Abstract
Slow Wallerian degeneration (WldS) encodes a chimeric Ube4b/nicotinamide mononucleotide adenylyl transferase 1 (Nmnat1) fusion protein that potently suppresses Wallerian degeneration, but the mechanistic action of WldS remains controversial. In this study, we characterize WldS-mediated axon protection in vivo using Drosophila melanogaster. We show that Nmnat1 can protect severed axons from autodestruction but at levels significantly lower than WldS, and enzyme-dead versions of Nmnat1 and WldS exhibit severely reduced axon-protective function. Interestingly, a 16–amino acid N-terminal domain of WldS (termed N16) accounts for the differences in axon-sparing activity between WldS and Nmnat1, and N16-dependent enhancement of Nmnat1-protective activity in WldS requires the N16-binding protein valosin-containing protein (VCP)/TER94. Thus, WldS-mediated suppression of Wallerian degeneration results from VCP–N16 interactions and Nmnat1 activity converging in vivo. Surprisingly, mouse Nmnat3, a mitochondrial Nmnat enzyme that localizes to the cytoplasm in Drosophila cells, protects severed axons at levels indistinguishable from WldS. Thus, nuclear Nmnat activity does not appear to be essential for WldS-like axon protection.
Introduction
Axons represent the key cellular outgrowth that permits connectivity between distant neurons or neurons and their nonneuronal targets (e.g., muscles). Axons can be extremely long, traversing great distances, and in most neurons, the axonal compartment composes the majority of the cellular volume. Because of the significant length and volume of the axon, its stabilization represents a major and ongoing challenge for neuronal cell types.
Wallerian degeneration serves as an extremely useful context in which to study molecular and cellular mechanisms that mediate axon integrity. Acute axotomy induces large-scale axon degeneration after a defined latent phase that bears striking resemblance to axon degeneration during developmental pruning (Luo and O'Leary, 2005) and in neurodegenerative disease (Coleman, 2005). Wallerian degeneration was long thought to represent a passive wasting away of the distal portion of the severed axons (Waller, 1850) caused by a lack of nutrients from the cell body, but a revolution in our understanding of axon biology came with the discovery of the spontaneous C57BL/slow Wallerian degeneration (WldS) mutant mouse. Amazingly, severed WldS mutant axons survived for weeks after axotomy (Lunn et al., 1989; Glass et al., 1993). This observation indicated that the WldS mutation somehow suppressed axon autodestruction and suggested for the first time that Wallerian degeneration might be an active process of axon autodestruction akin to apoptotic cell death (Coleman and Perry, 2002; Raff et al., 2002). Subsequent analysis of the WldS mutant has provided intriguing insights into the molecular relationships between axon degeneration in developmental, injury, and disease contexts. For example, the WldS mutation can suppress axon degeneration induced by chemical toxicity (Wang et al., 2001), nerve crush (Beirowski et al., 2005), and mouse models of neurodegenerative disease (Ferri et al., 2003; Coleman, 2005), suggesting that the underlying programs that drive axon autodestruction in these distinct degenerative contexts share common molecular features. However, WldS does not suppress axon degeneration during developmental axon pruning in Drosophila melanogaster or mammals (Hoopfer et al., 2006), arguing that WldS-independent molecular programs drive axon degeneration in developmental settings.
The WldS mutation was recently found to result in the fusion of two genes, the NAD+ biosynthetic molecule nicotinamide mononucleotide adenylyl transferase 1 (nmnat1) and Ube4b, an E4 ubiquitin ligase (Conforti et al., 2000; Mack et al., 2001). The protein encoded by this locus, termed WldS (Fig. 1 A), is composed of the N-terminal 70 amino acids of Ube4b (termed N70), a unique 18–amino acid domain generated during the gene fusion event (termed W18), and the full-length sequence of mouse nmnat1. Expression of this protein in mouse neurons fully recapitulates the WldS mutant phenotype (Mack et al., 2001) and can even protect severed axons from autodestruction in distant species like Drosophila (Hoopfer et al., 2006; MacDonald et al., 2006).
Figure 1.
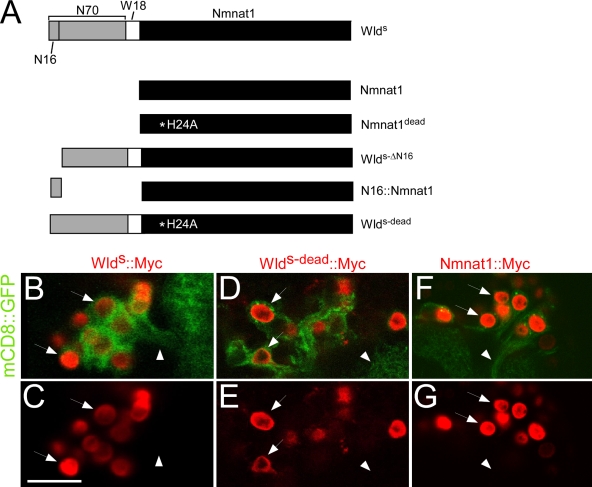
Constructs used to dissect WldS functional domains and localization in Drosophila neurons. (A) All constructs were generated from mouse WldS, which contains N70, the first 16 amino acids (N16) that encode the VCP-binding site, W18, an 18–amino acid linker, and full-length Nmnat1. Nmnat1dead harbors an H24A mutation; WldSΔN16 lacks the N16 domain; N16-Nmnat1 is N16 tethered to the N terminus of Nmnat1; WldS-dead is full-length WldS with an H24A mutation in Nmnat1. See Materials and methods for details. (B–G) UAS-WldS–Myc (B and C), UAS-WldS-dead–Myc (D and E), and UAS-Nmnat1–myc (F and G) were expressed with UAS-mCD8–GFP in projection neurons using GH146-Gal4. The arrows point to nuclei, and the arrowheads point to axons. Bar, 12.14 µm.
Three main models have been proposed to explain WldS-mediated axon protection. The first argues that Ube4b may have a dominant-negative effect on the ubiquitin proteasome (UPS) pathway. Although the N70 portion of Ube4b does not have ubiquitination activity on its own, it contains domains capable of binding Ube4b substrates, implying that N70 may bind key substrates in vivo, fail to target them for degradation, and, in turn, somehow stabilize severed axons. Consistent with this model, the UPS pathway has been shown to be essential in promoting axon autodestruction during Wallerian degeneration (Zhai et al., 2003) and developmental axon pruning (Watts et al., 2003), but a direct functional link between WldS and the UPS pathway is lacking. A second hypothesis is that Nmnat1, a NAD+ biosynthetic enzyme, acts to produce excess NAD+ in the axon after injury and thereby stabilize the axon. Two groups reported that application of NAD+ or its precursors to neuronal cultures partially suppressed axon degeneration in vitro (Araki et al., 2004; Wang et al., 2005); however, a third study found no enhancement of survival of severed axons from primary neuronal cultures with NAD+ application (Conforti et al., 2007). Moreover, transgenic mice expressing Nmnat1 in neurons failed to phenocopy WldS (Conforti et al., 2007). Finally, a third model proposes that Nmnat1 acts in the nucleus before injury through Sirt1, a member of the sirtuin family of NAD+-binding histone deacetylases, to change patterns of neuronal gene expression before injury, which protects severed axons after axotomy (Araki et al., 2004). Araki et al. (2004) provided in vitro data in support of this model showing that siRNAs directed toward sirt1 could partially suppress the neuroprotective effects of bath-applied NAD+; however, a null allele of sirt1 failed to suppress the ability of WldS to protect severed axons in vivo (Wang et al., 2005).
The aforementioned models remain open possibilities but have not been tested incisively in vivo. In this study, we explore the in vivo mechanistic action of WldS in Drosophila. We find that WldS-mediated protection requires two activities: Nmnat1 enzymatic function and the N-terminal 16 amino acids of N70 (termed N16). We show that N16 associates with TER94, the fly orthologue of the mouse N16-binding molecule valosin-containing protein (VCP; Laser et al., 2006), and that VCP is essential for WldS-like levels of axon protection. Thus, the converging activity of domains derived from both Ube4b and Nmnat1 are essential for WldS-like levels of axon protection in vivo. Surprisingly, we find that Nmnat3, a mouse mitochondrial Nmnat that localizes throughout the cytoplasm in Drosophila cells (but not to the nucleus), can protect severed axons as well as WldS. Thus, nuclear localization of Nmnat activity is not essential for robust axon protection, and these data raise the intriguing possibility that N16–VCP/Ter94 interactions may function to relocalize Nmnat1 outside of the nucleus, perhaps to mitochondria, where it exerts its neuroprotective effects.
