Abstract
Prohibitin ring complexes in the mitochondrial inner membrane regulate cell proliferation as well as the dynamics and function of mitochondria. Although prohibitins are essential in higher eukaryotes, prohibitin-deficient yeast cells are viable and exhibit a reduced replicative life span. Here, we define the genetic interactome of prohibitins in yeast using synthetic genetic arrays, and identify 35 genetic interactors of prohibitins (GEP genes) required for cell survival in the absence of prohibitins. Proteins encoded by these genes include members of a conserved protein family, Ups1 and Gep1, which affect the processing of the dynamin-like GTPase Mgm1 and thereby modulate cristae morphogenesis. We show that Ups1 and Gep1 regulate the levels of cardiolipin and phosphatidylethanolamine in mitochondria in a lipid-specific but coordinated manner. Lipid profiling by mass spectrometry of GEP-deficient mitochondria reveals a critical role of cardiolipin and phosphatidylethanolamine for survival of prohibitin-deficient cells. We propose that prohibitins control inner membrane organization and integrity by acting as protein and lipid scaffolds.
Introduction
Biological membranes are complex structures made up of a multiplicity of membrane lipids and proteins controlling essential cellular processes. This is exemplified by mitochondria, double-membrane bound organelles with essential roles in diverse metabolic and cellular signaling pathways (Chan, 2006; McBride et al., 2006). The mitochondrial inner membrane is considered to be the most protein-rich cellular membrane, whose functional impairment is associated with aging, myopathies, and neurological disorders in humans (Chan, 2006). It harbors the multisubunit complexes of the respiratory chain, key enzymes of many catabolic and anabolic pathways, and various protein translocases with crucial roles during mitochondrial biogenesis. Mitochondria form an interconnected, tubular network, which undergoes dynamic changes by balanced fusion and fission events (Hoppins et al., 2007). Mitochondrial biogenesis therefore requires adjusting and coordinating the assembly of proteins, encoded by both nuclear and mitochondrial genomes, and membrane lipids, some of which are synthesized within mitochondria. However, although protein transport to mitochondrial membranes has been extensively studied, mechanisms of lipid import into and within mitochondria are only poorly understood.
Prohibitins form an evolutionary conserved family of membrane proteins with a variety of suggested activities in different cellular compartments (Rajalingam et al., 2005; Kasashima et al., 2006; Wang et al., 2008). Cell proliferation depends on prohibitins targeted to mitochondria (Merkwirth et al., 2008), where two homologous subunits, Phb1 and Phb2, assemble into multimeric, high–molecular weight complexes in the inner membrane (Coates et al., 1997; Berger and Yaffe, 1998; Tatsuta et al., 2005). Prohibitins have been found to be part of mitochondrial nucleoids (Bogenhagen et al., 2003), to affect the organization of mitochondrial DNA (Kasashima et al., 2008), and to protect endothelial cells against reactive oxygen–induced senescence (Kasashima et al., 2008; Schleicher et al., 2008). Fibroblasts lacking prohibitins show aberrant mitochondrial cristae and exhibit a decreased resistance toward apoptosis (Merkwirth et al., 2008). These deficiencies are caused by an impaired processing of the dynamin-like GTPase OPA1 (Merkwirth et al., 2008), a core component of the mitochondrial fusion machinery (Hoppins et al., 2007), which is mutated in dominant optic atrophy (Alexander et al., 2000; Delettre et al., 2000).
Despite recent progress in understanding of cellular roles of prohibitins, their molecular activity remained largely elusive (Mishra et al., 2006). Prohibitin complexes assemble with m-AAA proteases in the mitochondrial inner membrane and thereby modulate the turnover of nonnative membrane proteins (Steglich et al., 1999). Therefore, they have been proposed to exert chaperone-like activity (Nijtmans et al., 2002). However, the ring structure of purified prohibitin complexes (Tatsuta et al., 2005) and the sequence similarity of prohibitins to lipid raft–associated proteins of the SPFH family (Browman et al., 2007) are also consistent with a scaffolding function of prohibitin complexes in the inner membrane.
Notably, although they are required for embryonic development in mice, Caenorhabditis elegans, and Drosophila melanogaster, deletion of prohibitin genes in yeast leads to premature ageing but does not affect cell survival (Coates et al., 1997; Artal-Sanz et al., 2003; Merkwirth et al., 2008). We therefore searched for yeast genes whose function is essential for cell growth in the absence of prohibitins. Functional studies on the uncovered genetic network identified novel modulators of the phospholipid composition of the inner membrane and link the function of prohibitin complexes to cardiolipin (CL) and phosphatidylethanolamine (PE), nonbilayer phospholipids critical for mitochondrial structure and integrity.
Results
Defining the genetic interactome of prohibitins
To identify genes genetically interacting with prohibitins, a synthetic genetic array (SGA) analysis was performed using Δphb1 cells and a collection of 4,850 yeast strains lacking nonessential genes. Moreover, we tested candidate genes and reexamined previously described genetic interactors of prohibitins in the same yeast strain background. In this way, we identified 35 GEP genes (for genetic interactors of prohibitins), whose deletion caused lethality or severely impaired growth of prohibitin-deficient yeast cells on glucose-containing media (Table I). Consistent with a mitochondrial function of prohibitin complexes, 31 genes code for mitochondrial proteins. Interestingly, 19 of the genetic interactors fall into two functional classes: genes with functions during the assembly of the respiratory chain, and genes required for the maintenance of mitochondrial morphology (Dimmer et al., 2002) and the assembly of β-barrel proteins in the outer membrane (Meisinger et al., 2007). The latter group includes the genes MMM1, MDM10, MDM12, MDM31, MDM32, and MDM34, which were previously found to genetically interact with each other (Dimmer et al., 2005). Diverse functions have been associated with another group of eight GEP genes, which include the synthesis of CL (by the CL synthase Crd1) and the synthesis of PE (by the phosphatidylserine [PS] decarboxylase Psd1). Finally, growth of prohibitin-deficient cells was found to depend on eight GEP genes of unknown function (Table I).
Table I.
GEP genes
| Functional group | Gene | ORF | Localization | Genetic interaction |
| Respiratory chain assembly | YTA10a | YER017c | IM | SL |
| YTA12a | YMR089c | IM | SL | |
| YME1 | YPR024w | IM | SL | |
| OXA1 | YER154w | IM | GD | |
| COX6 | YHR051w | IM | GD | |
| COX24 | YLR204w | IM | GD | |
| ATP7 | YKL016c | IM | GD | |
| ATP10a | YLR292w | IM | GD | |
| ATP17 | YDR377w | IM | GD | |
| ATP23a | YNR020c | IMS | GD | |
| COQ1 | YBR003w | MA | GD | |
| Mitochondrial morphology/β-barrel assembly | MMM1ab | YLL006w | IM/OM | SL |
| MDM10ab | YAL010c | OM | SL | |
| MDM12ab | YOL009c | OM | GD | |
| MDM31b | YHR194w | IM | GD | |
| MDM32b | YOR147w | IM | SL | |
| MDM34b | YGL219c | OM | SL | |
| MDM35 | YKL053c-A | IMS | SL | |
| UPS1b | YLR193c | IMS | SL | |
| Unknown | GEP1 | YLR168c | IMS | SL |
| GEP3 | YOR205c | M | GD | |
| GEP4 | YHR100c | M | SL | |
| GEP5 | YLR091w | M | GD | |
| GEP6 | YMR293c | M | SL | |
| GEP7 | YGL057c | M | GD | |
| GEP8 | YER093c-A | ND | SL | |
| GEP9 | YNL170w | ND | SL | |
| Diverse | PSD1a | YNL169c | IM | SL |
| CRD1b | YDL142c | IM | SL | |
| HMI1 | YOL095c | IM/MA | GD | |
| MRE11 | YMR224c | M, N, C | GD | |
| CTK3 | YML112w | N, C | GD | |
| PIF1 | YML061c | M, N | GD | |
| PIH1 | YHR034c | N | GD | |
| EMI1 | YDR512c | C | GD |
C, cytosol; GD, growth defect associated with double mutant; IM, inner membrane; IMS, intermembrane space; M, mitochondria; MA, matrix; N, nucleus; ND, not determined; OM, outer membrane; SL, synthetically lethal.
