Abstract
Male C57BL/6 mice were fed diets supplemented with either β-carotene (BC) or lycopene (LY) that were formulated for human consumption. Four weeks of dietary supplementations results in plasma and lung carotenoid (CAR) concentrations that approximated the levels detected in humans. Bioactivity of the CARs was determined by assaying their effects on the activity of the lung transcriptome (~8,500 mRNAs). Both CARs activated the cytochrome P450 1A1 gene but only BC induced the retinol dehydrogenase gene. The contrasting effects of the two CARs on the lung transcriptome were further uncovered in mice exposed to cigarette smoke (CS) for 3 days; only LY activated ~50 genes detected in the lungs of CS-exposed mice. These genes encoded inflammatory-immune proteins. Our data suggest that mice offer a viable in vivo model for studying bioactivities of dietary CARs and their modulatory effects on lung genomic expression in both health and after exposure to CS toxicants.
Keywords: β-Carotene, Lycopene, Cigarette smoke, Lung inflammation, Oligonucleotide arrays, Granulocytes, C57BL/6 mice
Introduction
Many epidemiologic studies have suggested an ameliorating role for carotenoids (CARs) and other dietary antioxidant micronutrients in cigarette smoke (CS)-induced diseases including lung cancer [46]. A recent study of 1,194 French subjects suggested that β-carotene (BC) protects against the decline in forced expiration volume (FEV1) over an 8-year period in the general population and that BC and vitamin E are protective against lung function loss in heavy smokers [19]. Paradoxically, results from the α-tocopherol, BC cancer prevention (ATBC) study, and the BC and retinol efficacy trial (CARET), two large investigations that focused on disease chemoprevention with nutritional supplements, suggested that smokers should avoid high-dose BC supplements because of an increased risk of lung cancer (e.g., 18 and 28% more lung cancers, respectively) [54].
These paradoxical and unexpected data have stimulated a number of in vivo studies to seek better understanding of the lung cellular and molecular pathways in which CARs could influence CS-induced lung pathobiology [18, 32, 35, 37, 38, 40, 41, 51, 56, 57, 61, 73, 75]. High doses of BC were shown to increase the activities of cytochrome P450s (CYPs) in rat lungs [57]. In the A/J mouse model of CS-induced lung tumorogenesis, dietary BC supplementation augmented lung BC concentrations but had no effect on lung tumors induced by either whole CS [51] or gas-phase of CS [75]. In a similar model, BC supplementation had no effect on CS carcinogen 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone-induced lung tumorogenesis [18].
Studies in ferrets have made important contributions in deciphering the in vivo effects of CARs, CS and their biological interactions [61]. In ferrets exposed to CS, accelerated levels of lung cell proliferation and squamous metaplasia were found in animals fed high doses of BC. This study suggested that diminished retinoid signaling pathways could represent a contributing mechanism related to the increased tumor incidence seen in human smokers [41]. Importantly, these investigators have also presented evidence that the beneficial and/or detrimental effects of BC supplementation could be related to the doses of BC administered [40, 41]. They have also shown that both low- and high-dose lycopene (LY) supplementations substantially inhibited CS-induced lung tissue squamous metaplasia and proliferating cellular nuclear antigen (PCNA) expression in ferrets [37], extending earlier observations that LY administration appears to decrease some markers of CS-induced lung bimolecular damages [1].
In a recent study A/J mice fed varying concentrations of BC were exposed to CS [32]. These investigators documented CS-induced effects on lung pathways related to drug metabolism, oxidative stress, inflammation, matrix degradation and apoptosis and demonstrated that BC itself had minimal changes in lung gene expression, but it decreased CS-related induction of inflammatory pathways. These investigators concluded that BC effects on CS-induced lung genomic profiles were negligible.
In the present study, we have explored CAR effects on the lung transcriptome of C57BL/6 mice and compared these effects with those in CAR (BC or LY) fed mice exposed to CS using a somewhat similar CAR administration and CS exposure strategy that was used previously in A/J mice [51, 76]. Although C57BL/6 mice have been less frequently used to study CS-related lung tumorogenesis [47, 76] and mice present a complex and somewhat different in vivo CAR digestion, absorption and metabolism profiles compared to humans [33], a rich array of genetically engineered mice on C57BL6 genetic background are available for the study of nutrient-environmental interactions in this mouse strain.
Methods
Protocols for humane treatment and utilization of mice described in this study were approved by the Institutional Animal Care and Use Committee of the University of California, Davis.
Diets and mice
The basal diet was the AIN-93G rodent diet which is optimized for rodent growth and health, and lacks added BC or LY [36]. The AIN-93G diet was supplemented with gelatin beadlets without CAR or gelatin beadlets with a crystalline form of synthetic CAR. The beadlets themselves consist of a starch-coated matrix of gelatin and sucrose stabilized with small amounts of tocopherol, sodium ascorbate and ascorbyl palmitate with tricalcium phosphate (BASF website). Gelatin beadlets contained either 100 g BC or 100 g LY (~77% all trans- and 23% total cis isomers) or no added CAR/kg gelatin beadlets provided by BASF (Ballerup, Denmark) and were developed for human consumption [83]. The final pelleted diets were prepared by DYETS, Bethleham, PA. These diets contained 5 g of beadlets/kg diet. The two CAR containing diets thus contained 0.5 g of CAR/kg diet, a supplementation several-fold higher than that used in human studies [54] but necessitated because of the poor systemic deliveries of ingested CARs in rodents [33]. It is likely that the pelleting process itself along with the chamber CS exposures affected the stability of the added BC and LY, causing some isomerization and oxidation of at least a portion of the two CARs.
Preliminary studies of dose ranging protocol for CAR administration
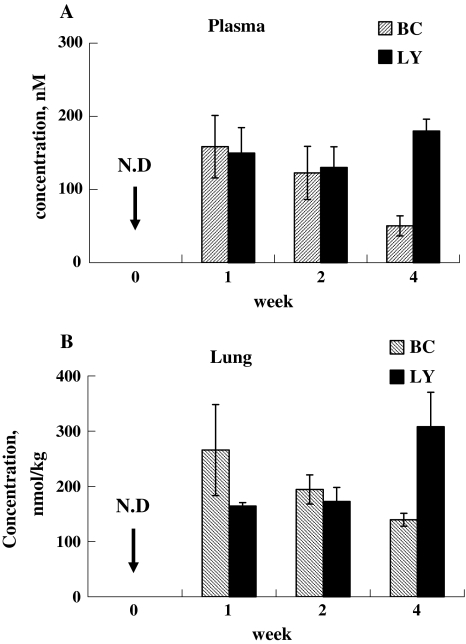
Male C57BL/6 mice (6 week old, 20–25 g) were obtained from Charles River Laboratories (Wilmington, MA). Twenty-four mice (6 groups of 4 mice per group) received the basal AIN-93 G rodent unsupplemented diet. They were housed in groups of 4 in polycarbonate cages with free access to tap water and diet. All mice received the basal diet for 2 weeks, reaching baseline low CAR concentrations in plasma and tissues. After 2 weeks of this acclimatization period, mice were subsequently fed with diets containing either 0.5 g BC/kg diet or 0.5 g LY/kg diet or the unsupplemented basal diet for 1, 2, and 4 weeks. At 1, 2, or 4 weeks mice were sacrificed and plasma and lung tissue were collected for analysis utilizing previously described HPLC methodologies [51, 52]. As depicted in Fig. 1, plasma and lung tissue BC peaked at 1 week, whereas LY levels remained more stable but were higher than BC levels at 4 weeks. The noted discrepancy in plasma and lung CAR levels with constant oral intakes is most likely secondary to differences in BC/LY cleavages by 15′, 15′ and 9′, 10′ monooxygenases, different BC/LY substrate and/or metabolite inductions of these metabolizing enzymes, and/or other less clarified biotransformations of the two CARs [24]. We empirically selected the 4-week feeding regimens for all the subsequent experiments designed to characterize CAR effects on global lung genomic responses at two widely divergent lung CAR concentrations, albeit with the realization that plasma CAR levels only reached low human levels for the two CARs [29, 63].
Fig. 1.
Plasma and lung CARs levels in air breathing mice fed BC (0.5 g/kg diet) or LY (0.5 g/kg diet) supplemented diet after 1, 2 and 4 weeks. a Plasma levels of BC and LY increased after 1 week. BC levels decreased after 2 and 4 weeks but LY levels were maintained for 4 weeks (n = 4 mice per group). b Lung BC levels (nmol/kg lung tissue) increased after 1 week and decreased after 2 and 4 weeks and appear to track the plasma BC levels. Lung LY levels increased after 1 week, and its levels slightly increased after 4 weeks (n = 4 mice per group) (N.D not detected)
Experimental protocol
Twenty-four mice were randomly assigned to one of three groups of eight mice. Each group was allowed to feed on either a basal diet or the BC or LY-supplemented diets as in the preliminary studies after receiving the basal diet for 2 weeks to allow mice to acclimatize to the diet containing the gelatin beadlets without added CARs. After 2 weeks the diets were changed to either the BC-supplemented diet (0.5 g BC/kg diet), the LY-supplemented diet (0.5 g LY/kg diet) or the basal diet lacking added CARs for four additional weeks. While continuing on the three assigned diets, four mice in each dietary group were then exposed to either CS or filtered air for three additional days. Body weight was measured before and after exposure to filtered air or CS. Mice had unrestricted access to water and their assigned diets during the filtered air or CS exposures.
CS exposure system
Mice were exposed to CS for 6 h/day for 3 days (8 am–2 pm) in chambers as previously described [51]. The chamber atmosphere was controlled with relative humidity of 41 ± 7% and temperature 21 ± 1°C. The nicotine content in the chamber during the CS exposure was 5.1 ± 0.4 mg/m3, carbon monoxide level 14 ± 1.0 ppm, and total suspended particulate concentration 60 ± 2.5 mg/m3. This level of CS exposure could be expected to result in carboxyhemoglobin (HbCO) levels, of approximately 5% [23]. Immediately after the last CS or filtered air exposure, all of the mice were euthanized by injection of beuthanasia (120 mg/kg body weight, i.p.). Lung parenchymal tissue was dissected away from extra-parenchymal airways and blood vessels and stored at −80°C until RNA extraction (within 4 weeks).
GeneChip analysis
A total of 24 mouse genome 430A 2.0 arrays, GeneChips, (Affymetrix, Santa Clara, CA), 4 GeneChips/group of 4 mice were utilized to obtain mRNA expression data. An aliquot of total RNA extracted from half-a-lung from each of the 24 mice was processed for GeneChip analysis as previously described using Affymetrix protocols [15].
RNA extraction and synthesis of biotin-labeled RNA
Lung tissue (~100 mg) from each mouse was homogenized in 1 mL of Trizol Reagent (Invitrogen, Carlsbad, CA) and total RNA was extracted and quantified as described by the manufacturer. An aliquot (20 μg/8 μL of water) of total RNA from each mouse lung from each of the three dietary groups exposed to either CS or filtered air was used for cDNA synthesis (first-strand and second-strand cDNA synthesis) followed by cleanup of double-stranded cDNA and synthesis of biotin-labeled cRNA. The biotin-labeled cRNA (40 μg) from each lung was used for fragmentation and hybridization to GeneChips.
