Abstract
Current concepts of cell migration were established in regular two-dimensional (2D) cell culture, but the roles of topography are poorly understood for cells migrating in an oriented 3D fibrillar extracellular matrix (ECM). We use a novel micropatterning technique termed microphotopatterning (μPP) to identify functions for 1D fibrillar patterns in 3D cell migration. In striking contrast to 2D, cell migration in both 1D and 3D is rapid, uniaxial, independent of ECM ligand density, and dependent on myosin II contractility and microtubules (MTs). 1D and 3D migration are also characterized by an anterior MT bundle with a posterior centrosome. We propose that cells migrate rapidly through 3D fibrillar matrices by a 1D migratory mechanism not mimicked by 2D matrices.
Introduction
Cell migration is a fundamental biological process in which cells often interact with ECM, which provides cells with physical and chemical signals that orchestrate their movement. Except for ameboid cells (Lammermann et al., 2008), migration of most cells consists of four basic sequential events: (1) protrusion at the leading edge, (2) adhesion to ECM, (3) cell body translocation, and (4) retraction of the trailing edge (Ridley et al., 2003). In routine 2D cell culture, the rate of cell migration depends on the coordination between these events as well as the concentration or density of the underlying ECM (DiMilla et al., 1993). The fastest migration rates of fibroblasts are achieved at an intermediate, optimal ligand density, with higher ECM ligand densities inhibiting protrusion and cell body movement by preventing retraction of the trailing edge.
Other physical aspects of ECM such as matrix stiffness can affect intracellular phosphorylation events (Pelham and Wang, 1997) and gene expression (Engler et al., 2006) as well as cell migration (Pelham and Wang, 1998; Zaman et al., 2006). In vivo, most cells have 3D interactions absent from 2D cell culture (Nelson and Bissell, 2006; Yamada and Cukierman, 2007), which can alter cell migration (Even-Ram and Yamada, 2005). For example, the protein concentration of 3D gel matrices affects ligand density, matrix stiffness, and pore size, which can physically impede cells and require activation of proteolytic pathways for migration (Zaman et al., 2006). The phenomenon of contact guidance described by Weiss and Garber (1952) more than 50 yr ago suggests that the structure of the surrounding ECM plays a role in cell orientation and migration. Oriented 3D matrix can provide directional cues that promote a characteristic aligned spindle morphology of cells in 3D tissue explants and directional migration of cells during gastrulation (Weiss and Garber, 1952; Nakatsuji and Johnson, 1984). Thus, the interplay between cells and ECM organization is vital for cell migration.
A major discrepancy with the known inhibitory effect of high ligand density in 2D is that fibroblasts readily migrate along ligand-dense ECM fibers in 3D at rates ∼1.5× faster than in 2D cell culture (Cukierman et al., 2001). We hypothesized that fibrillar ECM topography could provide a physical basis for this and other distinctive characteristics of 3D cell migration. In this report, we demonstrate that a single, nearly 1D micropatterned line can mimic many aspects of the phenotype induced by cell-derived oriented 3D matrices, including rapid uniaxial migration that is independent of ligand density. Additional contrasts to 2D migration include a dependence on myosin II contractility for rapid migration with a requirement for an anterior microtubule (MT) bundle for both 1D and 3D forms of ECM-directed migration.
Results and discussion
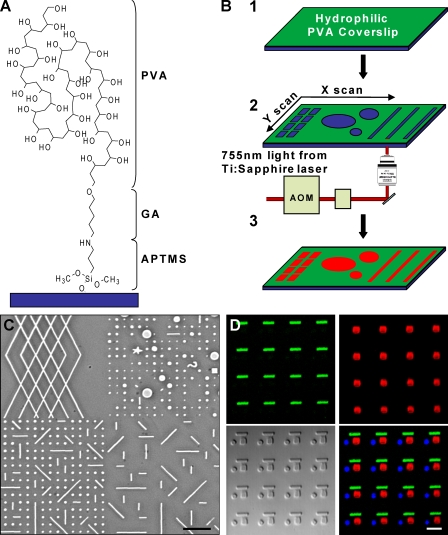
To investigate the role of ECM topography in cell migration, we developed a novel micropatterning technique we term microphotopatterning (μPP; Fig. 1 and Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1). We used polyvinyl alcohol (PVA) for its high hydrophilicity and ability to prevent protein adsorption and cell adhesion. We found that a thin (∼0.1 µm) macromolecular PVA monolayer was sufficient to block cell attachment. Many hydrogels derived from PVA, polyethylene glycol, and polyacrylamide are susceptible to photolytic ablation (Yamato et al., 2003). We used a two-photon confocal microscope to ablate precise patterns in the PVA film. Through the computer-controlled region of interest (ROI) function, we generated as many as several hundred ablation spots per field of view and stored them as templates (Fig. 1 and Fig. S2). These templates could be applied through AIM software to adjacent areas to generate a large array of a single or a combination of patterns (Fig. 1 C). To create smaller, more closely spaced patterns, we simply changed to a higher magnification objective or used a digital zoom through the software.
Figure 1.
Generation of micropatterned PVA films. (A) Schematic of a single PVA molecule conjugated to a glass coverslip through 3-(amino)propyl-trimethyloxysilane (APTMS) and glutaraldehyde (GA). (B) PVA-coated surfaces (1) are photoablated using an LSM 510 NLO META system (Carl Zeiss, Inc.). (2) ROI template generated by the software dictates the ablation pattern. (3) Protein (red) added to the dish adsorbs only to ablated sites. (C) Phase image of four different ROI templates used in close proximity. (D) Serial ablation with μPP to generate patterns of fibrinogen (green dashes), vitronectin (red squares), and FN (blue circles) within micrometers of each other. The DIC image is shown in the bottom left. Bars: (C) 20 µm; and (D) 5 µm.