Previously described synthetic lethal interactions (Berger and Yaffe, 1998; Steglich et al., 1999; Birner et al., 2003; Osman et al., 2007).
Genetic interactors identified by sporulation and tetrad dissection only.
Mitochondrial inner membrane integrity depends on Gep1 and prohibitins
The open reading frame YLR168c, which was termed GEP1, attracted our attention, as it showed a strong genetic interaction with prohibitin genes and belongs to a highly conserved gene family, including previously described UPS1/Preli1 genes (Fig. 1 A; Dee and Moffat, 2005; Sesaki et al., 2006). Similar to Ups1 and PRELI1, Gep1 has been identified in the mitochondrial proteome (Kumar et al., 2002). To assess mitochondrial defects in the absence of both Gep1 and Phb1, we generated Δgep1Δphb1 cells expressing Phb1 from a tetracycline-regulatable promoter ([PHB1]). These cells grew normally on both fermentable and nonfermentable carbon sources under nonrepressing conditions, whereas addition of the tetracycline analogue doxycycline shut off PHB1 expression and inhibited cell growth (Fig. 1 B). Immunoblot analysis of cell extracts revealed that outer membrane proteins accumulated normally in these cells (Fig. 1 C). However, the loss of Phb1 was accompanied by a decrease of mitochondrial inner membrane and matrix proteins, which suggests functional deficits in the inner membrane (Fig. 1 C). We therefore determined in Phb1-depleted cells the formation of the mitochondrial membrane potential, which is required for protein transport across and into the inner membrane and thereby essential for cell survival. Depletion of Phb1 decreased the membrane potential in Δphb1[PHB1] cells and led to its complete dissipation in Δgep1Δphb1[PHB1] cells (Fig. 1 D). These findings point to essential functions of both genes for the integrity of the inner membrane and provide an explanation for the synthetic lethal interaction of GEP1 and PHB1.
Figure 1.
Phb1 is required for mitochondrial inner membrane integrity in the absence of Gep1. (A) Multiple sequence alignment (score matrix: Blosum62) of Gep1, Gep2, Ups1, and human homologues. The conserved MSF1′/PRELI domain is depicted. Numbers refer to amino acids. Black highlighting indicates full conservation across all species; gray highlighting represents amino acids with similar properties conserved in at least three of the analyzed sequences. (B) Synthetic lethal interaction of Δgep1 and Δphb1. Fivefold serial dilutions of cell suspensions were spotted on glucose (Glc)- or galactose (Gal)-containing YP plates, which were supplemented with 2 µg/ml doxycycline (Dox) when indicated and incubated at 30°C. WT, wild type. (C) Steady-state levels of mitochondrial proteins in Δgep1Δphb1[PHB1] cells. Mitochondria were isolated from cells grown on glucose-containing media in the presence of doxycycline for different time periods. Mitochondrial proteins were analyzed by SDS-PAGE and immunoblotting. IM, inner membrane; OM, outer membrane; M, matrix; L, long isoform of Mgm1; S, short isoform of Mgm1. (D) Dissipation of the membrane potential in mitochondria lacking Gep1 and Phb1. Mitochondria were isolated from cells grown for 12 h on glucose-containing media in the presence or absence of doxycycline and stained with the potential-sensitive dye 3,3′-dipropylthiadicarbocyanine iodide (DiDC3(5)).
Requirement of Gep1 for mitochondrial cristae morphogenesis
To assess mitochondrial morphology in cells lacking Gep1 and prohibitins, we expressed mitochondrially targeted GFP variants in wild-type, Δgep1, or Δphb1 cells and Δgep1 or Δgep1Δphb1 cells harboring [PHB1]. An inspection of these cells by fluorescence microscopy revealed the accumulation of ball-shaped and clustered mitochondria in Δgep1Δphb1 cells upon down-regulation of Phb1 (Fig. 2 A). In the absence of doxycycline, mitochondrial morphology was slightly impaired in these cells, most likely reflecting a deleterious effect of Phb1 overexpression. Notably, the aberrant morphology of mitochondria was not a consequence of cell death, as mitochondria were inspected while at least 85% of the cells were still viable as determined by FUN-1 staining (unpublished data). Deletion of PHB1 alone did not interfere with the formation of tubular mitochondria (Fig. 2 A).
Figure 2.
Impaired mitochondrial cristae morphogenesis in cells lacking Gep1 and Phb1. (A, left) Wild-type (WT), Δphb1, Δgep1, Δphb1[PHB1], and Δgep1Δphb1[PHB1] cells expressing mitochondria-targeted GFP or DsRed were grown to log phase in YPD medium in the presence or absence of doxycycline, and analyzed by differential interference contrast (left) and fluorescence (right) microscopy. Bar, 5 µm. (right) The bar graph indicates the percentage of wild type–like (light gray), fragmented (dark gray), and ball-shaped (black) mitochondria. n ≥ 100; data represent mean values ± SD of three independent experiments. (B, left) Wild type, Δphb1, and Δgep1 or Δphb1[PHB1] and Δgep1Δphb1[PHB1] cells depleted of prohibitin were grown to log phase in YPD medium containing doxycycline and analyzed by transmission electron microscopy. Bar, 500 nm. (right) The bar graph indicates the percentage of wild type–like (light gray) or clustered mitochondria (black), and other mitochondrial phenotypes (dark gray). n = 100. Two representative micrographs of Δgep1Δphb1[PHB1] cells depleted of prohibitin are shown. (C) The cristae/contour ratio was determined as described in Materials and methods. The quantification is illustrated in the bar graph. n ≥ 100.
The analysis of mitochondrial ultrastructure by transmission electron microscopy revealed swollen and clustered mitochondria in ∼40% of Δgep1Δphb1[PHB1] but not Δphb1[PHB1] cells depleted of Phb1 (Fig. 2 B). We carefully quantified the surface of cristae membranes and related it to the surface of the mitochondrial contours in ultrathin sections of Gep1- and Phb1-deficient cells (Fig. 2 C). The surface of cristae membranes decreased only slightly in Δphb1[PHB1] cells but was reduced dramatically by ∼80% in Δgep1Δphb1[PHB1] cells upon down-regulation of Phb1 (Fig. 2 C), which demonstrates that Gep1 and Phb1 concomitantly affect mitochondrial cristae morphogenesis.
When cells were grown on glycerol-containing medium, ∼80% of Δgep1 and ∼60% of Δphb1 cells contained an at least partially fragmented mitochondrial network, which indicates that both proteins are crucial for mitochondrial morphology under conditions of increased mitochondrial demand (Fig. 3 A). Inner membrane fusion and the formation of cristae depend on the dynamin-like GTPase Mgm1 (Meeusen et al., 2006), which undergoes proteolytic processing by the rhomboid protease Pcp1 in the inner membrane. Cleavage results in the accumulation of two isoforms, L- and S-Mgm1, that functionally cooperate during inner membrane fusion (Sesaki et al., 2003). Interestingly, deletion of GEP1 significantly impaired the formation of S-Mgm1 in cells grown under respiring conditions (Fig. 3 B). These findings identify Gep1 as a novel modulator of Mgm1 processing in the inner membrane and suggest that the unbalanced accumulation of L- and S-Mgm1 causes deficiencies in cristae morphogenesis in Δgep1 mitochondria.
Figure 3.
Gep1 affects mitochondrial morphology and Mgm1 processing under respiring conditions. (A, left) Wild-type (WT), Δphb1, and Δgep1 cells expressing mitochondria-targeted GFP were grown to log phase in YP medium containing glycerol (YPG) and analyzed as in Fig. 2 A. Bar, 5 µm. (right) The bar graph indicates the percentage of wild type–like (light gray), fragmented (dark gray), and short tubular, partially fragmented (black) mitochondria. Data represent mean values ± SD of three independent experiments. (B) Decreased PE levels impair Mgm1 processing. Extracts of the indicated cells grown in YPG were analyzed by SDS-PAGE and immunoblotted using Mgm1- and Tom40-specific antisera. A quantification of the immunoblots is shown in the bottom panel. The percentage of S-Mgm1 was calculated from the ratio S-Mgm1/(S-Mgm1 + L-Mgm1). Data represent mean values ± SD of four independent experiments. *, P < 0.05; **, P < 0.01. L, long isoform of Mgm1; S, short isoform of Mgm1.