The scanned images of hybridized signals were analyzed with the Affymetrix GeneChip Operating Software (GCOS 1.0). When the P value for detection signal was ≤0.049 (range of P value 0.0002–0.049), the expression of the mRNA was classified as “present” (P). All mRNAs with the P value for detection ≥0.05 were considered “absent” (A). The signal intensities for transcripts classified as present ranged from 5 to 7,000 U. The .cel files from each scan were imported into dChip analyzer, a web based software package (http://www.dchip.org/) implementing model-based expression analysis of oligonucleotide arrays [34]. The dChip analyzer was used for obtaining statistically significant differences (P ≤ 0.05, fold change ≥2.0, and the difference in signal intensity ≥100) in the mRNA expression between the different treatment groups and for generating qualitative “heat-maps” of selected functionally related gene clusters. CAR and CS sensitive genes were further classified into functional clusters based on their annotations and descriptions of their functions in the literature.
Validation of GeneChip data
Many reports [16, 44, 66] have shown that the changes in the expression of mRNAs selected by the analysis of hybridization data as described above could be confirmed by independent analysis such as Northern, qualitative reverse transcriptase (RT) PCR, quantitative real-time RT-PCR (qRT-PCR), and in some cases by immunoblot analysis of the encoded proteins [22, 28]. In this study, selected differentially expressed genes that were relevant to the major new findings reported here were subjected to confirmation analysis by real-time PCR.
Quantitative real-time PCR (qRT-PCR)
An aliquot equivalent to 5 μg of total RNA extracted for GeneChip analysis was reverse-transcribed to obtain cDNA in a final volume of 20 μL reaction buffer consisting of oligo dT primer, DTT, dNTPs and Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). qRT-PCR method with SYBR green as fluorescent reporter was used to quantify the expression of selected genes identified by GeneChip assay. All the gene specific primers (Table 1) were designed with Primer Express 1.0 software (Applied Biosystems) using gene specific template obtained from Affymetrix probe set IDs. The reaction was carried out in 96-well optical well plate containing 6.25 ng RNA in each well. The applied RNA quantity was further normalized by amplifying cDNA samples simultaneously with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) specific primers. The transcript levels were measured by real-time RT-PCR using ABI PRISM 7700 Sequence detection system (PE Applied Biosystems, Foster City, CA). PCR amplification parameters were: initial denaturation step at 95°C for 10 min followed by 40 cycles, each at 95°C for 15 s (melting), 60°C for 1 min (annealing and extension). The 2-ΔΔCT method [43] was used to calculate relative changes in gene expression determined from real-time quantitative PCR experiments (Applied Biosystems User Bulletin No.2 (P/N4303859). The threshold cycle, Ct, which correlates inversely with the target mRNA levels, was measured as the cycle number at which the SYBR Green emission increases above a threshold level. Specific mRNA transcripts were expressed as fold difference in the expression of specific genes in RNA samples from lungs of mice fed basal diet compared to CAR fed diets or between CS-exposed and air-exposed lungs fed the assigned diets.
Table 1.
Primer sequences for real-time RT-PCR
| Gene | Primer sequence (5′–3′) |
|---|---|
| GAPDH | Sense-GCAACAGGGTGGTG |
| Antisense-GGATAGGGCCTCTC | |
| CYP1A1 | Sense-CAGATGATAAGGTCATCACGA |
| Antisense-TTGGGGATATAGAAGCCATTC | |
| S100A8 (calgranulin A) | Sense-GCATCTCACAAAGACAGCCACA |
| Antisense-AGCCCTAGGCCAGAAGCTCT | |
| S100A9 (calgranlin B) | Sense-GATGGCCAACAAAGCACCTT |
| Antisense-ATGATGGTGGTTATGCTGCG | |
| Slfn4 | Sense-ACAAGTGATGCCTGGAAAGG |
| Antisense-GACTGCCCTGGGAAATATGA | |
| IL1-beta | Sense-CCAAAAGATGAAGGGCTGCT |
| Antisense-TCATCTGGACAGCCCAGGTC | |
| CBR-3 | Sense-TACTTGGCTCTCCTGCCTCC |
| Antisense-GACTAGCTGGCCGTGAGGTT |
The oligonucleotide sequence for each primer was obtained by Primer Express software using gene specific sequence from Affymetrix probe set data base. The primers were custom prepared and used as described in Sect. “Methods”
Statistical analysis
Statistical evaluation of the CAR concentrations, qRT-PCR data, and the effects of CS exposure in the three dietary groups were done by Student’s t test using the statistical software GraphPad Prism 4.0. In all comparisons differences with P ≤ 0.05 were considered as significant. Results are expressed as mean ± SEM, the number of mice/group (n) and the P value.
Results
Effects of CARs and CS on body weight
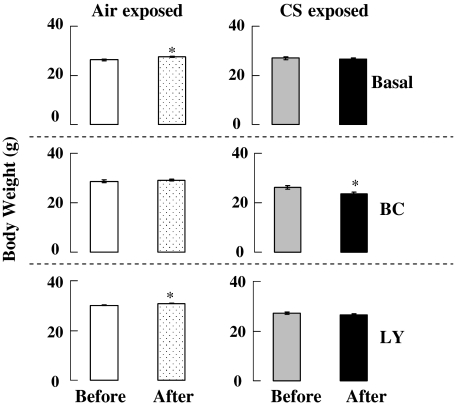
After the 2-week diet acclimation period, mice were fed on their assigned diets for 4 weeks. There were no significant differences in body weight between the groups of mice before CS exposure (Fig. 2). Half of the mice were then exposed to CS (6 h/day for 3 days) and the remaining half to filtered air. Air breathing mice on basal or LY-supplemented diets gained a small but significant amount of weight after 3 days. All CS-exposed mice showed a small decrease in body weight which was only statistically significant (P ≤ 0.05) in BC fed mice.
Fig. 2.
Effects of CARs on body weight in mice breathing air or after 3 days of CS exposure. Air breathing mice on basal or LY-supplemented diet gained small but significant weight. In the CS breathing groups of mice, only BC fed mice lost small but significant weight after CS exposure. Air air breathing for 3 days, CS 60 mg/m3 of total suspended particles for 6 h/day for 3 days, n = 4, * P < 0.05
CAR concentration
Plasma and lung concentrations of BC and LY in mice fed the three assigned diets for 4 weeks are shown in Table 2. Neither of the two CARs were detectable in plasma or lungs of mice fed the basal AIN-93G diet supplemented with gelatin beadlets (the carriers of synthetic crystalline CARs). After 4 weeks of feeding CAR supplemented diets, large and significant (P ≤ 0.001) increases in the plasma and lung CARs were detected. BC and LY concentrations in plasma were 50.3 ± 13.7 and 179.8 ± 16.2 nM, respectively. BC and LY concentrations in lungs were 139.6 ± 11.6 and 308.0 ± 62.4 nmol/kg wet weight of lung tissue, respectively. The data suggest that the lung tissue bioavailability of LY may exceed that of BC under similar basal diet composition and feeding conditions. However, as an unspecified amount of BC was probably converted to vitamin A in gut and/or liver tissue [29] or metabolized [24, 82], direct comparisons of overall absorption kinetics of the two CARs cannot be ascertained. The concentrations of lung CARs are within the range of those described for humans [63], the plasma LY concentrations being similar to those of 866 men [27].
Table 2.
Mice were fed either the basal diet or diets containing 500 mg/kg of either BC or LY for 4 weeks
| Control diet | β-carotene diet | Lycopene diet | |
|---|---|---|---|
| Plasma (nM) | |||
| β-carotene | <3.7 | 50.3 ± 13.7a | <3.7 |
| Lycopene | <3.7 | <3.7 | 179.9 ± 16.2a |
| Lung (nmol/kg) | |||
| β-carotene | <60 | 139.6 ± 11.6a | <60 |
| Lycopene | <60 | <60 | 308.1 ± 62.5a |
Concentration of BC and LY were measured in plasma (nM) or lung (nmol/kg) at time of sacrifice
aAll data are given as mean ± SEM, n = 4. BC and LY were below the limit of detection in plasma and lung of mice fed the basal diet. Four weeks of dietary supplementation resulted in a large and significant (P < 0.001) increase in plasma and lung concentrations of the two CARs
Bioactivity of CARs: CAR sensitive genes in mice breathing air
The bioactivity of the two CARs was assessed by evaluation of their effects on the lung transcriptome. High density oligonucleotide arrays containing 22,600 probe sets (mouse genome 430 A 2.0) that represent a large fraction of the mouse expressed genome were used. The total number of mRNAs detected in the lungs from the air and CS breathing mice fed the three assigned diets were ~15,000 and they were not significantly different from each other in the various groups of mice. The coefficient of variation of the total number of expressed genes detected in each group of 3–4 mice was <5%. Differential analysis of gene expression data showed that the elevated concentrations of CARs significantly affected the expression of a small number of genes (Table 3) which were a very small fraction (0.09 and 0.02%, respectively), of the ~15,000 lung genes that were reliably detected.
Table 3.
Summary of genome-wide responses of lungs to diets in air and cigarette smoke (CS) exposed mice
| Diet/treatment | Total number of genes affected | % of total number of genes affected | Up-regulated | Down-regulated |
|---|---|---|---|---|
| BC/AIR | 13 | 0.09 | 11 | 2 |
| LY/AIR | 3 | 0.02 | 1 | 2 |
| Basal/CS | 40 | 0.27 | 27 | 13 |
| BC/CS | 73 | 0.49 | 17 | 56 |
| LY/CS | 65 | 0.43 | 54 | 11 |
Effects of dietary β-carotene (BC) or lycopene (LY) on gene expression in the lungs of air breathing mice were obtained by comparing the entire list of genes from mice fed either BC- or LY-supplemented diet with that obtained from the lungs of air breathing mice fed the basal diet. The number differentially expressed, carotenoid sensitive genes (column 2) is also shown as the % of the total number of genes detected in the lungs from either BC- or LY-fed mice (column 3). CS sensitive genes in each dietary group were obtained by comparing the entire list of genes detected in the lungs of CS breathing mice with that expressed in air breathing mice. The total number of differentially expressed, CS sensitive genes (column 2) is shown also shown as % of the total genes detected (column 3) in the lungs of CS breathing mice
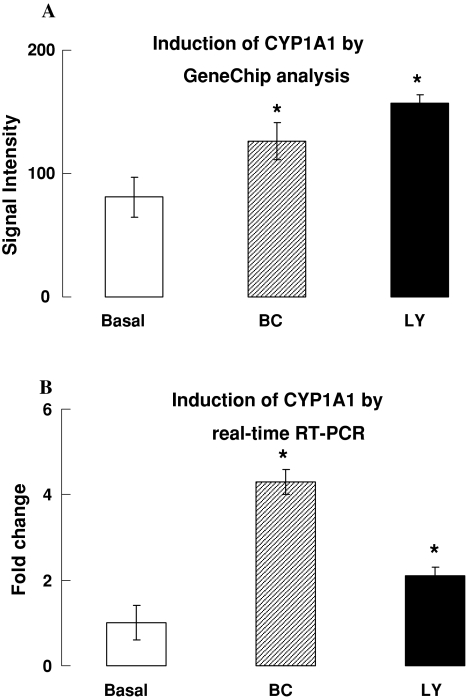
Global gene expression analysis identified qualitative and quantitative differences in the lungs response to diet induced CAR augmentations. Although the lung concentration of BC was lower than that of LY in air breathing mice, more genes were affected by BC than by LY when compared to the expression of genes in lungs of mice fed the basal diet (Tables 3, 4). The expression of cytochrome P450 1a1 (cyp1a1) was induced by ~twofold in the lungs of the CAR fed mice (Table 4; Fig. 3). The induction of cyp1a1 by BC and LY in air breathing lungs was also independently confirmed by qRT-PCR (Fig. 3b). The GeneChip data also suggest that another transcriptionally regulated gene that encodes D site albumin binding protein (Dbp) also responds like the cyp1a1 gene in the presence of BC or LY (Table 4).
