Abstract
Molecular methods allow the detection of pathogen nucleic acids (DNA and RNA) and, therefore, the detection of contamination in food is carried out with high selectivity and rapidity. In the last 2 decades molecular methods have accompanied traditional diagnostic methods in routine pathogen detection, and might replace them in the upcoming future. In this review the implementation in diagnostics of four of the most used molecular techniques (PCR, NASBA, microarray, LDR) are described and compared, highlighting advantages and limitations of each of them. Drawbacks of molecular methods with regard to traditional ones and the difficulties encountered in pathogen detection from food or clinical specimen are also discussed. Moreover, criteria for the choice of the target sequence for a secure detection and classification of pathogens and possible developments in molecular diagnostics are also proposed.
Keywords: Molecular biology, Food safety, Diagnostic, PCR, Microarray, NASBA, LDR
Introduction
Zoonoses and foodborne pathogens are health threatening agents that cause disease and death in humans, even in developed countries. In Europe as well as in the USA, pathogens such as Listeria, Escherichia coli, Salmonella, Campylobacter and Mycobacterium bovis are widespread and are occasionally the cause of disease outbreaks [54]. Traditional diagnostic methods identify a pathogen based on its phenotype: e.g. classification according to the ability to grow on a certain media, to metabolize a given chemical compound, etc. The exact classification of a serotype is achieved with the use of antibodies, generally directed against membrane proteins, or with serotype specific bacteriophages. The correct assessment of a clinical isolate can take 2–3 days or longer. Therefore, the development of rapid and secure methods to detect and trace the origin of pathogens and contaminants is urgently needed [7]. Faster and simpler methods would be a great advantage for many diagnostic purposes. Food safety could be greatly enhanced by the use of fast diagnostic methods allowing the immediate detection of pathogens [49].
Fast diagnostic methods include those based on the recognition and amplification of nucleic acids. As the same detection technique can be applied to identify nucleic acids from all organisms, the same strategies can be used in clinical diagnosis as for the detection of food-borne pathogens and GMOs. Methods for the amplification and detection of very small quantities of nucleic acids have been available for many years, but only in the last 10–15 years have been employed in diagnostics. Furthermore, in the last decade the amount of nucleic acid sequence data available for many organisms, including the whole genome sequence of a large number of pathogens has provided more support for DNA/RNA-based tests.
In this review, we describe some of the most commonly used nucleic acid-based methods for contamination detection and compare the advantages and limitations of these techniques.
Polymerase chain reaction
The Polymerase chain reaction (PCR) was the most important development for research in molecular biology [36, 41]. It is now the basic technique for the development of most molecular diagnostic methods for food safety and other fields [35].
In diagnostic PCR, specific primers directed against the DNA of the organism to be detected are used. The homology between primers and the target DNA confers specificity to the amplification. The presence of the amplification product at given reaction conditions reveals the presence of the organism in the tested sample.
The traditional method of visualizing the amplified product by ethidium bromide (EtBr) on an agarose gel has more recently been replaced by the less toxic and more sensitive SYBR GREEN, a dye that emits fluorescence upon intercalating into the double stranded DNA. SYBR GREEN can also be conveniently used in a real-time PCR machine.
The real-time PCR machine is a thermal cycler able to stimulate the fluorescent dye with a laser and to quantify the fluorescence of the reaction mix, and so the amplification product, after each cycle. The measurement of the amplified product in real-time allows to be quantified while the reaction is in the exponential phase and before plateaus. During the exponential phase, differences between samples are a simple function of the initial concentration of the target DNA and can be, therefore, immediately assessed. Moreover, the comparison with reference samples of known concentration allows the quantification of the initial concentration of the target DNA.
Nevertheless, the implementation of SYBR GREEN in real-time amplification experiments does not allow discriminating between specific target amplifications and co-produced PCR artefacts, such as non-specific amplifications or primer dimmers [24]. This could interfere with the detection and quantification of the target DNA, especially at low concentrations.
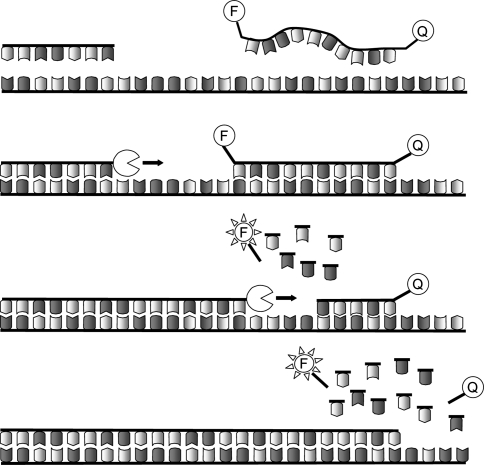
PCR reliability, in terms of specificity of pathogen detection and quantification, has been improved by the use of dye quenched probes [3, 39, 55]. TaqMan probes, which are the most commonly used dye quenched probes in diagnostics, are short DNA oligonucleotides (normally 10 bp long–10mer) specific to the target sequence between the two primers used in the PCR. TaqMan probes carry a fluorophore at one end and a quencher at the other, which prevents the fluorophore from being visible. During PCR cycling, the TaqMan probe specifically anneals to the single strand DNA target sequence and is degraded by the 3′–5′ exonuclease activity of the DNA polymerase. The fluorophore, separated from the quencher, then becomes visible (Fig. 1). The fluorescence, measured after each cycle in a real-time PCR machine, is proportional to the amount of the specific target amplification product and does not include PCR artefacts.
Fig. 1.
TaqMan PCR. Primers and TaqMan probe anneal to the target DNA. The Taq polymerase synthesises the new strand starting from the primer. The exonuclease activity of the Taq polymersase degrades the TaqMan probe to nucleotides, uncoupling the fluorophore (F) from the quencher (Q). The fluorescence emitted from the freed fluorophore is detected after each cycle
Moreover, the use of labelled probes allows the use of an internal positive control (IPC), to reveal the presence of PCR inhibitors. This feature is essential to assess the absence of amplification from a suitable sample, and to dispel the suspicion of false negative results. The IPC comprises a second TaqMan probe and of an artificial oligo carrying the annealing site for the IPC probe. The site is flanked by the two annealing sites of the same primers used in the amplification of the test sample, so that the artificial oligo can be amplified in the reaction. The IPC is added in the reaction mix and the reaction is performed in a real-time PCR machine equipped with two lasers capable of revealing and distinguishing the IPC and the sample signals. The IPC amplification demonstrates the absence of PCR inhibitors. Therefore, the concomitant absence of sample amplification can be considered as a valid result [27].