Gep1 regulates the accumulation of PE within mitochondria
To further define the role of Phb1 and Gep1 for mitochondrial morphogenesis and Mgm1 processing, we screened for genes whose overexpression promotes growth of cells lacking both genes. Δgep1Δphb1 cells expressing plasmid-borne PHB1 were transformed with a multicopy yeast library. After plasmid shuffling, the previously uncharacterized GEP2 gene (YDR185c), which codes for a homologue of Gep1, was identified as a multicopy suppressor of the synthetic lethal interaction of Δgep1 and Δphb1 (Fig. 4 A), indicating overlapping functions of members of this conserved protein family.
Figure 4.
The accumulation of PE in mitochondria depends on Gep1. (A) Overexpression of Gep2 or Cho1 allows growth of Δgep1Δphb1 cells. Δgep1Δphb1[PHB1] cells overexpressing Phb1, Gep2, or Cho1 were incubated at 30°C on media with or without 5′ fluoroorotic acid, which prevents growth of cells harboring the [PHB1] expression plasmid. (B) TLC analysis of mitochondrial phospholipids isolated from Δgep1 cells overexpressing Cho1, Ups1, or Gep2. The asterisk indicates an unidentified lipid species. (C) Gep1 and Psd2 interact genetically. Yeast cells were spotted on YP plates containing glucose (YPD) or glycerol (YPG) and incubated at 30°C or 37°C. (D) Synthetic lethal interaction of Δgep1 and Δcrd1. A diploid strain heterozygous for deletions of GEP1 and CRD1 was subjected to sporulation and tetrad dissection. Arrowheads indicate inviable double mutant progeny.
Strikingly, overexpression of the PS synthase Cho1 was also found to completely restore growth of Δgep1Δphb1 cells, linking functional defects in these cells to the cellular phospholipid metabolism (Fig. 4 A). PS, which is synthesized by Cho1 in the ER, is transported to mitochondria and decarboxylated to form PE by Psd1 in the inner membrane (Voelker, 2005). Psd1 is a bifunctional protein, which is required for both PE synthesis and the regulation of multidrug resistance (Gulshan et al., 2008). It has been observed to genetically interact with prohibitins (Table I; Birner et al., 2003), indicating a complex network of genetic interactions between prohibitins, Gep1, and the cellular phospholipid metabolism.
We therefore determined the mitochondrial phospholipid profile in Δgep1 cells by TLC. Whereas the majority of mitochondrial phospholipids accumulated at normal levels in the absence of Gep1, PE was present at significantly reduced levels in Δgep1 mitochondria (Fig. 4 B). Overexpression of Cho1 or Gep2, but not of Ups1, restored normal PE levels in Gep1-deficient mitochondria (Fig. 4 B). Further genetic experiments corroborated the role of Gep1 for the formation of cellular PE. Cell survival depends on PE, which is synthesized either by Psd1 within mitochondria or by Psd2 in the Golgi apparatus (Voelker, 2004). To examine which of these pathways is affected by Gep1, we deleted GEP1 in Δpsd1 and Δpsd2 cells and examined cell growth both on fermentable and nonfermentable carbon sources (Fig. 4 C). The absence of Gep1 did not impair growth of cells deficient of Psd1 at any temperature, which is consistent with an epistatic relationship of these genes (Fig. 4 C). However, growth of Δgep1Δpsd2 cells was inhibited on nonfermentable carbon sources at 37°C (Fig. 4 C), which suggests that Gep1 specifically affects mitochondrial PE. The synthesis of PE within mitochondria was previously found to be essential for the viability of yeast mutants lacking the CL synthase Crd1, which is localized in the mitochondrial inner membrane (Gohil et al., 2005). Consistent with the significantly reduced levels of PE in Δgep1 mitochondria, we did not obtain viable double mutant offspring after tetrad dissection of diploid cells heterozygous for deletions in GEP1 and CRD1 indicating synthetic lethality (Fig. 4 D). We conclude from these experiments that Gep1 is required for the accumulation of PE in mitochondria.
These findings raise the possibility that an altered phospholipid composition of the inner membrane impairs Mgm1 cleavage and causes an altered cristae morphogenesis in the absence of Gep1. We therefore assessed processing of Mgm1 in mitochondria lacking Psd1 or Crd1 (Fig. 3 B). Deletion of PSD1, as well as that of GEP1, impaired the formation of S-Mgm1, whereas Mgm1 cleavage was not affected in the absence of CRD1 under these conditions (Fig. 3 B). We conclude that the phospholipid composition, in particular the PE content, is crucial for efficient Mgm1 processing by rhomboid in the inner membrane.
Gep1 is dispensable for PE synthesis but controls its stability in mitochondria
To unravel the molecular basis of impaired PE accumulation in the absence of Gep1, we first determined the localization of Gep1 within mitochondria. A Gep1 variant carrying a C-terminal hemagglutinin tag was expressed in Δgep1 cells and in Δgep1Δphb1 cells. Expression of this variant promoted growth of Δgep1Δphb1 cells, demonstrating that the C-terminal extension did not interfere with Gep1 function (unpublished data). Mitochondria were isolated from Δgep1 cells and subjected to protease treatment. Gep1 was degraded only under hypotonic conditions, which resulted in osmotic disruption of the outer membrane, indicating that Gep1 is localized in the intermembrane space (Fig. 5 A). A hydropathy blot did not provide any evidence for the presence of a membrane-spanning segment in Gep1. However, Gep1 was found in the pellet fraction after carbonate extraction of mitochondrial membranes, which indicates a tight membrane association (Fig. 5 A).
Figure 5.
Gep1 is required for the stability of mitochondrial PE. (A) Localization of Gep1 in the mitochondrial intermembrane space. (left) Isolated mitochondria were treated with Na2CO3, pH 11.5 (T), and split into a soluble (S) and insoluble (P) fractions by ultracentrifugation. The asterisk indicates an unspecific cross-reaction of the antiserum. (right) Mitochondria or mitoplasts, generated by osmotic disruption of the outer membrane (SW), were treated with 50 µg/ml trypsin and analyzed by SDS-PAGE and immunoblotting. Tom70, an outer membrane protein; Yme1, an integral inner membrane protein exposed to the IMS; and Hsp60, a soluble matrix protein, served as controls. (B) Psd1 activity in the absence of Gep1. Mitoplasts of WT or Δgep1 cells were incubated with NBD-PS for indicated time periods. Phospholipids were fractionated by TLC and fluorescent lipids were quantified by fluor imaging. Equal loading was monitored by molybdenum blue staining. NBD-PE accumulating in wild-type mitochondria after 20 min was set to 100%. Data represent ± SD of four independent experiments. **, P < 0.01. (C) Mitochondrial PE synthesis in Gep1-deficient cells. Δpsd2 and Δpsd2Δgep1 cells were incubated with [3H]serine for 0, 20, 40, or 60 min. Phospholipids extracted from crude mitochondrial isolations were subjected to TLC. PE was recovered from the TLC plate and radioactivity was determined by liquid scintillation counting. Labeled PE accumulating in Δpsd2 cells after 60 min was set to 100%. Data represent mean values ± SD of five independent experiments. (D) PE stability is decreased in the absence of Gep1. Δpsd2 and Δpsd2Δgep1 cells were incubated with [3H]serine for 10 min, and, after addition of excess serine and further incubation for the time points indicated, PE was quantified as in C. Labeled PE accumulating in Δpsd2 cells at time point 0 was set to 100%. Data represent mean values ± SD of four independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As Gep1 may directly regulate Psd1 in the inner membrane, we assessed the enzymatic activity of Psd1 in wild-type and Δgep1 mitochondria. After osmotic disruption of the outer membrane, mitoplasts were incubated with fluorescently labeled PS (NBD-PS), which is converted to NBD-PE (Fig. 5 B). NBD-PE did not accumulate in Δpsd1 mitochondria, which demonstrates that NBD-PE is formed by Psd1. The synthesis of NBD-PE was not affected in the absence of Gep1 (Fig. 5 B). We observed a slightly but significantly increased rate of NBD-PE synthesis in Δgep1 mitochondria, which may indicate an alleviated product inhibition of Psd1 in these mitochondria.