Table 4.
β-carotene (BC) or lycopene (LY) sensitive genes in lungs of mice
| Diet | Name of gene | Up-/down-regulated |
|---|---|---|
| BC | Cytochrome P450, 1a1; Cyp1a1 | Up |
| D site albumin promoter binding protein, Dbp | Up | |
| Period homolog 2 (Drosophila), Per2 | Up | |
| Fibroblast growth factor 3, Fgf3 | Up | |
| Hepatic leukemia factor, Hlf | Up | |
| Immunoglobulin joining chain, Igj | Down | |
| Lecithin-retinol acyltransferase, Lrat | Up | |
| Aquaporin 3, Aqp3 | Up | |
| Sodium channel, voltage-gated, type VIII, alpha polypeptide, Scn8a | Down | |
| RIKEN cDNA A930004D23 gene | Up | |
| Expressed sequence AL033314 | Up | |
| RIO kinase 3 (yeast) | Up | |
| Unknown gene | Up | |
| Unknown gene | Up | |
| LY | Cytochrome P450, 1a1; Cyp1a1 | Up |
| D site albumin promoter binding protein, Dbp | Up | |
| RIKEN cDNA 5730454B08 gene | Down | |
| RIKEN cDNA 2210401K01 gene | Down |
Gene symbols are shown in bold figures
Fig. 3.
Induction of cytochrome P450 1a1 (cyp1a1) in lungs of mice fed BC or LY-supplemented diet. a Induction of cyp1a1 detected by GeneChip analysis, (n = 3–4, * P < 0.05 as compared to control); b induction of cyp1a1 detected by real-time quantitative RT-PCR (n = 3–4, * P < 0.05)
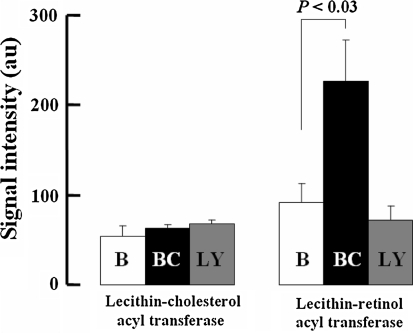
Several genes were modulated by BC but not LY (Table 4). These data suggest that the GeneChip assay can discriminate between the in vivo actions of the two CARs. The most noteworthy difference was the induction of the gene encoding lecithin-retinol acyltransferase (Lrat), which is a retinol esterifying enzyme. Fig. 4 shows GeneChip data for the expression of Lrat and a related acyltransferase to illustrate the specific induction of Lrat in the lungs of mice fed only the BC diet. Additional genes whose induction was similar to that of Lrat included genes encoding transcription factors such as period 2 (Per2) and hepatic leukemia factor (Hlf). BC diet supplementation also up-regulated the expression of aquaporin-3 (Aqp3) and down-regulated that of sodium channel (Scn8a) genes. Two genes of unknown functions were repressed by LY but unaffected by BC.
Fig. 4.
Selective induction of lecithin-retinol acyl transferase only in mice fed the BC-supplemented diet. The data obtained from GeneChip expression analysis show signal intensity for the two acyl transferases in the lungs of air breathing mice fed the 3 assigned diets
Cigarette smoke-related effects on lung gene expression and modification by CARs
A large number of genes were modulated in the lungs of mice breathing CS compared to those breathing filtered air. The analysis detected a higher number of CS sensitive genes in the lungs of mice fed both CAR supplemented diets compared to those of mice fed the CAR deficient, basal diet. The total number of genes affected was 40, 73, and 65 in the lungs of mice fed the basal, BC- or LY-supplemented diets, respectively (Table 3). Many of these CAR-modulated genes in CS-exposed lungs relate to genes involved in inflammatory-immune functions.
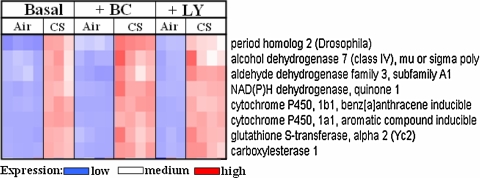
A robust activation of Phase I and Phase II response detected by the GeneChip assay in the lungs of CS-exposed mice was expected [14, 50] and further validated the 3-day (6 h/day) acute CS exposure paradigm and GeneChip analysis. Fig. 5 illustrates the qualitative relative expression of a cluster of eight co-regulated genes in the lungs of mice that were on the three assigned diets and were allowed to breathe either air or CS. Table 5 shows a robust expression of Phase I and II response genes as a result of 3 days of CS breathing in the mice fed the three diets. The CAR diets did not affect CS-induced responses of this cluster of genes. The induction of this family of genes in mouse lungs has previously been described [58] and further validates our GeneChip analysis. A number of genes encoding transcription factors which were induced by the BC-supplemented diet in air breathing mice were further increased after CS exposure. These genes included Dbp, Per2, Per3, Cry1 and Hlf (Fig. 4; Table 5).
Fig. 5.
“Heat-map” of cyp1a1-like cluster of 8 genes. Each column represents a mouse. Each row represents a gene. Lighter color shows low expression and darker color shows high expression of the gene. The expression of the 8 genes was low in the lungs of air breathing mice but was high in the lungs of CS breathing mice
Table 5.
Differential expression of genes modulated by carotenoids and cigarette smoke
| Gene name and description | Signal intensity | Accession no. | |||||
|---|---|---|---|---|---|---|---|
| CA | CCS | BA | BCS | LYA | LYCS | ||
| Xenobiotic/drug metabolism | |||||||
| Cytochrome P450, 1a1; Cyp1a1 | 81 ± 16 | 4,947 ± 730*** | 126 ± 15 | 3,985 ± 168*** | 157 ± 7 | 4,652 ± 326*** | NM_009992 |
| Cytochrome P450, f1b1; Cyp1b1 | 223 ± 61 | 2,150 ± 541** | 113 ± 7 | 1,657 ± 110*** | 165 ± 15 | 1,760 ± 1023*** | BI251808 |
| Cytochrome P450, family 2, a4///a5; Cyp2a4///Cyp2a5 | 938 ± 336 | 2,042 ± 477** | 942 ± 69 | 2,071 ± 127*** | 630 ± 113 | 2,195 ± 273** | NM_007812 |
| Aldehyde dehydrogenase family 3, a1; Aldh3a1 | 378 ± 91 | 1,072 ± 262** | 171 ± 12 | 836 ± 43*** | 312 ± 24 | 1,296 ± 61*** | NM_007436 |
| Alcohol dehydrogenase 7 (class IV); Adh7 | 269 ± 73 | 814 ± 182** | 151 ± 13 | 718 ± 33*** | 201 ± 18 | 859 ± 109** | NM_009626 |
| Aldo-keto reductase family 1, member B8; Akr1b8 | 623 ± 175 | 945 ± 242# | 343 ± 30 | 759 ± 36*** | 449 ± 3 | 1,095 ± 70*** | NM_008012 |
| Carboxylesterase 1; Ces1 | 93 ± 29 | 511 ± 146** | 68 ± 8 | 420 ± 35*** | 88 ± 7 | 575 ± 42*** | NM_021456 |
| NAD(P)H dehydrogenase, quinone 1; Nqo1 | 119 ± 23 | 403 ± 64** | 85 ± 3 | 366 ± 8*** | 124 ± 8 | 534 ± 10*** | AV158882 |
| Glutathione S-transferase, alpha 2; Gsta2 | 242 ± 60 | 569 ± 159# | 148 ± 13 | 485 ± 11*** | 245 ± 19 | 654 ± 39*** | NM_008182 |
| Glutathione S-transferase, alpha 3; Gsta3 | 349 ± 91 | 770 ± 139 | 294 ± 21 | 615 ± 58** | 364 ± 16 | 771 ± 42*** | AI172943 |
| Carbonyl reductase 3; Cbr3 | 604 ± 148 | 1,398 ± 304* | 284 ± 87 | 1,174 ± 29*** | 469 ± 23 | 1,472 ± 67*** | AK003232 |
| Glutathione reductase 1; Gsr | 277 ± 30 | 348 ± 29# | 492 ± 48 | 443 ± 18# | 356 ± 15 | 1,007 ± 176** | AK019177 |
| Glutamate-cysteine ligase, modifier subunit; Gclm | 402 ± 75 | 887 ± 173** | 320 ± 14 | 706 ± 32*** | 326 ± 35 | 1,105 ± 29*** | NM_008129 |
| Flavin containing monooxygenase 2; Fmo2 | 1,814 ± 583 | 2,284 ± 629# | 1,203 ± 188 | 1,688 ± 156# | 876 ± 115 | 2,056 ± 127*** | NM_018881 |
| Inflammatory-immune response | |||||||
| Paired-Ig-like receptor A1; Pira1 | 138 ± 38 | 80 ± 29# | 139 ± 20 | 69 ± 4** | 108 ± 5 | 310 ± 64** | NM_011087 |
| Paired immunoglobin-like type 2 receptor alpha; Pilra | 139 ± 42 | 104 ± 48# | 130 ± 10 | 73 ± 7** | 89 ± 2 | 262 ± 62* | BB775785 |
| Paired immunoglobin-like type 2 receptor beta; Pilrb | 248 ± 47 | 166 ± 46# | 254 ± 34 | 92 ± 3** | 186 ± 10 | 563 ± 131* | NM_133209 |
| EGF-like module containing, mucin-like, r4; Emr4 | 285 ± 99 | 170 ± 117# | 179 ± 13 | 45 ± 3*** | 191 ± 15 | 162 ± 22# | AF396935 |
| Killer cell lectin-like receptor, a3; Klra3 | 595 ± 188 | 307 ± 103# | 299 ± 21 | 131 ± 16*** | 401 ± 42 | 365 ± 48# | U49865 |
| Fibrinogen-like protein 2; Fgl2 | 269 ± 90 | 165 ± 58# | 240 ± 43 | 120 ± 9** | 171 ± 10 | 377 ± 63** | BF136544 |
| Protein tyrosine phosphatase, receptor type, C; Ptprc | 3,348 ± 987 | 1,540 ± 533# | 2,266 ± 156 | 960 ± 50*** | 2,287 ± 53 | 3,103 ± 391# | NM_011210 |
| Lymphocyte antigen 86; Ly86 | 825 ± 272 | 304 ± 86# | 433 ± 45 | 186 ± 14** | 599 ± 70 | 443 ± 9** | NM_010745 |
| Leukocyte specific transcript 1; Lst1 | 359 ± 101 | 127 ± 46* | 280 ± 54 | 49 ± 6** | 215 ± 20 | 460 ± 84* | U72644 |
| Coronin, actin binding protein 1A; Coro1a | 1,153 ± 319 | 574 ± 180# | 821 ± 91 | 416 ± 37** | 910 ± 113 | 1,415 ± 231# | BB740218 |