After local PVA ablation, ECM proteins were adsorbed to the etched surfaces (Fig. S1 C); adsorption was dependent on the extent of ablation from the total amount of light energy focused on a given region (Fig. S1 B). This method permits rapid design and testing of multiple patterns without needing the physical masks or stamps associated with microcontact printing (Mrksich et al., 1996). The high refractive index (>1.5) of PVA allows for live cell fluorescence techniques such as total internal reflection fluorescence (TIRF) microscopy, even through nonablated regions of the film, to observe dynamics of GFP-linked cytoskeletal components (e.g., see Fig. 3, E–G; and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1). Ablated regions of the PVA thin film are also visible by phase and differential interference contrast microscopy (Fig. 1 C). Because μPP does not require a physical stamp, multiple different protein patterns can be generated in close proximity. Fig. 1 D shows the result of three rounds of serial ablation, ECM protein attachment, washing, and blocking to deposit fibrinogen, vitronectin, and finally fibronectin (FN) within 2 µm of each other.
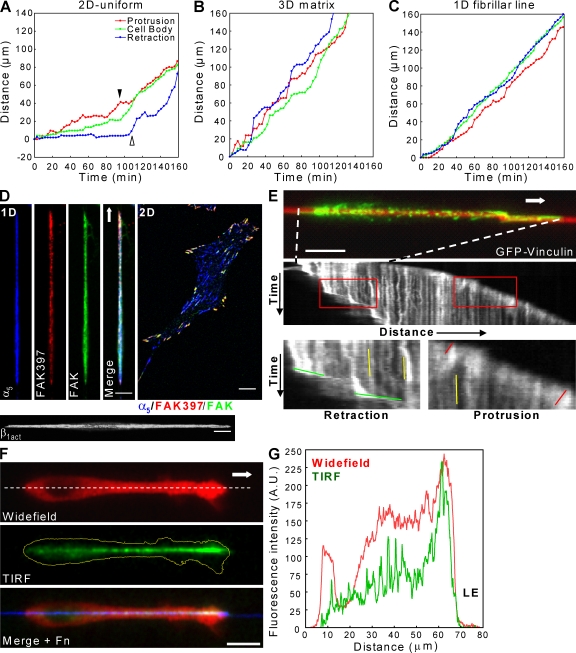
Figure 3.
Coordinated protrusion–retraction cycles and unique linear adhesions are associated with 1D migration. (A–C) Coordination between leading edge protrusion (red), cell body movement (green), and tail retraction (blue) in directionally migrating cells on 2D (A), in 3D matrix (B), and along a 1D line (C). In A, protrusion is reduced (filled arrowhead) until the tail retracts. As retraction begins (open arrowhead), protrusion resumes. Both 3D matrix and 1D migration display efficient protrusion–retraction cycles. (D) Confocal images showing localization of FAK, phospho-FAK397, α5-integrin, and activated β1-integrin during 1D fibrillar migration and a 2D control. (E, top) Adhesions containing GFP-vinculin using TIRF (green) are shown. Kymograph analysis (middle) of GFP-vinculin adhesions during 1D migration. Red boxes (bottom) show leading and trailing edges. Red, yellow, and green lines indicate slopes of vinculin movement within adhesions at the leading edge, under the cell body, and at the tail, respectively. Dashed lines indicate the cell’s initial position. (F) Widefield image of vinculin (top) shows elongated leading lamella of rapidly migrating cells. TIRF reveals vinculin in a front to rear gradient spanning leading to trailing edge. (G) Line scan (dashed line in F) of the cell in F shows the gradient pattern (green) and close proximity of adhesions to the leading edge (LE). Arrows indicate the direction of migration. Bars, 10 µm.
Using the flexibility of μPP, we generated multiple patterns to test roles of different ECM topographies in fibroblast migration. When attached to dot arrays (∼5-µm spacing; 3.4-µm2 area per ECM island), fibroblasts showed multiple points of protrusive activity (multiaxial) with slow migration (Fig. S2 B). In striking contrast, fibroblasts attaching to single 1.5-µm-wide fibrillar lines (termed 1D) demonstrated rapid spreading, polarization, motility, and uniaxial phenotype within 2–4 h, which is similar to previous observations of fibroblasts in cell-derived 3D matrices (Fig. 2, A–C; and Videos 2 and 3, available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1; Cukierman et al., 2001). These effects were independent of the ligand because the same phenotype was observed if fibrillar lines were coated with vitronectin or collagen IV (unpublished data).
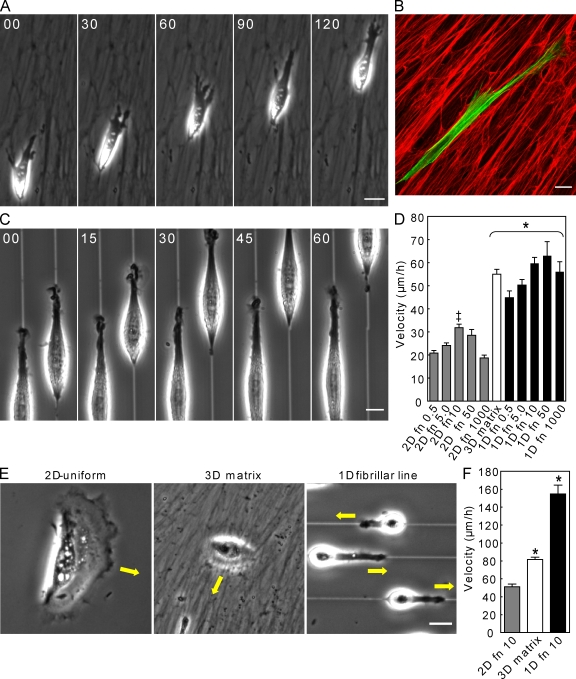
Figure 2.