These results were substantiated by pulse-labeling experiments in vivo, which were performed in a Δpsd2 strain background to exclude masking effects of nonmitochondrial PE synthesis (Fig. 5 C). We labeled Δpsd2 and Δpsd2Δgep1 cells with [3H]serine and monitored the incorporation of 3H in PE within mitochondria. Deletion of GEP1 did not affect mitochondrial PE synthesis by mitochondrial Psd1 (Fig. 5 C). Pulse chase experiments, however, revealed a significantly decreased stability of newly synthesized PE in the absence of Gep1 without a significant accumulation of other lipid species (Fig. 5 D). We conclude that the decreased PE concentration in the absence of Gep1 is not caused by an impaired Psd1 activity or uptake of its substrate PS but instead reflects a reduced stability of PE within Δgep1 mitochondria.
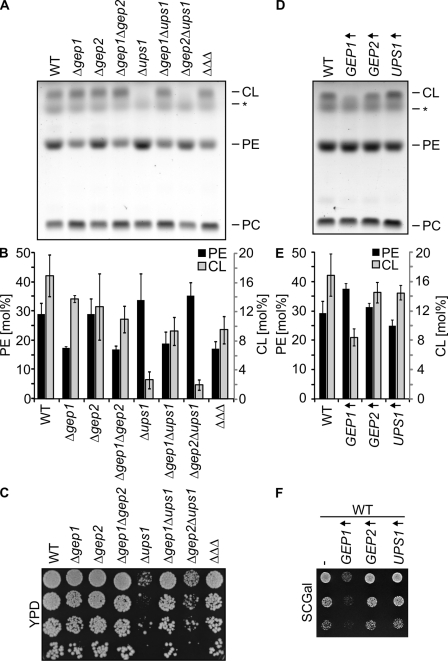
Lipid-specific functions of Gep1-like proteins for PE and CL
Several lines of evidence point to overlapping activities of Gep1 family members: first, both Δgep1 and Δups1 show a synthetic lethal interaction with prohibitin mutants (Table I); and second, overexpression of Gep2 promoted growth of Δgep1Δphb1 cells and restored PE accumulation in Gep1-deficient mitochondria (Fig. 4 A, B). We therefore determined the accumulation of PE in Δgep1, Δgep2, and Δups1 mitochondria both by TLC and by mass spectrometry (Fig. 6, A and B). In contrast to GEP1, deletion of GEP2 and UPS1 did not affect PE levels within mitochondria (Fig. 6, A and B). Our analysis revealed, however, a crucial role of Ups1 for CL levels. CL is a dimeric phosphoglycerolipid predominantly present in mitochondria, where it is synthesized by the CL synthase Crd1 in the inner membrane (Schlame, 2008). CL was decreased approximately sevenfold in Δups1 mitochondria but remained unaffected in the absence of Gep1 or Gep2 (Fig. 6, A and B). Growth of Δups1 cells was severely impaired on glucose-containing medium (Fig. 6 C), a phenotype reminiscent of yeast cells lacking Pgs1 that catalyzes the rate-limiting step of CL biosynthesis. Deletion of GEP1 and GEP2, however, did not affect cell growth under these conditions (Fig. 6 C). Similar to cells lacking Psd1, Δgep1 cells exhibited an increased tendency to lose mitochondrial DNA under these conditions (unpublished data). We conclude that the accumulation of CL in mitochondria depends on Ups1, whereas Gep1 controls mitochondrial PE.
Figure 6.
Gep1 and Ups1 regulate the phospholipid composition of mitochondrial membranes. (A and B) Phospholipid profile of mitochondria lacking Gep1-like proteins. Phospholipids were extracted from mitochondria isolated from the indicated strains (ΔΔΔ: Δgep1Δgep2Δups1) and analyzed by TLC (A) and mass spectrometry (B). Mean values ± SD obtained from at least two independent mitochondrial isolations; samples analyzed in duplicate are shown in B. Asterisks indicate unidentified lipid species. (C) Cell growth in the absence of Gep1-like proteins. Fivefold serial dilutions of the indicated cells were spotted on YPD plates. Strains were grown at 30°C. (D and E) Phospholipid profile of mitochondria in cells overexpressing Gep1-like proteins. Mitochondrial phospholipids were analyzed by TLC (D) and mass spectrometry (E). Mean values ± SD obtained from three mitochondrial isolations, each analyzed in duplicate, are shown in E. (F) Impaired cell growth upon Gep1 overexpression. Gep1-like proteins were expressed in wild-type cells from high-copy plasmids under the control of the GAL promoter. Fivefold serial dilutions of the cells were grown on synthetic media containing galactose as the carbon source (SCGal) at 30°C.
Strikingly, deletion of GEP1 restored normal CL levels in membranes of Δups1 mitochondria (Fig. 6, A and B) and suppressed growth defects associated with the loss of Ups1 (Fig. 6 C). In contrast, neither CL levels nor cell growth were affected upon deletion of GEP2 in Δups1 cells, although Gep1 and Gep2 share 55% identical amino acids (Fig. 6, A–C). PE, however,remained reduced in Δgep1Δups1 mitochondria and accumulated at similar levels as in Δgep1 mitochondria (Fig. 6, A and B), which demonstrates that the absence of Ups1 does not alleviate the requirement of Gep1 for the accumulation of PE. Thus, a complex functional network of Gep1-like proteins controls mitochondrial PE and CL.
To further define the functional interdependence of PE and CL pathways, we generated yeast strains overexpressing Gep1, Gep2, or Ups1 from galactose-inducible promoters. Overexpression of Gep1 impaired cell growth on galactose-containing medium, whereas increased cellular levels of Gep2 or Ups1 did not interfere with cell growth (Fig. 6 F). Mitochondria were isolated from these cells, and the phospholipid profile was determined by TLC and mass spectrometry (Fig. 6, D and E). In agreement with the observed growth defect, we noted a significantly reduced CL content of mitochondria containing overexpressed Gep1 (Fig. 6 D, E). Moreover, PE levels were increased, whereas other phospholipids were present at normal levels in these mitochondria (Fig. 6, D and E; and unpublished data). PE and CL were not altered in a statistically significant manner upon overexpression of Ups1 or Gep2 (Fig. 6, D and E).
Collectively, these experiments define lipid-specific activities of Gep1-like proteins for CL and PE and, at the same time, point to common steps in the regulation of both phospholipids within mitochondria. Both the observed restoration of CL levels in Δups1 cells upon deletion of GEP1 and the reduction of CL levels upon Gep1 overexpression suggest a competition between Gep1 and Ups1.
