Dox is usually administered intravenously over a brief period (e.g., 15 min) at a dose of 60∼75 mg/m2 in humans, with the peak plasma concentration ranging between 5 and 15 μM and an average half-life of ∼25 h (35, 41). Considering that ∼75% of Dox in the plasma is bound by plasma proteins independently of its plasma concentration up to 2 mM (12), the peak concentration of free Dox (available for acting on cells) is between 1.25 and 3.75 μM, which is below 5 μM and well within the dose range (0.1∼5 μM) that activates the proteasome in vitro. Therefore, a typical therapeutic dose of Dox should activate proteasomes in humans. In other words, the dose range of Dox that activates the proteasome is clinically relevant, whereas the concentration of Dox required to inhibit the proteasome appears to be too high to be clinically relevant.
Consistently with the findings from the cell-free systems, our tests with the cell culture (Figs. 1–5) have also convincingly demonstrated a significant enhancement of UPS-mediated protein degradation of both surrogate and endogenous substrates by Dox treatments. GFPu is a surrogate substrate for the proteasome (2). Dox (5 μM) effectively antagonized MG132-induced GFPu protein accumulation in 3T3 cells (Fig. 1). Dox (10 μM or lower) significantly destabilized GFPu but not desmin in cultured NRVMs (Fig. 2). More importantly, Dox at a dose up to 10 μM induced a dose-dependent reduction of endogenous ubiquitinated proteins in cultured NRVMs at baseline (Fig. 3A) and significantly attenuated the accumulation of Ub conjugates induced either by a pharmacological inhibitor (MG262; Fig. 3B) or by expression of a misfolded desmin protein (Fig. 3C). Furthermore, Dox (2.5 μM) significantly attenuated MG132-induced accumulation of β-catenin and c-Jun (Fig. 4, B and C), two bona fide proteasome substrate proteins in 3T3 cells (Fig. 4A). Similarly, Dox (1 μM) destabilized β-catenin and c-Jun in cultured cardiomyocytes (Fig. 5). Notably, Dox in these cell culture experiments displayed UPS enhancing effects at a dose ranging from 0.5 to 10 μM, which seems to be inconsistent with the proteasome-activating dose range (0.1∼5 μM) observed in the in vitro experiments using reconstituted UPS (Fig. 6) or purified 20S proteasomes (Fig. 8). This seeming “inconsistency” actually underscores the importance of Dox bioavailability to the proteasome. In the cell culture settings, a significant fraction of Dox molecules added to the culture medium, just like Dox intravenously administered to humans, will be bound by nonproteasome factors in the culture medium and inside the cell such that the actual amount of free Dox available for interacting directly with proteasomes in the cell is inevitably lower than the nominal concentration initially used in the culture media. Therefore, the nominal concentration of Dox in cell culture to enhance UPS proteolytic function can reasonably be higher than the highest concentration observed for Dox to directly activate purified proteasomes in the test tube.
Several previous reports by others have in fact provided indirect evidence that Dox enhances UPS-mediated protein degradation (17, 29, 38). IκB-α is a very important substrate of the UPS. In quiescent cells, the active form of NF-κB in the cytosol remains bound to its inhibitory molecule, IκB. Upon stimulation by proinflammatory cytokines or oxidants, IκB is first phosphorylated by upstream kinases and then polyubiquitinated and degraded by proteasomes (44), resulting in the release and translocation of the NF-κB complex into the nucleus, where NF-kB regulates the expression of its target genes, which mediate inflammatory responses, apoptosis, and carcinogenesis. Dox has been shown to induce a dose- and time-dependent activation of NF-κB in both bovine aortic endothelial cells and adult rat cardiomyocytes (48). p300 is a general transcriptional coactivator that controls many biological activities such as cellular growth, cellular differentiation, tumorigenesis, and apoptosis. p300 is essential for heart development, regulates cardiomyocyte-specific gene expression, and plays a critical role in cardiac hypertrophy (37). It has been reported that exposure of cardiac cells to Dox caused UPS-mediated depletion of p300 protein, although the enhanced p300 degradation was considered a specific effect of Dox (37, 38). Nuclear factors of activated T cells (NFATs) are important transcription factors in cardiomyocytes (16). A recent study (17) has shown that Dox induced depletion of NFAT5 by proteasome-mediated degradation. Here, we show that Dox enhances UPS-mediated degradation of two other important transcription regulators, β-catenin and c-Jun, in both cultured cells (Figs. 4 and 5) and test tubes (Fig. 4). Taken together, proteasome-mediated degradation of a variety of transcription factors, including ones critical to the heart, have proven to be significantly enhanced by Dox, which is consistent with the general enhancement of UPS proteolytic function by Dox that we observed previously and more definitively in the present study.
