Abstract
In yeast, three proteins are essential for mitochondrial fusion. Fzo1 and Mgm1 are conserved guanosine triphosphatases that reside in the outer and inner membranes, respectively. At each membrane, these conserved proteins are required for the distinct steps of membrane tethering and lipid mixing. The third essential component is Ugo1, an outer membrane protein in the mitochondrial transport protein family. We show that Ugo1 is a modified member of this family, containing three transmembrane domains and existing as a dimer, a structure that is critical for the fusion function of Ugo1. Our functional analysis of Ugo1 indicates that it is required distinctly for both outer and inner membrane fusion after membrane tethering, indicating that it operates at the lipid-mixing step of fusion. This role is distinct from the fusion dynamin-related proteins and thus demonstrates that at each membrane, a single fusion protein is not sufficient to drive the lipid-mixing step, but instead, this step requires a more complex assembly of proteins.
Introduction
Mitochondrial fusion is a conserved and functionally important process that evolved to create a more connected compartment that facilitates content exchange and access to mitochondrial DNA (Hoppins et al., 2007). Unlike other fusion events, mitochondrial fusion is not mediated by SNAREs but instead is driven by the action of dynamin-related proteins (DRPs). DRPs are GTPases that, through their ability to self-assemble, control a variety of membrane remodeling events. Through an analysis of mitochondrial fusion in vitro, we have demonstrated that two distinct DRPs are essential for fusion. Specifically, the transmembrane proteins Fzo1 (yeast)/Mfn1/2 (mammals) and Mgm1 (yeast)/Opa1 (mammals) drive outer and inner mitochondrial membrane fusion, respectively (Meeusen et al., 2004, 2006). Data suggest a model in which, in the initial stages of fusion, outer and inner membrane tethering is mediated by the self-assembly of mitochondrial fusion DRPs in trans via intermolecular coiled-coil interactions (Ishihara et al., 2004; Koshiba et al., 2004; Meeusen et al., 2004, 2006; Griffin and Chan, 2006). Analysis of mutant alleles of the fusion DRPs indicates that membrane tethering is separable from subsequent lipid content mixing, which completes the membrane fusion process, and that the mitochondrial fusion DRPs are essential at both stages (Meeusen et al., 2006).
To date, the only non-DRP essential for mitochondrial fusion is Ugo1 (Sesaki and Jensen, 2001). Although Ugo1 is localized to the outer membrane, it is classified as a member of the mitochondrial transport/carrier protein family by virtue of possessing signature energy transfer motifs (ETMs; Belenkiy et al., 2000). Mitochondrial transport proteins are typically inner membrane proteins composed of three homologous carrier repeats of ∼100 amino acids, which each contain two helical transmembrane domains (TMDs). Based on the structure of the mitochondrial ATP/ADP carrier, the ETMs are thought to act together to close the central pore formed by the six TMDs through an intramolecular salt bridge network (Pebay-Peyroula et al., 2003; Nury et al., 2006). Consistent with its classification, there are three ETMs present in Ugo1; however, the third motif lacks a critical and conserved charged residue. Mutational analysis of the first two ETMs in Ugo1 indicates that both are important for fusion but that the second motif is essential (Coonrod et al., 2007).
Hydropathy analysis of Ugo1 predicts that, like other transport proteins, there are six regions that may function as TMDs. However, protease protection analysis of Ugo1 has demonstrated that its C terminus is in the intermembrane space (IMS), and the N terminus is localized in the cytosol, constraining Ugo1 to an odd number of TMDs (Sesaki and Jensen, 2001). These findings were initially interpreted to indicate that Ugo1 contains a single TMD; however, more recent protease protection analysis indicates that Ugo1 contains either three or five membrane-spanning regions (Coonrod et al., 2007).
Interaction data indicate that Ugo1 potentially connects the outer and inner membrane fusion DRPs via two nonoverlapping, independent Fzo1 and Mgm1 interaction regions within the N-terminal and C-terminal halves of Ugo1, respectively (Sesaki et al., 2003; Wong et al., 2003; Sesaki and Jensen, 2004). A small C-terminal deletion of Fzo1 simultaneously abolishes the Fzo1–Ugo1 interaction and causes loss of mitochondrial fusion activity in vivo, suggesting that the Fzo1–Ugo1 interaction is mechanistically important (Sesaki and Jensen, 2004). However, the exact functional relevance of this interaction is not known. Indeed, the mechanistic role of Ugo1 in fusion is not known, specifically whether it is required for outer or inner membrane fusion or if it functions at either the membrane tethering or lipid-mixing steps. In this study, we have determined the role of Ugo1 in fusion by characterizing its structure and function in vitro, where we can resolve membrane tethering and lipid-mixing steps of outer and inner membrane fusion events. Our findings demonstrate that Ugo1 plays a unique, postmembrane tethering role in mitochondrial fusion. This role is distinct from the fusion DRPs and thus demonstrates that at each membrane, a single fusion protein is not sufficient to drive the lipid-mixing step, but instead, this stage requires a more complex assembly of proteins.