PCR implementation in diagnostics
The TaqMan PCR system has been recently used to detect bacterial, fungal, viral and nematode contamination from different matrices including food, human and animal specimens, environmental samples, etc. [21, 27, 43]. PCR is the most widely used molecular diagnostic technique due to its fast and easy to use protocol. Originally, 2–3 h were needed to complete a PCR, but nowadays more advanced PCR systems can deliver a result in a matter of minutes [57]. The high sensitivity of the PCR can be further improved with the application of reverse-transcription-PCR (RT-PCR). This requires the reverse-transcription of RNA by random primers and an RNA dependent DNA polymerase enzyme called reverse transcriptase. The resulting DNA copy of the RNA (cDNA) is used as template in the PCR. So far, RT-PCR has been mostly used to detect retroviruses, such as HIV, norovirus or avian influenza virus, as will be described later [42].
Numerous TaqMan PCR based tests were implemented in the detection of foodborne pathogens. For instance, Fratamico [15] showed the validity of this technique against traditional immunoassays in the detection of Salmonella sp. in naturally contaminated ground chicken, turkey and beef and [59] developed a test able to detect and discriminate pathogenic strains of Yersinia enterocolitica in pork meat. Elizaquível and colleagues [13] tested the sensitivity of TaqMan PCR in the detection of Leuconostoc sp. from meat products, setting the detection threshold to less than five genomic equivalents per reaction.
It is noteworthy that several PCR based tests have been approved by government agencies for food safety (e.g. Food and Drug Administration in USA), for routine food quality control and clinical diagnosis [63].
Nucleic acid sequence based amplification (NASBA)
Nucleic acid sequence based amplification (NASBA) is a technology developed in the early 90s to amplify nucleic acids without the use of a thermal cycler [9]. It is most often used to obtain many copies of RNA starting from a few RNA molecules.
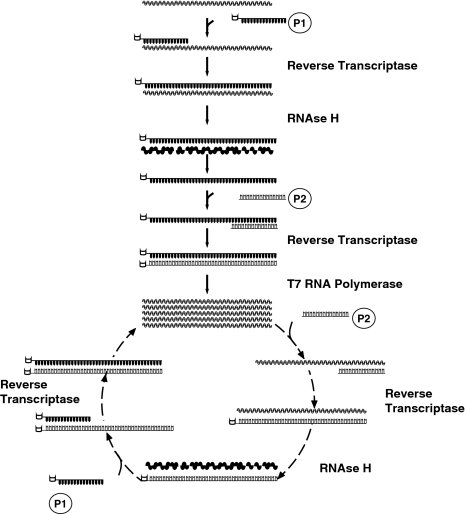
This approach employs two specific primers flanking the sequence to be amplified. The first primer (P1) carries at its 5′ end, the binding sequence for the T7 RNA polymerase and is used to initiate the RNA reverse-transcription (RT) reaction, catalyzed by a reverse-transcriptase. The RNAse H is then used to degrade the RNA strand in the RNA–DNA hybrid molecules that result from the RT reaction. The remaining cDNA is then accessible to the second primer (P2) which initiates the synthesis of the complementary strand. A third enzyme, the T7 RNA Polymerase, docks the double strand DNA on the sequence at the 5′ end of P1, transcribing many RNA copies of the gene. This process, i.e. the cycle of first strand synthesis/RNA hydrolysis/second strand synthesis and RNA transcription, is repeated indeterminately starting from the newly transcribed RNA (Fig. 2).
Fig. 2.
NASBA. The primer P1, carrying at the 5′ end the T7 RNA polymersase binding site (open box), anneals to the RNA strand and is extended by the reverse transcriptase to synthesize the cDNA. The RNA strand in the RNA/DNA duplex is degraded by the RNAse H. The cDNA is accessible to the second primer, P2 that allows the synthesis of the complementary strand. The resulting double strand DNA has a T7 RNA polymerase binding site where the enzyme can bind and produce many RNA copies. The cycle can be repeated indefinitely and the initial target strand is exponentially amplified
RNA and double strand cDNA accumulate exponentially and can be detected by EtBr/agarose gel electrophoresis. Recently, fluorescently labelled probes and a fluorescence scanner are employed to follow the NASBA real-time amplification of a viral RNA genome [28]. Bacteria detection can also take advantage quick amplification of expressed genes by NASBA, as described for the enterotoxin related genes from the pasteurization resistant Bacillus sp. in milk [18].
Compared with PCR, the lab equipment for NASBA is less expensive. NASBA does not require a thermal cycler, which is required to perform the PCR: the NASBA reaction, apart from the initial denaturation step, is carried out in isothermal conditions and a water bath or an isothermal block is sufficient. The use of NASBA could contribute to the development of an affordable, portable and easy to use pathogen detection kit, suitable even for less well equipped laboratories. For instance, NABSA amplification could be coupled to a dipstick, that is a simple and ready to use display method that relies on capillarity for nucleic acid migration and, like NASBA, does not require expensive lab equipment [11].
Oligonucleotide DNA microarray
DNA arrays represent an evolution of nucleic acid detection techniques. They were developed in the late 80s with the intent to directly and simultaneously assess gene expressions. The first versions were represented by collections of cDNA probes, each targeting a different gene, spotted on a nitrocellulose membrane and known as macroarrays. The development of sophisticated probe printing and image scanning technologies during the last 15 years has contributed to the miniaturization of the support and incremented the spot density, generating microarrays. Microarrays typically consist of a few squared centimetre support (e.g. a microscope glass slide), also known as chip, containing up to hundreds of thousands of probes representing for instance the whole genome or the whole transcriptome of the organisms under investigation. In certain cases, the use of this many probes is not necessary and the microarray is more conveniently divided into subarrays. For instance, each subarray can represent replicates of the same probe set and different experiments can be carried out in parallel on the same chip.
In spite to the evolution of the array, the typical experimental procedure conserved similar steps for macro and microarrays. First, the sample under investigation, which can be mRNA, cDNA, PCR products or genomic DNA, is labelled with a dye (e.g. a fluorescent or radioactive dye). The labelled sample is denaturated to produce single strand fragments and then hybridized to the array. Each DNA fragment will be captured by the corresponding DNA probe.
The excess of labelled nucleic acid is then washed away and the array is scanned to quantify the dye signal from each spot. The signal intensity corresponds to the concentration of each labelled nucleic acid fragment.
Oligo DNA microarray that have been extensively used in diagnosis, feature specific DNA oligonucleotides as probes. Each oligonucleotide probe can target a specific part of a gene, a fragment of genomic DNA or a universal Zip Code (see below).