Interestingly, enhanced proteasome peptidase activities have recently been reported in the genesis of pressure-overload cardiac hypertrophy and remodeling and during chaperone H11 kinase/HSP22 overexpression-induced cardiac growth, and pharmacologically induced proteasome inhibition has been shown to suppress the reactivation of the cardiomyocyte fetal gene program and pressure overload-induced cardiac hypertrophic growth and remodeling (7, 14, 15, 33, 42). These results suggest that increasing proteasome proteolytic function may be detrimental to the heart. Notably, it has also recently been shown that proteasome inhibition specifically abolishes Dox-induced DNA damage signals in H9C2 cardiomyocytes (29), implicating the participation of proteasome activation in the chronic cardiotoxicity of Dox.
Mechanisms by which Dox alters the activity of the proteasome.
Dox has been previously shown to be able to bind proteasomes in the cell, and proteasomes have been considered to be the carrier for Dox to enter the nucleus (20, 21). The binding of Dox to proteasomes was believed to inhibit proteasome function (10, 19). Indeed, we observed in the present study that a higher dose (e.g., 10 μM or higher) of Dox slowed down the proteasome-dependent in vitro degradation of GFPu by 3T3 cell extracts, failed to facilitate the in vitro degradation of GFPu and c-Jun by the reconstituted UPS (data not shown), and significantly decreased the peptidase activity of purified 20S proteasomes in test tubes (Fig. 8). However, we also discovered that the more clinically relevant lower doses of Dox not only facilitated UPS-mediated degradation of full-length proteins in cultured cells and in vitro (Figs. 3–7) but also increased the chymotrypsin-like activity of purified 20S proteasomes in test tubes (5.0 μM or less; Fig. 8). The mechanisms by which Dox alters proteasome activities remain to be elucidated. A direct oxidative modification on the proteasome does not appear to be the cause, because our in vitro tests using purified 20S proteasomes showed no significant effect of H2O2 (30∼150 μM) treatments on the activity, whereas the effects of Dox were readily detected (Fig. 8). It is nevertheless conceivable that the binding of a small amount of Dox to the 20S proteasome may stimulate the latter's activity by changing its conformation such that the catalytic sites work more efficiently while the binding of a larger amount of Dox may jam the proteasome and slow down the substrate flux through the chamber of the 20S proteasome. Dox and its metabolites induced posttranslational modifications to UPS substrates, and proteasome subunits may also play a role in its effects on UPS-mediated protein degradation. Tsimokha et al. (45) recently reported that Dox treatment can increase the phosphorylation of proteasome subunits and enhance the chymotrypsin-like activity of the 26S proteasome in cultured neoplastic cells.
Dox triggers posttranscriptional increases in CHIP protein expression.
Another interesting discovery of the present study is that Dox increased, whereas various proteasome inhibitors all remarkably decreased, CHIP protein levels in the cells (Fig. 7, A and B). In agreement with a recent report (39) showing that CHIP is the Ub E3 ligase for the degradation of HSP70, reciprocal changes in HSP70 protein levels were observed upon the treatments of proteasome inhibitors and Dox (Fig. 7B). As a potentially major Ub E3 ligase for removing abnormal proteins in the cytosol, CHIP can be upregulated by the overexpression of misfolded proteins, and the overexpression of CHIP, in turn, decreases the accumulation of misfolded proteins in the cell (8, 54). The decrease in CHIP protein levels by proteasome inhibition might conceivably be intended by the cell to stabilize HSP70 for dealing with non-native proteins accumulated by proteasome inhibition, but the increase of CHIP and the reciprocal decrease of HSP70 caused by Dox likely diminish the cell's ability to handle stress and may contribute to Dox cytotoxicity.