| GLI pathogenesis-related 2; Glipr2 | 146 ± 44 | 65 ± 24# | 148 ± 15 | 62 ± 2** | 132 ± 3 | 276 ± 68# | BM208214 |
| Matrix metalloproteinase 8; Mmp8 | 145 ± 33 | 86 ± 11# | 272 ± 83 | 78 ± 11** | 69 ± 6 | 2,465 ± 670** | NM_008611 |
| Matrix metalloproteinase 9; Mmp9 | 185 ± 33 | 84 ± 18** | 222 ± 66 | 68 ± 5** | 92 ± 3 | 830 ± 237** | NM_013599 |
| Interleukin 1 beta; Il1b | 534 ± 171 | 141 ± 39* | 763 ± 243 | 102 ± 12** | 179 ± 37 | 1,759 ± 464** | BC011437 |
| Interleukin 8 receptor, beta; Il8rb | 83 ± 29 | 19 ± 5* | 103 ± 37 | 14 ± 7** | 20 ± 6 | 256 ± 41** | NM_009909 |
| Phospholipase A2, group VII; Pla2g7 | 229 ± 66 | 129 ± 55# | 350 ± 77 | 129 ± 8** | 124 ± 14 | 975 ± 244** | AK005158 |
| Colony stimulating factor 3 receptor (granulocyte); Csf3r | 417 ± 90 | 163 ± 45** | 619 ± 200 | 133 ± 23** | 175 ± 30 | 1,322 ± 279** | NM_007782 |
| Formyl peptide receptor, related sequence 2; Fpr-rs2 | 475 ± 110 | 399 ± 165# | 383 ± 44 | 217 ± 7** | 273 ± 14 | 1,256 ± 282** | NM_008039 |
| Formyl peptide receptor 1; Fpr1 | 238 ± 63 | 154 ± 41# | 224 ± 11 | 126 ± 15** | 133 ± 19 | 572 ± 140** | NM_013521 |
| Secretory leukocyte protease inhibitor; Slpi | 721 ± 115 | 683 ± 193# | 735 ± 96 | 655 ± 89# | 461 ± 44 | 1,817 ± 331** | NM_011414 |
| Synuclein, alpha; Snca | 393 ± 87 | 601 ± 109# | 292 ± 17 | 388 ± 31** | 398 ± 25 | 954 ± 109** | NM_009221 |
| Fc receptor, IgG, low affinity III; Fcgr3 | 287 ± 57 | 278 ± 65# | 300 ± 35 | 201 ± 26* | 213 ± 21 | 730 ± 129** | NM_010188 |
| Triggering receptor expressed on myeloid cells 3; Trem3 | 131 ± 32 | 89 ± 31# | 108 ± 8 | 41 ± 2*** | 76 ± 4 | 315 ± 64** | NM_021407 |
| Cadherin 2; Cdh2 | 154 ± 39 | 101 ± 35# | 128 ± 15 | 95 ± 8# | 223 ± 12 | 166 ± 16** | BC022107 |
| Glycoprotein 49 A; Gp49a///Lilrb4 | 945 ± 247 | 580 ± 157# | 945 ± 193 | 413 ± 15** | 580 ± 12 | 1,564 ± 263** | U05264 |
| C-type lectin domain family 4, member d; Clec4d | 214 ± 42 | 121 ± 38# | 413 ± 128 | 103 ± 12** | 122 ± 12 | 1,226 ± 341** | NM_010819 |
| C-type lectin domain family 4, member e; Clec4e | 59 ± 19 | 44 ± 19# | 63 ± 18 | 22 ± 2* | 26 ± 4 | 226 ± 72* | NM_019948 |
| Metabolism and ion-channels | |||||||
| Argininosuccinate synthetase 1; Ass1 | 183 ± 42 | 253 ± 50# | 209 ± 78 | 174 ± 10# | 145 ± 8 | 186 ± 23# | NM_007494 |
| Histidine decarboxylase; Hdc | 1,075 ± 269 | 368 ± 127* | 624 ± 48 | 304 ± 14*** | 844 ± 97 | 862 ± 139# | BG072171 |
| Ubiquitin specific protease 2; Usp2 | 111 ± 33 | 214 ± 57# | 64 ± 2 | 162 ± 8*** | 95 ± 6 | 153 ± 7** | AI553394 |
| Lysyl oxidase-like 1; Loxl1 | 1,666 ± 319 | 642 ± 181** | 1,000 ± 74 | 454 ± 48*** | 1573 ± 55 | 874 ± 141** | AF357006 |
| Sodium channel, voltage-gated, 3b; Scn3b | 1,220 ± 201 | 681 ± 170# | 753 ± 55 | 348 ± 28*** | 1,221 ± 139 | 910 ± 136# | BE951842 |
| Stearoyl-Coenzyme A desaturase 1; Scd1 | 917 ± 265 | 617 ± 124# | 801 ± 82 | 341 ± 59** | 1,192 ± 292 | 937 ± 195# | NM_009127 |
| Serine (or cysteine) proteinase inhibitor, clade H1; Serpinh1 | 1,377 ± 40 | 802 ± 74*** | 1,574 ± 153 | 799 ± 76** | 1,915 ± 230 | 1,253 ± 188* | BB329489 |
| Solute carrier family 7a10; Slc7a10 | 415 ± 46 | 270 ± 42* | 381 ± 12 | 214 ± 18*** | 533 ± 35 | 414 ± 57# | NM_017394 |
| Solute carrier family 28a2; Slc28a2 | 146 ± 41 | 51 ± 13* | 128 ± 21 | 37 ± 6** | 125 ± 19 | 274 ± 66# | NM_021520 |
| Topoisomerase (DNA) II alpha; Top2a | 183 ± 43 | 117 ± 30# | 127 ± 8 | 74 ± 8** | 62 ± 32 | 165 ± 20# | BM211413 |
| Solute carrier family 40 (iron-regulated transporter)1; Slc40a1 | 387 ± 103 | 310 ± 77# | 334 ± 88 | 284 ± 28# | 284 ± 16 | 1,006 ± 147** | AF226613 |
| Solute carrier family 38, member 5; Slc38a5 | 462 ± 85 | 231 ± 55** | 285 ± 27 | 211 ± 26# | 472 ± 9 | 298 ± 44** | BG066984 |
| Guanine deaminase; Gda | 92 ± 22 | 73 ± 23# | 105 ± 19 | 52 ± 4** | 61 ± 5 | 308 ± 65** | NM_010266 |
| Peptidyl arginine deiminase, type IV; Padi4 | 43 ± 10 | 20 ± 4# | 64 ± 15 | 17 ± 3** | 40 ± 4 | 249 ± 78* | NM_011061 |
| Arachidonate 5-lipoxygenase activating protein; Alox5ap | 1,428 ± 246 | 1,050 ± 171# | 1,226 ± 185 | 867 ± 27# | 1,212 ± 75 | 2,748 ± 434** | BC026209 |
| Amiloride binding protein 1; Abp1 | 355 ± 100 | 116 ± 43* | 155 ± 24 | 106 ± 8# | 230 ± 35 | 94 ± 17** | BC021880 |
| Apoptosis | |||||||
| Granzyme A; Gzma | 1,176 ± 392 | 531 ± 194# | 633 ± 44 | 219 ± 20*** | 688 ± 92 | 801 ± 85# | NM_010370 |
| Insulin-like growth factor binding protein 3; Igfbp3 | 757 ± 122 | 350 ± 95** | 468 ± 38 | 357 ± 21** | 794 ± 23 | 345 ± 37*** | AV175389 |
| Chloride channel calcium activated 1; Clca1 | 122 ± 24 | 147 ± 39# | 127 ± 22 | 181 ± 9* | 103 ± 4 | 261 ± 44** | AF047838 |
| Receptor | |||||||
| Angiotensin receptor-like 1; Agtrl1 | 251 ± 36 | 138 ± 17** | 169 ± 13 | 135 ± 19# | 2,934 ± 210 | 1,485 ± 453** | BB533323 |
| Membrane-spanning 4-domains, a1; Ms4a1 | 259 ± 86 | 95 ± 31# | 120 ± 16 | 55 ± 5** | 201 ± 17 | 159 ± 30# | BB236617 |
| Membrane-spanning 4-domains, a4b; Ms4a4b | 655 ± 197 | 312 ± 126# | 342 ± 35 | 145 ± 11** | 481 ± 44 | 422 ± 62# | BB199001 |
| Membrane-spanning 4-domains, a4c; Ms4a4c | 408 ± 145 | 78 ± 33* | 216 ± 51 | 38 ± 2** | 236 ± 47 | 295 ± 62# | NM_029499 |
| Membrane-spanning 4-domains, a6b; Ms4a6b | 1,031 ± 305 | 549 ± 162# | 703 ± 56 | 332 ± 24*** | 701 ± 43 | 805 ± 98# | NM_027209 |
| Cell proliferation | |||||||
| tetraspanin 4; Tspan4 | 376 ± 64 | 1,387 ± 243** | 532 ± 32 | 1,145 ± 99*** | 622 ± 13 | 1,180 ± 160** | NM_053082 |
| Placenta-specific 8; Plac8 | 2,254 ± 538 | 913 ± 292* | 1,601 ± 144 | 485 ± 41*** | 1,415 ± 114 | 2,077 ± 200** | AF263458 |
| Unknown function | |||||||
| Macrophage activation 2 like; Mpa2 l | 154 ± 33 | 68 ± 20* | 82 ± 10 | 41 ± 3** | 130 ± 10 | 117 ± 15# | BM241485 |
| Histone 1, H2ad; Hist1h2ad | 319 ± 111 | 230 ± 93# | 231 ± 45 | 132 ± 7* | 112 ± 17 | 351 ± 30** | W91024 |
| Apoptosis | |||||||
| Granzyme A; Gzma | 1,176 ± 392 | 531 ± 194# | 633 ± 44 | 219 ± 20*** | 688 ± 92 | 801 ± 85# | NM_010370 |
| Insulin-like growth factor binding protein 3; Igfbp3 | 757 ± 122 | 350 ± 95** | 468 ± 38 | 357 ± 21** | 794 ± 23 | 345 ± 37*** | AV175389 |
| Chloride channel calcium activated 1; Clca1 | 122 ± 24 | 147 ± 39# | 127 ± 22 | 181 ± 9* | 103 ± 4 | 261 ± 44** | AF047838 |
| Receptor | |||||||
| Angiotensin receptor-like 1; Agtrl1 | 251 ± 36 | 138 ± 17** | 169 ± 13 | 135 ± 19# | 2,934 ± 210 | 1,485 ± 453** | BB533323 |
| Membrane-spanning 4-domains, a1; Ms4a1 | 259 ± 86 | 95 ± 31# | 120 ± 16 | 55 ± 5** | 201 ± 17 | 159 ± 30# | BB236617 |
| Membrane-spanning 4-domains, a4b; Ms4a4b | 655 ± 197 | 312 ± 126# | 342 ± 35 | 145 ± 11** | 481 ± 44 | 422 ± 62# | BB199001 |
| Membrane-spanning 4-domains, a4c; Ms4a4c | 408 ± 145 | 78 ± 33* | 216 ± 51 | 38 ± 2** | 236 ± 47 | 295 ± 62# | NM_029499 |
| Membrane-spanning 4-domains, a6b; Ms4a6b | 1,031 ± 305 | 549 ± 162# | 703 ± 56 | 332 ± 24*** | 701 ± 43 | 805 ± 98# | NM_027209 |
| Cell proliferation | |||||||
| Tetraspanin 4; Tspan4 | 376 ± 64 | 1,387 ± 243** | 532 ± 32 | 1,145 ± 99*** | 622 ± 13 | 1,180 ± 160** | NM_053082 |
| Placenta-specific 8; Plac8 | 2,254 ± 538 | 913 ± 292* | 1,601 ± 144 | 485 ± 41*** | 1,415 ± 114 | 2,077 ± 200** | AF263458 |
| Unknown function | |||||||
| Macrophage activation 2 like; Mpa2 l | 154 ± 33 | 68 ± 20* | 82 ± 10 | 41 ± 3** | 130 ± 10 | 117 ± 15# | BM241485 |
| Histone 1, H2ad; Hist1h2ad | 319 ± 111 | 230 ± 93# | 231 ± 45 | 132 ± 7* | 112 ± 17 | 351 ± 30** | W91024 |
| Transcription and growth factors | |||||||
| Period homolog 2 (Drosophila); Per2 | 175 ± 51 | 983 ± 221** | 296 ± 16 | 857 ± 48*** | 313 ± 17 | 926 ± 53*** | AF035830 |
| Period homolog 3 (Drosophila); Per3 | 75 ± 42 | 384 ± 51** | 224 ± 22 | 438 ± 24*** | 282 ± 11 | 409 ± 26 | NM_011067 |
| D site albumin promoter binding protein; Dbp | 222 ± 63 | 613 ± 110** | 505 ± 58 | 530 ± 80# | 509 ± 44 | 446 ± 91 | BC018323 |
| Cryptochrome 1; Cry1 | 143 ± 45 | 260 ± 91# | 61 ± 8 | 222 ± 25*** | 88 ± 6 | 281 ± 42** | BG069864 |