Results
Ugo1 contains three TMDs
Determination of Ugo1 topology is critical to understand its function in mitochondrial fusion, specifically in evaluating its tertiary and quaternary structure and its interactions with the outer and inner membrane fusion DRPs, Fzo1 and Mgm1. To determine the exact topology of Ugo1, we generated anti-Ugo1 antibodies and created functional, internally epitope-tagged Ugo1 alleles for use in protease protection experiments.
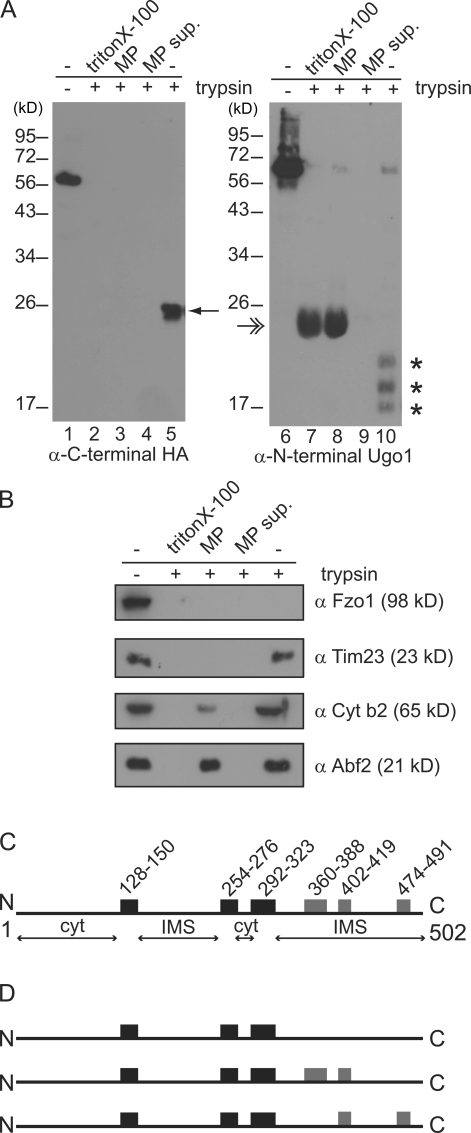
We examined the topology of Ugo1 by treating isolated mitochondria with the exogenous protease trypsin. Mitochondria were isolated from a strain expressing a functional C-terminal influenza HA epitope-tagged Ugo1 (Wong et al., 2003). Consistent with published observations, in intact mitochondria, we detected a protected C-terminal fragment of Ugo1 of ∼23 kD, which indicates that the C terminus resides in an internal compartment (Fig. 1 A, compare lane 1 with lane 5; Sesaki and Jensen, 2001). To further explore Ugo1 topology, we used a polyclonal antibody to the N-terminal 125 amino acids of Ugo1 and observed that several Ugo1 fragments in the range of 17–20 kD were protected from trypsin digestion in intact mitochondria (Fig. 1 A, compare lane 6 with lane 10). Under these conditions, the outer membrane marker Fzo1 was digested completely, indicating that the cytosolic face of the outer membrane was fully accessible to the protease (Fig. 1 B). In addition, both IMS (cytochrome b2 and the C-terminal soluble domain of Tim23) and matrix (Abf2) marker proteins were protected from proteolysis, confirming that the mitochondrial outer and inner membranes were intact (Fig. 1 B). However, when both inner and outer mitochondrial membranes were disrupted by treatment with detergent, complete proteolysis of Ugo1 was observed, as detected by both polyclonal and anti-HA antibodies and marker proteins in all mitochondrial compartments (Fig. 1, A and B).
Figure 1.