Increasing CHIP protein levels in the cell by Dox appears to be mediated by a posttranscriptional mechanism. This is because the transcript level of CHIP in the cell was not significantly increased by Dox treatments (Fig. 7, C and D). To the contrary, Dox at 5 μM significantly decreased the transcript levels of both CHIP and HSP70. Similarly, the downregulation of CHIP protein by proteasome inhibition also seems to be mediated by a transcription-independent mechanism because the steady-state transcript level of CHIP was not significantly altered, although HSP70 transcript levels were markedly increased, by the treatment of MG132 at a concentration (0.5 μM) sufficient to decrease the abundance of CHIP proteins (Fig. 7, C and D). Although Dox at 1 μM did not change HSP70 transcript levels, Dox at 5 μM significantly reduced HSP70 transcript levels in 3T3 cells. Hence, the downregulation of HSP70 protein in Dox-treated cells may possibly result from both decreased synthesis and increased degradation, especially when Dox is used at the higher end of the therapeutic dosage. It will be interesting to further decipher the posttranscriptional mechanisms underlying the opposing effects on CHIP protein expression between Dox treatment and proteasome inhibition.
Clinical implications.
With rare exceptions, virtually all studies on the involvement of the proteasome in pathology and/or pharmacology to date have emphasized the significance of proteasome functional insufficiency in many pathophysiological processes (3, 40, 46). Dox appears to be the only exception in that it can enhance UPS proteolytic function in degrading both surrogate and endogenous substrates. Although a causal link between the UPS-activating effects of Dox and its cardiotoxicity remains to be established, the present study unveils that Dox is likely the first pharmacological agent that possesses an intrinsic proteasome activation property. Transcription factors whose degradation is enhanced by Dox, such as β-catenin, c-Jun, and NFAT, play essential roles in maintaining the homeostasis in the heart under both physiological and pathological conditions. Therefore, UPS activation by Dox is likely detrimental to the heart, and it is very tempting to consider proteasome inhibition as a strategy to subside the side effect, especially in light of the fact that proteasome inhibitors have already been used clinically to treat certain types of malignancy that are treated by Dox (40). However, cardiac toxicities have been observed in patients with chronic use of the proteasome inhibitor bortezomib (Velcade) (1, 9, 47). Nevertheless, chemotherapies combining Dox and proteasome inhibitors seem to show improved clinical outcome in clinical trials (6, 46). Hence, it is hopeful that studies like the present one will stimulate very interesting and important investigations into whether chemotherapies combining Dox and proteasome inhibition display either less or more severe cardiotoxicity than those using either alone.
GRANTS
This work was supported in part by National High Technology Research and Development Program of China Grant 2006AA02Z4B5, National Natural Science Foundation of China Grant 30670827, the Scientific Research Foundation for the Returned Overseas Chinese Scholars of State Education Ministry of China, and Guangdong Provincial Natural Science Foundation of China Grant 6104746 (to J. Liu). X. Wang is an Established Investigator of the American Heart Association (AHA). This work was also supported in part by National Heart, Lung, and Blood Institute Grants HL-072166 and HL-085629 (to X. Wang) and AHA Grants 0740025N (to X. Wang) and 0620032Z (to H. Zheng).
Acknowledgments
Present address of H. Zheng: Department of Anesthesia and Medicine, Brigham and Women's Hospital, Boston, MA 02115.
Present address of M. Tang: Cardiovascular Research Center and Department of Physiology, Temple University School of Medicine, Philadelphia, PA 19140.