Results
Constructs generated for in vivo functional dissection of WldS
The adult Drosophila olfactory system offers an excellent opportunity to study Wallerian degeneration and WldS function in a well-controlled in vivo setting: odorant receptor (OR)–Gal4 driver lines can be used to reproducibly label specific subsets of axons with membrane-tethered GFP (mCD8-GFP); nonlethal surgical ablation of antennae or maxillary palps induces Wallerian degeneration of olfactory receptor neuron (ORN) axons; severed axon degeneration can be monitored in the intact brain for months after injury; and neuroprotective molecules like WldS can be expressed in these axons at standardized levels to quantitatively compare neuroprotective activity (MacDonald et al., 2006). In this study, we use this system to define the functional domains in WldS that are essential for protection of severed axons in vivo.
We constructed several transgenic Drosophila lines carrying variants of the WldS molecule (Fig. 1 A): (a) upstream activating sequence (UAS)–nmnat1, full-length mouse nmnat1; (b) UAS-nmnat1dead, full-length mouse nmnat1 with an H24A substitution, which has been previously shown to allow folding of Nmnat and binding of its substrate but largely to abolish substrate conversion to NAD+ (Saridakis et al., 2001); (c) UAS-WldS-ΔN16, WldS with a deletion of the first 16 amino acids (termed N16), which was previously shown to bind VCP (Laser et al., 2006); (d) UAS-N16–nmnat1, a fusion of N16 to full-length Nmnat1; and (e) UAS-WldS-dead, full-length WldS with an H24A substitution in the Nmnat1 portion of the molecule. To determine the subcellular localization of WldS and the aforementioned variants, we also generated C-terminally Myc epitope–tagged versions of WldS, WldS-dead, and Nmnat1 (UAS-WldS–Myc, UAS-WldS-dead–Myc, and UAS-Nmnat1–Myc). When expressed in Drosophila, antennal lobe projection neurons in the central brain of all of these molecules localized exclusively to the nucleus (Fig. 1, B–G), indicating that the H24A mutations that abolish NAD+ biosynthetic activity do not alter the subcellular localization of WldS. We further note the conspicuous absence of WldS+ nuclear puncta in Drosophila neurons (Fig. 1, B and C) and S2 cells (see Fig. 5, A and B) that have been observed when WldS is expressed in mammalian cell lines (Laser et al., 2006). Thus, WldS localizes to the nucleus in Drosophila neurons, and these data argue that the presence of WldS+ nuclear puncta is not essential for WldS to exert its neuroprotective effects.
Figure 5.
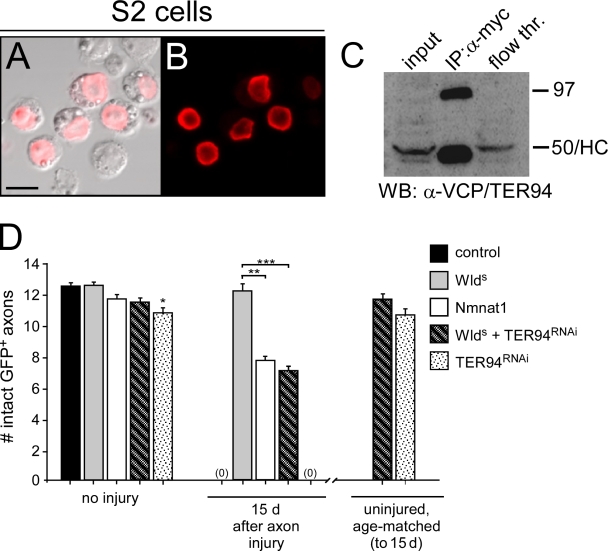
WldS localizes to the nucleus in Drosophila S2 cells and associates with TER94, and TER94 is essential for WldS-like levels of axon protection. (A and B) S2 cells were transfected with WldS-myc and stained with α-Myc antibodies, and α-Myc immunoreactivity localized to the nucleus. (C) Immunoprecipitation (IP) from WldS-Myc–transfected cell lysates with α-Myc antibodies led to the purification of a 97-kD isoform of Ter94. HC, heavy chain from α-Myc IgG; WB, Western blot. (D) UAS-Ter94RNAi was driven in OR22a+ ORNs in the presence or absence of WldS. n ≥ 10 for all genotypes; *, P < 0.05; **, P < 0.01; ***, P < 0.001. Error bars represent SEM. Bar, 13.56 µm.
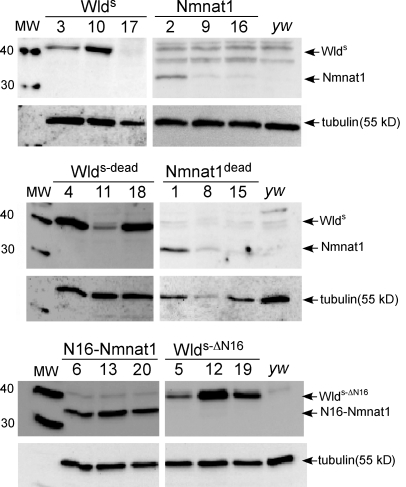
Multiple transgenic lines were established for each construct, and at least three lines bearing independently isolated transgene insertions were assayed for protein expression levels (Fig. 2) and functional protection (see following section). The efficiency of protein expression from each transgene was assayed by crossing individual UAS lines to the panneuronal driver elav-Gal4, preparing protein extracts from head, and assaying protein levels with α-Nmnat1 antibodies on Western blots. All WldS variants carry full-length Nmnat1, and thus, this antibody should recognize each with equal affinity. In some cases, each of the three transgenic lines for a given construct expressed very similar levels of protein (e.g., N16-Nmnat1 and WldS-ΔN16); for others, the total expression level of each protein varied among transgenic lines (e.g., WldS, Nmnat1, Nmnat1dead, and WldS-dead). Despite this fact, the in vivo axon-protective function of each molecule was in most cases strikingly similar among transgenic lines carrying different insertions of the same transgene (see following section).
Figure 2.
WldS variant proteins are stably expressed in Drosophila neurons. Three independent UAS-driven transgene insertion lines for each WldS variant molecule were crossed to elav-Gal4, and protein extracts from heads were assayed for expression using α-Nmnat1 antibodies. Multiple lines for each construct were found to express detectable levels of protein. The expression of each of these molecules in ORNs with OR22a-Gal4 did not lead to expression of proteins at levels detectable by Western blot analysis (not depicted). Molecular masses are shown in kilodaltons. MW, molecular weight.
Wallerian degeneration is potently suppressed by WldS and partially blocked by Nmnat1, and axonal protection requires Nmnat1 enzymatic activity
To compare the protective effects of the aforementioned transgenes, we crossed each to the ORN-specific driver OR22a-Gal4, severed ORN axons, and determined the number of remaining intact axonal fibers present at 5, 10, 15, 20, and 30 d after injury (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200808042/DC1). Each neuroprotective molecule showed a unique pattern of protection, but this was indistinguishable among the other transgenic lines carrying the same transgene (Fig. S2), and therefore, axon protection data were pooled for each transgene (Fig. 3).
Figure 3.
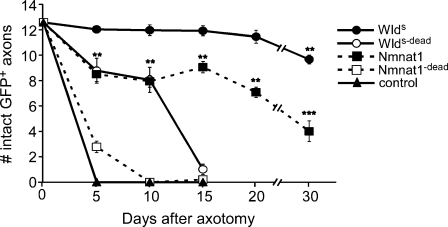
WldS requires N70 and Nmnat1 biosynthetic activity for maximal protection of severed axons in vivo. UAS-regulated versions of WldS variants were expressed in OR22a+ ORNs with OR22a-Gal4, axons were severed, and the number of remaining intact GFP+ axons was scored at the time points indicated. Two to four independent insertion lines were tested for each UAS-regulated WldS variant molecule (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200808042/DC1); n ≥ 10 for individual lines; data were subsequently pooled and are presented here. For Nmnat1, **, P < 0.01 and ***, P < 0.001 (Nmnat1 vs. WldS at corresponding time points). For WldS, **, P < 0.01 (day 0 vs. day 30). Error bars represent SEM.
We first asked whether Nmnat1 could protect severed axons in vivo at a level similar to WldS and whether Nmnat1 enzymatic activity was essential for axonal protection. Previously, we reported that mouse WldS could robustly suppress Wallerian degeneration in Drosophila for at least 20 d after injury (MacDonald et al., 2006). We have now confirmed and extended this finding to 30 d after injury, with the majority (77%) of WldS-expressing axons remaining intact for this period (Fig. 3). Interestingly, we found that expression of mouse Nmnat1 in ORNs also partially suppressed Wallerian degeneration; however, Nmnat1 was significantly less effective than WldS at protecting severed axons. For example, the majority of severed axons in animals expressing WldS remained intact even 30 d after injury (77%), whereas a minority of axons expressing Nmnat1 (32%) remained intact at this time point. Thus, Nmnat1 is indeed a neuroprotective molecule in vivo and likely represents an important component of the severed axon-protecting activity of WldS. However, the fact that Nmnat1 provided significantly reduced protection of severed axons compared with WldS argues that N70 and/or W18 enhances the neuroprotective nature of Nmnat1 in important ways to provide WldS-like levels of axon protection.