1D topography functionally mimics 3D fibrillar matrix. (A) Rapid migration of NIH-3T3 fibroblasts through a 3D cell-derived matrix. (B) Cytoskeletal alignment (green, actin) along oriented FN fibers (red). (C) Fibroblasts plated on single ∼1.5-µm lines mimic this 3D phenotype and migrate continuously. (D) Cell migration rates on 2D surfaces and 1D fibrillar lines at different FN ligand densities versus 3D cell-derived matrix. (E) HK migration over 2D, 3D matrix, and 1D fibrillar lines. (F) HK migration rates on substrates in E. Bars: (A, B, and E) 20 µm; and (C) 10 µm. *, P < 0.01 versus 2D; ‡, P < 0.05 versus 0.5 and 1,000 FN. Error bars indicate SEM.
We tested the role of ligand density in cell migration rate by varying FN concentrations for coating 1D lines and 2D surfaces from 0.5 µg/ml to the near-saturating level of 1,000 µg/ml, mimicking the high ligand density of 3D fibrillar matrix (Fig. S2, C–E). On 2D surfaces, varying ligand density produced the expected biphasic effects on migration rate, whereas 1D migration was significantly elevated above 2D velocities at all concentrations (Fig. 2 D). Above 0.5 µg/ml, 1D fibrillar migration rates remained at an elevated plateau in contradiction to the well-established effect of increased ligand density on 2D cell migration.
To test whether epithelial cells also display rapid 1D migration, we compared human epidermal keratinocyte migration. Although unable to elongate on or invade into 3D cell-derived matrices, human keratinocytes (HKs) often followed the linearity of the matrix; migration velocity on top of matrices increased ∼1.6-fold in 3D versus 2D. On 1D fibrillar lines, HKs acquired a uniaxial phenotype similar to fibroblasts (Fig. 2, E and F; and Video 4, available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1) with a remarkable threefold faster migration than in 2D. These data suggest that epithelial cell migration on fibrillar ECM structures is rapid and mimicked by 1D lines.
We further compared 1D fibrillar, 2D, and 3D cell migration by quantifying the coordination between three key steps of fibroblast migration: protrusion, cell body translocation, and tail retraction. As expected, fibroblasts migrating on 2D uniform surfaces demonstrated the hindrance of leading edge protrusion by inefficient tail retraction described previously (Fig. 3 A; Chen, 1979, 1981).
In contrast, both 3D and 1D fibrillar migration at 10 and 50 µg/ml FN displayed highly efficient coordination between all three processes according to correlation analyses, with little reduction in leading edge protrusion as a result of inefficient tail retraction (Table I and Fig. 3, B and C). Interestingly, the degree of coordination between leading edge protrusion and tail retraction on 1D fibrillar lines did decrease at high ligand densities (1,000 µg/ml FN) caused by protrusive activity at the rear, resulting in tail elongation, yet this effect did not prevent either leading edge protrusion or cell body translocation.
Table I.
Correlation coefficients between protrusion, cell body translocation, and tail retraction of fibroblasts migrating under 2D, 3D, and 1D conditions
| Correlations | 2D (50 FN) | 3D matrix | 1D (10 FN) | 1D (50 FN) | 1D (1,000 FN) |
| Protrusion–retraction | 0.45 | 0.87 | 0.77 | 0.88 | 0.63 |
| Protrusion–cell body | 0.63 | 0.84 | 0.83 | 0.91 | 0.91 |
| Cell body–retraction | 0.73 | 0.96 | 0.93 | 0.94 | 0.69 |
Spatiotemporal feedback occurs between the actomyosin cytoskeleton and focal adhesion components to regulate adhesion strength, which can in turn regulate cell migration (Gupton and Waterman-Storer, 2006). Thus, cytoskeletal and adhesion organization in 1D or 3D systems might be important for rapid migration. In striking contrast to its localization on 2D substrates, a primary component of fibrillar and 3D matrix adhesions, α5-integrin, was present in a single, long adhesion spanning the entire length of fibroblasts, as was activated β1-integrin, as shown by 9EG7 (Fig. 3 D). The focal adhesion proteins FAK, vinculin, and paxillin localized in long linear adhesions along the entire cell axis, resembling 3D matrix adhesions (Cukierman et al., 2001). In addition, phospho-FAK397 staining was concentrated at leading and often trailing edges in a heterogeneous pattern, suggesting localized signal regulation. No significant change in adhesion staining was observed at higher ECM concentrations (unpublished data).
Because adhesion assembly and disassembly mechanisms influence cell migration, we performed time-lapse TIRF imaging of GFP vinculin on 1D fibrillar patterns. Kinematic data revealed that adhesions formed within 1 µm of the leading edge (Fig. 3, F and G), and their assembly rate matched the rate of leading edge protrusion (Fig. 3 E and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1). In addition, once vinculin was incorporated into the adhesion at the front, adhesion disassembly was observed only at the trailing edge with rates similar to adhesion assembly. Consequently, adhesions beneath the cell may serve as a stable anchor, whereas the main sites of adhesion regulation are at the leading and trailing edges.