| Hepatic leukemia factor; Hlf | 150 ± 52 | 425 ± 129# | 173 ± 25 | 383 ± 25*** | 188 ± 14 | 405 ± 76 | BB744589 |
| DNA-damage-inducible transcript 4; Ddit4 | 983 ± 346 | 1,470 ± 434# | 637 ± 110 | 1,136 ± 122** | 967 ± 60 | 815 ± 92 | AK017926 |
| Thyrotroph embryonic factor; Tef | 1,545 ± 282 | 3,447 ± 565** | 1,883 ± 124 | 3,151 ± 182*** | 2,138 ± 81 | 2,836 ± 233 | BC017689 |
| v-maf musculoaponeurotic fibrosarcoma oncogene family F; Maff | 164 ± 31 | 453 ± 61** | 239 ± 35 | 421 ± 18*** | 214 ± 18 | 359 ± 29 | BC022952 |
| FK506 binding protein 5; Fkbp5 | 136 ± 23 | 377 ± 78** | 169 ± 6 | 397 ± 26*** | 116 ± 19 | 327 ± 45** | U16959 |
| Cold inducible RNA binding protein; Cirbp | 373 ± 68 | 502 ± 55# | 231 ± 35 | 551 ± 44*** | 629 ± 29 | 610 ± 113# | NM_007705 |
| Epithelial stromal interaction 1 (breast); Epsti1 | 421 ± 83 | 274 ± 75# | 299 ± 15 | 149 ± 3*** | 424 ± 56 | 399 ± 18# | BF020640 |
| SAM domain, SH3 domain & nuclear localisation signals 1; Samsn1 | 185 ± 56 | 104 ± 39# | 119 ± 4 | 53 ± 2*** | 106 ± 6 | 197 ± 45# | NM_023380 |
| Neutrophil cytosolic factor 4; Ncf4 | 226 ± 54 | 125 ± 44# | 190 ± 30 | 77 ± 4** | 168 ± 14 | 362 ± 66* | NM_008677 |
| Tripartite motif protein 30///tripartite motif protein 30-like; Trim30 | 646 ± 216 | 942 ± 598# | 398 ± 51 | 206 ± 22** | 376 ± 38 | 593 ± 64** | BG068242 |
| Nuclear factor, erythroid derived 2; Nfe2 | 106 ± 18 | 71 ± 13# | 133 ± 14 | 78 ± 4** | 108 ± 5 | 325 ± 79* | L09600 |
| POU domain, class 2, associating factor 1; Pou2af1 | 418 ± 10 | 201 ± 97# | 222 ± 38 | 112 ± 9** | 346 ± 21 | 266 ± 71# | NM_011136 |
| Deafness, autosomal dominant 5 homolog (human); Dfna5 h | 47 ± 16 | 31 ± 6# | 43 ± 7 | 19 ± 4** | 28 ± 4 | 172 ± 45** | NM_018769 |
| Zinc finger and BTB domain containing 16; Zbtb16 | 101 ± 24 | 152 ± 33# | 113 ± 20 | 129 ± 14# | 74 ± 1 | 162 ± 21** | Z47205 |
| Erythroid differentiation regulator 1; Erdr1 | 712 ± 133 | 642 ± 121# | 525 ± 60 | 442 ± 157# | 366 ± 131 | 870 ± 107** | AJ007909 |
| Nuclear receptor subfamily 1, d1; Nr1d1 | 415 ± 106 | 245 ± 29# | 406 ± 20 | 317 ± 33* | 504 ± 20 | 243 ± 40** | W13191 |
| Neuronatin; Nnat | 205 ± 57 | 138 ± 13# | 132 ± 12 | 109 ± 7# | 198 ± 24 | 159 ± 16# | AV218841 |
| Pleckstrin homology domain containing, a6; Plekha6 | 109 ± 21 | 98 ± 28# | 91 ± 10 | 72 ± 8# | 180 ± 9 | 99 ± 12** | BB486127 |
| Unknown gene | |||||||
| RIKEN cDNA 9030611N15 gene | 1,085 ± 293 | 2,188 ± 530# | 7,12.3 ± 90.3 | 2,028 ± 102*** | 788 ± 44 | 1,899 ± 155*** | NM_134072 |
| Expressed sequence AI447904 | 885 ± 283 | 364 ± 137# | 421 ± 23.7 | 171 ± 13*** | 585 ± 53 | 515 ± 42# | BM241008 |
| DNA segment, Chr 7, Wayne State University 130, expressed | 42.2 ± 12.6 | 124 ± 45# | 37 ± 10 | 95 ± 17** | 40 ± 4 | 94 ± 9** | AI480533 |
| – | 289 ± 104 | 591 ± 176# | 252 ± 44 | 437 ± 59** | 364 ± 45 | 359 ± 56# | BC003855 |
| – | 89 ± 37 | 162 ± 46# | 74 ± 13 | 113 ± 12* | 93 ± 25 | 118 ± 33# | M11024 |
| RIKEN cDNA 1700012B18 gene | 138 ± 37 | 329 ± 81* | 112 ± 8 | 259 ± 18*** | 145 ± 24 | 374 ± 37** | BC022135 |
| RIKEN cDNA C130076O07 gene | 231 ± 48 | 81 ± 16** | 184 ± 12 | 70 ± 6*** | 191 ± 18 | 97 ± 20** | BB202655 |
| RIKEN cDNA 6330500D04 gene | 482 ± 140 | 239 ± 68# | 328 ± 47 | 149 ± 4** | 371 ± 34 | 610 ± 102# | BM242294 |
| – | 518 ± 133 | 202 ± 48** | 228 ± 34 | 121 ± 8** | 368 ± 58 | 309 ± 27# | BE686052 |
| RIKEN cDNA 1100001G20 gene | 208 ± 30 | 202 ± 50# | 311 ± 42 | 168 ± 16** | 153 ± 15 | 1,268 ± 394* | AV006463 |
| RIKEN cDNA 2300003P22 gene | 61 ± 13 | 33 ± 7# | 139 ± 51 | 96 ± 60# | 33 ± 12 | 842 ± 278* | AK004007 |
| CD300 antigen like family member F | 328 ± 93 | 186 ± 41# | 290 ± 78 | 162 ± 7# | 171 ± 7 | 699 ± 156** | BM230330 |
Gene symbols are shown in bold figures
*** P < 0.001, ** P < 0.05, #P < 0.5
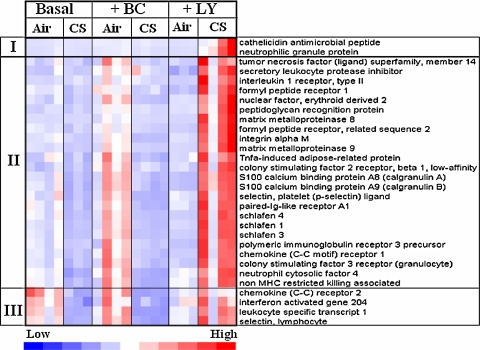
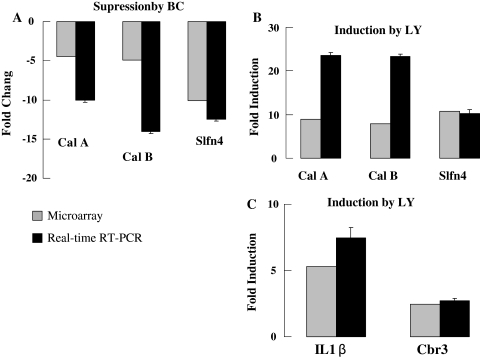
The major novel discovery of this in vivo genome-wide screen is the identification of a large cluster of inflammatory-immune genes that are induced by dietary LY but not BC in the lungs of CS-exposed mice (Fig. 6). Three distinct clusters of genes could be identified in this group of inflammatory-immune genes. Cluster I (2 genes) showed low expression in the lungs of all mice except those that were fed LY-supplemented diet and exposed to CS; these two genes are frequently used as markers of neutrophils [8, 70]. A large sub-group of 24 genes, cluster II, Fig. 6, was moderately expressed in the lungs of mice fed the basal diet or the BC-supplemented diet. The lungs of BC-supplemented mice breathing air showed higher expression of this cluster of genes when compared to that in the lungs of other groups of mice breathing air. Their expressions were suppressed by exposure to CS. In contrast to the suppressing effects of CS on these cluster II genes in mice fed the basal or the BC diet, the LY fed mice showed obvious augmentation of their expression. Inductions of Cal A, Cal B, Slfn4, IL1β, and carbonyl reductase 3 (Cbr3) genes in CS breathing lungs of mice fed LY-supplemented diet and suppression of these genes in the CS breathing lungs of mice fed BC-supplemented diet were independently confirmed by qRT- PCR (Fig. 7a–c). Genes in cluster III behave like those in cluster II except that they were highly expressed in the lungs of air breathing mice fed the basal or the BC diets.
Fig. 6.
“Heat-map” inflammation-immune related genes in the lungs of mice fed the 3 assigned diets. Each column represents a mouse and each row a gene. Three distinct patterns of expression could be identified. Cluster I shows high expression only in the CS-exposed lungs of LY fed mice. Cluster II shows medium-expression of genes in air breathing, BC fed mice and the expression was suppressed by CS. The expression of the same cluster of genes was low in air breathing mice fed LY but was induced when the mice were exposed to CS. Cluster III shows similar expression in mice fed the basal or the BC-supplemented diet but the expression is reversed in the mice fed the LY-supplemented diet
Fig. 7.
a Confirmation of selected inflammatory genes by real-time quantitative RT-PCR. Suppression of Cal A, Cal B, slfn 4, gene expression in BC + CS-exposed group. b Induction of Cal A, Cal B, slfn 4, gene expression in LY + CS-exposed group. c Induction of IL1β and Cbr3 gene expression in LY + CS-exposed group (n = 4)
Discussion
Concentrations of CARS in mouse plasma and lungs
β-carotene and LY supplementations were approximately equivalent to 125 mg/kg body weight/per day for a human. A large amount of dietary CARs are necessary in rodents to obtain increases in tissue CAR concentrations comparable to levels seen in humans, explained in part by active CAR cleavage enzyme(s) in the rodent intestine [33]. These high CAR supplementations were well tolerated by mice as suggested by the lack of a significant decrease in whole body weights in air breathing CAR supplemented mice (Fig. 2). Plasma BC concentrations in this study were ~50% of those reported previously in A/J mice fed similar CAR concentrations [51] and may reflect strain specific differences in absorption and metabolism of BC or differences in dietary constituents. In addition, the lipid content of the rodent diet also appears to be an important contributor to the bioavailability of CARs. This is suggested by the detection of higher serum BC levels in A/J mice whose BC-supplemented diets included corn oil (5%) and sodium cholate (0.25%) [32]. Such modified rodent diets containing bile salts and increased vegetable oils have not systematically been compared with the well characterized AIN-93G diet optimized for rodent growth and development [36]. Plasma and lung concentrations of BC (50 nM and 140 nmol/kg, lung wet weight, respectively) approximated levels described for human plasma and lung tissue, respectively [29, 63].