Differences in oligonucleotide DNA microarrays regard mainly the technique used to print the spots and the oligo length. Manufacturers like Affymetrix use a photolithography strategy to synthesize the oligo directly on the chip, producing 25mer oligonucleotides [15, 51]. This technique allows strict control over number, concentration and spacing of each probe, enabling the production of high density microarray (500 k unique 25mer, each contained in an 18 μm2 spot; [34]).
Other microarray manufacturers take advantage of a spotter. The spotter, either with a pin or with an “ink jet” method, deposit droplets of oligo DNA (normally 25–70mers) that bind to the chip surface [26]. Although microarrays produced by photolithography have better performance in terms of data reproducibility and higher level of miniaturization, spotted microarrays can be constructed “in house” by a research laboratory and are generally cheaper. Nevertheless, cross contamination of probes and pin clogging are problems often encountered, resulting in non-specific signals and missing spots [12].
The length of the oligos varies from 25 to 70 bps. Generally, long oligos have higher sensitivity, but short oligos offer a higher power to recognise unspecific hybridisations [2, 61]. The higher selectivity of 25mer microarrays is achieved with the probe pair system, first introduced by Affymetrix: each probe (that is a “perfect match”, PM) is accompanied by another “mismatched” (MM) probe. The MM probe differs from the PM probe only for a single nucleotide located in the middle of the 25mer oligo. In absence of cross hybridization, the signal from the PM spot is always stronger than that of the MM spot. If the situation is reversed, i.e. the signal from the mismatched probe is not significantly lower than the PM probe, both PM and MM signals are most likely due to unspecific hybridization. This methodology not only allows the detection of cross-hybridizations, but also with some modifications can be used for genotyping, i.e. in the determination of the presence of specific alleles, with a single nucleotide resolution. In this case, four probes are used that differ for the oligonucleotide in the middle position, that is in turn either and A or a G or a C or a T. The probe that perfectly matches the sample DNA shows the highest signal. By comparing the probe signals is, therefore, possible to determine the allelic composition of the investigated DNA sample.
Microarray-based diagnostics
In food quality control as well as in veterinary and medical diagnostics, it is crucial to quickly identify the pathogens causing certain symptoms or contaminations. The accuracy of pathogen detection is also very important as some closely related innocuous and harmful bacteria can coexist in the same matrix or product. If the two kinds are not distinguished, detection might result in false positives or false negatives. The wide range of pathogens that can contaminate a given product might delay the right pathogen diagnosis.
The possibility, offered by microarrays, to miniaturize many different pathogen specific probes on a single support well accompanies their high sensitivity and specificity. In this scenario, it is easy to imagine the opportunities microarrays can offer, making them the technique of choice for pathogen detection and diagnostics in the upcoming future. Recently, Lin et al. [30] set up a diagnostic test for the detection of a total of 23 bacteria and viruses. The test implements Affymetrix microarrays on which pathogen specific sequences are synthesized that are redundantly informative, to guarantee high detection quality. The system incorporates the PM/MM method to discriminate between real and artefacts.
Wang et al. [62] presented an array-based assay for the identification of 23 foodborne pathogens. Their array consists of 20–30mers spotted on a glass slide, and is designed to hybridize to the 16S gene of target pathogens. Authors found a cross reactivity, expected from theoretical prediction, among strains. Nevertheless, this cross reactivity does not hamper the correct classification of each isolate, relying on the pathogen specific hybridization pattern, i.e. on its “molecular signature”.
Other microarray-based tests also focused on important food-borne pathogens. For instance, Shigella sp. and Escherichia coli received a lot of attention and much effort has been put into the development of a test able to detect and determine the serotype of pathogenic strains [29, 31, 32]. The identification of a specific serotype is particularly important with E. coli, as pathogenicity varies among different strains, ranging from the harmless K-12 to the life threatening O157:H7. Another approach exploits over 400 long DNA oligos (70mers) to detect and classify 11 different E. coli pathotypes, i.e. groups of strains hosting similar virulence factor and causing similar clinical symptoms [44]. The assay relies on pathotype-specific genes as well as E. coli core sequences. The system shows some non-specificity, meaning that several probes cross-react also with pathotypes different from those to which they should be specific. Again, the correct assessment of each strain is based on the molecular signature of the pathotype. The lower stringency of long probes allows DNA from previously uncharacterized pathotypes to hybridise even in absence of a full identity with the probes. This can result in a new hybridisation pattern and can serve to define a new molecular signature for the strain under examination.
Food safety along with traceability took advantage of array-based tests like CarnoCheck® (greiner bio-one) that allow the simultaneous identification of eight different animal species to detect food fraudulent manipulations.
Until recently the high costs of reagents (chip and labelling kit) and of instruments (scanner and hybridization chamber) limited the use of diagnostic microarrays mainly to the medical field. It is not unreasonable though to believe that in the upcoming future the widespread use of this technology will make microarray-based diagnostics more affordable and will diffuse this technology to the routine use in food safety.
Ligation detection reaction-universal arrays (LDR-UA)
The ligation detection reaction (LDR) combined with universal arrays (UA) was introduced in the late 90s by Gerry et al. [16]. The LDR-UA technique was developed to uncouple the probe/target recognition and annealing from the microarray hybridization, reducing costs and conferring higher flexibility and selectivity [5].
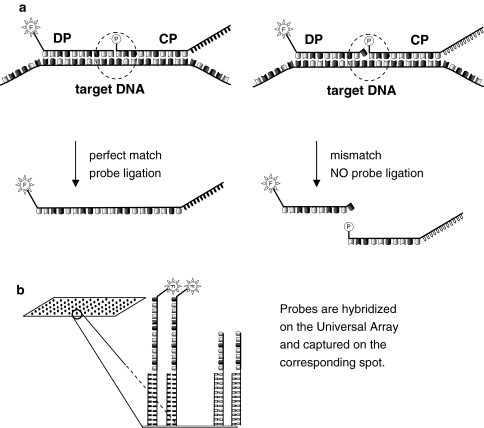
The LDR takes advantage of probe sets composed of two probes: a discriminating probe (DP) and a common probe (CP). DP and CP are designed to anneal juxtaposed on the target sequence, with the DP positioned at the 5′ end of the CP and no gaps between them. The DP is modified at its 5′ end with a fluorophore, whereas the CP is modified at its 5′ end with a phosphate and it is extended at the 3′ end with a unique 25 bp sequence named Zip Code, that characterize the probe set. The target sequence is typically a PCR product that is denatured to allow the CP and DP to anneal one next to each other. A thermostable ligase is then used to ligate the two probes together, bringing the fluorophore and the cZip Code together on the same resulting molecule (Fig. 3).