The costs of publication of this article were defrayed in part by the payment of page charges. The article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
REFERENCES
- 1.Appelman YE, Doevendans PA. Proteasome inhibition and stress compromise the heart in chemotherapy. Cardiovasc Res 79: 547–548, 2008. [DOI] [PubMed] [Google Scholar]
- 2.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science 292: 1552–1555, 2001. [DOI] [PubMed] [Google Scholar]
- 3.Birks EJ, Latif N, Enesa K, Folkvang T, Luong LA, Sarathchandra P, Khan M, Ovaa H, Terracciano CM, Barton PJ, Yacoub MH, Evans PC. Elevated p53 expression is associated with dysregulation of the ubiquitin-proteasome system in dilated cardiomyopathy. Cardiovasc Res 79: 472–480, 2008. [DOI] [PubMed] [Google Scholar]
- 4.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, Li F, Gerdes AM, Wawrousek EF, Wang X. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circ Res 97: 1018–1028, 2005. [DOI] [PubMed] [Google Scholar]
- 5.Ciftci O, Ullrich O, Schmidt CA, Diestel A, Hass R. Regulation of the nuclear proteasome activity in myelomonocytic human leukemia cells after adriamycin treatment. Blood 97: 2830–2838, 2001. [DOI] [PubMed] [Google Scholar]
- 6.Ciolli S, Leoni F, Casini C, Breschi C, Santini V, Bosi A. The addition of liposomal doxorubicin to bortezomib, thalidomide and dexamethasone significantly improves clinical outcome of advanced multiple myeloma. Br J Haematol 141: 814–819, 2008. [DOI] [PubMed] [Google Scholar]
- 7.Depre C, Wang Q, Yan L, Hedhli N, Peter P, Chen L, Hong C, Hittinger L, Ghaleh B, Sadoshima J, Vatner DE, Vatner SF, Madura K. Activation of the cardiac proteasome during pressure overload promotes ventricular hypertrophy. Circulation 114: 1821–1828, 2006. [DOI] [PubMed] [Google Scholar]
- 8.Dikshit P, Jana NR. The co-chaperone CHIP is induced in various stresses and confers protection to cells. Biochem Biophys Res Commun 357: 761–765, 2007. [DOI] [PubMed] [Google Scholar]
- 9.Enrico O, Gabriele B, Nadia C, Sara G, Daniele V, Giulia C, Antonio S, Mario P. Unexpected cardiotoxicity in haematological bortezomib treated patients. Br J Haematol 138: 396–397, 2007. [DOI] [PubMed] [Google Scholar]
- 10.Fekete MR, McBride WH, Pajonk F. Anthracyclines, proteasome activity and multi-drug-resistance. BMC Cancer 5: 114, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fielitz J, Kim MS, Shelton JM, Latif S, Spencer JA, Glass DJ, Richardson JA, Bassel-Duby R, Olson EN. Myosin accumulation and striated muscle myopathy result from the loss of muscle RING finger 1 and 3. J Clin Invest 117: 2486–2495, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Greene RF, Collins JM, Jenkins JF, Speyer JL, Myers CE. Plasma pharmacokinetics of adriamycin and adriamycinol: implications for the design of in vitro experiments and treatment protocols. Cancer Res 43: 3417–3421, 1983. [PubMed] [Google Scholar]
- 13.Haq S, Michael A, Andreucci M, Bhattacharya K, Dotto P, Walters B, Woodgett J, Kilter H, Force T. Stabilization of beta-catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc Natl Acad Sci USA 100: 4610–4615, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hedhli N, Lizano P, Hong C, Fritzky LF, Dhar SK, Liu H, Tian Y, Gao S, Madura K, Vatner SF, Depre C. Proteasome inhibition decreases cardiac remodeling after initiation of pressure overload. Am J Physiol Heart Circ Physiol 295: H1385–H1393, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedhli N, Wang L, Wang Q, Rashed E, Tian Y, Sui X, Madura K, Depre C. Proteasome activation during cardiac hypertrophy by the chaperone H11 kinase/Hsp22. Cardiovasc Res 77: 497–505, 2008. [DOI] [PubMed] [Google Scholar]
- 16.Heineke J, Molkentin JD. Regulation of cardiac hypertrophy by intracellular signalling pathways. Nat Rev Mol Cell Biol 7: 589–600, 2006. [DOI] [PubMed] [Google Scholar]