Next, we assayed the ability of enzyme-dead variants of WldS and Nmnat1 to protect severed axons. Strikingly, blocking the enzymatic activity of Nmnat1 with an H24A mutation almost completely eliminated the ability of Nmnat1 to protect severed axons. In Nmnat1dead-expressing animals, ∼100% of all axons had undergone Wallerian degeneration within 10 d after injury (Fig. 3). Thus, Nmnat1-dependent axon protection requires the NAD+ biosynthetic activity of Nmnat1. Consistent with a role for Nmnat1 enzymatic activity in promoting WldS-dependent axon protection, we found that WldS-dead also exhibited strongly reduced neuroprotection: in WldS-dead-expressing animals, 92% of axons had degenerated within 15 d after injury (Fig. 3). Interestingly, and in contrast to Nmnat1dead, WldS-dead exhibited intermediate levels of protection until 10 d after injury, indicating that WldS-dead retains a moderate level of axon-sparing activity despite having reduced Nmnat1 enzymatic activity. These data provide further strong evidence that N70 or W18 domains of WldS are important for WldS-dependent protection of severed axons and indicate that Nmnat1 enzymatic activity is essential for sustained WldS-dependent protection of axons.
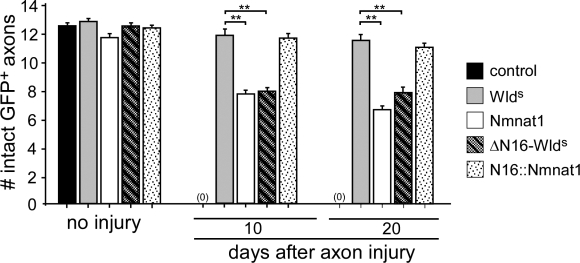
The N16 domain of WldS is essential for maximal axon protection and accounts for differences in neuroprotection observed between WldS and Nmnat1
Which portion of N70 or W18 is responsible for the increased neuroprotection of WldS relative to that observed with Nmnat1? Previous biochemical experiments identified N16 as a binding site for association of WldS with VCP (Laser et al., 2006). We sought to determine the functional requirements for the N16 domain in WldS-mediated protection of severed axons in vivo. Therefore, we generated two transgenic constructs, WldS-ΔN16 (harboring a specific deletion of the N16 domain) and N16-Nmnat1 (in which only the N16 domain is directly fused to the N terminus of Nmnat1; Fig. 1 A) and assayed axon-protective effects. In striking contrast to the protection observed with WldS, WldS-ΔN16 provided significantly reduced protection of severed axons, with 25% and 35% of axons degenerating by day 10 and day 20 after injury, respectively (Fig. 4). WldS-ΔN16-mediated protection was indistinguishable from that afforded by Nmnat1, suggesting that N16 is the key domain responsible for the increased protection we observed in axons expressing WldS compared with Nmnat1. Reciprocally, N16-Nmnat1 expression in axons provided significantly increased protection of severed axons relative to Nmnat1 alone, with >90% of axons surviving as long as 20 d after injury (Fig. 4). Therefore, N16-Nmnat1–mediated protection of severed axons was indistinguishable from the protection provided by WldS. In summary, we find that N16 is essential for WldS-like neuroprotective activity and is sufficient when tethered to Nmnat1 to provide WldS-like levels of severed axon protection in vivo. Together, the aforementioned data indicate that WldS-mediated axon protection results from two synergizing activities: (1) Nmnat1 enzymatic activity and (2) N16-dependent protein–protein interactions.
Figure 4.
N16 is the key domain in the N terminus of WldS that is essential for WldS-like axon protection. WldS, Nmnat1, ΔN16-WldS, and N16-Nmnat1 were expressed in ORNs using OR22a-Gal4, axons were severed, and the number of remaining intact GFP+ severed axons were scored at the time points indicated. n ≥ 10 for all genotypes; **, P < 0.01. Error bars represent SEM.
VCP/TER94 binds to N16 and is essential for WldS-like neuroprotection
The only protein known to physically interact with WldS is VCP, and it does so through the N16 domain (Laser et al., 2006), but its role in WldS-mediated axon protection remains unclear. To determine whether Drosophila VCP, known as TER94 (Pinter et al., 1998), associates with mouse WldS, we transfected Drosophila S2 cells with WldS-Myc and performed coimmunoprecipitation experiments with α-Myc antibodies. We found that WldS-Myc was stably expressed in Drosophila S2 cells and localized primarily to the nucleus (Fig. 5, A and B). Two isoforms of TER94 are present in Drosophila, one of 50 kD (isoform A) and a second of 97 kD (isoform B). We found that S2 cells normally express the 50-kD isoform and lower levels of the 97-kD isoform. Immunoprecipitations with α-Myc antibodies from WldS-Myc–transfected S2 cells led to robust copurification of the 97-kD isoform (which is >80% identical to mouse VCP; Fig. 5 C), indicating that WldS-Myc can associate with TER94. Whether WldS-Myc associates with the 50-kD isoform of TER94 remains unclear because this protein runs at the same size as the mouse IgG heavy chain used for WldS-Myc pull-downs. Thus, TER94, like VCP in mammals, can physically associate with WldS in Drosophila.
Next, we sought to determine whether loss of TER94 function could suppress the ability of WldS to protect severed axons. Because deletion of N16 from WldS suppresses the axon-protective function of WldS to levels equivalent to Nmnat1 (Fig. 4), we predicted that blocking TER94 function in WldS-expressing neurons should phenocopy the protective effects of Nmnat1 and WldS-ΔN16. We first assayed whether null alleles of TER94 could dominantly suppress the ability of WldS to protect severed axons, but WldS-expressing animals lacking a single copy of TER94 showed the normally high levels of axon protection we observe in WldS-expressing axons (unpublished data). We were unable to assay TER94 function in WldS-mediated axon protection in a null TER94 background because animals homozygous for strong loss of function alleles of TER94 are homozygous lethal (Ruden et al., 2000). Therefore, we turned to RNAi methods to knock down TER94 function specifically in WldS-expressing axons. Strikingly, we found that knocking down TER94 in WldS-expressing axons with UAS-TER94RNAi suppressed the axon-protective function of WldS to levels indistinguishable from Nmnat1 and WldS-ΔN16 (Fig. 5 D). Uninjured age-matched controls for WldS/TER94RNAi and TER94RNAi animals were indistinguishable from day 0 controls, and thus, the axon loss observed in WldS/TER94 animals was dependent on axotomy. We note that RNAi for TER94 alone did result in a slight but significant reduction in the number of axons present in uninjured animals in the absence of WldS (Fig. 5 D), suggesting that TER94RNAi can induce low level axon degeneration. However, the vast majority of axons in these animals were intact before injury and in age-matched controls, indicating that TER94RNAi treatment does not cause widespread degeneration of axons on its own. In summary, we conclude that TER94 is autonomously required in WldS-expressing neurons in vivo for WldS to maximally protect severed axons from autodestruction. Furthermore, we propose that the key neuroprotective portion of the N70/W18 domain of WldS is N16 and that its neuroprotective effects are mediated by a TER94/VCP-dependent mechanism.
A major cellular role for TER94/VCP is targeting proteins for refolding, or destruction as part of the UPS degradation pathway. TER94/VCP sits at a pivotal point in the cascade, having the ability to direct refolding or degradation of substrates based on its binding partners. When associated with ubiquitinated proteins, VCP can either bind Npl4 and Ufd2 and target proteins for proteasomal degradation or bind Ufd1 and Plap and release refolded proteins into the cytoplasm (for review see Halawani and Latterich, 2006). In an attempt to identify additional molecules that interact with TER94/VCP to modulate axon autodestruction or WldS-mediated neuroprotection, we assayed the requirements of the following Drosophila genes in Wallerian degeneration and WldS function: CG4673, encoding the Drosophila orthologue of npl4; ufd2; ufd1-like; and plap, encoding the Drosophila orthologue of Ufd3. For each molecule, we used UAS-regulated RNAi constructs to knock down their function in neurons and assayed the ability of these molecules to (a) induce spontaneous axon degeneration, (b) suppress axon autodestruction 15 d after injury, and (c) suppress the ability of WldS to protect severed axons. We found that knockdown of each of these genes failed to dramatically suppress normal axon development, axon stability, degeneration after axotomy, or the ability of WldS to protect severed axons (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200808042/DC1). These data argue that additional components of the TER94/VCP complex are not involved in Wallerian degeneration or WldS-mediated protection of severed axons.
A lack of evidence for a requirement for Sir2 function in WldS-mediated axon protection
Sirt1, a member of the mammalian family of Sir2-like NAD+-binding histone deacetylases (the sirtuins), has been implicated in NAD+-mediated axon protection through a lentiviral-expressing siRNA study in mammalian tissue culture. Specifically, siRNA knockdown of Sirt1, but not the six other mammalian sirtuins (Sirt2–7), partially suppressed the ability of NAD+ application to protect axons in dorsal root ganglion explant cultures (Araki et al., 2004). Surprisingly, in a subsequent study in which a sirt1-null mutation was crossed into the WldS background, loss of sirt1 failed to suppress the ability of WldS to protect severed axons in vivo (Wang et al., 2005), calling into question the role of Sirt1 in executing WldS function. Importantly, it remains an open possibility that the remaining mammalian sirtuins (Sirt2–7) might functionally compensate for Sirt1 in this context, and thus, the functional requirements for mammalian sirtuins in WldS function remain unclear.