A potential explanation for the persistent high velocities we observed with 1D fibrillar migration could be physical limitation of lateral cell spreading and lateral lamellae. This proposal is supported by analysis of cell adhesion area as measured by vinculin staining and total spread area, both of which were significantly reduced in 1D compared with 2D conditions (adhesion area, 54.6 ± 22.1 vs. 89.9 ± 37.4; total spread area, 370.5 ± 183 vs. 1,414 ± 630; P < 0.05). To test this hypothesis further, we generated fiberlike patterns with widths of 1–40 µm (Fig. 4 A). With increasing fibril width, cells generally continued to migrate in one direction but demonstrated increased cell spreading, loss of uniaxial morphology above 5-µm fibril widths, and decreased migration rate (Fig. 4 B). These data indicate that loss of the uniaxial phenotype in conjunction with increased lateral cell spreading on wider fibrils is detrimental to rapid cell migration.
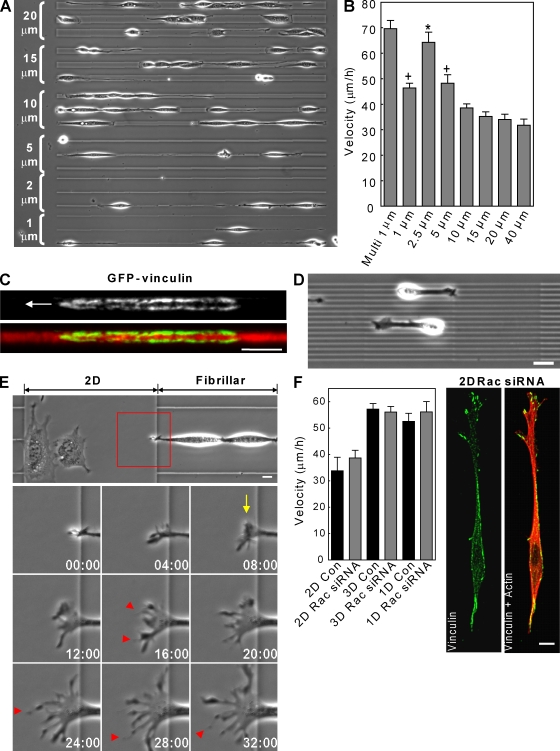
Figure 4.
Regulation of cell migration by fiber width and dimensionality. (A) Triplets (bracketed) of 1, 2.5, 5, 15, and 20 µm-wide lines used for cell migration tracking. (B) Biphasic effect of fiber width on fibrillar migration rate. (C) TIRF images of GFP-vinculin containing adhesions on a 2.5-µm line show two parallel sets of adhesions. The arrow indicates the direction of migration. (D) Fibroblasts migrating with a 3D/1D phenotype along multiple 1-µm lines. (E) 1D to 2D ECM transition induces spreading of the leading edge (arrow) and formation of many cell processes (arrowheads). The red box indicates ROI in time lapse below. (F) Rac siRNA knockdown in 2D induces a uniaxial phenotype lacking the single linear adhesion seen in 1D and does not increase migration rate. *, P < 0.05 versus all conditions except multilines; +, P < 0.05 versus 15–40 µm. Con, control. Error bars indicate SEM. Bars, 10 µm.
Interestingly, cells on fibrillar lines below 1.5-µm wide showed a reduced migration rate compared with 2.5-µm widths. We hypothesized that this reduced migration could be a result of insufficient ECM area available for forming new adhesions. As a test, we generated multiple 1-µm-wide lines spaced ∼5 µm apart and found that migration rates increased to the levels on 2.5-µm fibrils (Fig. 4, B and D). Moreover, vinculin-containing adhesions were localized to either side of the 2.5-µm fibrils, suggesting that fibroblasts treat this width as two smaller parallel fibers analogous to railroad tracks (Fig. 4 C).
The importance of topography to migration was particularly apparent when cells migrating on fibrillar patterns (single or multiple lines) confronted either a perpendicular fibrillar pattern or a 2D region. In both cases, the uniaxial lamella transformed into several multiaxial lamellae, leading to a spread cell morphology and slowing of migration (Fig. 4 E and Video 6, available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1). The opposite transformation also occurred from 2D to 1D mode of migration, although less frequently, accompanied by an increase in velocity (Video 7). Therefore, 1D fibrillar patterns reduce lateral spreading of the lamella, thereby increasing the efficiency by which cells migrate along a single axis.
We tested whether the uniaxial phenotype with reduced spread area associated with fibrillar migration is sufficient to promote rapid migration by manipulating the small GTPase Rac. We previously reported that Rac activity is lower in 3D than 2D and that RNAi knockdown of Rac in 2D induces a similar uniaxial phenotype and directional cell migration (Pankov et al., 2005). Knockdown of Rac to ∼25% of control levels (Fig. 4 F and Fig. S3 A, available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1) reduced cell spreading in 2D and mimicked the uniaxial phenotype closely in terms of cell shape, yet migration rates did not increase under any condition. Immunostaining for vinculin revealed that adhesions on the 2D ECM remain spatially separated. These data indicate that the uniaxial phenotype alone is not sufficient to promote rapid migration and that ECM topography can affect the spatial distribution of cell adhesions.