Fig. 3.
LDR-UA. a Discriminating probe (DP) and common probe (CP) anneal juxtaposed to the target DNA. DP is labelled with a fluorophore. CP is phosphorylated at the 5′ end and is linked to its 3′ end a Zip Code. The probe/target perfect match allows the thermostable ligase to bind the two probes together. One single mismatch at the 3′ end of the DP prevents the ligation of the probes. b The ligation reaction is hybridized to a universal array. Each CP is captured on a spot corresponding to a complementary of a Zip Code (cZIP Code). The successful ligation is detected by array scanning
The ligation product is hybridized to the UA. The UA is an oligonucleotide DNA microarray with a different “cZip Code” (the complementary sequence of the ZIP Code) in each spot. Each CP is captured by the corresponding cZip Code. If CP and DP have been ligated together in the LDR, i.e. if the captured molecule carries a fluorophore, then the array spot will be visible.
Normally Taq or Pfu ligases are used as they have the highest performance at 65°C. This allows designing probes with high melting temperature, thus minimizing the cross reactivity and increasing specificity. Moreover, due to the thermostable properties of the ligase, the denaturation/annealing/ligation process can be repeated many times with a theoretical linear amplification of the target (30-fold for 30 cycles). The LDR reaction is also very sensitive to mismatches: a single base pair substitution located at the 3′ end of the DP prevents the ligation, resulting in no signal at the corresponding spot of the UA. This confers to the LDR-UA detection system a very high discriminatory power that finds a good application in diagnostics. By using LDR-UA it is possible to safely identify and distinguish related species that differ by only one base pair in a considered DNA sequence.
The use of multiple CP/DP sets targeting one or more PCR fragments is possible. The detection of DNA from different species can be achieved by implementing multiplex PCR products and a mixture of CP/DP sets. The array hybridization pattern will describe the content of the DNA pool under investigation. The possibility to spot a large number of cZip Codes onto a single slide gives the opportunity to detect as many DNA species at the same time.
The same Zip Code can be associated with probe sets labelled with fluorophores of different colours. This approach can be exploited to increase the number of DNA species detectable simultaneously without altering the number of available spots on the array.
An alternative to UA hybridization is the use of capillary electrophoresis (CE). The LDR products are separated in capillaries and the ligation occurrence is visible due to the band-shift. The use of various probe sets simultaneously is still possible and the ligated probes can be separated with a one base pair resolution [17, 46]. This approach can be cost effective, but when the simultaneous visualization of many different DNA species or a higher sensitivity is needed UAs are more convenient.
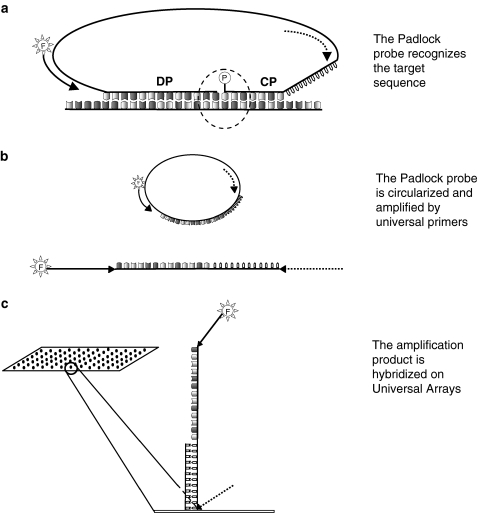
Recently, the use of padlock probes allowed the detection of genomic DNA without an intermediate PCR amplification step. In this approach, the two CP/DP probes are at the extremities of one long (~100 bp) DNA oligo (padlock probe, Fig. 4). Within the padlock probe two primer annealing sites and a Zip Code are included. The parts of the padlock probe corresponding to the two probes anneal to the DNA target and are ligated together upon sequence recognition, resulting in the circularization of the molecule. The molecular ring is then amplified by universal primers and labelled with a fluorophore. The amplicon, that must include the Zip Code, is then hybridized to an UA [52].
Fig. 4.
LDR-UA with padlock probes. a Padlock probes carry discriminating probe (DP) and common probe (CP) on a single molecule. The CP is normally shorter than DP to increase specificity. The padlock probe also contains two annealing sites for universal primers (indicated as an arrow and a dashed arrow), and a cZIP Code (jagged line). The padlock probe is circularized upon target recognition and ligation mediated by a ligase. b Circularized molecules are amplified with universal primers, one of which carries a fluorophore. c Amplification product is hybridized to a universal array
LDR-UA in diagnostics
The sensitivity and flexibility offered by the LDR-UA system creates many possibilities in various fields, from genotyping to diagnosis. Although the full potential of this technique has not been exploited yet, some authors report interesting applications.
Food safety has been the concern of LDR-UA based kit developers, as at least two tests have been published that contribute to this research field. First, the LDR-UA technology has been used for the detection and classification of water borne pathogens such as cyanobacteria [6]. The authors used a mix of 19 probe sets able to detect over 300 cyanobacteria strains and classify them in 14 phylogenetic lineages. Second, this technology was implemented to detect five different genetically modified organism (GMO) DNAs simultaneously [4]. The LDR, followed by UA hybridization, was performed on PCR products obtained from samples containing as little as 0.5% GMO DNA, producing unambiguous results.
Finally, a LDR-UA based kit was also developed to detect mutations of bovine milk protein genes with the aim of improving the composition and quality of milk and milk derived products [8].
Molecular detection techniques comparison
Many factors guide the choice of which molecular technique is to be implemented. The available budget, the response time, the number of species to be detected simultaneously, and the required resolution (genera, species, pathotype, serotype), are some of the features to be considered (Table 1).
Table 1.
Limitations and advantages of molecular diagnostic techniques
| CONS | PROS | Application examples | |
|---|---|---|---|
| NASBA | Needs the bacteria to be viable | Cheap | Detection of enterotoxin related genes from Bacillus sp. in milk [18] |
| Needs the gene to be expressed RNA handling might be troublesome |
Sensitive | ||
| TaqMan | Cost effective for a limited number of DNA species | Easy to use | Detection of Salmonella sp. in ground meat [14] |
| The number of simultaneous DNA species detectable is limited by the fluorophores SNP detection not always reliable |
Quick result | ||
| High density microarrays | Expensive | Detects the highest number of DNA species simultaneously | Assessment of E. Coli pathotype [44] |
| Slower than NASBA and TaqMan PCR | |||
| LDR-UA | Slower than NASBA and TaqMan PCR | Highest performance in SNP detection | Detection of GMO contamination in food and cyanobacteria in water [4, 6] |
| Highly sensitive | |||
| Very flexible |
The most popular approach is TaqMan PCR that is considered a good compromise of the features listed above. This system can provide results in a matter of hours and has also been used to discriminate sequences that differ only for one base pair. This is done by using two competing probes corresponding to the two genotypes under investigation [50]. Nevertheless, other methods have been shown to perform better in single nucleotide polymorphism (SNP) identification (see below and [22]).