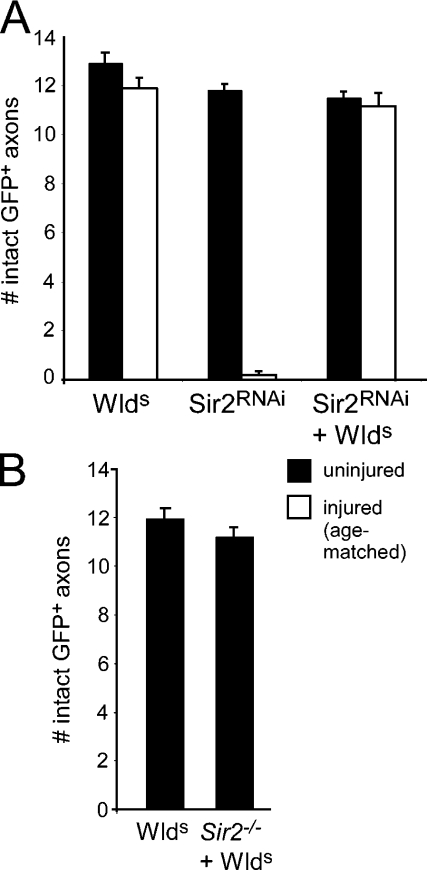
In Drosophila, there is a single gene that bears strong sequence similarity to the mammalian sirtuins, Sir2; Sir2 is 55.9% identical to the Sirt1 at the amino acid level. The simplicity of the Drosophila sirtuin family offers the opportunity to directly reassess the requirements for sirtuins in WldS function in vivo. We first assayed the requirements for Drosophila Sir2 in WldS-mediated protection of severed axons using a UAS-regulated RNAi construct targeted to Sir2. We found that expression of Sir2RNAi in axons did not induce spontaneous degeneration and that ORN axons appeared to develop normally (unpublished data). Axotomized Sir2RNAi-expressing axons exhibited normal axon degeneration, arguing that Sir2 function is not required for Wallerian degeneration (Fig. 6 A). Moreover, expression of Sir2RNAi in WldS-expressing axons did not suppress the ability of WldS to protect severed axons (Fig. 6 A). Next, we confirmed our observation with Sir2RNAi genetically by crossing WldS into a Sir2-null mutant background. Loss of function alleles of Sir2 are not available; however, removal of the Sir2 gene can be accomplished using a heteroallelic combination of the two partially overlapping deletions in sir25.26 and sir24.5 mutants (Newman et al., 2002). As with our Sir2RNAi experiments, we found that deletion of the Sir2 gene in a WldS background failed to suppress the ability of WldS to protect severed axons from degeneration (Fig. 6 B). These data argue strongly that WldS does not require the NAD+-binding histone deacetylase Sir2 to protect severed axons from autodestruction.
Figure 6.
Sir2 is not required for WldS-mediated protection of severed axons, and membrane-tethered WldS fails to suppress axon degeneration. (A) UAS-Sir2RNAi was driven in OR22a+ ORNs with OR22a-Gal4 in the presence or absence of WldS, and axons were assayed 15 d after injury. (B) The requirements for Sir2 in WldS-mediated axon protection were assayed 15 d after injury in WldS-expressing axons and WldS axons that lacked sir2 (sir24.5/sir25.26). WldS, n = 10; Sir2−/− + WldS, n = 6. Error bars represent SEM.
Wallerian degeneration is strongly suppressed by mouse mitochondrial Nmnat3, but NAD+ synthase (NADS), membrane-tethered WldS, and cytoplasmic Nmnat2 fail to protect severed axons
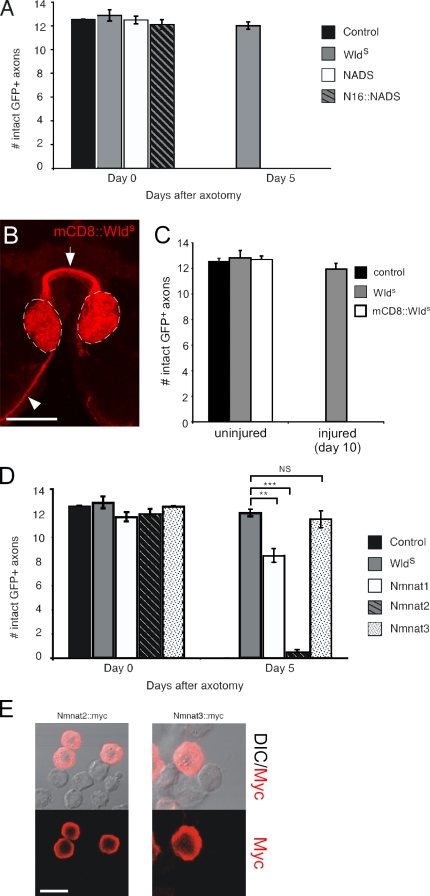
Do all NAD+ biosynthetic enzymes protect axons from autodestruction? The Drosophila genome encodes one additional NAD+ biosynthetic enzyme called NADS that by primary sequence is not related to Nmnat and in yeast localizes throughout the cell (Suda et al., 2003). We generated transgenic flies carrying a UAS-NADS construct, drove expression in ORN axons as described in the first section of Results, and assayed axon-protective function 5 d after injury (Fig. 7 A). Surprisingly, NADS provided no protection of axons. To determine whether NADS might require VCP/TER94 interactions through N16 for detectable axon-protective activity, we also generated flies bearing an N16-NADS fusion protein but found that this molecule also completely failed to suppress axon autodestruction (Fig. 7 A).
Figure 7.
Axon-protective function of NAD+-producing enzymes is specific to Nmnat1 and -3, and Nmnat3 protects as well as WldS. (A) NADS and N16-NADS were expressed in ORNs using OR22a-Gal4, and axons were severed and analyzed for protection 5 d after axotomy. (B) Localization of mCD8-WldS was assayed using α-CD8 antibodies, and expression of mCD8-WldS was driven in ORNs using the OR22a-Gal4 driver. Axons are indicated projecting to glomeruli (arrowhead) and across the midline (arrow), and OR22a+ ORN-innervated glomeruli are shown with dashed lines. (C) Membrane-tethered WldS (mCD8-WldS) was assayed for axon-protective function 10 d after axotomy. n ≥ 10 for all genotypes. (D) Nmnat2 and -3 were driven by OR22a-Gal4 in ORNs to assay for protective function 5 d after axotomy. n ≥ 10 for all genotypes; **, P < 0.01; ***, P < 0.001. (E) S2 cells were transfected with Nmnat2-myc and Nmnat3-myc and stained with α-myc. In both cases, α-myc immunoreactivity was localized to the cytoplasm but not the nucleus. DIC, differential interference contrast. Error bars represent SEM. Bars: (B) 27.47 µm; (E) 14.71 µm.
WldS is normally localized in the nucleus, but can WldS protect severed axons from degeneration if it is forced to localize elsewhere in the cell? We used two approaches to address this question. First, we generated a membrane-tethered version of WldS. In brief, we fused the extracellular and transmembrane domains of the mouse CD8 surface receptor (Lee and Luo, 2001) to the N terminus of WldS (termed mCD8-WldS) and drove the expression of this molecule in 22a+ axons. We found that adult ORNs stably expressed high levels of mCD8-WldS in the axonal compartment (Fig. 7 B). However, we found that mCD8-WldS completely failed to suppress the degeneration of severed axons; 10 d after axotomy, 100% of mCD8-WldS+ axons degenerated and were cleared from the central nervous system (Fig. 7 C). Thus, tethering WldS to the cell surface in vivo is sufficient to block the ability of WldS to protect severed axons.
As a second approach, we assayed the protective function of mammalian Nmnat molecules that show unique patterns of subcellular localization. In mammals, there are three genes encoding Nmnat molecules: Nmnat1, which, as described in the first section of Results, localizes to the nucleus; Nmnat2, which is cytoplasmic; and Nmnat3, which localizes primarily to mitochondria (Sasaki et al., 2006). Nmnat2 localized to the cytoplasm in Drosophila S2 cells (Fig. 7 E) but completely failed to suppress axon degeneration (Fig. 7 D). In striking contrast, Nmnat3, which also appeared to localize largely to the cytoplasm in S2 cells (Fig. 7 E), provided levels of axon protection that were indistinguishable from those afforded by WldS (Fig. 7 D).