- 17.Ito T, Fujio Y, Takahashi K, Azuma J. Degradation of NFAT5, a transcriptional regulator of osmotic stress-related genes, is a critical event for doxorubicin-induced cytotoxicity in cardiac myocytes. J Biol Chem 282: 1152–1160, 2007. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem 276: 42938–42944, 2001. [DOI] [PubMed] [Google Scholar]
- 19.Kiyomiya K, Kurebe M, Nakagawa H, Matsuo S. The role of the proteasome in apoptosis induced by anthracycline anticancer agents. Int J Oncol 20: 1205–1209, 2002. [PubMed] [Google Scholar]
- 20.Kiyomiya K, Matsuo S, Kurebe M. Proteasome is a carrier to translocate doxorubicin from cytoplasm into nucleus. Life Sci 62: 1853–1860, 1998. [DOI] [PubMed] [Google Scholar]
- 21.Kiyomiya K, Satoh J, Horie H, Kurebe M, Nakagawa H, Matsuo S. Correlation between nuclear action of anthracycline anticancer agents and their binding affinity to the proteasome. Int J Oncol 21: 1081–1085, 2002. [PubMed] [Google Scholar]
- 22.Kobayashi S, Mao K, Zheng H, Wang X, Patterson C, O'Connell TD, Liang Q. Diminished GATA4 protein levels contribute to hyperglycemia-induced cardiomyocyte injury. J Biol Chem 282: 21945–21952, 2007. [DOI] [PubMed] [Google Scholar]
- 23.Koegl M, Hoppe T, Schlenker S, Ulrich HD, Mayer TU, Jentsch S. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96: 635–644, 1999. [DOI] [PubMed] [Google Scholar]
- 24.Kumarapeli RKA, Horak KM, Glasford JW, Li J, Chen Q, Liu J, Zheng H, Wang X. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J 19: 2051–2053, 2005. [DOI] [PubMed] [Google Scholar]
- 25.Li HH, Willis MS, Lockyer P, Miller N, McDonough H, Glass DJ, Patterson C. Atrogin-1 inhibits Akt-dependent cardiac hypertrophy in mice via ubiquitin-dependent coactivation of forkhead proteins. J Clin Invest 117: 3211–3223, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, Wang X. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J 20: 362–364, 2006. [DOI] [PubMed] [Google Scholar]
- 27.Liu J, Tang M, Mestril R, Wang X. Aberrant protein aggregation is essential for a mutant desmin to impair the proteolytic function of the ubiquitin-proteasome system in cardiomyocytes. J Mol Cell Cardiol 40: 451–454, 2006. [DOI] [PubMed] [Google Scholar]
- 28.Liu X, Chua CC, Gao J, Chen Z, Landy CL, Hamdy R, Chua BH. Pifithrin-α protects against doxorubicin-induced apoptosis and acute cardiotoxicity in mice. Am J Physiol Heart Circ Physiol 286: H933–H939, 2004. [DOI] [PubMed] [Google Scholar]
- 29.Lyu YL, Kerrigan JE, Lin CP, Azarova AM, Tsai YC, Ban Y, Liu LF. Topoisomerase IIbeta mediated DNA double-strand breaks: implications in doxorubicin cardiotoxicity and prevention by dexrazoxane. Cancer Res 67: 8839–8846, 2007. [DOI] [PubMed] [Google Scholar]
- 30.Marques C, Guo W, Pereira P, Taylor A, Patterson C, Evans PC, Shang F. The triage of damaged proteins: degradation by the ubiquitin-proteasome pathway or repair by molecular chaperones. FASEB J 20: 741–743, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones 8: 303–308, 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mearini G, Schlossarek S, Willis MS, Carrier L. The ubiquitin-proteasome system in cardiac dysfunction. Biochim Biophys Acta. In press. [DOI] [PubMed]
- 33.Meiners S, Dreger H, Fechner M, Bieler S, Rother W, Gunther C, Baumann G, Stangl V, Stangl K. Suppression of cardiomyocyte hypertrophy by inhibition of the ubiquitin-proteasome system. Hypertension 51: 302–308, 2008. [DOI] [PubMed] [Google Scholar]
- 34.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev 56: 185–229, 2004. [DOI] [PubMed] [Google Scholar]
- 35.Mross K, Maessen P, van der Vijgh WJ, Gall H, Boven E, Pinedo HM. Pharmacokinetics and metabolism of epidoxorubicin and doxorubicin in humans. J Clin Oncol 6: 517–526, 1988. [DOI] [PubMed] [Google Scholar]