Comparing the performance of NASBA and PCR on the same number of bacteria cells, NASBA is generally more sensitive than PCR performed on genomic DNA, possibly because the amplification starts from the many mRNA copies of the gene under investigation, in contrast to its single copy in the genome [58]. The NASBA-based diagnostic assay for the detection of Plasmodium falciparum proved to provide results in less time than the RT-PCR equivalent [48]. RNA based techniques, such as NASBA or RT-PCR, have been mainly applied to the detection of retroviruses, leaving the diagnosis of bacteria to genomic DNA based systems. This choice is probably due to the difficulty in handling RNA, the need for the gene used as marker to be constitutively expressed and for the cell to be viable.
The great advantage of miniaturized systems like microarrays resides in the possibility to visualize many different DNA sequences at the same time. High density microarray chips can contain hundreds of thousand of probes and allow the detection of a large range of pathogens, as well as undesired food components (e.g. GMOs). The high costs of microarrays are justified if their potential is fully exploited and are used with samples, where a wide spectrum of contaminants is expected to be found.
Differently from the TaqMan PCR, where the detection occurs as the reaction runs, the use of microarrays requires further steps: the purification and labelling of the amplified DNA and its hybridization to the chip. The recommended hybridization time for short oligo chips is 14–16 h at 45°C (Affymetrix).
With regard to the LDR-UA system, the LDR itself adds an additional 3–4 h to the process. This time consuming step is largely compensated by a reduced microarray hybridization period. Since the Zip Codes are designed to have an optimal melting temperature, incubation at 65°C for 1–2 h is sufficient, and the cross reactivity is minimal.
The LDR-UA has proved to be very sensitive, being able to detect target PCR product up to 50 pM, a concentration certainly not visible with an EtBr agarose gel or with a conventional microarray [6]. The high sensitivity of LDR-UA is mainly due to the linear amplification performed during the reaction and to the concentration of the fluorescent probes on the UA spot. The PCR–LDR–CE variant too gives good performance, with the detection limit of about 120 bacterial genome equivalents in the initial PCR.
The tight control on the probe ligation due to the thermostable ligase results in no non-specific signal in case of probe/target mismatch and consequently unambiguous interpretation of the results. Moreover, this system allows the most reliable SNP detection and the safe discrimination of species or serotypes that differ even by as little as one nucleotide.
The universality of the array resides in the cZip Code that has the lowest possible homology to any other sequence. The advantage of this system is that the probe sets can be changed without printing a new microarray. In this way, the replacement of a malfunctioning probe or the addition of new probe sets is easily and quickly done at the cost of the new probes only. Furthermore, the probe/target annealing temperature is not constrained to a certain value, as it happens during regular array hybridization: the LDR can be performed separately according to each probe optimum temperature and the ligation products can be pooled together before hybridizing on the UA [8].
Choice of the target gene
Regardless the detection technique chosen for the development of a diagnostic method, the selection of the target sequence must be done carefully and a few important points must be considered.
First, the huge amount of sequence data available from many online databases offers a powerful resource for the design of primers and probes to be implemented in the detection of pathogens. Nevertheless, isolates from field samples or clinical specimens can significantly deviate from the reference strain sequence reported in the database, as the reference strains represent single isolates. For this reason the largest number of sequences should be considered in the design of the molecular probes, and if needed in house sequencing should be performed. Moreover, to add robustness to the molecular test and to be reasonably sure to detect all strains, the target sequence should be well conserved in the various species to be included in the assay. Similarly, if the intent is to distinguish a species from a closely related one, reliable polymorphisms should be chosen. Non synonymous polymorphisms, i.e. those that alter the deduced amino acid sequence of a coding region, are normally considered to be more reliably species-specific than the non-conservative ones, as they result in a phenotypic change that might be specific of the strain.
Secondly, the detection of pathogenic genes located in plasmids (e.g. the shiga-like toxin encoding genes in the O157 plasmid of E. coli) might increase the sensitivity of the test with respect to genomic sequences, being plasmids present in multiple copies in each cell. However, the use of these sequences as target could lead to the incorrect classification of the detected bacteria, due to the high ratio of horizontal plasmid transfer, even between different species [56].
Thirdly, one should consider whether to target a protein coding sequence or any other conserved regions, e.g. those encoding the 16S or 18S ribosomal RNA. The latter are very conserved sequences and are normally present in many copies in the genome. These features elected 16S or 18S the target of choice for many diagnostic purposes and a databases have been created to collect the 16S sequences from a huge number of living forms (RDPII database, over 706 k entries in October 2008; http://rdp.cme.msu.edu/).
However, some authors chose genes that are more strongly related to virulence, hence considered more pathogen specific. For instance, Volokhov and colleagues [60] developed a Listeria diagnostic microarray-based on genes essential for the bacteria virulence but found in all Listeria strains, such as the invasion associated protein (iap), and in pathogenic strain-specific genes, such as hly.
The use of virulence genes as well as other species-specific sequences has an additional advantage: they are normally less conserved than the ribosomal RNA encoding genes; therefore, they permit discrimination with a higher resolution. In this way, the correct classification of very closely related species, such as E. coli and Shigella sp. is possible. For instance, targeting various virulence and non-virulence genes, a PCR based assay was developed to perform a highly discriminative classification of E. coli distinguishing the most frequently isolated serotypes [45].
Molecular diagnostic limitations
Molecular diagnostic methods offer a faster and more sensitive alternative to traditional immunoassays and culture techniques. Nevertheless, in spite of the unquestionable advantages a quick diagnosis brings to food safety, so far they have replaced traditional methods only in a limited number of analyses. There are still some factors that hinder the adoption of diagnostic assays based on the pathogen’s nucleic acids, rather than on its phenotype.
One major factor is the abundance of false positives and false negatives results. The presence of DNA in the environment, in the laboratory and even in the instruments used to prepare the reaction mix often results in the false detection of a pathogen. Differently from living cell contaminations that can be easily wiped out from surfaces and lab equipment, DNA is less easily removable.