In summary, these data indicate that only a subset of NAD+ biosynthetic enzymes can provide significant protection of severed axons, that membrane-tethered WldS is not sufficient to protect axons, and that Nmnat3, an enzyme that localizes primarily to mitochondria in mammalian cells, provides WldS-like levels of axon protection. Thus, nuclear localization appears to be nonessential for WldS-like axon protection, and Nmnat3 and WldS appear to have unique activities or localizations relative to Nmnat2 and mCD8-WldS that allow for robust axon-protective function in vivo.
Discussion
The enzymatic activity of Nmnat1 is a critical feature of WldS
Whether Nmnat1 and its enzymatic product NAD+ underlie the protective activity of WldS has remained a point of great controversy. Two studies found that transfection of nmnat1 into dorsal root ganglia neurons or direct application of NAD+ or its biosynthetic precursors to dorsal root ganglia cultures was sufficient to prolong the survival of severed axons in vitro (Araki et al., 2004; Wang et al., 2005). In contrast, a subsequent study reported a lack of protection of axons by direct expression of Nmnat1 in neurons or application of NAD+ in vitro, and transgenic mice expressing Nmnat1 failed to exhibit significant increased axon protection in vivo (Conforti et al., 2007). We find that expression of Nmnat1 in Drosophila axons provides significant levels of protection for severed axons, with the majority of axons surviving for longer than 2 wk. Thus, Nmnat1 is indeed strongly neuroprotective on its own, although the levels of protection provided by Nmnat1 are significantly reduced compared with WldS. This observation argues that Nmnat1 is a critical component of WldS but indicates that the protection afforded by WldS and Nmnat1 is not equivalent. As discussed in the following section, we propose that the increased efficiency of protection provided by WldS maps to the N16 domain of WldS, which interacts with VCP/TER94. Why doesn't Nmnat1 protect severed axons in transgenic mice? We can think of at least two explanations for the difference observed between mice and flies with respect to in vivo axon protection by Nmnat1. First, we express nmnat1 in Drosophila using the Gal4/UAS binary expression system, which can drive very high levels of expression in ORN neurons. It is possible that we have increased the relative amount of Nmnat1 in neurons to a point exceeding that necessary for Nmnat1-dependent protection of severed axons, whereas expression levels of Nmnat1 in mouse sciatic nerve using the β-actin promoter (Conforti et al., 2007) remain below this critical threshold. Second, the axons of Drosophila ORNs are much shorter than mammalian axons in the sciatic nerve and may thus be more easily protected relative to lengthy mammalian axons.
The protection of severed axons by Nmnat1 critically depends on its enzymatic activity. To generate an Nmnat1 molecule that fails to synthesize NAD+, we engineered an H24A substitution in Nmnat1. Based on analysis of the crystal structure of Methanobacterium thermoautotrophicum NMNAT, the corresponding histidine (H19) resides in the binding site for ATP. Mutational analysis of the importance of this histidine with an H19A substitution demonstrated that mutation of this residue reduced NAD+ biosynthetic activity to <1%, did not significantly affect protein folding (determined by solving the crystal structure of the H19A mutant version), and led to the copurification of NMNAT with its substrate nicotinamide mononucleotide trapped in the active site rather than its product NAD+, which copurifies with wild-type NMNAT (Saridakis et al., 2001). This suggests a failure of NMNAT to convert nicotinamide mononucleotide to NAD+. We find that an H24A substitution in Nmnat1 completely suppresses the neuroprotective effects of Nmnat1, with close to 100% of the axons from Nmnat1dead-expressing neurons degenerating within 5 d after injury. WldS-dead, harboring the same H24A mutation in Nmnat1, also shows dramatically reduced efficacy in the protection of severed axons. Together, these data demonstrate a requirement for Nmnat1 enzymatic activity in WldS-mediated protection of severed axons in vivo. In turn, along with the aforementioned studies, they suggest that the effects of Nmnat1 are mediated by its biosynthetic products. NAD+ is certainly the best-known product of Nmnat1 enzymatic activity and likely plays a key role in axon protection; however, we note that we cannot exclude the formal possibility that in the context of axon protection, Nmnat1 may generate additional biosynthetic products beyond NAD+ that might be neuroprotective.
Interestingly, recent work on Drosophila Nmnat (dNmnat) has revealed a role for dNmnat1 in the suppression of activity- or spinocerebellar ataxia 1 (Sca1)–induced degeneration in the visual system (Zhai et al., 2006, 2008). Surprisingly, the dNmnat-dependent protection of neurons in the visual system is independent of its enzymatic activity, and dNmnat appears capable of acting as a chaperone-like molecule in an enzymatic activity–independent manner (Zhai et al., 2008). Is this chaperone-like activity contributing to WldS-dependent neuroprotection? Although this remains an open possibility, we suspect it is not a major contributing force in the mechanistic action of WldS in severed axons for two reasons. First, WldS and Nmnat1 both require their enzymatic activity to protect severed axons in vivo, whereas dNmnat can protect against Sca1-induced neurodegeneration in the absence of enzyme function. Second, dNmnat is induced in response to the formation of Sca1 aggregates and localizes to sites of protein aggregation, whereas WldS and Nmnat1 show no such localization. A more thorough delineation of the mechanistic action of WldS, Nmnat1, and dNmnat will be essential to resolve these issues.
N70 contributes to WldS neuroprotective function through interactions with VCP/TER94
How N70 or W18 could contribute to WldS function has remained enigmatic. The portion of Ube4b found in WldS, N70, lacks the U box found in Ube4b that is essential for E4 ubiquitination activity (Hatakeyama et al., 2001; Hatakeyama and Nakayama, 2003) and thus does not have ubiquitin-conjugating activity on its own. However, a deletion study of Ube4b has shown that this N-terminal domain is indeed essential for Ube4b ubiquitination activity (Mahoney et al., 2002). Although specific domains necessary for ubiquitination activity of mouse Ube4b have not been identified in N70, the first 16 amino acids of N70 (termed N16) have recently been shown to bind VCP/p97 (Laser et al., 2006). Through N16, WldS can redistribute VCP into the nucleus (Dai and Li, 2001). VCP, in turn, could result in the association of WldS with a variety of cellular proteins because VCP is capable of binding an array of oligonucleotide-ubiquitinated proteins or misfolded proteins and can act to drive their polyubiquitination and proteasomal degradation or, alternatively, deubiquitination and release of refolded proteins into the cytoplasm (for review see Halawani and Latterich, 2006). Consistent with a VCP-dependent recruitment of polyubiquitinated proteins to WldS, α-ubiquitin immunoreactivity appears to be enriched with WldS in the nucleus in vitro (Laser et al., 2006).
The aforementioned data provide an exciting link between WldS and VCP/polyubiquitin, but a requirement for WldS–VCP interactions or N16 in vivo in protecting severed axons has not been demonstrated. In this study, we have shown that (a) deletion of the VCP-binding domain N16 from WldS reduces the axon-protective ability of WldS to levels indistinguishable from Nmnat1; (b) tethering N16 to Nmnat1 is sufficient to provide levels of axon protection equivalent to those observed with WldS; (c) WldS associates with Drosophila TER94, the fly orthologue of VCP; and (d) knocking down TER94 function in WldS-expressing neurons suppresses the axon-protective effects of WldS to levels indistinguishable from those observed with Nmnat1. Together, these data argue strongly for a role for N16–VCP interactions and VCP function in mediating the ability of WldS to protect severed axons.
How do N16–VCP interactions help WldS protect axons? A simple model is that N16–VCP interactions function to redistribute nuclear Nmnat1 activity in the cell to a compartment in which its axon-protective effect is maximal. As discussed in the following section, an interesting possibility is that this redistribution mimics the localization of Nmnat3 (a mitochondrial Nmnat), which appears to protect Drosophila axons in vivo at a level indistinguishable from WldS. Alternatively, N16 may bring Nmnat1 together with VCP-associating proteins that mediate protective effects. For example, perhaps VCP/TER94 binds an unidentified ubiquitinated protein that promotes axon destruction. Because WldS redistributes VCP/TER94 to the nucleus, this molecule would in turn be titrated out of axons. This effect, combined with the NAD+ biosynthetic capacity of Nmnat1, which is partially neuroprotective on its own, could combine for robust protection of axons. Alternatively, VCP/TER94 may bring a polyubiquitinated NAD+-sensitive molecule into close proximity with Nmnat1, which in turn promotes robust axon protection. Identified VCP-binding proteins that mediate refolding versus degradative outputs of the VCP complex may not be involved in protecting the axons of WldS-expressing neurons. We assayed the ability of several VCP-interacting molecules that are essential to promote proteasomal degradation of polyubiquitinated proteins (Ufd2 and Npl4/CG4673) or refolding and cytoplasmic release of misfolded proteins (Ufd1-like and Plap). Knockdown of these molecules in neurons failed to induce widespread spontaneous axon degeneration, suppress axon degeneration after axotomy, or reduce the ability of WldS to protect severed axons. Although we cannot exclude the possibility that RNAi knockdown of these molecules was incomplete, these data begin to argue that VCP/TER94 interactions with N16 do not require these molecules for the execution of WldS-mediated axon protection. Further work aimed at defining the site of action of WldS and identifying molecules interacting with WldS–VCP/TER94 complexes in vivo will be essential to clarify these issues.