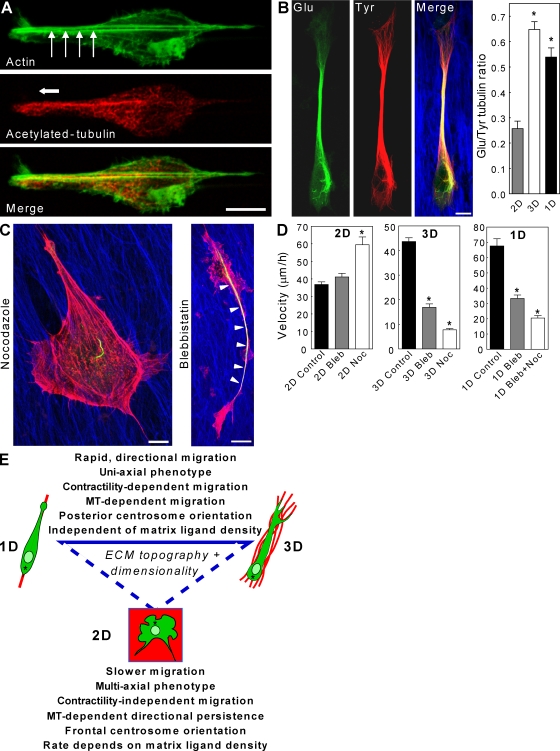
Immunostaining of actin and MT cytoskeletal networks in 1D revealed linear organization (Fig. 5 A). On fibrillar patterns, stress fibers stretched from leading to trailing edge parallel to the matrix, whereas MTs, implicated in establishing cell polarity, became localized in parallel arrays extending into the lamellipodium. Staining for detyrosinated Glu-tubulin, a posttranslational modification of tubulin associated with MT stabilization, revealed large quantities of stabilized MTs throughout 1D- and 3D-migrating cells and accumulated in an anterior bundle (Fig. 5 B), as shown by an elevated Glu/Tyr-tubulin ratio in both. Western blotting further confirmed this difference between 2D and 3D (Fig. S3 B). This distribution resembles the stabilized MTs of neurons required for axon maintenance (Shea, 1999), suggesting that it may help maintain uniaxial polarity and migration along a matrix fibril. Surprisingly, the centrosome as marked by pericentrin was oriented toward the rear of fibroblasts migrating in 3D and 1D (Fig. S3 C), as was recently discovered for epithelial cells and in zebrafish (Pouthas et al., 2008). Similar but less pronounced results were obtained for Golgi orientation (unpublished data). This inconsistency with frontal centrosome and Golgi orientation in 2D wound-healing assays (Gundersen and Bulinski, 1988; Etienne-Manneville and Hall, 2001) suggests distinct mechanisms of polarity in single cell migration on 1D and 3D ECM fibrils.
Figure 5.
Cytoskeletal regulation by ECM dimensionality. (A) Actin (green) stress fibers (arrows) extend from the leading to the trailing edge, whereas stabilized acetylated tubulin (red) is tightly compacted toward the front of the cell. (B) Cells in 3D matrix (blue) show high levels of stabilized Glu-tubulin (green) directed toward the leading edge, which contains mostly Tyr-tubulin (red). The graph shows the Glu/Tyr tubulin ratio in 2D, 3D, and 1D. (C) After blebbistatin treatment, Glu-tubulin remains concentrated in an axonlike structure (arrowheads) polarized along the matrix, whereas nocodazole disrupts this architecture. (D) Comparison of effects of blebbistatin (Bleb) and nocodazole (Noc) on cell migration in 2D, 3D, and 1D conditions. (E) Summary of the effects of ECM topography/dimensionality on cell morphology and migration. Asterisks indicate centrosomes. (B and D) *, P < 0.05 versus 2D. Error bars indicate SEM. Bars, 10 µm.
We tested whether the cross talk between MTs and the myosin II contractile apparatus reported for cells in 2D (Even-Ram et al., 2007) occurred in 1D and 3D. Inhibition of myosin II ATPase activity with blebbistatin produced the opposite effect on fibroblast motility in 2D versus both 3D and 1D conditions. Although blocking myosin II–dependent contractility in 2D led to a small increase in migration rate (+13%), fibroblasts migrating in 3D matrices or along 1D fibrils showed 61% and 55% reductions, respectively (Fig. 5 D). Moreover, unlike 2D, 3D and 1D conditions showed no significant increases in cell protrusion; fibroblasts still followed the FN fibrils/patterns, suggesting that contractility is not involved in establishing cell polarity on fibrillar structures.
The MT inhibitor nocodazole caused a loss of cell polarity and detachment from the 1D matrix. To reduce this hypercontractile response, we pretreated cells with a low dose of blebbistatin (5 µM) before nocodazole addition. This led to uncontrolled protrusion in all directions and reduced migration in 1D and 3D conditions, which is in contrast to increased migration in 2D (Fig. 5 D). Inhibitor treatments showed that the uniaxial phenotype is maintained after loss of contractility (blebbistatin treatment), whereas cells lose the ability to orient along ECM fibrils in the absence of MTs (nocodazole treatment; Fig. 5 C). These results highlight the importance of MTs in 1D and 3D rapid oriented migration compared with their previously characterized role in maintaining directionality of 2D migration.
Conclusions
Our data indicate that ECM topography can regulate cell migration rate and cellular phenotype regardless of ligand density. 1D topography can mimic many characteristics of the rapid, uniaxial migration phenotype observed in fibrillar 3D cell-derived matrices, and both 1D and 3D migration differ from 2D migration with respect to morphology, cytoskeleton organization, and function (Fig. 5 E). The similarities between 3D matrix and 1D fibrillar migration establish that external physical stimuli in the form of the linear topographical cues shown in this report can regulate cell behavior. In fact, previous research has suggested that aligned ECM fibers can be an important environmental factor directing cell migration in embryonic development (Nakatsuji and Johnson, 1984) as well as in mouse tumor (Provenzano et al., 2008) and in vivo metastasis models (Sidani et al., 2006). We propose that oriented 3D fibrillar matrices provide an avenue by which fibroblasts can align, polarize, and migrate directionally using 1D fibril regulation of actomyosin and MT networks to modify cellular axial phenotype and promote proficient directional protrusion. 1D fibrillar migration may provide a simpler and more representative system than 2D systems for understanding 3D fibrillar matrix regulation of the dynamics of cell adhesions, cytoskeleton, and signaling during cell migration.