Analysis of Ugo1 topology. (A and B) Mitochondria were left intact (lanes 5 and 10), converted to mitoplasts (lanes 3 and 8; mitoplast supernatant, lanes 4 and 9), or solubilized (lanes 2 and 7) before treatment with (+) or without (−) trypsin (lanes 1 and 6) and were analyzed by SDS-PAGE and immunoblotting with the indicated antisera. The 23-kD protected C-terminal Ugo1 fragment is indicated by a single arrow, and the 17–20-kD protected N-terminal fragments are indicated by asterisks. The double arrow indicates a nonspecific interaction of the secondary antibody with trypsin (HRP-α rabbit). (C) Schematic of Ugo1 structure. Black boxes designate hydrophobic regions predicted to be bona fide TMDs based on data in A, and gray boxes represent additional possible TMDs. Amino acid positions are listed above the boxes. Predicted cytosolic (cyt) and IMS regions are also indicated. (D) Schematic representations of possible Ugo1 structures. MP, mitoplast; MP sup, mitoplast supernatant. N, N terminus; C, C terminus.
We converted intact mitochondria to mitoplasts by selectively rupturing the mitochondrial outer membrane by hypoosmotic shock. In mitoplasts, the matrix marker Abf2 was protected from trypsin digestion, indicating that the inner membrane was intact, whereas, as expected, the IMS protein cytochrome b2 and the C-terminal IMS domain of Tim23 were both degraded (Fig. 1 B). Under these conditions, the HA-tagged C terminus of Ugo1 behaved exactly like the Tim23 IMS marker and was also sensitive to digestion in mitoplasts, which is in agreement with published data (Fig. 1 A, compare lane 3 with lane 5; Sesaki and Jensen, 2001). This result indicates that the C terminus of Ugo1 resides in the IMS. Significantly, the N-terminal Ugo1 species detected using polyclonal anti-Ugo1 antibodies were accessible to trypsin digestion in outer membrane–permeabilized mitoplasts (Fig. 1 A, compare lane 8 with lane 10). Ugo1 was readily proteolyzed in detergent-solubilized mitochondria, and no Ugo1 species were detected in the supernatant of the mitoplast fractions, indicating that they are not intrinsically protease resistant and instead are protected in mitochondria from proteolysis by an intact outer membrane (Fig. 1 A, lanes 2 and 7 and lanes 4 and 9).
Our polyclonal antibodies directed to the first Ugo1 125 residues revealed an IMS region that was previously undetected because prior analyses relied on an N-terminal epitope-tagged version of Ugo1 that was accessible to protease in intact mitochondria (Sesaki and Jensen, 2001). Thus, our data indicate that there are at least two regions of Ugo1 localized to IMS (Fig. 1 C). In combination with hydropathy plot analysis, these protease protection data predict that there is an N-terminal IMS region between the first and second predicted TMDs in Ugo1 of ∼100 amino acids. These data are consistent with three models of Ugo1 topology in which Ugo1 contains three or five TMDs (Fig. 1 D).
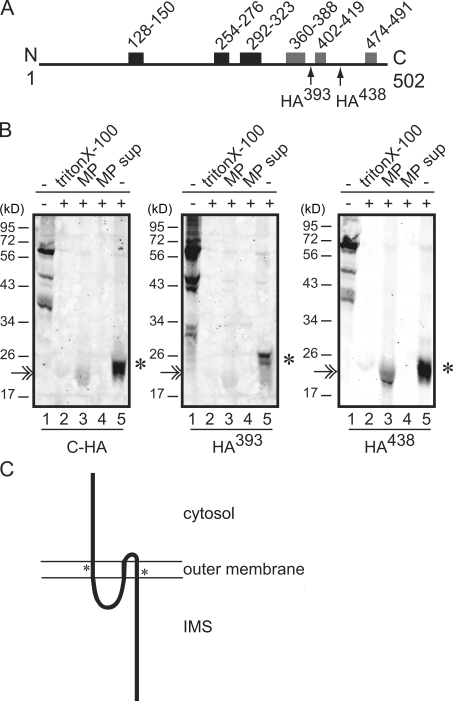
To distinguish between these models, we generated versions of Ugo1 with internal HA tags located in predicted loop regions that follow potential TMDs ending at amino acid 393 (Ugo1HA393) or 438 (Ugo1HA438; Fig. 2 A). Both Ugo1HA393 and Ugo1HA438 were functional as assessed by their ability to rescue the mitochondrial DNA loss and glycerol growth defects of Δugo1 cells (unpublished data), indicating that they are correctly targeted to the outer membrane and are suitable for topology analysis. Protease protection analysis of intact mitochondria isolated from yeast strains expressing Ugo1HA393 and Ugo1HA438 revealed that the HA epitope in both versions is protease resistant but becomes sensitive to trypsin proteolysis in outer membrane–permeabilized mitoplasts (Fig. 2 B, lanes 1, 3, and 5). These data indicate that regions of Ugo1 at amino acid 393 and 438 are localized to the IMS. Consistently, these regions were readily proteolyzed in detergent-solubilized mitochondria and were not detected in the supernatant of the mitoplast fractions, indicating that they are protected from proteolysis by an intact outer membrane (Fig. 2 B, lanes 2 and 4). We conclude from these data that there are no TMDs in the C-terminal half of Ugo1.