Finally, we note that we have defined the minimal domains essential for WldS-like levels of neuroprotection as N16-Nmnat1. W18 is not normally a domain found in any mouse protein, being generated by the spontaneous fusion event in WldS mice, but a role for this domain in axon protection remained an open possibility. The deletion of amino acids 17–70 of N70 and W18 in the N16-Nmnat1 construct demonstrates that these domains are dispensable for WldS-mediated axon protection.
Severed axon autodestruction: the role of nuclear WldS, NAD+, and Nmnat activity
The strong localization of WldS to the nucleus in mice and Drosophila provides a compelling argument for a nuclear role for WldS in protecting severed axons. Impressively, even expression of high levels of WldS in fly neurons with the Gal4/UAS binary system failed to result in detectable axonal localization of WldS-Myc in vivo. Despite its clear localization to the nucleus, it has remained unclear whether nuclear localization is essential for WldS to protect severed axons or whether its presence in the nucleus points to an underlying role in modifying gene expression patterns and thereby axon stability. A role for the NAD+-binding histone deacetylase Sirt1 in WldS-mediated axon protection seems increasingly unlikely; previous work has shown that crossing null alleles of Sirt1 into the WldS background fails to suppress the ability of WldS to protect axons (Wang et al., 2005), and we show that loss of the Drosophila orthologue of Sirt1, Sir2, does not affect the ability of WldS to protect severed axons in vivo. Interestingly, we found that tethering WldS to the membrane by fusing it with mCD8 completely suppressed the ability of WldS to protect severed axons; therefore, the presence of membrane-tethered (likely) monomers of WldS is insufficient for any level of axon protection, suggesting that localization or free solubility of WldS is important to its function.
Are changes in NAD+ production responsible for the axon-protective effects of WldS? To further explore the role of NAD+ biosynthesis in suppressing axon autodestruction, we assayed the ability of various Drosophila and mammalian NAD+ biosynthetic enzymes to block Wallerian degeneration. Surprisingly, of the four NAD+ biosynthetic molecules we tested, only two, Nmnat1 and Nmnat3, protected severed axons from autodestruction. Nmnat2, which localizes to the cytoplasm in mammalian and Drosophila cells, exhibited no protective effect. Likewise, NADS, an NAD+ biosynthetic molecule that generates NAD+ from a different pool of precursors than Nmnat (Rongvaux et al., 2003), failed to protect severed axons from degeneration. In striking contrast, Nmnat3, which localizes to mitochondria in mammals and throughout the cytoplasm in Drosophila cells, provided robust protection of axons at levels indistinguishable from those afforded by WldS 5 d after injury. These observations raise several important points: (a) not all NAD+ biosynthetic enzymes are capable of suppressing Wallerian degeneration, which argues that Nmnat1 and Nmnat3 are somehow unique in localization, protein–protein interactions, or enzymatic activity, which results in axon-protective activity; (b) the fact that multiple NAD+ biosynthetic enzymes protect severed axons may indicate that changes in NAD+ are a key factor in suppression of Wallerian degeneration; (c) Nmnat3 is not expected to bind VCP/TER94 but can protect axons at levels equivalent to WldS; thus, N16–VCP interactions do not appear to be essential for WldS-like neuroprotection by Nmnat molecules; and (d) Nmnat3 localizes outside the nucleus in Drosophila and mammals, arguing that nuclear localization of Nmnat activity is not essential for robust axon protection. Interestingly, it was recently found that Nmnat1 and Nmnat3 can also suppress degeneration induced by rotenone, an activator of mitochondrial oxidative stress (Press and Milbrandt, 2008). This observation, coupled with the robust protection of axons we observe with Nmnat3, may point to a role for WldS/Nmnat activity in mitochondria for axon protection. In the future, it will be critical to define the site of action of WldS and Nmnat3 and determine whether they are protecting axons by similar mechanisms.
The WldS mutation protects axons through gain of function mechanisms. Is WldS telling us that axons initiate an active axon autodestruction pathway akin to cell death, as has been previously speculated (Coleman and Perry, 2002; Raff et al., 2002), or is WldS an aberration that fortuitously blocks Wallerian degeneration? Based on work in other invertebrates, we favor the notion that axons in mice, Drosophila, and many other species indeed initiate an active process of autodestruction after axotomy. A considerable literature exists supporting the ability of severed axons in both vertebrates and invertebrates to exhibit long-term survival (Bittner, 1991). A striking example comes from two species of crickets, Teleogryllus commodus and Gryllus bimaculatus. In these animals, behavioral rhythmicity is governed by a small number of pigment-dispersing factor–positive neurons whose cell bodies reside in the optic lobes. The removal of cell bodies fails to induce degeneration of severed pigment-dispersing factor–positive axons, which can survive for up to 90 d after axotomy, and, impressively, behaviors subject to circadian control remain robust, indicating that severed surviving axonal stumps remain functionally intact for many weeks after axotomy (Stengl, 1995). Why some axons spontaneously degenerate after injury whereas others do not is unknown, but defining the molecular pathways that account for these striking differences in the axonal survival and precisely how WldS impinges upon these pathways will be an extremely important future goal to further our understanding of the basic cellular mechanisms by which neurons maintain the functional integrity of axons. Our findings point to the importance of defining molecules downstream of WldS, perhaps outside the nucleus, that interact with VCP or NAD+, and we suggest that these will be key modulators of Wallerian degeneration.
Materials and methods
Drosophila stocks and transgenics
The following stocks were used: OR22a-Gal4 (a gift from J. Carlson, Yale University, New Haven, CT); pUAST-mCD8–GFP, GH146-Gal4, and elav-Gal4 (a gift from T. Lee, University of Massachusetts Medical School, Worcester, MA); Sir24.5 and Sir25.26 (a gift from S. Smolik, Oregon Health and Science University, Portland, Oregon); UAS-Ter94RNAi, UAS-CG4673RNAi, UAS-CG9934RNAi, UAS-PlapRNAi, and UAS-Ufd1–likeRNAi (Vienna Drosophila RNAi Center); and UAS-Sir2-RNAi (National Institute of Genetics Stock Center).
pUAST-WldS–myc was created by PCR amplifying a 1.1-kb fragment from pUAST-WldS and fusing six tandem Myc tags onto the C terminus of WldS using the primers 5′-GATCAGATCTCAAAACATGGAGGAGCTGAGCGCTGAC-3′ and 5′-ATAATCCGCGGCAGAGTGGAATGGTTGTGCTTG-3′ and subsequently subcloning this fragment into pUAST with BglII and SacII. pUAST-Nmnat1 was created by subcloning mouse nmnat1 (858 bp from Mus musculus) from pHSVPrPUC (a gift from Z. He, Harvard University, Cambridge, MA) into pBluescript-KS with BamHI and EcoRI. It was then subcloned from pBluescript-KS into pUAST with BamHI and XhoI cut sites and was subsequently subcloned into pUAST using BglII and XhoI cut sites. Nmnat1dead was subcloned in the same manner but carries the H24A mutation (a gift from Z. He).
pUAST-WldS-dead was created by subcloning WldS from pUAST (a gift from L. Luo, Stanford University, Stanford, CA) to pBluescript-KS using EcoRI and XbaI, and then a 245-bp C-terminal fragment including the H24A mutation was used to replace the corresponding region of nmnat1. The resulting 1.1-kb fragment (WldS-dead) was then subcloned from pBluescript-KS into pUAST. pUAST-WldS-ΔN16 was created by cloning amino acids 17–373 of WldS into pUAST with BglII and XhoI and adding a 5′ Met-encoding codon using the primers 5′-GATCAGATCTCAAAACATGCTTGCTGGTGGACAGACCTCCC-3′ and 5′-TATACTCGAGTCACAGAGTGGAATGGTTGTGCTTGGCC-3′.
pUAST-N16–nmnat1 was created by subcloning N16 into pUAST and subsequently amplifying an 850-bp fragment of M. musculus nmnat1 using the primers 5′-GAATGCTAGCATGGACTCATCCAAGAAGACAGAGGTGG-3′ and 5′-TATACTCGAGTCACAGAGTGGAATGGTTGTGCTTGGCC-3′ and then subcloning this fragment into pUAST-N16 with NheI and XhoI. pUAST-mCD8–WldS was created by amplifying a 672-bp fragment encoding the extracellular and transmembrane domains of mCD8 from pUAST-mCD8–GFP (Lee and Luo, 2001) and subcloning it into pBluescript-KS using BglII and XbaI. The WldS open reading frame was then subcloned into this plasmid using engineered BglII and XbaI sites to generate pBluescript-KS–mCD8–WldS. mCD8–WldS was then subcloned into pUAST using BglII and XbaI to generate pUAST-mCD8–WldS.