Materials and methods
Cells culture and transfection
NIH-3T3 fibroblasts (American Type Culture Collection) were cultured in phenol red–free DME (Hyclone) containing 10% bovine calf serum (Hyclone), 100 U/ml penicillin/streptomycin (Invitrogen), and 2 mM l-glutamine (Invitrogen) at 37°C with 10% CO2. Human foreskin fibroblasts were provided by S. Yamada (National Institute of Dental and Craniofacial Research, Bethesda, MD) and were cultured in the same medium except using 10% fetal bovine serum (Hyclone). Human epidermal keratinocytes were cultured in keratinocyte KGM-2 supplemented medium (Lonza). 3D FN-containing matrices were prepared from human foreskin fibroblasts as described previously (Cukierman et al., 2001). Plasmids were transfected into NIH-3T3 fibroblasts by electroporation using a Gene Pulser (Bio-Rad Laboratories) at 170 V and 960 μFd (micro farad) with external capacitance and a time constant of 17–22 µs in 0.4-cm gap cuvettes.
siRNA
Rac1 siRNAs were obtained as SMARTpool plus preparations and single duplexes from Thermo Fisher Scientific. siRNA transfections were performed as previously described (Pankov et al., 2005).
Reagents
The following sources were used: mouse antiacetylated tubulin (Sigma-Aldrich), rat antityrosinated tubulin (Abcam), rabbit antidetyrosinated Glu-tubulin (Millipore), mouse antivinculin (Sigma-Aldrich), mouse antiactin (Sigma-Aldrich), rabbit anti-Rac (Millipore), Alexa Fluor 488 and rhodamine phalloidins (Invitrogen), rabbit antipericentrin (Covance), blebbistatin (−/−; Toronto Chemicals or EMD), and nocodazole (EMD); rabbit anti-FN R745 and goat anti-FN 1213 were generated in our laboratory.
Immunofluorescence staining
For staining of MTs, cells were permeabilized and fixed in 0.5% glutaraldehyde, 3% paraformaldehyde, 0.5% Triton X-100, and 5% sucrose in PHEM buffer (60 mM Pipes, 2 mM Hepes, 10 mM EGTA, and 2 mM MgCl2, pH 6.9) for 5 min, rinsed once with PHEM + 100 mM glycine, and fixed again with 4% paraformaldehyde in PHEM buffer for 20 min. For all other immunostaining, the glutaraldehyde was omitted, and the paraformaldehyde concentration was 4%. To quench glutaraldehyde autofluorescence, samples were incubated with 0.5% NaBH4 in PBS for 5 min. Nonspecific sites were blocked with 20% donkey serum (Jackson ImmunoResearch Laboratories) together with mouse on mouse reagent (M.O.M.; Vector Laboratories) in PHEM buffer for 1 h. All other steps were performed as described previously (Even-Ram et al., 2007). IgG secondary antibodies were obtained from Jackson ImmunoResearch Laboratories.
Coverslip preparation
Glass-bottom dishes (MatTek) were washed in 50% HNO3 for 30 min and rinsed overnight in H2O. Glass surfaces were treated with 200 mM NaOH for 15 min, rinsed with H2O, and silanized with 1% 3-(amino)propyl-trimethoxysilane (Sigma-Aldrich) for 10 min. After rinsing twice, surfaces were blown dry and cured at 65°C for 2 h. Dishes were activated with 0.5% glutaraldehyde for 30 min and rinsed three times for >30 min with H2O. Surfaces were again blown dry and stored desiccated at 4°C.
PVA film synthesis
PVA (molecular weight ∼98,000; 98% hydrolyzed; Sigma-Aldrich) was diluted in H2O to a 5.6% stock solution. This mixture was solubilized at ∼90°C in a water bath and was immediately 0.2-µm filtered to remove impurities. 1,124 µl 2 N HCl was added to 8,876 µl of the PVA solution. Activated glass dishes were covered with this mixture and incubated for 4–5 min at room temperature. Dishes were spin coated for 40 s at 7,000 rpm with an acceleration of 550 rps using a spin coater (Laurel Technologies). PVA-coated dishes were allowed to cure overnight at 4°C. As a result of our ability to observe GFP fluorescence in nonablated areas of the PVA film during TIRF microscopy, we estimate its thickness to be <0.2 µm.
Photoablation (μPP)
A confocal microscope (LSM 510 NLO META; Carl Zeiss, Inc.) equipped with a two-photon laser (1.5 W total power; Ti:Sapphire Chameleon; Coherent) was used for μPP. The two photon was tuned to 755 nm, which provides an excitation wavelength similar to 365 nm, and used at ∼90% power. The PVA solution has high ultraviolet absorbance between 200 and 380 nm, suggesting that the ablation is likely a result of two-photon absorption in the 350–380-nm range. ROIs were generated using the AIM software (Carl Zeiss, Inc.). A 63× NA 1.4 oil immersion objective (Carl Zeiss, Inc.) was used during ablation. Scan speed was set to four and averaged 2–4 times per field of view to provide a laser dwell time of ∼25.6 µs/pixel. The digital zoom function was set to 1.6× to mimic a 100× objective to increase laser dwell time. Scanning was performed in both directions for a mean of ∼15 s per field of view. The hydrogel surface was located using a dichroic (KP 700/488) and 1% power on the He-Ne2 633-nm laser with a long pass 560 emission filter to generate a reflected light image (Fig. S1). Unless otherwise specified, components were associated with the confocal system.