Survival of prohibitin-deficient cells depends on the lipid composition of mitochondrial membranes
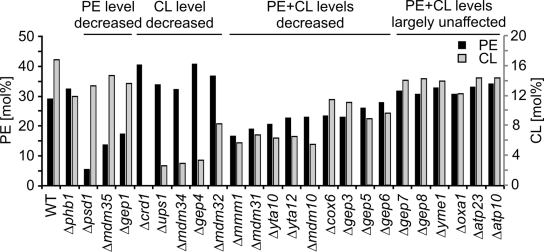
The regulation of the phospholipid composition of the inner membrane by Gep1 and Ups1 proteins together with their synthetic lethal interaction with prohibitins suggests that reduced levels of PE and CL are deleterious for inner membrane integrity in prohibitin-deficient cells. Accordingly, other GEP genes may affect the PE and CL levels in mitochondrial membranes as well. We therefore isolated mitochondria from various yeast strains lacking Phb1 or GEP genes, extracted membrane lipids, and determined PE and CL levels by mass spectrometry (Fig. 7). Strikingly, mitochondrial PE and/or CL were affected in the majority of 23 examined strains (Fig. 7). Only some strains showed normal PE and almost unaltered CL levels in mitochondria (Fig. 7). The latter group included cells lacking assembly factors of the FO particle of the F1FO ATP-synthase, like Atp10 and Atp23, that have been previously found to interact genetically with prohibitins (Osman et al., 2007), or Oxa1, for which a function during FO assembly was recently described (Jia et al., 2007). Deletion of PSD1 or CRD1 resulted in the expected drastic reduction of the PE or CL content of mitochondrial membranes, respectively (Fig. 7). Notably, we observed increased PE levels in cells showing a severely reduced CL content. This is in agreement with previous findings describing an increase in mitochondrial PE in Δcrd1 cells, and suggested a coordinated regulation of both phospholipids (Zhong et al., 2004). Moreover, membranes isolated from Phb1-deficient mitochondria contained reduced amounts of CL and slightly increased PE levels. Strikingly, the loss of a large number of GEP genes genetically interacting with prohibitins led to strongly reduced levels of PE and/or CL (Fig. 7). These included genes with functions for mitochondrial morphology and the assembly of β-barrel proteins (MDM10, MMM1, MDM31, MDM32, MDM34, and MDM35; Merz et al., 2007; Bolender et al., 2008), genes associated with the assembly of respiratory chain complexes (COX6, YTA10, and YTA12), and several uncharacterized open reading frames (GEP3-6). Our findings link the function of these genes to the mitochondrial lipid metabolism and point to a critical role of the PE and CL content of the inner membrane for the survival of prohibitin-deficient cells.
Figure 7.
Lipid profile of mitochondria lacking GEP genes. CL and PE levels were determined by mass spectrometry in mitochondria isolated from wild type (WT) and Δphb1 cells, and cells lacking various GEP genes grown on galactose-containing media. Mean values of two mitochondrial lipid extracts are shown. Effects of Yme1 and Oxa1 on mitochondrial PE levels have been previously observed in cells grown on glucose-containing media (Nebauer et al., 2007).
Discussion
The network of prohibitin-interacting genes unraveled by our SGA analysis demonstrates an intimate functional relationship of prohibitins to the lipid composition of mitochondrial membranes (Fig. 8 A). While they are dispensable for yeast cell growth under normal conditions, prohibitins are essential for the integrity of the inner membrane and cell survival if membranes are deficient for CL or PE. Both CL and PE have similar physical properties and cluster into nonbilayer, hexagonal phase structures in lipid membranes (de Kruijff, 1997). Disturbances in both biosynthetic pathways are synthetic lethal in yeast (Gohil et al., 2005) and bacteria, illustrating that the physical similarities of CL and PE are of functional relevance in vivo. Consistently, clusters of PE and CL have been detected in bacterial membranes (Matsumoto et al., 2006). It is therefore conceivable that defined lipid clusters with specific functions exist in the inner membrane of mitochondria. Contact sites between inner and outer mitochondrial membrane, at which import of nuclear-encoded mitochondrial proteins occurs (Reichert and Neupert, 2002) and which have been linked to phospholipid transport processes (Simbeni et al., 1990), were found to be enriched in PE and CL, and may represent such specialized membrane domains. Ringlike prohibitin complexes may serve as membrane organizers and affect the distribution of CL and PE in the membrane bilayer (Fig. 8 B). If CL and PE are present only at low concentrations, this function may become essential for inner membrane integrity and membrane-associated processes. Such an activity of prohibitins is in perfect accordance with the predicted function of prohibitins as protein scaffolds, and may ensure the recruitment of membrane proteins to a specific lipid environment. This includes m-AAA proteases that assemble with prohibitins into large supercomplexes in the inner membrane (Steglich et al., 1999). It should be noted that a fencelike activity of prohibitin ring complexes could also ensure the formation of protein-free domains in the mitochondrial inner membrane, which is considered to be the most protein-rich cellular membrane. These proposed functions of prohibitins as membrane organizers are reminiscent of other SPFH proteins like flotillins/reggies, which are distantly related to prohibitins and were found to induce microdomains in the plasma membrane and to modulate the assembly of signaling complexes (Frick et al., 2007; Langhorst et al., 2008).
Figure 8.
Prohibitins and mitochondrial inner membrane organization. (A) Genetic interaction of prohibitins with PE and CL biosynthetic pathways. Synthetic lethal interactions between CHO1 and PGS1 and between PSD1 and CRD1 have been described previously (Janitor et al., 1996; Gohil et al., 2005). (B) Hypothetical model for the role of prohibitins as membrane organizers. The maintenance of putative functional membrane domains containing CL and PE (gray dots) depends on prohibitin ring complexes or a high level of CL and PE in the inner membrane.
Altered levels of CL or PE compromise mitochondrial activities and are associated with many pathophysiological states, but the mechanisms that determine the phospholipid composition of mitochondrial membranes are poorly understood. Our findings identify the conserved family of Gep1-like proteins as novel membrane-associated regulators of CL and PE in mitochondrial membranes. Gep1 is essential for the accumulation of PE, whereas Ups1 is required for the accumulation of CL in mitochondrial membranes. Strikingly, deletion of GEP1 in Δups1 cells restored CL levels in mitochondria, which suggests competition between Gep1 and Ups1. Consistently, overexpression of Gep1 reduces CL in mitochondria. Thus, the phospholipid content of mitochondrial membranes critically depends on the level of Gep1. The competitive action of Gep1 and Ups1 allows the adjustment of relative PE and CL levels simply by modulating the amounts or the availability of the regulatory proteins. These findings demonstrate that mitochondrial levels of PE and CL are regulated coordinately by related proteins and are therefore in agreement with previous notions that cells may require a critical amount of these nonbilayer-forming phospholipids (Zhong et al., 2004; Gohil et al., 2005).
Gep1-like proteins likely exert a regulatory role during membrane biogenesis, as the mitochondrial phospholipid profile is only modestly altered in cells lacking all Gep1-like proteins. The decrease of the PE content of mitochondrial membranes in the absence of Gep1 is not caused by an impaired PE synthesis. Rather, PE does not accumulate stably in Gep1-deficient mitochondria, suggesting that Gep1 inhibits either a PE-specific lipase or the export of PE from mitochondria. Accordingly, the competition of Gep1 and Ups1 may determine the specificity of mitochondrial phospholipases or lipid transport processes. In agreement with an inhibitory role of Gep1 for PE export from mitochondria, we observed in the absence of Gep1 a slight increase of PC, which is generated by methyl transferases in ER membranes using mitochondrial PE (Kodaki and Yamashita, 1989). Contact sites between inner and outer mitochondrial membranes have been discussed as sites of phospholipid transport (Simbeni et al., 1990). It will be therefore of interest to examine whether Gep1-like proteins locate to these sites.
We identify the morphogenesis of cristae as one process critically dependent on mitochondrial PE. Decreased PE levels in Gep1- or Psd1-deficient cells compromise the processing of the dynamin-like GTPase Mgm1, which is required for membrane fusion and cristae formation (Meeusen et al., 2006). Under these conditions, prohibitins are essential to maintain inner membrane integrity. Thus, residual Mgm1 processing sufficient to maintain mitochondrial cristae at decreased PE levels appears to critically depend on prohibitins. Notably, we have recently identified the processing of the mammalian Mgm1 homologue OPA1 as the central process controlled by prohibitins in mouse fibroblasts (Merkwirth et al., 2008), which indicates that the same processes depend on prohibitin function in evolutionary distant organisms. Therefore, phenotypic differences associated with the loss of prohibitins in yeast and mammals likely reflect differences in the phospholipid profile of mitochondrial membranes or the lipid dependence of Mgm1/OPA1 processing itself.