pUAST-Nmnat1–myc was created by amplifying Nmnat1 from the pUAST vector (as described for pUAST-Nmnat1) with the primers 5′-GGCGAGATCTCAAAACATGGACTCATCCAAGAAGACAGAGGTGG-3′ and 5′-TATTAATCCGCGGCAGAGTGGAATGGTTGTGCTTGG-3′ and was subsequently cloned into the C-terminal myc pUAST vector with BglII and SacII. pUAST-Nmnat2 was amplified from mouse cDNA clone BC098007 (Open Biosystems) using the primers 5′-AATTGATTGCGGCCGCAAAACATGACCGAGACCACAAAGACC-3′ and 5′-AATTGTCTAGACTAGCCCGAGGCGTTGATGTACAGC-3′ and subsequently cloned into pUAST with NotI and XbaI. pUAST-Nmnat2–myc was amplified from the same clone as Nmnat2 with the primers 5′-AATTGATTGCGGCCGCAAAACATGACCGAGACCACAAAGACC-3′ and 5′-ATTAATATTAGCGGCCGCGTGCCCGAGGCGTTGATGTACAGCTGACTCTTGAG-3′ and subsequently cloned into C-terminal myc pUAST vector with NotI.
pUAST-Nmnat3 was amplified from mouse cDNA clone BC092086 (Open Biosystems) using the primers 5′-ACTTATATGCGGCCGCAAAACATGAAGAACCGAATTCCCTGTGGTGC-3′ and 5′-AGCAGTCTAGACTAGCCAGTCTTTCCTTTCCC-3′ and was subsequently cloned into pUAST with NotI and XbaI. pUAST-Nmnat3–myc was amplified from the same clone as Nmnat3 using the primers 5′-ACTTATATGCGGCCGCAAAACATGAAGAACCGAATTCCCTGTGGTGC-3′ and 5′-TATAATTAGCGGCCGCGTGCCAGTCTTTCCTTTCCCTTTCCAGGAACCGTCATTGATG-3′ and was subsequently cloned into C-terminal myc pUAST vector with NotI.
pUAST-NADS was amplified from cDNA clone LD11409 (Berkeley Drosophila Genome Project) using the primers 5′-GATCGAATTCCAAAACATGGGACGCAAGGTGACCGTAGCGG-3′ and 5′-GATCCTCGAGCTAGACGGGAATACCGGTGCGA-3′ and was subsequently cloned into pUAST with EcoRI and XhoI. pUAST-N16–NADS was amplified from the same clone as NADS with the primers 5′-GATCGAATTCCAAAACATGGAAGAACTGAGCGCGGATGAAATTCGCCGCCGCCGCCTGGCGCGCATGGGACGCAAGGTGACCGTAGCGG-3′ and 5′-GATCCTCGAGCTAGACGGGAATACCGGTGCGA-3′ and subsequently cloned into pUAST with EcoRI and XhoI.
Axon injury protocol
Antennal ablations were performed on animals carrying OR22a-Gal4, UAS-mCD8–GFP, and the indicated constructs. 1 wk after eclosion, both third antennal segments were surgically ablated with tweezers, and the flies were kept at 25°C until dissection (5, 10, 15, 20, or 30 d), as indicated in Figs. 3–7 and Figs. S2 and S3. Uninjured flies were dissected 1 wk after eclosion unless denoted in legends as age matched. Adult heads were fixed with 4% formaldehyde (Sigma-Aldrich) in 1× PBS and 0.1% Triton X-100 (PTx; Sigma-Aldrich) for 20 min before dissection and washed five times with PTx. Dissected brains were subsequently fixed in 4% formaldehyde in PTx for 10 min and washed five times with PTx and three times with 1× PBS/0.1% Triton X-100/1% BSA (PBT; Thermo Fisher Scientific). Brains were rocked for 30 min in PBT at room temperature, and primary antibodies were added in PBT and rocked overnight at 4°C. Brains were washed 10 times over 1.5 h with PBT, and secondary antibodies were added in PBT. Brains were rocked for 3 h in secondary antibody at room temperature and subsequently washed 10 times over 1.5 h with PBT. Brains were stored at 4°C in antifade reagent (Bio-Rad Laboratories). In the case of animals expressing GFP, no antibodies were used to enhance the GFP signal.
Immunolabeling, confocal microscopy, and quantification of axon protection
Standard methods were used for dissection, fixation, and antibody labeling of the Drosophila adult brain (MacDonald et al., 2006). Primary antibodies were used at the following dilutions: 1:1,000 for immunolabeling of mouse tissue with α-Myc (9E10; Covance), 1:500 for Western blot analysis of α-VCP (BD), and 1:250 for rabbit α-Nmnat1 (a gift from M. Coleman, Babraham Institute, Cambridge, England, UK). Secondary antibodies (Cy3; Jackson ImmunoResearch Laboratories) for immunolabeling of tissues were used at 1:200. Specimens were mounted in a 1:1 mixture of antifade reagent and 75% glycerol. Confocal z stacks of antennal lobes and S2 cells were collected on a confocal microscope (Axioskop2 LSM 5 Pascal; Carl Zeiss, Inc.) with a Plan Apochromat 63× NA 1.4 objective lens (Carl Zeiss, Inc.), which was also equipped with a transmitted light detector used for acquisition of differential interference contrast images of S2 cells. All confocal microscopy was performed at room temperature (21°C) using acquisition software (LSM 5 Pascal; Carl Zeiss, Inc.). Confocal images were compressed with ImageJ software (National Institutes of Health), and intact GFP+ axons were counted individually at the indicated time points. At least two lines (in most cases three to four) were assayed for each construct, results among lines carrying the same construct were indistinguishable from one another, and data were subsequently pooled.
Immunoprecipitations from S2 cell extracts
Drosophila S2 cells were counted and plated at 1 × 106 cells/ml, incubated overnight at 30°C, transfected with pUAST-WldS–myc and pAC-Gal4 according to the instructions in the Transfectene kit (QIAGEN), and incubated for 48 h. Protein G–Sepharose fast-flow beads (GE Healthcare) were washed in extraction buffer (150 mM NaCl, 1% Igepal CA630, 50 mM Tris, pH 7.5, and 1× EDTA-free complete protease inhibitor) and bound with α-Myc antibody for 3 h at 4°C. Transfected and untransfected cells were isolated and centrifuged at 3,000 rpm for 5 min at 4°C. Cells were resuspended in extraction buffer and placed in ethanol/dry ice bath for 5 min, thawed in a cold ice bath, homogenized, and centrifuged at 11,000 rpm for 10 min at 4°C. A fraction of the supernatant was reserved as input, and remaining samples were precleared with equilibrated protein G–Sepharose beads for 2 h at 4°C, added to α-myc–conjugated protein G–Sepharose beads, and incubated at 4°C overnight. The samples were then centrifuged at 2,000 rpm for 1 min. A portion of the supernatant was reserved as the flow-through sample, and the remaining supernatant was discarded; beads were washed five times with extraction buffer, and bound protein was eluted from beads with SDS-PAGE loading buffer.
S2 cell transfection and immunolabeling
Drosophila S2 cells were counted and plated at 1 × 106 cells/ml, incubated overnight at 30°C, and transfected with pUAST-WldS–myc and pAC-Gal4 according to the instructions in the Transfectene kit. Cells were then fixed in PEM (Pipes, EGTA, and MgCl2 buffer)/4% formaldehyde and washed with PBT. Primary antibody (α-myc) was added to the cells at 1:1,000, cells were washed, and secondary α-mouse antibodies were added at 1:200. Immunolabeling was then viewed with confocal microscopy.
Online supplemental material
Fig. S1 shows how the quantification of intact axons was counted. Fig. S2 compares the various transgenes of each construct to show that expression level variations did not affect the protection level. Fig. S3 shows RNAi experiments of proteins thought to complex with VCP/Ter94, none of which appear to have a role in WldS-mediated protection. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200808042/DC1.
Acknowledgments
We wish to thank L. Conforti and M. Coleman for helpful discussions and sharing unpublished data. We thank Z. He and L. Luo for DNA constructs and J. Carlson and S. Smolik for Drosophila stocks. We thank all members of the Freeman laboratory and U. Homberg for helpful discussions.
This work was supported by National Institutes of Health (grants NS053538 and NS059991 to M.R. Freeman), the Christopher and Dana Reeves Foundation, and the Worcester Foundation for Biomedical Research. M.R. Freeman is an Alfred P. Sloan Research Fellow.
Footnotes
Abbreviations used in this paper: dNmnat, Drosophila Nmnat; NADS, NAD+ synthase; Nmnat, nicotinamide mononucleotide adenylyl transferase; OR, odorant receptor; ORN, olfactory receptor neuron; UAS, upstream activating sequence; UPS, ubiquitin proteasome; VCP, valosin-containing protein; WldS, slow Wallerian degeneration.