Post-μPP protein attachment
After glutaraldehyde quenching, samples were rinsed three times with PBS. Fluorescently labeled proteins were diluted in PBS containing 0.1% Pluronic F-127 (Invitrogen) and added to coverslips for 1 h at 37°C. This adsorption was followed by two washes with PBS and rinsing with 2 M NaCl. Dishes were blocked with 1% heat-denatured BSA for 1 h at 37°C. Finally, dishes were rinsed twice with PBS and kept in PBS until use.
Fluorescent labeling of ECM proteins
All proteins were conjugated with Alexa Fluor–based dyes (488, 568, and 647; Invitrogen) or DyLight 649 succinimidyl ester (Thermo Fisher Scientific). Conjugation of all proteins with Alexa Fluor followed protocols from Invitrogen. DyLight 649 was first adjusted to 1 mg/ml in DMSO, and 100 μl/1 ml of 5 mg/ml protein (PBS, pH 9.0, with 1 N NaOH) was added and incubated on ice for 1 h.
Microscopy
Confocal microscopy images were obtained using a confocal microscope (LSM 510 NLO META) equipped with a Plan-Apochromat 63× 1.4 NA or Plan-Neofluar 40× 1.3 NA objective (Carl Zeiss, Inc.). A 488-nm argon, 543-nm He-Ne1, and 633-nm He-Ne2 (Carl Zeiss, Inc.) were used to excite Cy2, Cy3, and Cy5 fluorophores, respectively. The pinholes for each laser line were aligned for optimal confocality. For DAPI illumination, the two-photon laser was tuned to 730 nm and used at ∼3–8% power.
TIRF imaging was performed using a microscope (IX-71; Olympus) using either a Plan Apo N 60× 1.45 NA (TIRFM UIS2; Olympus) or UApo 150× 1.45 NA objective (TIRFM UIS2; Olympus). Images were acquired with an electron multiplication charge-coupled device camera (9100–12 or 9100–09; Hamamatsu Photonics). Excitation was provided by a laser launch (Prairie Technologies) equipped with a multiline argon (488 nm) and krypton-argon (568/647 nm) gas lasers. Single emission filters (520, 605, and 690 nm) and triple dichroic (488/568/647 nm) mirrors were obtained from Chroma Technology Corp. MetaMorph imaging software (MDS Analytical Technologies) was used to acquire images and control all hardware.
Time-lapse videos were recorded on a microscope (Axiovert 135TV; Carl Zeiss, Inc.) fitted with a motorized xy- and z-stage focus drive (Ludl Electronic Products Ltd.) using an enhanced contrast Plan-Neofluar 10× 0.3 NA, a long-distance Plan-Neofluar Korr 20× 0.4 NA, or a long-distance Plan-Neofluar Korr 40× 0.6 NA phase objective (Carl Zeiss, Inc.). Images were acquired with a charge-coupled device camera (ORCA II ER; Hamamatsu Photonics). MetaMorph imaging software was used to acquire images and control all hardware. A custom environmental chamber (Lucite) enclosed both time-lapse and TIRF microscopes and maintained cells at 37°C with 10% CO2.
Image processing
Image processing was performed using custom-made smoothing, sharpening, and convolution filters using MetaMorph software. To flatten background in images, either a rolling ball averaging filter (size of 10) was used or one standard deviation above the mean fluorescence was subtracted from images. The following filters were used: a 7 × 7 tall Gaussian, a 7 × 7 low pass filter, a 7 × 7 unsharpen mask (scaling factor of 0.6), and a 5 × 5 sharpening hat filter. Filter kernel settings will be provided on request. For cells imaged in 3D cell-derived matrix, z stacks of each wavelength were either averaged or summed together to generate z-projected/2D images. Chemical structures were created using ChemSketch (version 10.0; Advanced Chemistry Development Inc.).
Statistics
Prism 4 software (GraphPad Software, Inc.) was used for all graphs and statistical analyses. One-way analysis of variance using a Tukey post-test for more than two datasets and Mann-Whitney t tests were used to establish significant differences (P < 0.05). All graphs display mean and error bars as standard error.
Online supplemental material
Fig. S1 is a summary of the process of μPP. Fig. S2 shows an example of a large-patterned array, migration over dot matrices, and quantification of ligand density on 2D and 1D surfaces. Fig. S3 demonstrates the posterior centrosome orientation in 1D and 3D and shows siRNA knockdown of Rac. Video 1 demonstrates adhesion formation and cell spreading of NIH-3T3 fibroblasts expressing GFP-vinculin using TIRF imaging. Videos 2 and 3 show fibroblasts undergoing 1D fibrillar and 3D migration. Video 4 shows regulation of human epidermal keratinocyte migration rate by ECM topography. Video 5 demonstrates adhesion assembly, anchorage, and disassembly during 1D fibrillar migration of NIH-3T3 fibroblasts expressing GFP-vinculin. Videos 6 and 7 illustrate the effects of ECM topography on cell spreading, phenotype, and migration. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810041/DC1.
Acknowledgments
We thank M. dos Santos, A. Green, A. Berrier, R. Petrie, and J. Harunaga for helpful suggestions.
Support was provided by the intramural research program of the National Institute of Dental and Craniofacial Research at the National Institutes of Health.
Footnotes
Abbreviations used in this paper: FN, fibronectin; HK, human keratinocyte; MT, microtubule; μPP, microphotopatterning; PVA, polyvinyl alcohol; ROI, region of interest; TIRF, total internal reflection fluorescence.