False negative results can be caused by the presence of inhibitors. It often happens that the sample to be analyzed is a complex matrix, such as cheese or salami that can contain chemical compounds able to interfere with the enzyme activity. Enzyme inhibition can thus bring about a false negative result. Therefore, some food matrices require nucleic acid extraction protocols that are ad hoc studied to remove inhibitors. Furthermore, the introduction of positive controls like the IPC described for the TaqMan PCR is necessary to bring validity to the test.
In many cases the genomic DNA of the host organism itself, especially from ground or processed meat or from complex food products, can interfere with the detection procedure by competing for the probe and primer annealing. The big size of vertebrate or plant genomes, on average 103–104 fold bigger than that of bacteria, renders this phenomenon not negligible even with relatively few host cells. The reduction of DNA complexity, for instance by eliminating genomic repetitive sequences, can in many cases help solve this problem [30]. In general, the presence of abundant inhibitors or competitors makes the bacteria enrichment via culture a necessary step [37, 53]. Enrichment is typically achieved with a stomacher device that mechanically disrupts the food matrix in a cultivation broth. The homogenized sample is then filtered and incubated at the proper growth temperature for 1 or 2 days.
In case of bacteria and fungi with a thick cell wall, an aggressive protocol that can involve both chemical and enzymatic reagents, is required for efficient wall lysis prior to the extraction of genomic DNA and the amplification reaction.
Moreover, in most cases, the detection of contaminations using molecular methods requires a priori knowledge of the target sequence and the occasional mutation in the genomic DNA can compromise the detection. This problem can be overcome with the use of redundant probes, or primers that make extremely unlikely the simultaneous variation of all target sequences, or with the implementation of long probes and less stringent hybridization conditions.
Finally, the DNA based assays are able to detect both viable and non viable pathogens. While this can be an advantage with non-cultivable pathogens, it can also have some drawbacks. In fact, the detection of death pathogenic organisms, whose genomic DNA is still contained in the sample, can bring about a false positive result. In these cases a molecular backup is offered by RNA based techniques, like NASBA or RT-PCR, since the RNA is less stable than DNA.
A possible solution to some limitations of the molecular methods could come from microfluidics, multidisciplinary technologies aimed to develop sub-millimetre scale devices. Microfluidics-based “Lab-on-chips” includes devices that perform reactions such as DNA extraction, PCR and microarray hybridization in one single piece of equipment and with minimal human intervention. Moreover, all these reactions can be combined and performed in a succession, reducing contamination risks [23]. Lab-on-chips have been designed to perform PCR from genomic DNA immediately followed by LDR. The hybridization on universal microarray is also possible and the entire process is completed in less than 1 h, instead of several hours [19, 20, 64].
Microfluidics allows the use of small volumes and sample amounts, increasing sensitivity and improving experiment throughput. More importantly, the sample carryover is very limited, reducing the risk of contamination. Furthermore, as lab-on-chip includes all steps in one device, procedures are more easily standardized. A major benefit of miniaturization and standardization will be portable devices for the on site detection of pathogens, thus limiting the laboratory work, sample handling and transportation.
References
- 1.Anjum MF, Mafura M, Slickers P, Ballmer K, Kuhnert P, Woodwart MJ, Ehricht R (2007) Pathotyping Escherchia coli by using miniaturized DNA microarrays. Appl Environ Microbiol 73:5692–5697 [DOI] [PMC free article] [PubMed]
- 2.Bates SR, Baldwin DA, Channing A, Gifford LK, Hsu A, Lu P (2005) Cooperativity of paired oligonucleotide probes for microarray hybridization assays. Anal Biochem 342:59–68 [DOI] [PubMed]
- 3.Bonnet G, Tyagi S, Libchaber A, Kramer FR (1999) Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc Natl Acad Sci USA 96:6171–6176 [DOI] [PMC free article] [PubMed]
- 4.Bordoni R, Germini A, Mezzelani A, MarchelliG De Bellis R (2005) A microarray platform for parallel detection of five transgenic events in foods: a combined polymerase chain reaction-ligation detection reaction-universal array method. J Agric Food Chem 53:912–918 [DOI] [PubMed]
- 5.Busti E, Bordoni R, Castiglioni B, Monciardini P, Sosio M, Donadio S, Consolandi C, Rossi Bernardi L, Battaglia C, De Bellis G (2002) Bacterial discrimination by means of a universal array approach mediated by LDR (ligase detection reaction). BMC Microbiol 2:27 [DOI] [PMC free article] [PubMed]
- 6.Castiglioni B, Rizzi E, Frosini A, Sivonen K, Rajaniemi P, Rantala A, Mugnai MA, Ventura S, Wilmotte A, Boutte C, Grubisic S, Balthasart P, Consolandi C, Bordoni R, Mezzelani A, Battaglia C, De Bellis G (2004) Development of a universal microarray based on the ligation detection reaction and 16S rrna gene polymorphism to target diversity of cyanobacteria. Appl Environ Microbiol 70:7161–7172 [DOI] [PMC free article] [PubMed]
- 7.CDC Health Protection Research Guide 2006–2015 (2005)
- 8.Chessa S, Chiatti F, Ceriotti G, Caroli A, Consolandi C, Pagnacco G, Castiglioni B (2007) Development of a single nucleotide polymorphism genotyping microarray platform for the identification of bovine milk protein genetic polymorphisms. J Dairy Sci 90:451–464 [DOI] [PubMed]
- 9.Compton J (1991) Nucleic acid sequence-based amplification. Nature 350:91–92 [DOI] [PubMed]