Although the absence of CL did not inhibit Mgm1 processing in our experiments, we do not exclude a role of CL for proteolytic cleavage under certain growth conditions in yeast or in other organisms. Variations of the PE content of the inner membrane may mask the dependence of Mgm1 processing on CL. Accordingly, differences in the relative content of PE and CL may explain why the loss of Ups1 was observed previously to inhibit Mgm1 processing (Sesaki et al., 2006). It is therefore an intriguing possibility that impaired processing of the mammalian Mgm1 homologue OPA1 causes the disturbed formation of mitochondrial cristae, which was observed in lymphoblasts of Barth syndrome patients or yeast cells lacking the CL transacylase tafazzin (Acehan et al., 2007; Claypool et al., 2008). The identification of Gep1-like proteins as regulators of CL and PE now allows for the direct examination of the role of an altered phospholipid composition for mitochondrial dysfunction in disease.
Gep1-like proteins and prohibitins acting as membrane organizers may affect various membrane-associated processes, which are known to depend on nonbilayer phospholipids. Mitochondrial fusion requires phospholipase D, which hydrolyzes CL in the outer membrane and generates the fusogenic lipid phosphatidic acid (Choi et al., 2006). CL and PE affect also the insertion and oligomerization of the proapoptotic Bcl2-family member Bax in the outer membrane (Lucken-Ardjomande et al., 2008). A CL deficiency in the inner membrane impairs the activity of mitochondrial enzymes (Jiang et al., 2000), decreases the stability of respiratory chain supercomplexes and of mitochondrial DNA (Pfeiffer et al., 2003), and accelerates apoptosis by facilitating the release of cytochrome c from the intramitochondrial storage compartments (Choi et al., 2007). It is conceivable that the assembly of the F1FO ATP-synthase and respiratory complexes is yet another process dependent on defined functional domains within mitochondrial membranes. This is suggested by the synthetic interaction of prohibitins with genes coding for assembly factors of inner membrane complexes (Osman et al., 2007), which do not drastically affect mitochondrial PE or CL levels when deleted.
Interestingly, mutations in the majority of GEP genes, which show a synthetic interaction with prohibitins, cause decreased levels of nonbilayer lipids. This includes MDM10 and MMM1, which were originally identified as genes required for mitochondrial inheritance and DNA stability (Boldogh et al., 1998) but were found more recently to function in the assembly of β-barrel proteins in the outer membrane (Meisinger et al., 2007). Accordingly, an impaired insertion of an outer membrane protein affecting mitochondrial lipid biosynthesis may explain the reduced levels of PE and CL in the absence of Mdm10 or Mmm1. However, it cannot be excluded that both proteins play a direct role for the mitochondrial lipid metabolism and that an altered lipid composition causes deficiencies in the assembly of outer membrane proteins. Similarly, it is an attractive possibility that at least some of the GEP genes identified in this study, including previously uncharacterized open reading frames, regulate directly the biosynthesis of PE and CL in mitochondria and affect the activity of biosynthetic enzymes or intramitochondrial lipid transport processes. The identification of components mediating lipid import from the ER, transbilayer flipping across mitochondrial membranes, or lipid transport across the intermembrane space of mitochondria has yet be accomplished.
Materials and methods
Yeast genetic procedures
The SGA using Δphb1 cells was performed as described previously (Tong et al., 2001). Synthetic genetic interactions with Δphb1 were confirmed by sporulation and tetrad dissection. In agreement with the functional interdependence of Phb1 and Phb2, the same interactors were identified in SGAs using Δphb2 cells. To identify multicopy suppressors of synthetic lethal interactions, Δphb1Δgep1[PHB1] cells were transformed with a genomic Yep13 high-copy library. After growth for 1 d at 30°C on SC-Leu, plates were replicated on plates containing 5′ fluoroorotic acid (1 mg/ml). Clones were analyzed after 2 d, and the suppressor gene was identified by subcloning. The genotypes of yeast strains used are described in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200810189/DC1).
Microscopy
Fluorescence and electron microscopy was performed as described previously (Dürr et al., 2006) using a microscope (Axioplan 2; Carl Zeiss, Inc.) equipped with a Plan-Neofluar 100×/1.30 NA Ph3 oil objective lens (Carl Zeiss, Inc.). Images were recorded with a monochrome camera (Evolution VF Mono Cooled; Intas) and processed with Image-Pro Plus 5.0 and Scope Pro4.5 software (Media Cybernetics). For the determination of the cristae/mitochondrial contour ratio, ultrathin sections were viewed in a transmission electron microscope (JEM-2100; JEOL Ltd.) and images were taken with a digital camera (Erlangshen ES500W, model 782; Gatan Inc.). Approximately 100 micrographs of mitochondria were taken for each strain at the same magnification. For each image, the numbers of cristae were counted and the contour of each mitochondrion was measured in micrometers by using the measure function of ImageJ. For each strain the numbers of cristae were put into relation to the sum of the mitochondrial contours. Processing of Mgm1 was monitored by immunoblotting using Mgm1-specific antiserum, provided by A. Reichert (University of Frankfurt, Frankfurt, Germany).
Lipid analysis
TLC analysis.
Mitochondria were isolated from cells grown on yeast-peptone (YP) medium containing 2% galactose and 0.5% lactate. The mitochondrial fraction was washed, resuspended in buffer A (0.6 M sorbitol and 5 mM MES, pH 6), and loaded on a continuous sucrose gradient (20–50% in buffer A). Mitochondria were harvested from the lower third of the gradient and diluted 1:5 in buffer A, pelleted, washed once in SEM buffer (250 mM sucrose, 10 mM MOPS, pH 7.2, and 1 mM EDTA), and finally resuspended in SEM buffer. The purity of the mitochondrial fraction and absence of contaminations by vacuolar and ER membranes was assessed by immunoblotting using Sec61- and Vac8-specific antisera, provided by T. Sommer (Max-Delbrück-Centre, Berlin, Germany) and C. Ungermann (University of Osnabrück, Osnabrück, Germany), respectively. Purified mitochondria (1 mg) were mixed with 1.5 ml chloroform/methanol (1:1, vol/vol) and vigorously shaken for 60 min. 300 µl H2O was added and samples were vortexed for 60 s. After centrifugation (1,000 rpm, 5 min), the aqueous phase was removed and the solvent phase was washed with 250 µl methanol/H2O (1:1, vol/vol). Samples were then dried under a constant stream of air. Lipids were dissolved in chloroform, phosphate concentration was determined (Rouser et al., 1970), and samples were subjected to TLC analysis. TLC plates (HPTLC; Merck & Co., Inc.) were developed with chloroform/methanol/25% ammonia (50:50:3, vol/vol/vol) when not stated otherwise, allowing the separation of PC, PE, and CL from other mitochondrial phospholipids (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200810189/DC1). TLC plates were stained with 470 mM CuSO4 in 8.5% o-phosphoric acid and subsequently incubated for 10 min at 180°C.
Quantification of PE and CL by mass spectrometry.
Mitochondrial lipids were extracted and phosphate concentration was determined according to Rouser et al. (1970). For mass spectrometric analysis, 1.5-nmol phospholipids of mitochondrial fractions were extracted in the presence of CL (CL56:0; Avanti Polar Lipids, Inc.) and PE standard (50 pmol each) as described previously (Brügger et al., 2006). Dried lipids were redissolved in 10 mM ammonium acetate in methanol.
Quantification of PE was performed by neutral-loss scanning, selecting for a neutral loss of 141 D as described previously (Brügger et al., 2006). Quantification of CL was performed in negative ion mode on a quadrupole time-of-flight mass spectrometer (QStar Elite, Applied Biosystems). 10 µl of lipid extracts was diluted 1:2 with 0.1% piperidine in methanol and automatically infused (Triversa Nanomate; Advion Biosciences). The ionization voltage was set to −0.95 kV and gas pressure was set to 0.5 psi. CLs were detected as single charged molecules. CL species (all combinations of fatty acids 16:0, 16:1, 16:2, 18:0, 18:1, and 18:2) were analyzed by targeted product ion scanning. The peak areas of CL-derived fatty acid fragments were extracted from the respective product ion spectra via the “Extract Fragments” script (Analyst QS 2.0; Applied Biosystems). Isotope correction for M+2 ions was performed manually, and values were corrected for response factors of standards.