The plasma concentrations of LY detected in this study were lower than those reported in rat serum [66, 81]. The latter observations suggest significant difference between C57BL/6 mice and F334 rats in the digestion, uptake and/or metabolism of LY since the basal diets, without added CARs used in this study were very similar to those that were used in F344 rats [81]. As for the above-mentioned case for BC, the lipid composition of the LY-supplemented diet is likely to affect the serum LY concentration, as evidenced by the finding of a higher LY concentration in the serum of Copenhagen rats compared to F344 rats fed the AIN-93G diet; the diets of Copenhagen rats were supplemented with coconut fat (6%w/w) [66].
Bioreactivity of CARs in lungs of air breathing mice
In spite of the somewhat lower concentrations of the two CARs in mouse lungs compared to those in human and rat lungs, the two CARs showed significant bioactivity, as determined by the GeneChip assay.
Previous studies have shown the induction of cypla1 by CARs [30]. The present study also detected the induction of cypla1 (Fig. 3). A twofold increase in the activity of cyp1a1 gene in mouse lungs by CAR concentrations that were lower than those described in previous studies in mice or in human lungs suggests that the mouse lung genome is sensitive to dietary CARs and further validates that the C57BL/6 mouse offers a useful in vivo model to study CAR metabolism [30]. The cyp1a1 gene encodes a transcriptionally regulated enzyme important in the metabolism of carcinogens [31]. It is driven by the aromatic hydrocarbon receptor (AhR), a ligand activated transcription factor of the nuclear receptor superfamily [74]. The role of AhR in normal physiology remains unclear, but appears to relate to both environmental pollutants and numerous host responses including inflammation [11, 68, 69]. Dietary factors appear to activate cyp1a1 by AhR pathways [10], although the role of other members of ligand activated nuclear receptors such as RARs cannot be excluded [9]. The induction of cyp1a1 activity by dietary BC has previously been reported in rats [57] and in ferrets [39] and has been implicated in the carcinogenic effects of BC [21], although it cannot be assumed that cyplal itself is necessarily procarcinogenic [26, 45].
The expression of Lrat was induced in the lungs of BC fed mice (Fig 4). Lrat is transcriptionally regulated by retinol [84] which can be generated from BC but not LY. Lrat plays a major role in the metabolism and storage of vitamin A in different cell types including epithelial cells. BC is a precursor of vitamin A which is oxidized to retinoic acid, a known diet derived factor for the regulation of lung growth and development [79], and regulation of surfactant proteins in lung type II cell [3, 80]. The basal expression of Lrat in mouse lungs is similar to that in the liver [42]. Our GeneChip data suggest that the Lrat gene is further activated by diet induced augmentation of lung BC.
The gene encoding Dbp (D site albumin promoter binding protein), a transcription factor implicated in the regulation of circadian rhythm [53] was seen to be induced by CARs. Dbp is the founding member of the PAR family of basic leucine zipper (bZip) transcription factors [48] and the gene encoding Hlf (hepatic leukemia factor), another member of the same family [25], was also seen to be up-regulated by CAR supplementation. Dbp and Hlf were reported to be sensitive to xenobiotics and stimulate transcription of genes against xenobiotics and oxidative stress and hence might play a role in modulating the toxicity of such compounds [13]. Modulation of members of ion- and water-channel genes by dietary BC and not LY is another novel finding in this study and warrants further characterization. BC fed mice showed increased expression of Aqp3 gene encoding, a water channel which has been shown to be regulated by growth factors, inflammation, and osmotic stress [4].
GeneChip data from this in vivo study, focused on lungs, are also noteworthy for the lack of induction of Phase II genes by CARs in air breathing mice which may in part be due to lower CAR levels in lung tissues compared to levels used in in vitro studies or the dramatic difference in extracellular milieu between the in vivo and in vitro experimental conditions [2]. CAR-dependent induction of Phase II response has been shown in various cell lines of liver and intestinal origins [64], and absence of this response in lungs in vivo may be attributed to differences in cellular origins and assay conditions including intracellular concentrations of the CARs and their metabolites. Some of the Phase II response genes are driven by the redox sensitive transcription factor Nrf2 which binds to antioxidant response elements (ARE) and up-regulates protective detoxifying enzymes related to oxidative stress [49]. BC and LY are singlet oxygen scavengers; therefore, lack of any modulation of “classical” antioxidant response genes such as heme-oxygenase 1 (HO-1), superoxide dismutases (Sod) and glutamate-cysteine ligase (Gclc) is also noted. We also noted a lack of induction in the expression of HO-1, Sod and Gclc in the lungs of mice with severe α-tocopherol deficiency imposed either by dietary depletion or by the deletion of the α-tocopherol transfer protein gene [17, 55].
Modulation of CS-induced transcriptomes by CARs
A large induction of cyp1a1 gene identified by the GeneChip assay in the CS-exposed lungs has previously been demonstrated [14, 39, 71]. As discussed above, the induction of Cyp1a1 is driven by AhR and the encoded protein is expressed in airway epithelia and lung parenchyma [77]. In the present experiments both CARs induced cyp1a1 in the lungs of air breathing mice; however, neither of them affected CS-caused induction of cyp1a1.
The genes of Phase II response encode detoxification and antioxidant enzymes which are induced by CS and their expression was unaffected by dietary CARs. The redox sensitive transcription factor, Nrf2, plays an important role in the expression of these genes [4, 58]. The co-ordinated induction of all but Hlf in this cluster of genes is noteworthy, because they are all associated with the regulation of circadian rhythm and are expressed in lung cells [5, 12, 59, 62]. Dbp, Per2 and Hlf are the members of the proline and acidic amino acid-rich basic leucine zipper (PAR bZIP) transcription factor family. The suggestion that this cluster of genes may regulate the cell cycle and cell proliferation [6] raises the possibility that dietary CARs and CS interactions may affect cellular homeostasis through their actions on major regulators of the circadian rhythm. In contrast to the effects of CS on the “clock-genes”, the expressions of several immune-response genes such as Spon2, Eln, Igh-6 and Igh-j558 were down-regulated in the CS-exposed lungs of mice fed the BC-supplemented diet. The functional implications of the reciprocal relationship between the “clock-genes” that may affect cell cycle, and immune related genes in response to dietary CARs and CS remains to be explored.
Carotenoid modulate immune-inflammation related genes in CS-exposed lungs
The major novel discovery of this in vivo genome-wide screen is the identification of a large cluster of inflammatory-immune genes that are induced by dietary LY but not by BC in the lungs of CS-exposed mice (Fig. 6). Review of the literature suggests that some of the immune-inflammatory genes, for example, calgranulins A and B, also designated calprotectins or S100 A8 and A9, and matrix metalloproteinase 8 and 9 are co-expressed in granulocytes (derived from myeloid cells). Hence, it is possible that dietary CARs affect homeostasis of lung specific granulocyte population in mice challenged with CS. A large induction of calgranulins by CS in LY fed mice is suggestive of an increased recruitment of circulatory phagocytes to the lung, possible via activation of AhR [68], although discordant data exist [69]. Cal B is suggested to be a chemotactic factor for leukocytes [20] and increased expression of these genes in lung tissues may further recruit circulating leukocytes to respiratory epithelium, propagating a cycle of granulocyte transmigration to the lung. In addition, members of calgranulin family may also be ligands for receptor for advanced glycation end products (RAGE) [67]. RAGE is abundantly expressed in mouse lungs [15] and its activation by ligands such as calgranulins is suspected to activate molecular signaling pathways that culminate in mitogenesis, growth inhibition, and apoptosis [67]. The expression of MMP 8 and MMP 9 which are produced by macrophages, were also increased in lungs of LY fed mice exposed to CS. Macrophage proliferation and activation are implicated in various types of pulmonary pathology. One important process associated with pulmonary fibrosis is injury to basement membranes by MMPs [72]. Expression of MMPs induced by CS exposure may play a role in the pathogenesis of COPD [78].
A recent study has addressed the effects of BC supplementation and 2-week CS exposure on transcriptomic responses of lungs in A/J mice [32]. The A/J mouse strain, in contrast to C57BL/6 used in our study, is susceptible to spontaneous age-related lung tumors. Furthermore, the tumor multiplicity of these mice is augmented during the recovery-phase after chronic (>6 month) exposure to CS [76]. Although there appears to be a robust Nrf2 driven response to CS in both the strains of mice, BC-supplementations appear to suggest different transcriptomic responses in the two strains of mice. We suggest that these differences are primarily attributed to the differences in the lung transcriptomes due to distinct genetic backgrounds of these two strains of mice. Our preliminary data suggest that ~2,000 genes are differentially expressed between the A/J and C57BL/6 mice, and many of the differentially expressed genes belong to inflammatory gene class. Additional variables such as the mode of CS exposure whole-body (our study) versus nose only [32], the exposure chamber concentration of CS of 60 mg/m3 (in our study) vs 141.2 mg/m3 [32] and the duration of exposure, 6 h/day for 3 days (our study) versus 4 h/day, 5 days/week for the first week and 7 days/week the second week [32] are likely to contribute differences in the BC-induced changes in CS-related lung gene expressions.
Pathobiologic implications and limitations
How relevant are these findings for humans? The studies described here are acute studies and thus represents only a “snap-shot” of genomic responses of lungs to dietary CAR supplements and their interactions with genomic responses of lungs to short-term CS exposure. A single dose of CAR and a single time interval of CS exposure were used to obtain the present data. The CAR intake and CS exposures were based on previous studies in mice [51, 75]. The study supports the concept that CAR supplements are capable of modulating lung gene expression and even more interestingly, modulate the effects of CS, and might be expected to modulate lung responses to other inhaled environmental toxicants. Although C57BL/6 mice are not the optimal strain for the study of CS-induced lung carcinogenesis [76], they offer the advantage of genetic manipulation to insert or delete genes implicated in human lung disease related to environmental toxins and dietary factors formulated to design and test chemopreventive strategies. The present studies were not designed to specifically focus on CAR modulations of CS-related lung carcinogenic processes.
Some limitations have to be considered; the effects of CARs and CS on lung specific cellular constituencies were not addressed. There is considerable heterogeneity in the cellular composition and response of different compartments of respiratory tract tissues to environmental pollutants, as exemplified by cell AhR system responses [11]. The CAR formulations used in the present study were similar to those described in a recently published human study [83]. It should be recognized that considerable differential susceptibilities to oxidant stimuli (such as that posed by CS) exist between different mouse strains [7, 76]. Lastly, it is highly possible that some of the observed changes in gene profiles are produced by the actions of non-enzymatic and enzymatically generated metabolites of the ingested CARs, including their metabolic and oxidative products. These CAR metabolic species were not measured in the present study, but are known to possess significant bioactivities [24, 65] and need to be addressed in future studies.