Figure 2.
Ugo1 contains three membrane-spanning domains. (A) A schematic of Ugo1 as in Fig. 1 with arrows indicating the locations of internal triple HA tags. (B) Protease protection analysis of internal HA-tagged versions of Ugo1, which are schematically depicted in A. The protected C-terminal fragments are indicated by asterisks. The double arrow indicates a nonspecific interaction of the antibody with trypsin (IRdye-α rabbit). (C) Model of Ugo1 topology in the outer mitochondrial membrane. The location of the putative carrier domains are each indicated with an asterisk. MP, mitoplast; MP sup, mitoplast supernatant. N, N terminus; C, C terminus.
As schematically depicted in Fig. 2 C, our protease protection data predict that Ugo1 possesses only three TMDs positioned in the N-terminal half of the protein, which is in contrast to a recently proposed topology of Ugo1 in which the last hydrophobic region (amino acids 474–491) was suggested to be a TMD (Coonrod et al., 2007). Our data place the N-terminal 14-kD portion of Ugo1 and the loop between the second and third TMD in the cytosol and the loop between the first and second TMD and the 23-kD C-terminal region in IMS (Fig. 2 C). This topology is consistent with the ability of Ugo1 to independently interact with the outer and inner membrane fusion DRPs, Fzo1 and Mgm1, as significant portions that likely represent interaction regions are positioned appropriately (Sesaki and Jensen, 2004).
Dimerization of Ugo1 creates a modified version of a typical transport protein
In contrast to inner membrane carrier proteins, we have determined that Ugo1 possesses only three TMDs. However, similar to transport proteins, the first and third TMDs of Ugo1 possess ETMs that form critical intramolecular salt bridges (Fig. 2 C). It has been shown that the charged residues in ETM in the third TMD and not in the first TMD of Ugo1 are important for mitochondrial fusion (Coonrod et al., 2007). Given the divergence of Ugo1, we considered the possibility that ETMs in Ugo1 form intermolecular rather than intramolecular salt bridges. In this case, ETMs would participate in the formation of a six TMD Ugo1 dimer, which would be similar to the mitochondrial transport family topology.
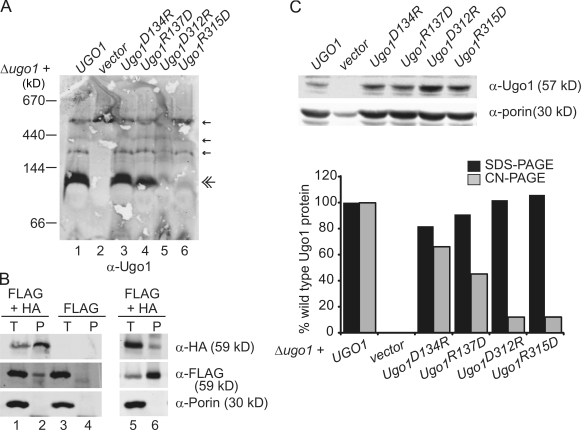
To test this possibility, we focused our analysis on the quaternary structure of Ugo1. Specifically, we determined the native size of Ugo1 in yeast mitochondria by high resolution clear native PAGE (hrCN-PAGE; Wittig et al., 2007). After solubilization with digitonin, mitochondrial proteins were subjected to hrCN-PAGE that was capable of resolving 70–700-kD-sized protein species. By Western analysis with polyclonal antibodies directed against the N terminus of Ugo1, we observed that the most abundant form of Ugo1 in wild-type mitochondria was a species with an apparent molecular mass of 115 kD, which is consistent with the size of a Ugo1 dimer (Ugo1 Mr 57,470; Fig. 3 A, lane 1). This species was not detected with antibodies directed against either Fzo1 or Mgm1 and was present in mitochondria lacking Fzo1 or Mgm1 but was absent in mitochondria isolated from a Δugo1 strain, which is consistent with the interpretation that this 115-kD species represents a Ugo1 homodimer (Fig. 3 A, lane 2; and not depicted). To further test that Ugo1 exists as dimer, we coexpressed Ugo1-HA and Ugo1-Flag in cells and asked whether they interact in mitochondria by immunoprecipitation analysis. We observed that anti-HA antibodies immunoprecipitated Ugo1-HA and Ugo1-Flag from chemically cross-linked mitochondrial extracts as detected by Western analysis (Fig. 3 B). This interaction was specific as judged by the fact that anti-HA immunoprecipitation of Ugo1-Flag was dependent on the HA-tagged form of Ugo1 (Fig. 3 B, compare lane 2 with lane 4). In addition, analysis of the reciprocal immunoprecipitation using anti-Flag antibodies detected a Ugo1-Flag–Ugo1-HA interaction, further supporting our conclusion that Ugo1 forms a dimer in the mitochondrial outer membrane (Fig. 3 B, lane 6).