- 10.Consolandi C, Palmieri L, Doveri S, Maestri E, Marmiroli N, Reale S, Lee D, Baldoni L, Tosti N, Severgnini M, De Bellis G, Castiglioni B (2007) Olive variety identification by ligation detection reaction in a universal array format. J Biotechnol 129:565–574 [DOI] [PubMed]
- 11.Deborggraeve S, Claes F, Laurent T, Mertens P, Leclipteux T, Dujardin JC, Herdewijn P, Buscher P (2006) Molecular dipstick test for diagnosis of sleeping sickness. J Clin Microbiol 44:2884–2889 [DOI] [PMC free article] [PubMed]
- 12.Ehrenreich A (2006) DNA microarray technology for the microbiologist: an overview. Appl Microbiol Biotechnol 73:255–273 [DOI] [PubMed]
- 13.Elizaquível P, Chenoll E, Aznar R (2007) A TaqMan-based real-time PCR assay for the specific detection and quantification of Leuconostoc mesenteroides in meat products. FEMS Microbiol Lett 278:62–71 [DOI] [PubMed]
- 14.Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D (1991) Light-directed, spatially addressable parallel chemical synthesis. Science 251:767–773 [DOI] [PubMed]
- 15.Fratamico PM (2003) Comparison of culture, polymerase chain reaction (PCR), Taqman salmonella, and transia card salmonella assays for detection of Salmonella sp. in naturally contaminated ground chicken, ground turkey, and ground beef. Mol Cell Probes 17:215–221 [DOI] [PubMed]
- 16.Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F (1999) Universal DNA microarray method for multiplex detection of low abundance point mutations. J Mol Biol 292:251–262 [DOI] [PubMed]
- 17.Gogol EB, Cummings CA, Burns RC, Relman DA (2007) Phase variation and microevolution at homopolymeric tracts in Bordetella pertussis. BMC Genomics 8:122 [DOI] [PMC free article] [PubMed]
- 18.Gore HM, Wakeman CA, Hull RM, McKillip JL (2003) Real-time molecular beacon NASBA reveals hblC expression from Bacillus sp. in milk. Biochem Biophys Res Commun 311:386–390 [DOI] [PubMed]
- 19.Hashimoto M, Barany F, Soper SA (2006) Polymerase chain reaction/ligase detection reaction/hybridization assays using flow-through microfluidic devices for the detection of low-abundant DNA point mutations. Biosens Bioelectron 21:1915–1923 [DOI] [PubMed]
- 20.Hashimoto M, Barany F, Xu F, Soper SA (2007) Serial processing of biological reactions using flow-through microfluidic devices: coupled PCR/LDR for the detection of low-abundant DNA point mutations. Analyst 132:913–921 [DOI] [PubMed]
- 21.Hoffmaster AR, Meyer RF, Bowen MD, Marston CK, Weyant RS, Thurman K, Messenger SL, Minor EE, Winchell JM, Rassmussen MV, Newton BR, Parker JT, Morrill WE, McKinney N, Barnett GA, Sejvar JJ, Jernigan JA, Perkins BA, Popovic T (2002) Evaluation and validation of a real-time polymerase chain reaction assay for rapid identification of Bacillus anthracis. Emerg Infect Dis 8:1178–1182 [DOI] [PMC free article] [PubMed]
- 22.Isler JA, Vesterqvist OE, Burczynski ME (2007) Analytical validation of genotyping assays in the biomarker laboratory. Pharmacogenomics 8:353–368 [DOI] [PubMed]
- 23.Jain KK (2007) Applications of nanobiotechnology in clinical diagnostics. Clin Chem 53:2002–2009 [DOI] [PubMed]
- 24.Jamnikar Ciglenecki U, Grom J, Toplak I, Jemersic L, Barlic-Maganja D (2008) Real-time RT-PCR assay for rapid and specific detection of classical swine fever virus: comparison of SYBR green and TaqMan MGB detection methods using novel MGB probes. J Virol Methods 147:257–264 [DOI] [PubMed]
- 25.Kay S, Van den Eede G (2001) The limits of GMO detection. Nat Biotechnol 19:405 [DOI] [PubMed]
- 26.Lausted C, Dahl T, Warren C, King K, Smith K, Johnson M, Saleem R, Aitchison J, Hood L, Lasky SR (2004) POSaM: a fast, flexible, open-source, inkjet oligonucleotide synthesizer and microarrayer. Genome Biol 5:R58 [DOI] [PMC free article] [PubMed]
- 27.Lehmann LE, Hunfeld KP, Emrich T, Haberhausen G, Wissing H, Hoeft A, Stuber F (2007) A multiplex real-time PCR assay for rapid detection and differentiation of 25 bacterial and fungal pathogens from whole blood samples. Med Microbiol Immunol 197:313–324 [DOI] [PubMed]
- 28.Leone G, van Schijndel H, van Gemen B, Kramer FR, Schoen CD (1998) Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res 26:2150–2155 [DOI] [PMC free article] [PubMed]
- 29.Li Y, Liu D, Cao B, Han W, Liu Y, Liu F, Guo X, Bastin DA, Feng L, Wang L (2006) Development of a serotype-specific DNA microarray for identification of some Shigella and pathogenic Escherichia coli strains. J Clin Microbiol 44:4376–4383 [DOI] [PMC free article] [PubMed]
- 30.Lin B, Wang Z, Vora GJ, Thornton JA, Schnur JM, Thach DC, Blaney KM, Ligler AG, Malanoski AP, Santiago J, Walter EA, Agan BK, Metzgar D, Seto D, Daum LT, Kruzelock R, Rowley RK, Hanson EH, Tibbetts C, Stenger DA (2006) Broad-spectrum respiratory tract pathogen identification using resequencing DNA microarrays. Genome Res 16:527–535 [DOI] [PMC free article] [PubMed]
- 31.Liu D (2006) Identification, subtyping and virulence determination of Listeria monocytogenes, an important foodborne pathogen. J Med Microbiol 55:645–659 [DOI] [PubMed]
- 32.Liu Y, Fratamico P (2006) Escherichia coli O antigen typing using DNA microarrays. Mol Cell Probes 20:239–244 [DOI] [PubMed]
- 33.Lopez-Campos G, Coiras M, Sanchez-Merino JP, Lopez-Huertas MR, Spiteri I, Martin-Sanchez F, Perez-Brena P (2007) Oligonucleotide microarray design for detection and serotyping of human respiratory adenoviruses by using a virtual amplicon retrieval software. J Virol Methods 145:127–136 [DOI] [PubMed]
- 34.Matsuzaki H, Loi H, Dong S, Tsai YY, Fang J, Law J, Di X, Liu WM, Yang G, Liu G, Huang J, Kennedy GC, Ryder TB, Marcus GA, Walsh PS, Shriver MD, Puck JM, Jones KW, Mei R (2004) Parallel genotyping of over 10,000 SNPs using a one-primer assay on a high-density oligonucleotide array. Genome Res 14:414–425 [DOI] [PMC free article] [PubMed]
- 35.Maurer J (2006) PCR methods in foods (food microbiology and food safety). Springer, Berlin
- 36.McPherson MJ, Moller S (2000) PCR: the basics. BIOS, Oxford