We conclude that C57BL/6 mice offer a viable in vivo model to study bioavailability and bioactivity of synthetic and natural CARs and their possible role in modulating environmentally induced lung pathobiologies. As there is substantial evidence that dietary supplementations are capable of modifying biologic responses to air pollutants [60], and as the C57BL/6 mouse has been genetically engineered to manipulate multiple pathways related to both air pollution and diet-related transcriptionally-related pathways, this mouse model should prove useful for the interrogation of key pathways of environment-nutrient interactions in lung tissues.
Acknowledgments
Support for this work was received from NIH grant #ES011985, USDA grant # 35200-134546, UC Davis Clinical Nutrition Research Unit, UC Davis Center for Human Nutrition Research, BASF and Pharmanex.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Arab L, Steck-Scott S, Fleishauer AT (2002) Lycopene and the lung. Exp Biol Med (Maywood) 227:894–899 [DOI] [PubMed]
- 2.Ben-Dor A, Steiner M, Gheber L, Danilenko M, Dubi N, Linnewiel K, Zick A, Sharoni Y, Levy J (2005) Carotenoids activate the antioxidant response element transcription system. Mol Cancer Ther 4:177–186 [PubMed]
- 3.Bogue CW, Jacobs HC, Dynia DW, Wilson CM, Gross I (1996) Retinoic acid increases surfactant protein mRNA in fetal rat lung in culture. Am J Physiol 271:L862–L868 [DOI] [PubMed]
- 4.Borok Z, Verkman AS (2002) Lung edema clearance: 20 years of progress: invited review: role of aquaporin water channels in fluid transport in lung and airways. J Appl Physiol 93:2199–2206 [DOI] [PubMed]
- 5.Burioka N, Takata M, Okano Y, Ohdo S, Fukuoka Y, Miyata M, Takane H, Endo M, Suyama H, Shimizu E (2005) Dexamethasone influences human clock gene expression in bronchial epithelium and peripheral blood mononuclear cells in vitro. Chronobiol Int 22:585–590 [DOI] [PubMed]
- 6.Canaple L, Kakizawa T, Laudet V (2003) The days and nights of cancer cells. Cancer Res 63:7545–7552 [PubMed]
- 7.Cho HY, Kleeberger SR (2007) Genetic mechanisms of susceptibility to oxidative lung injury in mice. Free Radic Biol Med 42:433–445 [DOI] [PubMed]
- 8.Cowland JB, Johnsen AH, Borregaard N (1995) hCAP-18, a cathelin/pro-bactenecin-like protein of human neutrophil specific granules. FEBS Lett 368:173–176 [DOI] [PubMed]
- 9.Delescluse C, Lemaire G, de Sousa G, Rahmani R (2000) Is CYP1A1 induction always related to AHR signaling pathway? Toxicology 153:73–82 [DOI] [PubMed]
- 10.Denison MS, Nagy SR (2003) Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol 43:309–334 [DOI] [PubMed]
- 11.Frericks M, Meissner M, Esser C (2007) Microarray analysis of the AHR system: tissue-specific flexibility in signal and target genes. Toxicol Appl Pharmacol 220:320–332 [DOI] [PubMed]
- 12.Fu Z, Inaba M, Noguchi T, Kato H (2002) Molecular cloning and circadian regulation of cryptochrome genes in Japanese quail (Coturnix coturnix japonica). J Biol Rhythms 17:14–27 [DOI] [PubMed]
- 13.Gachon F, Olela FF, Schaad O, Descombes P, Schibler U (2006) The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab 4:25–36 [DOI] [PubMed]
- 14.Gebel S, Gerstmayer B, Bosio A, Haussmann HJ, Van Miert E, Muller T (2004) Gene expression profiling in respiratory tissues from rats exposed to mainstream cigarette smoke. Carcinogenesis 25:169–178 [DOI] [PubMed]
- 15.Gohil K, Cross CE, Last JA (2003) Ozone-induced disruptions of lung transcriptomes. Biochem Biophys Res Commun 305:719–728 [DOI] [PubMed]
- 16.Gohil K, Moy RK, Farzin S, Maguire JJ, Packer L (2000) mRNA expression profile of a human cancer cell line in response to Ginkgo biloba extract: induction of antioxidant response and the Golgi system. Free Radic Res 33:831–849 [DOI] [PubMed]
- 17.Gohil K, Oommen S, Vasu VT, Aung HH, Cross CE (2007) Tocopherol transfer protein deficiency modifies nuclear receptor transcriptional networks in lungs: modulation by cigarette smoke in vivo. Mol Aspects Med 28:453–480 [DOI] [PubMed]
- 18.Goralczyk R, Wertz K, Lenz B, Riss G, Buchwald Hunziker P, Geatrix B, Aebischer CP, Bachmann H (2005) Beta-carotene interaction with NNK in the AJ-mouse model: effects on cell proliferation, tumor formation and retinoic acid responsive genes. Biochim Biophys Acta 1740:179–188 [DOI] [PubMed]
- 19.Guenegou A, Leynaert B, Pin I, Le Moel G, Zureik M, Neukirch F (2006) Serum carotenoids, vitamins A and E, and 8-year lung function decline in a general population. Thorax 61:320–326 [DOI] [PMC free article] [PubMed]
- 20.Harrison CA, Raftery MJ, Walsh J, Alewood P, Iismaa SE, Thliveris S, Geczy CL (1999) Oxidation regulates the inflammatory properties of the murine S100 protein S100A8. J Biol Chem 274:8561–8569 [DOI] [PubMed]
- 21.Hecht SS (1999) Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst 91:1194–1210 [DOI] [PubMed]
- 22.Herzog A, Siler U, Spitzer V, Seifert N, Denelavas A, Hunziker PB, Hunziker W, Goralczyk R, Wertz K (2005) Lycopene reduced gene expression of steroid targets and inflammatory markers in normal rat prostate. FASEB J 19:272–274 [DOI] [PubMed]
- 23.Hodge-Bell KC, Lee KM, Renne RA, Gideon KM, Harbo SJ, McKinney WJ (2007) Pulmonary inflammation in mice exposed to mainstream cigarette smoke. Inhal Toxicol 19:361–376 [DOI] [PubMed]
- 24.Hu KQ, Liu C, Ernst H, Krinsky NI, Russell RM, Wang XD (2006) The biochemical characterization of ferret carotene–9′,10′-monooxygenase catalyzing cleavage of carotenoids in vitro and in vivo. J Biol Chem 281:19327–19338 [DOI] [PMC free article] [PubMed]
- 25.Inaba T, Roberts WM, Shapiro LH, Jolly KW, Raimondi SC, Smith SD, Look AT (1992) Fusion of the leucine zipper gene HLF to the E2A gene in human acute B-lineage leukemia. Science 257:531–534 [DOI] [PubMed]
- 26.Ito S, Chen C, Satoh J, Yim S, Gonzalez FJ (2007) Dietary phytochemicals regulate whole-body CYP1A1 expression through an arylhydrocarbon receptor nuclear translocator-dependent system in gut. J Clin Invest 117:1940–1950 [DOI] [PMC free article] [PubMed]
- 27.Ito Y, Wakai K, Suzuki K, Ozasa K, Watanabe Y, Seki N, Ando M, Nishino Y, Kondo T, Ohno Y, Tamakoshi A (2005) Lung cancer mortality and serum levels of carotenoids, retinol, tocopherols, and folic acid in men and women: a case-control study nested in the JACC Study. J Epidemiol 15(Suppl 2):S140–S149 [DOI] [PMC free article] [PubMed]
- 28.Izzotti A, Cartiglia C, Longobardi M, Balansky RM, D’Agostini F, Lubet RA, De Flora S (2004) Alterations of gene expression in skin and lung of mice exposed to light and cigarette smoke. FASEB J 18:1559–1561 [DOI] [PubMed]
- 29.Khachik F, Carvalho L, Bernstein PS, Muir GJ, Zhao DY, Katz NB (2002) Chemistry, distribution, and metabolism of tomato carotenoids and their impact on human health. Exp Biol Med (Maywood) 227:845–851 [DOI] [PubMed]
- 30.Khachik F, Telang N, Enjo F, Lipkin M (2003) Development of a mixture of dietary carotenoids as cancer chemopreventive agents: C57BL/6 mice as a useful animal model for efficacy studies with carotenoids. In: Meskin MS, Bidlack WR, Davies AJ, Lewis DS, Randolph RK (eds) Phytochemicals: mechanisms of action. CRC Press, Boca Raton, pp 163–176
- 31.Kim JH, Sherman ME, Curriero FC, Guengerich FP, Strickland PT, Sutter TR (2004) Expression of cytochromes P450 1A1 and 1B1 in human lung from smokers, non-smokers, and ex-smokers. Toxicol Appl Pharmacol 199:210–219 [DOI] [PubMed]
- 32.Kuntz E, Borlak J, Riss G, Aebischer CP, Bachmann H, Seifert N, Hunziker PB, Solle D, Hunziker W, Goralczyk R, Wertz K (2007) Transcriptomics does not show adverse effects of beta-carotene in A/J mice exposed to smoke for 2 weeks. Arch Biochem Biophys 465:336–346 [DOI] [PubMed]
- 33.Lee CM, Boileau AC, Boileau TW, Williams AW, Swanson KS, Heintz KA, Erdman JW Jr (1999) Review of animal models in carotenoid research. J Nutr 129:2271–2277 [DOI] [PubMed]
- 34.Li C, Wong WH (2001) Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci USA 98:31–36 [DOI] [PMC free article] [PubMed]
- 35.Li T, Molteni A, Latkovich P, Castellani W, Baybutt RC (2003) Vitamin A depletion induced by cigarette smoke is associated with the development of emphysema in rats. J Nutr 133:2629–2634 [DOI] [PubMed]
- 36.Lien EL, Boyle FG, Wrenn JM, Perry RW, Thompson CA, Borzelleca JF (2001) Comparison of AIN-76A and AIN-93G diets: a 13-week study in rats. Food Chem Toxicol 39:385–392 [DOI] [PubMed]
- 37.Liu C, Lian F, Smith DE, Russell RM, Wang XD (2003) Lycopene supplementation inhibits lung squamous metaplasia and induces apoptosis via up-regulating insulin-like growth factor-binding protein 3 in cigarette smoke-exposed ferrets. Cancer Res 63:3138–3144 [PubMed]
- 38.Liu C, Russell RM, Wang XD (2004) Alpha-tocopherol and ascorbic acid decrease the production of beta-apo-carotenals and increase the formation of retinoids from beta-carotene in the lung tissues of cigarette smoke-exposed ferrets in vitro. J Nutr 134:426–430 [DOI] [PubMed]
- 39.Liu C, Russell RM, Wang XD (2003) Exposing ferrets to cigarette smoke and a pharmacological dose of beta-carotene supplementation enhance in vitro retinoic acid catabolism in lungs via induction of cytochrome P450 enzymes. J Nutr 133:173–179 [DOI] [PubMed]
- 40.Liu C, Russell RM, Wang XD (2004) Low dose beta-carotene supplementation of ferrets attenuates smoke-induced lung phosphorylation of JNK, p38 MAPK, and p53 proteins. J Nutr 134:2705–2710 [DOI] [PubMed]