Figure 3.
Native Ugo1 forms a 115-kD homooligomer that requires the conserved ETMs. (A) Mitochondria isolated from Δugo1 plus empty vector or Δugo1 expressing either UGO1, Ugo1D134R, Ugo1R137D, Ugo1D312R, or Ugo1R315D were solubilized in 1.5% digitonin and analyzed by hrCN-PAGE and immunodetection with Ugo1 polyclonal antiserum. Small amounts of nonspecific bands detected by the sera are visible on the blots (single arrows) in addition to the Ugo1 species (double arrow). (B) Immunoprecipitations were performed with either α-HA monoclonal antibodies (left) or α-Flag monoclonal antibodies (right) on 125 µg chemically cross-linked mitochondria isolated from a UGO1-Flag strain or UGO1-Flag strain expressing Ugo1-HA. Proteins were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. Total (T) represents 2.5% of the input, and pellet (P) represents 25% of the immunoprecipitated protein. (C) To examine protein stability, the digitonin-solubilized mitochondria in A (10 µg) were separated by SDS-PAGE and visualized by Western analysis with α-Ugo1 or α-porin. The graph represents the quantification of digitonin lysate Ugo1 total protein level normalized to porin as detected by SDS-PAGE analysis versus that detected in the 115-kD Ugo1 species resolved by hrCN-PAGE analysis. Quantification was performed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
To test the possibility that the ETMs participate in the formation of the 115-kD Ugo1-containing complex, we examined Ugo1 native structure in four charge reversal Ugo1 point mutants: D134R, R137D, D312R, and R315D. To examine Ugo1 quaternary structure, mitochondria from strains expressing Ugo1D134R, Ugo1R137D, Ugo1D312R, or Ugo1R315D were solubilized and analyzed by hrCN-PAGE and Western blotting with anti-Ugo1 polyclonal antibodies. This analysis revealed that, as compared with Ugo1 in wild-type mitochondria, all four mutations cause a decrease in the abundance of 115-kD Ugo1 species (Fig. 3 A, lanes 3–6). Mutations in the first ETM, Ugo1D134R and Ugo1R137D, caused a less severe decrease in the abundance of the 115-kD Ugo1 species compared with the equivalent mutations in the second motif, Ugo1D312R and Ugo1R315D, which is consistent with the report that fusion activity in vivo is more dependent on the second ETM (Fig. 3 A, compare lanes 3 and 4 with lanes 5 and 6; Coonrod et al., 2007). To rule out the possibility that mutations in ETMs simply caused a decrease in the steady-state level of Ugo1, the same samples that were analyzed by hrCN-PAGE were also analyzed by SDS-PAGE to resolve Ugo1 monomers. In contrast to our observations with hrCN-PAGE, Western analysis of SDS-PAGE indicates that all mutant Ugo1 protein levels were comparable with those observed in wild-type cells (Fig. 3 C). To further test our conclusion that the charge reversal mutations in the second ETM destabilized the Ugo1 dimer, we examined the hydrodynamic properties of digitonin-solubilized mitochondrial extracts by sucrose gradient centrifugation. This technique is likely to be less stringent than hrCN-PAGE; thus, we compared wild type to Ugo1D312R/R315D, which contains both charge reversals in the critical second ETM and cannot support fusion in vivo and is thus more severe than the single charge reversals (Coonrod et al., 2007). As expected, similar to the Ugo1D312R and Ugo1R315D single mutants, analysis of Ugo1D312R/R315D mitochondria by hrCN-PAGE demonstrated that a 115-kD Ugo1 species is absent. Consistently, analysis of the hydrodynamic properties of Ugo1 by sucrose gradient centrifugation indicated that the sedimentation profile of Ugo1D312R/R315D is distinct from wild type as is the calculated sedimentation coefficient for Ugo1D312R/R315D (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200809099/DC1). Specifically, we observed that the sedimentation coefficient calculated using standards for Ugo1D312R/R315D was smaller than wild-type Ugo1, whereas the sedimentation coefficients for the internal control porin were comparable from extracts prepared from Ugo1D312R/R315D and Ugo1 mitochondria (Fig. S1). Collectively, these data are consistent with a model in which Ugo1 exists as a dimer by virtue of intermolecular salt bridges formed by charged residues in ETMs in its first and third TMDs.