Determination of Psd1 activity.
Mitochondria were resuspended in assay buffer B (0.1 M Tris/HCl, pH 7.4, 10 mM EDTA, and 2 µM PS-C6-NBD [Avanti Polar Lipids]) to a final concentration of 5 mg/ml and incubated at 25°C. At the time points indicated, mitochondria (500 µg) were removed from the reaction mixture, and phospholipids were extracted and analyzed by TLC (developing solvent: chloroform/methanol/H2O/triethylamine; 30:35:7:35 vol/vol/vol/vol). NBD signals were detected and quantified by fluor imaging (TyphoonTrio; GE Healthcare).
In vivo labeling of phospholipids.
Logarithmically growing yeast cells were harvested and resuspended in medium supplemented with [3H]serine (12.5 µCi/ml) to a final OD600 of 5. After incubation at 30°C for 0, 20, 40, or 60 min, cells corresponding to 10 OD600 were harvested and stored in liquid nitrogen. Phospholipids were extracted and analyzed by TLC. PE spots were recovered from the TLC plates and mixed with 400 µl H2O and 8 ml of scintillation cocktail. Labeled PE was quantified by liquid scintillation counting. The stability of PE was monitored in pulse chase experiments. Yeast cells were labeled for 10 min as described above. Cells were then harvested, resuspended in medium containing 20 mM of unlabeled serine, and incubated at 30°C for 0, 30, 60, 90, 120, or 150 min before PE was quantified.
Online supplemental material
Fig. S1 documents the resolution of the TLC system applied in this study. Table S1 lists yeast strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810189/DC1.
Acknowledgments
We thank Stefan Geimer and Rita Grotjahn for help with electron microscopy, Martine Collart and Justus Ackermann for their support at different stages of this project, and Mark Dürr for helpful discussions. We would like to thank Iris Leibrecht and Timo Sachsenheimer for technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to T. Langer (SFB635/C4) and B. Westermann (WE2174/4-1), and support from the German-Israeli Project (DIP grant F.5.1) and the European Research Counsel to T. Langer.
Footnotes
Abbreviations used in this paper: CL, cardiolipin; PE, phosphatidylethanolamine; PS, phosphatidylserine; SGA, synthetic genetic array.
References
- Acehan D., Xu Y., Stokes D.L., Schlame M. 2007. Comparison of lymphoblast mitochondria from normal subjects and patients with Barth syndrome using electron microscopic tomography.Lab. Invest. 87:40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G., et al. 2000. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28.Nat. Genet. 26:211–215 [DOI] [PubMed] [Google Scholar]
- Artal-Sanz M., Tsang W.Y., Willems E.M., Grivell L.A., Lemire B.D., van der Spek H., Nijtmans L.G., Sanz M.A. 2003. The mitochondrial prohibitin complex is essential for embryonic viability and germline function in Caenorhabditis elegans.J. Biol. Chem. 278:32091–32099 [DOI] [PubMed] [Google Scholar]
- Berger K.H., Yaffe M.P. 1998. Prohibitin family members interact genetically with mitochondrial inheritance components in Saccharomyces cerevisiae.Mol. Cell. Biol. 18:4043–4052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birner R., Nebauer R., Schneiter R., Daum G. 2003. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine biosynthetic machinery with the prohibitin complex of Saccharomyces cerevisiae.Mol. Biol. Cell. 14:370–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D.F., Wang Y., Shen E.L., Kobayashi R. 2003. Protein components of mitochondrial DNA nucleoids in higher eukaryotes.Mol. Cell. Proteomics. 2:1205–1216 [DOI] [PubMed] [Google Scholar]
- Boldogh I., Vojtov N., Karmon S., Pon L.A. 1998. Interaction between mitochondria and the actin cytoskeleton in budding yeast requires two integral mitochondrial outer membrane proteins, Mmm1p and Mdm10p.J. Cell Biol. 141:1371–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolender N., Sickmann A., Wagner R., Meisinger C., Pfanner N. 2008. Multiple pathways for sorting mitochondrial precursor proteins.EMBO Rep. 9:42–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browman D.T., Hoegg M.B., Robbins S.M. 2007. The SPFH domain-containing proteins: more than lipid raft markers.Trends Cell Biol. 17:394–402 [DOI] [PubMed] [Google Scholar]
- Brügger B., Glass B., Haberkant P., Leibrecht I., Wieland F.T., Krausslich H.G. 2006. The HIV lipidome: a raft with an unusual composition.Proc. Natl. Acad. Sci. USA. 103:2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan D.C. 2006. Mitochondria: dynamic organelles in disease, aging, and development.Cell. 125:1241–1252 [DOI] [PubMed] [Google Scholar]
- Choi S.Y., Huang P., Jenkins G.M., Chan D.C., Schiller J., Frohman M.A. 2006. A common lipid links Mfn-mediated mitochondrial fusion and SNARE-regulated exocytosis.Nat. Cell Biol. 8:1255–1262 [DOI] [PubMed] [Google Scholar]
- Choi S.Y., Gonzalvez F., Jenkins G.M., Slomianny C., Chretien D., Arnoult D., Petit P.X., Frohman M.A. 2007. Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli-elicited apoptosis.Cell Death Differ. 14:597–606 [DOI] [PubMed] [Google Scholar]
- Claypool S.M., Boontheung P., McCaffery J.M., Loo J.A., Koehler C.M. 2008. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome.Mol. Biol. Cell. 19:5143–5155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coates P.J., Jamieson D.J., Smart K., Prescott A.R., Hall P.A. 1997. The prohibitin family of mitochondrial proteins regulate replicative lifespan.Curr. Biol. 7:607–610 [DOI] [PubMed] [Google Scholar]
- de Kruijff B. 1997. Lipid polymorphism and biomembrane function.Curr. Opin. Chem. Biol. 1:564–569 [DOI] [PubMed] [Google Scholar]
- Dee C.T., Moffat K.G. 2005. A novel family of mitochondrial proteins is represented by the Drosophila genes slmo, preli-like and real-time.Dev. Genes Evol. 215:248–254 [DOI] [PubMed] [Google Scholar]
- Delettre C., Lenaers G., Griffoin J.M., Gigarel N., Lorenzo C., Belenguer P., Pelloquin L., Grosgeorge J., Turc-Carel C., Perret E., et al. 2000. Nuclear gene OPA1, encoding a mitochondrial dynamin-related protein, is mutated in dominant optic atrophy.Nat. Genet. 26:207–210 [DOI] [PubMed] [Google Scholar]
- Dimmer K.S., Fritz S., Fuchs F., Messerschmitt M., Weinbach N., Neupert W., Westermann B. 2002. Genetic basis of mitochondrial function and morphology in Saccharomyces cerevisiae.Mol. Biol. Cell. 13:847–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmer K.S., Jakobs S., Vogel F., Altmann K., Westermann B. 2005. Mdm31 and Mdm32 are inner membrane proteins required for maintenance of mitochondrial shape and stability of mitochondrial DNA nucleoids in yeast.J. Cell Biol. 168:103–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr M., Escobar-Henriques M., Merz S., Geimer S., Langer T., Westermann B. 2006. Nonredundant roles of mitochondria-associated F-box proteins Mfb1 and Mdm30 in maintenance of mitochondrial morphology in yeast.Mol. Biol. Cell. 17:3745–3755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M., Bright N.A., Riento K., Bray A., Merrified C., Nichols B.J. 2007. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding.Curr. Biol. 17:1151–1156 [DOI] [PubMed] [Google Scholar]
- Gohil V.M., Thompson M.N., Greenberg M.L. 2005. Synthetic lethal interaction of the mitochondrial phosphatidylethanolamine and cardiolipin biosynthetic pathways in Saccharomyces cerevisiae.J. Biol. Chem. 280:35410–35416 [DOI] [PubMed] [Google Scholar]