References
- Chen W.T. 1979. Induction of spreading during fibroblast movement.J. Cell Biol. 81:684–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W.T. 1981. Mechanism of retraction of the trailing edge during fibroblast movement.J. Cell Biol. 90:187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cukierman E., Pankov R., Stevens D.R., Yamada K.M. 2001. Taking cell-matrix adhesions to the third dimension.Science. 294:1708–1712 [DOI] [PubMed] [Google Scholar]
- DiMilla P.A., Stone J.A., Quinn J.A., Albelda S.M., Lauffenburger D.A. 1993. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength.J. Cell Biol. 122:729–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler A.J., Sen S., Sweeney H.L., Discher D.E. 2006. Matrix elasticity directs stem cell lineage specification.Cell. 126:677–689 [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. 2001. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta.Cell. 106:489–498 [DOI] [PubMed] [Google Scholar]
- Even-Ram S., Yamada K.M. 2005. Cell migration in 3D matrix.Curr. Opin. Cell Biol. 17:524–532 [DOI] [PubMed] [Google Scholar]
- Even-Ram S., Doyle A.D., Conti M.A., Matsumoto K., Adelstein R.S., Yamada K.M. 2007. Myosin IIA regulates cell motility and actomyosin-microtubule crosstalk.Nat. Cell Biol. 9:299–309 [DOI] [PubMed] [Google Scholar]
- Gundersen G.G., Bulinski J.C. 1988. Selective stabilization of microtubules oriented toward the direction of cell migration.Proc. Natl. Acad. Sci. USA. 85:5946–5950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S.L., Waterman-Storer C.M. 2006. Spatiotemporal feedback between actomyosin and focal-adhesion systems optimizes rapid cell migration.Cell. 125:1361–1374 [DOI] [PubMed] [Google Scholar]
- Lammermann T., Bader B.L., Monkley S.J., Worbs T., Wedlich-Soldner R., Hirsch K., Keller M., Forster R., Critchley D.R., Fassler R., Sixt M. 2008. Rapid leukocyte migration by integrin-independent flowing and squeezing.Nature. 453:51–55 [DOI] [PubMed] [Google Scholar]
- Mrksich M., Chen C.S., Xia Y., Dike L.E., Ingber D.E., Whitesides G.M. 1996. Controlling cell attachment on contoured surfaces with self-assembled monolayers of alkanethiolates on gold.Proc. Natl. Acad. Sci. USA. 93:10775–10778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji N., Johnson K.E. 1984. Experimental manipulation of a contact guidance system in amphibian gastrulation by mechanical tension.Nature. 307:453–455 [DOI] [PubMed] [Google Scholar]
- Nelson C.M., Bissell M.J. 2006. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer.Annu. Rev. Cell Dev. Biol. 22:287–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankov R., Endo Y., Even-Ram S., Araki M., Clark K., Cukierman E., Matsumoto K., Yamada K.M. 2005. A Rac switch regulates random versus directionally persistent cell migration.J. Cell Biol. 170:793–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R.J., Jr., Wang Y. 1997. Cell locomotion and focal adhesions are regulated by substrate flexibility.Proc. Natl. Acad. Sci. USA. 94:13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R.J., Jr., Wang Y.L. 1998. Cell locomotion and focal adhesions are regulated by the mechanical properties of the substrate.Biol. Bull. 194:348–349 [DOI] [PubMed] [Google Scholar]
- Pouthas F., Girard P., Lecaudey V., Ly T.B., Gilmour D., Boulin C., Pepperkok R., Reynaud E.G. 2008. In migrating cells, the Golgi complex and the position of the centrosome depend on geometrical constraints of the substratum.J. Cell Sci. 121:2406–2414 [DOI] [PubMed] [Google Scholar]
- Provenzano P.P., Inman D.R., Eliceiri K.W., Trier S.M., Keely P.J. 2008. Contact guidance mediated three-dimensional cell migration is regulated by Rho/ROCK-dependent matrix reorganization.Biophys. J. 95:5374–5384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley A.J., Schwartz M.A., Burridge K., Firtel R.A., Ginsberg M.H., Borisy G., Parsons J.T., Horwitz A.R. 2003. Cell migration: integrating signals from front to back.Science. 302:1704–1709 [DOI] [PubMed] [Google Scholar]
- Shea T.B. 1999. Selective stabilization of microtubules within the proximal region of developing axonal neurites.Brain Res. Bull. 48:255–261 [DOI] [PubMed] [Google Scholar]
- Sidani M., Wyckoff J., Xue C., Segall J.E., Condeelis J. 2006. Probing the microenvironment of mammary tumors using multiphoton microscopy.J. Mammary Gland Biol. Neoplasia. 11:151–163 [DOI] [PubMed] [Google Scholar]
- Weiss P., Garber B. 1952. Shape and movement of mesenchyme cells as functions of the physical structure of the medium: contributions to a quantitative morphology.Proc. Natl. Acad. Sci. USA. 38:264–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K.M., Cukierman E. 2007. Modeling tissue morphogenesis and cancer in 3D.Cell. 130:601–610 [DOI] [PubMed] [Google Scholar]
- Yamato M., Konno C., Koike S., Isoi Y., Shimizu T., Kikuchi A., Makino K., Okano T. 2003. Nanofabrication for micropatterned cell arrays by combining electron beam-irradiated polymer grafting and localized laser ablation.J. Biomed. Mater. Res. A. 67:1065–1071 [DOI] [PubMed] [Google Scholar]
- Zaman M.H., Trapani L.M., Sieminski A.L., Mackellar D., Gong H., Kamm R.D., Wells A., Lauffenburger D.A., Matsudaira P. 2006. Migration of tumor cells in 3D matrices is governed by matrix stiffness along with cell-matrix adhesion and proteolysis.Proc. Natl. Acad. Sci. USA. 103:10889–10894 [DOI] [PMC free article] [PubMed] [Google Scholar]