Ugo1 is distinctly required for both outer and inner mitochondrial membrane fusion
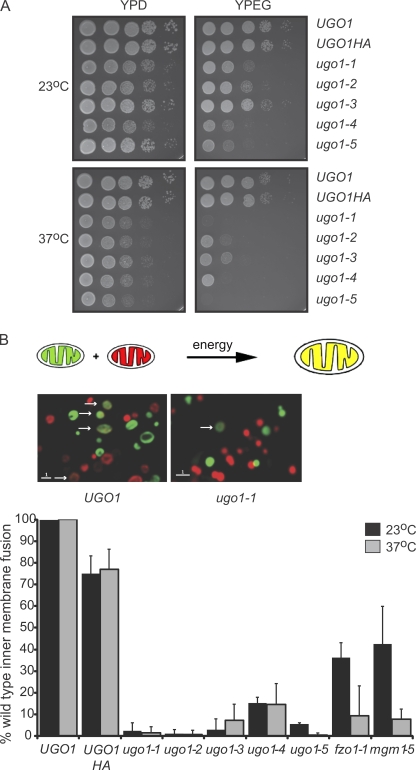
To determine whether Ugo1 plays a direct role in mitochondrial fusion, conditional temperature-sensitive alleles of UGO1 were isolated. To create and screen for ugo1ts alleles, we amplified the UGO1-HA gene by PCR under low fidelity DNA polymerase conditions and integrated mutagenized ugo1-HA into the genome of the wild-type yeast strain W303 at the UGO1 locus. To identify ugo1-HAts strains, transformants were screened for specific growth defects on the fermentable carbon source glycerol at 37°C. Five independent ugo1-HAts strains, displaying the largest differences in growth rate on glycerol at permissive and nonpermissive temperatures, were chosen for further characterization (Fig. 4 A, ugo1-1–ugo1-5). We performed sequence analysis of the UGO1 locus in each of these strains and discovered that each ugo1 allele contains between four and six missense mutations (Table I). Western analysis of SDS-PAGE of total cellular extracts or isolated mitochondria from each ugo1-HAts strain using either anti-HA or anti-Ugo1 polyclonal antibodies indicated that Ugo1 is expressed at levels comparable with wild type in each ugo1 mutant (unpublished data). In addition, analysis of mitochondrial morphology using mitochondria GFP in ugo1ts strains indicated that at the permissive temperature mitochondrial morphology was reticular, which is similar to that observed in UGO1-HA control cells (unpublished data). Upon shifting to the nonpermissive temperature, ugo1ts cells contained mitochondria that were fragmented compared with UGO1-HA control cells, which is consistent with the reported essential role of Ugo1 in mitochondrial fusion (Sesaki and Jensen, 2001; and unpublished data).
Figure 4.
Characterization of temperature-sensitive alleles of Ugo1. (A) Wild-type (UGO1 and UGO1-HA) and ugo1 temperature-sensitive cells (ugo1-1–ugo1-5) were plated on YPD and YPEG plates and incubated at the permissive (23°C) or nonpermissive temperature (37°C). (B, top) A schematic representation of the in vitro inner membrane fusion assay. (middle) Fluorescent images of in vitro mitochondrial fusion reactions with mitochondria isolated from UGO1 and ugo1-1. Arrows indicate fusion events. (bottom) A comparison of mitochondrial in vitro fusion efficiency in UGO1, ugots, fzo1-1, and mgm1-5 mitochondria at the permissive (23°C) and nonpermissive temperature (37°C). Fusion efficiency of ugo1ts mitochondria is expressed as a percentage of wild-type fusion, which averaged 8.5% of total mitochondria. Data are represented as mean ± SEM. Bars, 1 µm.
Table I.