- 37.Melo SC, Pungartnik C, Cascardo JC, Brendel M (2006) Rapid and efficient protocol for DNA extraction and molecular identification of the basidiomycete Crinipellis perniciosa. Genet Mol Res 5:851–855 [PubMed]
- 38.Monis PT, Giglio S (2006) Nucleic acid amplification-based techniques for pathogen detection and identification. Infect Genet Evol 6:2–12 [DOI] [PMC free article] [PubMed]
- 39.Morris T, Robertson B, Gallagher M (1996) Rapid reverse transcription-PCR detection of hepatitis C virus RNA in serum by using the TaqMan fluorogenic detection system. J Clin Microbiol 34:2933–2936 [DOI] [PMC free article] [PubMed]
- 40.Morshed MG, Lee MK, Jorgensen D, Isaac-Renton JL (2007) Molecular methods used in clinical laboratory: prospects and pitfalls. FEMS Immunol Med Microbiol 49:184–191 [DOI] [PubMed]
- 41.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H (1987) Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 51(Pt 1):263–273 [DOI] [PubMed]
- 42.Ngazoa ES, Fliss I, Jean J (2008) Quantitative study of persistence of human norovirus genome in water using TaqMan real-time RT-PCR. J Appl Microbiol 104:707–715 [DOI] [PubMed]
- 43.Nielsen MK, Peterson DS, Monrad J, Thamsborg SM, Olsen SN, Kaplan RM (2007) Detection and semi-quantification of Strongylus vulgaris DNA in equine faeces by real-time quantitative PCR. Int J Parasitol 38:443–453 [DOI] [PubMed]
- 44.Palaniappan RU, Zhang Y, Chiu D, Torres A, Debroy C, Whittam TS, Chang YF (2006) Differentiation of Escherichia coli pathotypes by oligonucleotide spotted array. J Clin Microbiol 44:1495–1501 [DOI] [PMC free article] [PubMed]
- 45.Perelle S, Dilasser F, Grout J, Fach P (2004) Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world’s most frequent clinical cases. Mol Cell Probes 18:185–192 [DOI] [PubMed]
- 46.Pingle MR, Granger K, Feinberg P, Shatsky R, Sterling B, Rundell M, Spitzer E, Larone D, Golightly L, Barany F (2007) Multiplexed identification of blood-borne bacterial pathogens by use of a novel 16S rRNA gene PCR-ligase detection reaction-capillary electrophoresis assay. J Clin Microbiol 45:1927–1935 [DOI] [PMC free article] [PubMed]
- 47.Reithinger R, Dujardin JC (2007) Molecular diagnosis of leishmaniasis: current status and future applications. J Clin Microbiol 45:21–25 [DOI] [PMC free article] [PubMed]
- 48.Schneider P, Wolters L, Schoone G, Schallig H, Sillekens P, Hermsen R, Sauerwein R (2005) Real-time nucleic acid sequence-based amplification is more convenient than real-time PCR for quantification of Plasmodium falciparum. J Clin Microbiol 43:402–405 [DOI] [PMC free article] [PubMed]
- 49.Schrenzel J (2007) Clinical relevance of new diagnostic methods for bloodstream infections. Int J Antimicrob Agents 30(Suppl 1):S2–S6 [DOI] [PubMed]
- 50.Shi MM, Myrand SP, Bleavins MR, de la Iglesia FA (1999) High throughput genotyping for the detection of a single nucleotide polymorphism in NAD(P)H quinone oxidoreductase (DT diaphorase) using TaqMan probes. Mol Pathol 52:295–299 [DOI] [PMC free article] [PubMed]
- 51.Singh-Gasson S, Green RD, Yue Y, Nelson C, Blattner F, Sussman MR, Cerrina F (2000) Maskless fabrication of light-directed oigonucleotide microarrays using a digital micromirror array. Nat Biotechnol 17:974–978 [DOI] [PubMed]
- 52.Szemes M, Bonants P, de Weerdt M, Baner J, Landegren U, Schoen CD (2005) Diagnostic application of padlock probes-multiplex detection of plant pathogens using universal microarrays. Nucleic Acids Res 33:e70 [DOI] [PMC free article] [PubMed]
- 53.Taylor GM, Worth DR, Palmer S, Jahans K, Hewinson RG (2007) Rapid detection of Mycobacterium bovis DNA in cattle lymph nodes with visible lesions using PCR. BMC Vet Res 3:12 [DOI] [PMC free article] [PubMed]
- 54.The community summary report on trends and sources of zoonoses, zoonotic agents, antimicrobial resistance and foodborne outbreaks in the European union in 2006 (2007) The EFSA Journal 130
- 55.Thelwell N, Millington S, Solinas A, Booth J, Brown T (2000) Mode of action and application of Scorpion primers to mutation detection. Nucleic Acids Res 28:3752–3761 [DOI] [PMC free article] [PubMed]
- 56.Thomas CM, Nielsen KM (2005) Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat Rev Microbiol 3:711–721 [DOI] [PubMed]
- 57.Tomlinson JA, Barker I, Boonham N (2007) Faster, simpler, more-specific methods for improved molecular detection of Phytophthora ramorum in the field. Appl Environ Microbiol 73:4040–4047 [DOI] [PMC free article] [PubMed]
- 58.Van der Meide W, Guerra J, Schoone G, Farenhorst M, Coelho L, Faber W, Peekel I, Schallig H (2008) Comparison between quantitative nucleic acid sequence-based amplification, real-time reverse transcriptase PCR, and real-time PCR for quantification of Leishmania parasites. J Clin Microbiol 46:73–78 [DOI] [PMC free article] [PubMed]
- 59.Vishnubhatla A, Fung DY, Oberst RD, Hays MP, Nagaraja TG, Flood SJ (2000) A rapid 5′ nuclease (TaqMan) assay for detection of virulent strains of Yersinia enterocolitica. Appl Environ Microbiol 66:4131–4135 [DOI] [PMC free article] [PubMed]
- 60.Volokhov D, Rasooly A, Chumako K, Chizhikov vV (2002) Identification of Listeria species by microarray-based assay. J Clin Microbiol 40:4720–4728 [DOI] [PMC free article] [PubMed]
- 61.Wiesinger-Mayr H, Vierlinger K, Pichler R, Kriegner A, Hirschl AM, Presterl E, Bodrossy L, Noehammer C (2007) Identification of human pathogens isolated from blood using microarray hybridisation and signal pattern recognition. BMC Microbiol 7:78 [DOI] [PMC free article] [PubMed]
- 62.Wang XW, Zhang L, Jin LQ, Jin M, Shen ZQ, An S, Chao FH, Li JW (2007) Development and application of an oligonucleotide microarray for the detection of food-borne bacterial pathogens. Appl Microbiol Biotechnol 76:225–233 [DOI] [PubMed]
- 63.Yang SR, Rothman E (2004) PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis 4:337–348 [DOI] [PMC free article] [PubMed]
- 64.Yi C, Zhang Q, Li CW, Yang J, Zhao J, Yang M (2006) Optical and electrochemical detection techniques for cell-based microfluidic systems. Anal Bioanal Chem 384:1259–1268 [DOI] [PubMed]