- Gulshan K., Schmidt J.A., Shahi P., Moye-Rowley W.S. 2008. Evidence for the bifunctional nature of mitochondrial phosphatidylserine decarboxylase: role in Pdr3-dependent retrograde regulation of PDR5 expression.Mol. Cell. Biol. 28:5851–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria.Annu. Rev. Biochem. 76:751–780 [DOI] [PubMed] [Google Scholar]
- Janitor M., Obernauerova M., Kohlwein S.D., Subik J. 1996. The pel1 mutant of Saccharomyces cerevisiae is deficient in cardiolipin and does not survive the disruption of the CHO1 gene encoding phosphatidylserine synthase.FEMS Microbiol. Lett. 140:43–47 [DOI] [PubMed] [Google Scholar]
- Jia L., Dienhart M.K., Stuart R.A. 2007. Oxa1 directly interacts with Atp9 and mediates its assembly into the mitochondrial F1Fo-ATP synthase complex.Mol. Biol. Cell. 18:1897–1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F., Ryan M.T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M.L. 2000. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function.J. Biol. Chem. 275:22387–22394 [DOI] [PubMed] [Google Scholar]
- Kasashima K., Ohta E., Kagawa Y., Endo H. 2006. Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2.J. Biol. Chem. 281:36401–36410 [DOI] [PubMed] [Google Scholar]
- Kasashima K., Sumitani M., Satoh M., Endo H. 2008. Human prohibitin 1 maintains the organization and stability of the mitochondrial nucleoids.Exp. Cell Res. 314:988–996 [DOI] [PubMed] [Google Scholar]
- Kodaki T., Yamashita S. 1989. Characterization of the methyltransferases in the yeast phosphatidylethanolamine methylation pathway by selective gene disruption.Eur. J. Biochem. 185:243–251 [DOI] [PubMed] [Google Scholar]
- Kumar A., Agarwal S., Heyman J.A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., et al. 2002. Subcellular localization of the yeast proteome.Genes Dev. 16:707–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langhorst M.F., Reuter A., Jaeger F.A., Wippich F.M., Luxenhofer G., Plattner H., Stuermer C.A. 2008. Trafficking of the microdomain scaffolding protein reggie-1/flotillin-2.Eur. J. Cell Biol. 87:211–226 [DOI] [PubMed] [Google Scholar]
- Lucken-Ardjomande S., Montessuit S., Martinou J.C. 2008. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane.Cell Death Differ. 15:929–937 [DOI] [PubMed] [Google Scholar]
- Matsumoto K., Kusaka J., Nishibori A., Hara H. 2006. Lipid domains in bacterial membranes.Mol. Microbiol. 61:1110–1117 [DOI] [PubMed] [Google Scholar]
- McBride H.M., Neuspiel M., Wasiak S. 2006. Mitochondria: more than just a powerhouse.Curr. Biol. 16:R551–R560 [DOI] [PubMed] [Google Scholar]
- Meeusen S., DeVay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1.Cell. 127:383–395 [DOI] [PubMed] [Google Scholar]
- Meisinger C., Pfannschmidt S., Rissler M., Milenkovic D., Becker T., Stojanovski D., Youngman M.J., Jensen R.E., Chacinska A., Guiard B., et al. 2007. The morphology proteins Mdm12/Mmm1 function in the major beta-barrel assembly pathway of mitochondria.EMBO J. 26:2229–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkwirth C., Dargazanli S., Tatsuta T., Geimer S., Lower B., Wunderlich F.T., von Kleist-Retzow J.C., Waisman A., Westermann B., Langer T. 2008. Prohibitins control cell proliferation and apoptosis by regulating OPA1-dependent cristae morphogenesis in mitochondria.Genes Dev. 22:476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merz S., Hammermeister M., Altmann K., Dürr M., Westermann B. 2007. Molecular machinery of mitochondrial dynamics in yeast.Biol. Chem. 388:917–926 [DOI] [PubMed] [Google Scholar]
- Mishra S., Murphy L.C., Murphy L.J. 2006. The prohibitins: emerging roles in diverse functions.J. Cell. Mol. Med. 10:353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nebauer R., Schuiki I., Kulterer B., Trajanoski Z., Daum G. 2007. The phosphatidylethanolamine level of yeast mitochondria is affected by the mitochondrial components Oxa1p and Yme1p.FEBS J. 274:6180–6190 [DOI] [PubMed] [Google Scholar]
- Nijtmans L.G., Artal S.M., Grivell L.A., Coates P.J. 2002. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease.Cell. Mol. Life Sci. 59:143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C., Wilmes C., Tatsuta T., Langer T. 2007. Prohibitins interact genetically with Atp23, a novel processing peptidase and chaperone for the F1Fo-ATP synthase.Mol. Biol. Cell. 18:627–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer K., Gohil V., Stuart R.A., Hunte C., Brandt U., Greenberg M.L., Schagger H. 2003. Cardiolipin stabilizes respiratory chain supercomplexes.J. Biol. Chem. 278:52873–52880 [DOI] [PubMed] [Google Scholar]
- Rajalingam K., Wunder C., Brinkmann V., Churin Y., Hekman M., Sievers C., Rapp U.R., Rudel T. 2005. Prohibitin is required for Ras-induced Raf-MEK-ERK activation and epithelial cell migration.Nat. Cell Biol. 7:837–843 [DOI] [PubMed] [Google Scholar]
- Reichert A.S., Neupert W. 2002. Contact sites between the outer and inner membrane of mitochondria-role in protein transport.Biochim. Biophys. Acta. 1592:41–49 [DOI] [PubMed] [Google Scholar]
- Rouser G., Fkeischer S., Yamamoto A. 1970. Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots.Lipids. 5:494–496 [DOI] [PubMed] [Google Scholar]
- Schlame M. 2008. Cardiolipin synthesis for the assembly of bacterial and mitochondrial membranes.J. Lipid Res. 49:1607–1620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleicher M., Shepherd B.R., Suarez Y., Fernandez-Hernando C., Yu J., Pan Y., Acevedo L.M., Shadel G.S., Sessa W.C. 2008. Prohibitin-1 maintains the angiogenic capacity of endothelial cells by regulating mitochondrial function and senescence.J. Cell Biol. 180:101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H., Southard S.M., Hobbs A.E., Jensen R.E. 2003. Cells lacking Pcp1p/Ugo2p, a rhomboid-like protease required for Mgm1p processing, lose mtDNA and mitochondrial structure in a Dnm1p-dependent manner, but remain competent for mitochondrial fusion.Biochem. Biophys. Res. Commun. 308:276–283 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Dunn C.D., Iijima M., Shepard K.A., Yaffe M.P., Machamer C.E., Jensen R.E. 2006. Ups1p, a conserved intermembrane space protein, regulates mitochondrial shape and alternative topogenesis of Mgm1p.J. Cell Biol. 173:651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simbeni R., Paltauf F., Daum G. 1990. Intramitochondrial transfer of phospholipids in the yeast, Saccharomyces cerevisiae.J. Biol. Chem. 265:281–285 [PubMed] [Google Scholar]
- Steglich G., Neupert W., Langer T. 1999. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria.Mol. Cell. Biol. 19:3435–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T., Model K., Langer T. 2005. Formation of membrane-bound ring complexes by prohibitins in mitochondria.Mol. Biol. Cell. 16:248–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Evangelista M., Parsons A.B., Xu H., Bader G.D., Page N., Robinson M., Raghibizadeh S., Hogue C.W., Bussey H., et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants.Science. 294:2364–2368 [DOI] [PubMed] [Google Scholar]
- Voelker D.R. 2004. Lipid synthesis and transport in mitochondrial biogenesis. Topics in Current Genetics, vol. 8 Springer, Berlin/Heidelberg; 267–291 [Google Scholar]
- Voelker D.R. 2005. Bridging gaps in phospholipid transport.Trends Biochem. Sci. 30:396–404 [DOI] [PubMed] [Google Scholar]
- Wang X., Zuo X., Kucejova B., Chen X.J. 2008. Reduced cytosolic protein synthesis suppresses mitochondrial degeneration.Nat. Cell Biol. 10:1090–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q., Gohil V.M., Ma L., Greenberg M.L. 2004. Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56.J. Biol. Chem. 279:32294–32300 [DOI] [PubMed] [Google Scholar]