References
- Araki T., Sasaki Y., Milbrandt J. 2004. Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration.Science. 305:1010–1013 [DOI] [PubMed] [Google Scholar]
- Beirowski B., Adalbert R., Wagner D., Grumme D.S., Addicks K., Ribchester R.R., Coleman M.P. 2005. The progressive nature of Wallerian degeneration in wild-type and slow Wallerian degeneration (WldS) nerves.BMC Neurosci. 6:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner G.D. 1991. Long-term survival of anucleate axons and its implications for nerve regeneration.Trends Neurosci. 14:188–193 [DOI] [PubMed] [Google Scholar]
- Coleman M. 2005. Axon degeneration mechanisms: commonality amid diversity.Nat. Rev. Neurosci. 6:889–898 [DOI] [PubMed] [Google Scholar]
- Coleman M.P., Perry V.H. 2002. Axon pathology in neurological disease: a neglected therapeutic target.Trends Neurosci. 25:532–537 [DOI] [PubMed] [Google Scholar]
- Conforti L., Tarlton A., Mack T.G., Mi W., Buckmaster E.A., Wagner D., Perry V.H., Coleman M.P. 2000. A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse.Proc. Natl. Acad. Sci. USA. 97:11377–11382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti L., Fang G., Beirowski B., Wang M.S., Sorci L., Asress S., Adalbert R., Silva A., Bridge K., Huang X.P., et al. 2007. NAD(+) and axon degeneration revisited: Nmnat1 cannot substitute for Wld(S) to delay Wallerian degeneration.Cell Death Differ. 14:116–127 [DOI] [PubMed] [Google Scholar]
- Dai R.M., Li C.C. 2001. Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation.Nat. Cell Biol. 3:740–744 [DOI] [PubMed] [Google Scholar]
- Ferri A., Sanes J.R., Coleman M.P., Cunningham J.M., Kato A.C. 2003. Inhibiting axon degeneration and synapse loss attenuates apoptosis and disease progression in a mouse model of motoneuron disease.Curr. Biol. 13:669–673 [DOI] [PubMed] [Google Scholar]
- Glass J.D., Brushart T.M., George E.B., Griffin J.W. 1993. Prolonged survival of transected nerve fibres in C57BL/Ola mice is an intrinsic characteristic of the axon.J. Neurocytol. 22:311–321 [DOI] [PubMed] [Google Scholar]
- Halawani D., Latterich M. 2006. p97: the cell's molecular purgatory? Mol. Cell. 22:713–717 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Nakayama K.I. 2003. Ubiquitylation as a quality control system for intracellular proteins.J. Biochem. 134:1–8 [DOI] [PubMed] [Google Scholar]
- Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K.I. 2001. U box proteins as a new family of ubiquitin-protein ligases.J. Biol. Chem. 276:33111–33120 [DOI] [PubMed] [Google Scholar]
- Hoopfer E.D., McLaughlin T., Watts R.J., Schuldiner O., O'Leary D.D., Luo L. 2006. Wlds protection distinguishes axon degeneration following injury from naturally occurring developmental pruning.Neuron. 50:883–895 [DOI] [PubMed] [Google Scholar]
- Laser H., Conforti L., Morreale G., Mack T.G., Heyer M., Haley J.E., Wishart T.M., Beirowski B., Walker S.A., Haase G., et al. 2006. The slow Wallerian degeneration protein, WldS, binds directly to VCP/p97 and partially redistributes it within the nucleus.Mol. Biol. Cell. 17:1075–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Luo L. 2001. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development.Trends Neurosci. 24:251–254 [DOI] [PubMed] [Google Scholar]
- Lunn E.R., Perry V.H., Brown M.C., Rosen H., Gordon S. 1989. Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve.Eur. J. Neurosci. 1:27–33 [DOI] [PubMed] [Google Scholar]
- Luo L., O'Leary D.D. 2005. Axon retraction and degeneration in development and disease.Annu. Rev. Neurosci. 28:127–156 [DOI] [PubMed] [Google Scholar]
- MacDonald J.M., Beach M.G., Porpiglia E., Sheehan A.E., Watts R.J., Freeman M.R. 2006. The Drosophila cell corpse engulfment receptor Draper mediates glial clearance of severed axons.Neuron. 50:869–881 [DOI] [PubMed] [Google Scholar]
- Mack T.G., Reiner M., Beirowski B., Mi W., Emanuelli M., Wagner D., Thomson D., Gillingwater T., Court F., Conforti L., et al. 2001. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene.Nat. Neurosci. 4:1199–1206 [DOI] [PubMed] [Google Scholar]
- Mahoney J.A., Odin J.A., White S.M., Shaffer D., Koff A., Casciola-Rosen L., Rosen A. 2002. The human homologue of the yeast polyubiquitination factor Ufd2p is cleaved by caspase 6 and granzyme B during apoptosis.Biochem. J. 361:587–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman B.L., Lundblad J.R., Chen Y., Smolik S.M. 2002. A Drosophila homologue of Sir2 modifies position-effect variegation but does not affect life span.Genetics. 162:1675–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinter M., Jekely G., Szepesi R.J., Farkas A., Theopold U., Meyer H.E., Lindholm D., Nassel D.R., Hultmark D., Friedrich P. 1998. TER94, a Drosophila homolog of the membrane fusion protein CDC48/p97, is accumulated in nonproliferating cells: in the reproductive organs and in the brain of the imago.Insect Biochem. Mol. Biol. 28:91–98 [DOI] [PubMed] [Google Scholar]
- Press C., Milbrandt J. 2008. Nmnat delays axonal degeneration caused by mitochondrial and oxidative stress.J. Neurosci. 28:4861–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff M.C., Whitmore A.V., Finn J.T. 2002. Axonal self-destruction and neurodegeneration.Science. 296:868–871 [DOI] [PubMed] [Google Scholar]
- Rongvaux A., Andris F., Van Gool R., Leo O. 2003. Reconstructing eukaryotic NAD metabolism.Bioessays. 25:683–690 [DOI] [PubMed] [Google Scholar]
- Ruden D.M., Sollars V., Wang X., Mori D., Alterman M., Lu X. 2000. Membrane fusion proteins are required for oskar mRNA localization in the Drosophila egg chamber.Dev. Biol. 218:314–325 [DOI] [PubMed] [Google Scholar]
- Saridakis V., Christendat D., Kimber M.S., Dharamsi A., Edwards A.M., Pai E.F. 2001. Insights into ligand binding and catalysis of a central step in NAD+ synthesis: structures of Methanobacterium thermoautotrophicum NMN adenylyltransferase complexes.J. Biol. Chem. 276:7225–7232 [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Araki T., Milbrandt J. 2006. Stimulation of nicotinamide adenine dinucleotide biosynthetic pathways delays axonal degeneration after axotomy.J. Neurosci. 26:8484–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengl M. 1995. Pigment-dispersing hormone-immunoreactive fibers persist in crickets which remain rhythmic after bilateral transection of the optic stalks.J. Comp. Physiol. [A]. 176:217–228 [Google Scholar]
- Suda Y., Tachikawa H., Yokota A., Nakanishi H., Yamashita N., Miura Y., Takahashi N. 2003. Saccharomyces cerevisiae QNS1 codes for NAD(+) synthetase that is functionally conserved in mammals.Yeast. 20:995–1005 [DOI] [PubMed] [Google Scholar]
- Waller A. 1850. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observation of the alterations produced thereby in the structure of their primitive fibres.Philos. Trans. R. Soc. Lond. (1776–1886). 140:423–429 [Google Scholar]
- Wang J., Zhai Q., Chen Y., Lin E., Gu W., McBurney M.W., He Z. 2005. A local mechanism mediates NAD-dependent protection of axon degeneration.J. Cell Biol. 170:349–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Wu Y., Culver D.G., Glass J.D. 2001. The gene for slow Wallerian degeneration (Wld(s)) is also protective against vincristine neuropathy.Neurobiol. Dis. 8:155–161 [DOI] [PubMed] [Google Scholar]
- Watts R.J., Hoopfer E.D., Luo L. 2003. Axon pruning during Drosophila metamorphosis: evidence for local degeneration and requirement of the ubiquitin-proteasome system.Neuron. 38:871–885 [DOI] [PubMed] [Google Scholar]
- Zhai Q., Wang J., Kim A., Liu Q., Watts R., Hoopfer E., Mitchison T., Luo L., He Z. 2003. Involvement of the ubiquitin-proteasome system in the early stages of Wallerian degeneration.Neuron. 39:217–225 [DOI] [PubMed] [Google Scholar]
- Zhai R.G., Cao Y., Hiesinger P.R., Zhou Y., Mehta S.Q., Schulze K.L., Verstreken P., Bellen H.J. 2006. Drosophila NMNAT maintains neural integrity independent of its NAD synthesis activity.PLoS Biol. 4:e416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai R.G., Zhang F., Hiesinger P.R., Cao Y., Haueter C.M., Bellen H.J. 2008. NAD synthase NMNAT acts as a chaperone to protect against neurodegeneration.Nature. 452:887–891 [DOI] [PMC free article] [PubMed] [Google Scholar]