Mutations identified at the UGO1 locus in ugo1ts alleles
| Allele | Mutations identified |
| ugo1-1 | Ser76Pro, Asp153Gly, Asp263Gly, Ile314Thr, Lys448Arg |
| ugo1-2 | Asp263Gly, Phe266Ile, Phe323Ser, Val384Asp, Phe389Ser |
| ugo1-3 | Tyr28Phe, Leu95Pro, Leu298Val, Asn420Asp |
| ugo1-4 | Ile112Thr, The154Ala, Ile262Met, Asn420Tyr |
| ugo1-5 | Glu183Asp, Tyr255His, Lys415Met, Leu418Ser, Asn428Lys |
To pinpoint the function of Ugo1 in mitochondrial fusion, mitochondria isolated from the ugo1ts allele strains were assayed for fusion activity using an in vitro mitochondrial fusion assay (Meeusen et al., 2004, 2006). Specifically, mitochondria were labeled with either matrix-targeted GFP or matrix-targeted Discosoma sp. red fluorescent protein (m-dsRed) and isolated for in vitro analysis as previously described (Meeusen et al., 2004). Outer and inner membrane fusion events were measured by matrix content mixing of the fluorophores as indicated by the colocalization of matrix-targeted GFP and m-dsRed in a single matrix compartment (Fig. 4 B).
As a control, we tested the function of mitochondria containing UGO1-HA in the in vitro fusion assay. We observed only a slight decrease in fusion activity in homoallelic reactions with mitochondria isolated from the UGO1-HA strain as compared with mitochondria isolated from wild-type UGO1 cells, which is consistent with the fact that we detect no apparent fusion defect in UGO1-HA cells in vivo (Fig. 4, A and B; and not depicted). In contrast, in homoallelic in vitro reactions of mitochondria isolated from ugo1ts strains, we observed that matrix content mixing was severely compromised both at permissive and nonpermissive temperatures compared with reactions containing either wild-type or UGO1-HA mitochondria (Fig. 4 B). These observations demonstrate that Ugo1 function is directly required for either mitochondrial outer and/or inner membrane fusion events.
We have previously demonstrated that outer and inner membrane fusion events are separable and have distinct mechanistic and energetic requirements (Meeusen et al., 2004). To separate and stage outer and inner membrane fusion events, we have taken advantage of the fact that mitochondrial fusion in vitro requires two distinct steps: a mixing, centrifugation, and incubation step that supports time-dependent outer membrane fusion only (stage 1 [S1]) and a resuspension step in a buffer solution containing exogenous energy that supports inner membrane fusion and results in complete mitochondrial content mixing (stage 2 [S2]). In S1 and S2 assays with mitochondria isolated from temperature-sensitive mutants of fusion DRPs, we have shown that Fzo1 and Mgm1 are required for outer and inner membrane fusion, respectively. Specifically, the fzo1ts mitochondria are defective for outer membrane fusion (Meeusen et al., 2004). In contrast, outer membrane fusion of mgm1ts mitochondria is unaffected as compared with wild-type mitochondria, but inner membrane fusion is blocked (Meeusen et al., 2006). Outer membrane fusion activity of mitochondria can also be directly assessed using fluorescence microscopy by labeling one population of mitochondria with matrix blue fluorescent protein (BFP) and a second population of mitochondria with m-dsRed and outer membrane GFP (Fig. 5 A). In the fluorescent microscope, outer membrane–fused mitochondria are structures that have nonoverlapping, adjacent red and blue matrices demarcated by a continuous green outer membrane.
Figure 5.
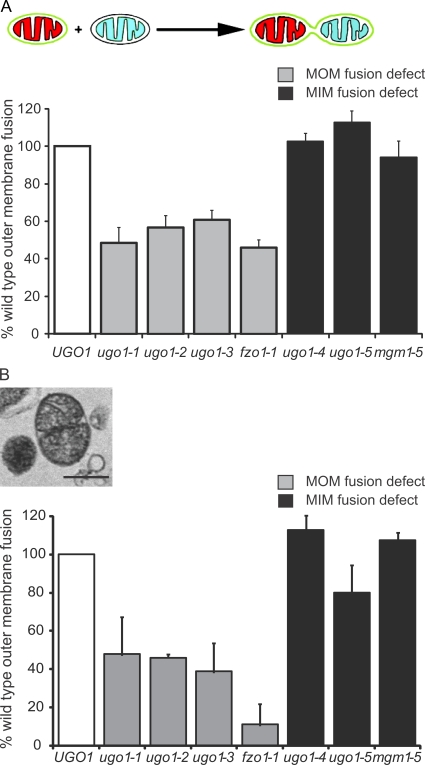
Ugo1 is distinctly required for both mitochondrial outer and inner membrane fusion. (A) A schematic representation of the in vitro mitochondrial outer membrane fusion assay is shown. The graph is a comparison of outer membrane in vitro fusion efficiency at the nonpermissive temperature (37°C) of mitochondria isolated from UGO1, ugo1ts, fzo1-1, and mgm1-5 strains. The two classes of fusion mutants are represented by gray and black bars for mitochondrial outer membrane (MOM) and mitochondrial inner membrane (MIM) fusion deficient, respectively. The absolute values of the mean of three independent experiments (percentage of total mitochondria, n > 350) are UGO1, 5.2; ugo1-1, 2.5; ugo1-2, 2.9; ugo1-3, 3.2; ugo1-4, 5.3; ugo1-5, 5.9; fzo1-1, 2.4; and mgm1-5, 4.9. (B) Representative electron micrograph of outer membrane–fused mitochondria observed in samples subjected to in vitro fusion at the nonpermissive temperature. The graph compares the abundance of outer membrane–fused mitochondria observed in electron micrographs. The mitochondria were isolated from UGO1, ugo1ts, fzo1-1, and mgm1-5 strains and subjected to mitochondrial outer membrane (S1) in vitro fusion conditions at the nonpermissive temperature (37°C). The absolute values of the mean of three independent experiments (percentage of total mitochondria, n > 350) are UGO1, 7; ugo1-1, 3.3; ugo1-2, 3.2; ugo1-3, 2.7; ugo1-4, 7.9; ugo1-5, 5.6; fzo1-1, 0.8; and mgm1-5, 7.5. White bars show wild-type control strains. Data are represented as mean ± SEM. Bar, 0.5 µm.