- 36.Patterson C, Ike C, Willis PWt Stouffer GA, Willis MS. The bitter end: the ubiquitin-proteasome system and cardiac dysfunction. Circulation 115: 1456–1463, 2007. [DOI] [PubMed] [Google Scholar]
- 37.Poizat C, Puri PL, Bai Y, Kedes L. Phosphorylation-dependent degradation of p300 by doxorubicin-activated p38 mitogen-activated protein kinase in cardiac cells. Mol Cell Biol 25: 2673–2687, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poizat C, Sartorelli V, Chung G, Kloner RA, Kedes L. Proteasome-mediated degradation of the coactivator p300 impairs cardiac transcription. Mol Cell Biol 20: 8643–8654, 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature 440: 551–555, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richardson PG, Mitsiades C, Hideshima T, Anderson KC. Proteasome inhibition in the treatment of cancer. Cell Cycle 4: 290–296, 2005. [PubMed] [Google Scholar]
- 41.Robert J, Vrignaud P, Nguyen-Ngoc T, Iliadis A, Mauriac L, Hurteloup P. Comparative pharmacokinetics and metabolism of doxorubicin and epirubicin in patients with metastatic breast cancer. Cancer Treat Rep 69: 633–640, 1985. [PubMed] [Google Scholar]
- 42.Stansfield WE, Tang RH, Moss NC, Baldwin AS, Willis MS, Selzman CH. Proteasome inhibition promotes regression of left ventricular hypertrophy. Am J Physiol Heart Circ Physiol 294: H645–H650, 2008. [DOI] [PubMed] [Google Scholar]
- 43.Su H, Huang W, Wang X. The COP9 signalosome negatively regulates proteasome proteolytic function and is essential to transcription. Int J Biochem Cell Biol. In press. [DOI] [PMC free article] [PubMed]
- 44.Tanaka K, Kawakami T, Tateishi K, Yashiroda H, Chiba T. Control of IkappaBalpha proteolysis by the ubiquitin-proteasome pathway. Biochimie 83: 351–356, 2001. [DOI] [PubMed] [Google Scholar]
- 45.Tsimokha AS, Mittenberg AG, Kulichkova VA, Kozhukharova IV, Gause LN, Konstantinova IM. Changes in composition and activities of 26S proteasomes under the action of doxorubicin–apoptosis inductor of erythroleukemic K562 cells. Cell Biol Int 31: 338–348, 2007. [DOI] [PubMed] [Google Scholar]
- 46.Voorhees PM, Orlowski RZ. Emerging data on the use of anthracyclines in combination with bortezomib in multiple myeloma. Clin Lymphoma Myeloma 7, Suppl 4: S156–S162, 2007. [DOI] [PubMed] [Google Scholar]
- 47.Voortman J, Giaccone G. Severe reversible cardiac failure after bortezomib treatment combined with chemotherapy in a non-small cell lung cancer patient: a case report. BMC Cancer 6: 129, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang S, Kotamraju S, Konorev E, Kalivendi S, Joseph J, Kalyanaraman B. Activation of nuclear factor-kappaB during doxorubicin-induced apoptosis in endothelial cells and myocytes is pro-apoptotic: the role of hydrogen peroxide. Biochem J 367: 729–740, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang X, Gerdes AM. Chronic pressure overload cardiac hypertrophy and failure in guinea pigs: III. Intercalated disc remodeling. J Mol Cell Cardiol 31: 333–343, 1999. [DOI] [PubMed] [Google Scholar]
- 50.Wang X, Su H, Ranek MJ. Protein quality control and degradation in cardiomyocytes. J Mol Cell Cardiol 45: 11–27, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Willis MS, Ike C, Li L, Wang DZ, Glass DJ, Patterson C. Muscle ring finger 1, but not muscle ring finger 2, regulates cardiac hypertrophy in vivo. Circ Res 100: 456–459, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yi C, Deng XW. COP1–from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 15: 618–625, 2005. [DOI] [PubMed] [Google Scholar]
- 53.Young GW, Wang Y, Ping P. Understanding proteasome assembly and regulation: importance to cardiovascular medicine. Trends Cardiovasc Med 18: 93–98, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell 126: 571–582, 2006. [DOI] [PubMed] [Google Scholar]