- 41.Liu C, Wang XD, Bronson RT, Smith DE, Krinsky NI, Russell RM (2000) Effects of physiological versus pharmacological beta-carotene supplementation on cell proliferation and histopathological changes in the lungs of cigarette smoke-exposed ferrets. Carcinogenesis 21:2245–2253 [DOI] [PubMed]
- 42.Liu L, Gudas LJ (2005) Disruption of the lecithin:retinol acyltransferase gene makes mice more susceptible to vitamin A deficiency. J Biol Chem 280:40226–40234 [DOI] [PubMed]
- 43.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408 [DOI] [PubMed]
- 44.Lockhart DJ, Dong H, Byrne MC, Follettie MT, Gallo MV, Chee MS, Mittmann M, Wang C, Kobayashi M, Horton H, Brown EL (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 14:1675–1680 [DOI] [PubMed]
- 45.Ma Q, Lu AY (2007) CYP1A induction and human risk assessment: an evolving tale of in vitro and in vivo studies. Drug Metab Dispos 35:1009–1016 [DOI] [PubMed]
- 46.Mannisto S, Smith-Warner SA, Spiegelman D, Albanes D, Anderson K, van den Brandt PA, Cerhan JR, Colditz G, Feskanich D, Freudenheim JL, Giovannucci E, Goldbohm RA, Graham S, Miller AB, Rohan TE, Virtamo J, Willett WC, Hunter DJ (2004) Dietary carotenoids and risk of lung cancer in a pooled analysis of seven cohort studies. Cancer Epidemiol Biomarkers Prev 13:40–48 [DOI] [PubMed]
- 47.Meyer AM, Dwyer-Nield LD, Hurteau GJ, Keith RL, O’Leary E, You M, Bonventre JV, Nemenoff RA, Malkinson AM (2004) Decreased lung tumorigenesis in mice genetically deficient in cytosolic phospholipase A2. Carcinogenesis 25:1517–1524 [DOI] [PubMed]
- 48.Mueller CR, Maire P, Schibler U (1990) DBP, a liver-enriched transcriptional activator, is expressed late in ontogeny and its tissue specificity is determined posttranscriptionally. Cell 61:279–291 [DOI] [PubMed]
- 49.Nguyen T, Yang CS, Pickett CB (2004) The pathways and molecular mechanisms regulating Nrf2 activation in response to chemical stress. Free Radic Biol Med 37:433–441 [DOI] [PubMed]
- 50.Nishikawa A, Mori Y, Lee IS, Tanaka T, Hirose M (2004) Cigarette smoking, metabolic activation and carcinogenesis. Curr Drug Metab 5:363–373 [DOI] [PubMed]
- 51.Obermueller-Jevic UC, Espiritu I, Corbacho AM, Cross CE, Witschi H (2002) Lung tumor development in mice exposed to tobacco smoke and fed beta-carotene diets. Toxicol Sci 69:23–29 [DOI] [PubMed]
- 52.Obermuller-Jevic UC, Olano-Martin E, Corbacho AM, Eiserich JP, van der Vliet A, Valacchi G, Cross CE, Packer L (2003) Lycopene inhibits the growth of normal human prostate epithelial cells in vitro. J Nutr 133:3356–3360 [DOI] [PubMed]
- 53.Oishi K, Kasamatsu M, Ishida N (2004) Gene- and tissue-specific alterations of circadian clock gene expression in streptozotocin-induced diabetic mice under restricted feeding. Biochem Biophys Res Commun 317:330–334 [DOI] [PubMed]
- 54.Omenn GS, Goodman G, Thornquist M, Grizzle J, Rosenstock L, Barnhart S, Balmes J, Cherniack MG, Cullen MR, Glass A et al (1994) The beta-carotene and retinol efficacy trial (CARET) for chemoprevention of lung cancer in high risk populations: smokers and asbestos-exposed workers. Cancer Res 54:2038s–2043s [PubMed]
- 55.Oommen S, Vasu VT, Leonard SW, Traber MG, Cross CE, Gohil K (2007) Genome wide responses of murine lungs to dietary alpha-tocopherol. Free Radic Res 41:98–109 [DOI] [PMC free article] [PubMed]
- 56.Pang B, Wang C, Weng X, Tang X, Zhang H, Niu S, Mao Y, Xin P, Huang X, Zhu J (2003) Beta-carotene protects rats against bronchitis induced by cigarette smoking. Chin Med J (Engl) 116:514–516 [PubMed]
- 57.Paolini M, Cantelli-Forti G, Perocco P, Pedulli GF, Abdel-Rahman SZ, Legator MS (1999) Co-carcinogenic effect of beta-carotene. Nature 398:760–761 [DOI] [PubMed]
- 58.Rangasamy T, Cho CY, Thimmulappa RK, Zhen L, Srisuma SS, Kensler TW, Yamamoto M, Petrache I, Tuder RM, Biswal S (2004) Genetic ablation of Nrf2 enhances susceptibility to cigarette smoke-induced emphysema in mice. J Clin Invest 114:1248–1259 [DOI] [PMC free article] [PubMed]
- 59.Richter HG, Torres-Farfan C, Rojas-Garcia PP, Campino C, Torrealba F, Seron-Ferre M (2004) The circadian timing system: making sense of day/night gene expression. Biol Res 37:11–28 [DOI] [PubMed]
- 60.Romieu I, Castro-Giner F, Kunzli N, Sunyer J (2008) Air pollution, oxidative stress and dietary supplementation: a review. Eur Respir J 31:179–197 [DOI] [PubMed]
- 61.Russell RM (2004) The enigma of beta-carotene in carcinogenesis: what can be learned from animal studies. J Nutr 134:262S–268S [DOI] [PubMed]
- 62.Sakamoto K, Nagase T, Fukui H, Horikawa K, Okada T, Tanaka H, Sato K, Miyake Y, Ohara O, Kako K, Ishida N (1998) Multitissue circadian expression of rat period homolog (rPer2) mRNA is governed by the mammalian circadian clock, the suprachiasmatic nucleus in the brain. J Biol Chem 273:27039–27042 [DOI] [PubMed]
- 63.Schmitz HH, Poor CL, Gugger ET, Erdman JW Jr (1993) Analysis of carotenoids in human and animal tissues. Methods Enzymol 214:102–116 [DOI] [PubMed]
- 64.Sharoni Y, Danilenko M, Dubi N, Ben-Dor A, Levy J (2004) Carotenoids and transcription. Arch Biochem Biophys 430:89–96 [DOI] [PubMed]
- 65.Siems W, Wiswedel I, Salerno C, Crifo C, Augustin W, Schild L, Langhans CD, Sommerburg O (2005) Beta-carotene breakdown products may impair mitochondrial functions—potential side effects of high-dose beta-carotene supplementation. J Nutr Biochem 16:385–397 [DOI] [PubMed]
- 66.Siler U, Barella L, Spitzer V, Schnorr J, Lein M, Goralczyk R, Wertz K (2004) Lycopene and vitamin E interfere with autocrine/paracrine loops in the Dunning prostate cancer model. FASEB J 18:1019–1021 [DOI] [PubMed]
- 67.Stern DM, Yan SD, Yan SF, Schmidt AM (2002) Receptor for advanced glycation endproducts (RAGE) and the complications of diabetes. Ageing Res Rev 1:1–15 [DOI] [PubMed]
- 68.Teske S, Bohn AA, Regal JF, Neumiller JJ, Lawrence BP (2005) Activation of the aryl hydrocarbon receptor increases pulmonary neutrophilia and diminishes host resistance to influenza A virus. Am J Physiol Lung Cell Mol Physiol 289:L111–L124 [DOI] [PubMed]
- 69.Thatcher TH, Maggirwar SB, Baglole CJ, Lakatos HF, Gasiewicz TA, Phipps RP, Sime PJ (2007) Aryl hydrocarbon receptor-deficient mice develop heightened inflammatory responses to cigarette smoke and endotoxin associated with rapid loss of the nuclear factor-kappaB component RelB. Am J Pathol 170:855–864 [DOI] [PMC free article] [PubMed]
- 70.Tomasinsig L, Scocchi M, Di Loreto C, Artico D, Zanetti M (2002) Inducible expression of an antimicrobial peptide of the innate immunity in polymorphonuclear leukocytes. J Leukoc Biol 72:1003–1010 [PubMed]
- 71.Villard PH, Seree EM, Re JL, De Meo M, Barra Y, Attolini L, Dumenil G, Catalin J, Durand A, Lacarelle B (1998) Effects of tobacco smoke on the gene expression of the Cyp1a, Cyp2b, Cyp2e, and Cyp3a subfamilies in mouse liver and lung: relation to single strand breaks of DNA. Toxicol Appl Pharmacol 148:195–204 [DOI] [PubMed]
- 72.Wang L, Antonini JM, Rojanasakul Y, Castranova V, Scabilloni JF, Mercer RR (2003) Potential role of apoptotic macrophages in pulmonary inflammation and fibrosis. J Cell Physiol 194:215–224 [DOI] [PubMed]
- 73.Wang XD, Liu C, Bronson RT, Smith DE, Krinsky NI, Russell M (1999) Retinoid signaling and activator protein-1 expression in ferrets given beta-carotene supplements and exposed to tobacco smoke. J Natl Cancer Inst 91:60–66 [DOI] [PubMed]
- 74.Whitlock JP Jr (1999) Induction of cytochrome P4501A1. Annu Rev Pharmacol Toxicol 39:103–125 [DOI] [PubMed]
- 75.Witschi H (2005) Carcinogenic activity of cigarette smoke gas phase and its modulation by beta-carotene and N-acetylcysteine. Toxicol Sci 84:81–87 [DOI] [PubMed]
- 76.Witschi H (2005) A/J mouse as a model for lung tumorigenesis caused by tobacco smoke: strengths and weaknesses. Exp Lung Res 31:3–18 [DOI] [PubMed]
- 77.Witschi H, Espiritu I, Peake JL, Wu K, Maronpot RR, Pinkerton KE (1997) The carcinogenicity of environmental tobacco smoke. Carcinogenesis 18:575–586 [DOI] [PubMed]
- 78.Yoshida T, Tuder RM (2007) Pathobiology of cigarette smoke-induced chronic obstructive pulmonary disease. Physiol Rev 87:1047–1082 [DOI] [PubMed]
- 79.Zachman RD (1995) Role of vitamin A in lung development. J Nutr 125:1634S–1638S [DOI] [PubMed]
- 80.Zachman RD, Grummer MA (1998) Effect of maternal/fetal vitamin A deficiency on fetal rat lung surfactant protein expression and the response to prenatal dexamethasone. Pediatr Res 43:178–183 [DOI] [PubMed]
- 81.Zaripheh S, Boileau TW, Lila MA, Erdman JW Jr (2003) [14C]-lycopene and [14C]-labeled polar products are differentially distributed in tissues of F344 rats prefed lycopene. J Nutr 133:4189–4195 [DOI] [PubMed]
- 82.Zaripheh S, Nara TY, Nakamura MT, Erdman JW Jr (2006) Dietary lycopene downregulates carotenoid 15,15′-monooxygenase and PPAR-gamma in selected rat tissues. J Nutr 136:932–938 [DOI] [PubMed]
- 83.Zhao X, Aldini G, Johnson EJ, Rasmussen H, Kraemer K, Woolf H, Musaeus N, Krinsky NI, Russell RM, Yeum KJ (2006) Modification of lymphocyte DNA damage by carotenoid supplementation in postmenopausal women. Am J Clin Nutr 83:163–169 [DOI] [PubMed]
- 84.Zolfaghari R, Cifelli CJ, Banta MD, Ross AC (2001) Fatty acid delta(5)-desaturase mRNA is regulated by dietary vitamin A and exogenous retinoic acid in liver of adult rats. Arch Biochem Biophys 391:8–15 [DOI] [PubMed]