To determine the specific fusion defect in ugo1ts mitochondria, outer membrane fusion was assessed using our fluorescent-based assay under S1 conditions. Significantly, and in contrast to the distinct outer and inner membrane defects associated with fzo1 and mgm1 mutant mitochondria, respectively, our analysis revealed that ugo1ts mitochondria are defective for either outer or inner membrane fusion in an allele-specific manner. Specifically, we observed that ugo1-1, ugo1-2, and ugo1-3 mitochondria are defective for outer membrane fusion to a similar extent as that observed for fzo1-1 mitochondria (Fig. 5 A, gray bars). In contrast, we observed that ugo1-4 and ugo1-5 mitochondria are competent for outer membrane fusion but are specifically blocked for inner membrane fusion, which is similar to that observed for mgm1-5 mitochondria (Fig. 5 A, black bars).
To further substantiate these observations, EM analysis was performed on mitochondria from ugo1ts fusion reactions. We have previously observed S1 reactions by EM analysis and can distinguish outer membrane–fused mitochondria (Meeusen et al., 2004, 2006). These structures contain two or more matrices whose inner membranes are tightly aligned along an interface, surrounded by a single outer membrane that is also tightly aligned with adjacent inner membrane (Fig. 5 B). Analysis of S1 ugo1ts mitochondria by EM confirmed our observations from the fluorescent outer membrane fusion assay. Specifically, the amount of outer membrane–fused structures observed in ugo1-4 and ugo1-5 reactions was comparable with the amount observed in wild-type and mgm1-5 reactions, indicating that outer membrane fusion is unaffected (Fig. 5 B, black bars). In contrast, the amount of outer membrane–fused structures in ugo1-1, ugo1-2, and ugo1-3 reactions was similar to that observed for fzo1-1 reactions and was significantly less than the amount of outer membrane–fused structures observed in wild-type reactions (Fig. 5 B, gray bars). Therefore, unlike the DRP fusion proteins Fzo1 and Mgm1, which are required for either outer or inner membrane fusion, Ugo1 function is distinctly required for both outer and inner membrane fusion events.
Ugo1 is required for lipid mixing but not outer or inner membrane tethering
Homotypic trans-interactions of fusion DRPs are thought to be important for both outer and inner membrane tethering events that precede membrane fusion (Hoppins et al., 2007). Under S1 assay conditions, we have previously observed Mgm1-dependent inner membrane tethering by EM analysis (Meeusen et al., 2006). Specifically, we found that in outer membrane–fused pairs of some mgm1 mutants, inner membranes from separate matrices were grossly dissociated rather than tightly aligned as observed in wild-type mitochondria.
Trans-interactions between Fzo1 or its mammalian orthologues Mfn1 and Mfn2 are also likely important for outer membrane tethering, but outer membrane–tethered intermediates of wild-type mitochondria have not been observed. We have previously shown in vitro that Fzo1 is required on both mitochondrial partners for efficient outer membrane fusion (Meeusen et al., 2004). Also, mutations in the C-terminal heptad repeat of Mfn2 block mitochondrial fusion and cause an accumulation of aggregated mitochondria in mammalian cells (Koshiba et al., 2004). The spacing between mitochondria within these aggregates is comparable with the dimensions of the antiparallel dimer structure formed by two C-terminal heptad repeat regions in vitro, which is consistent with the hypothesis that they represent stalled Mfn1/2-tethered organelles (Koshiba et al., 2004).
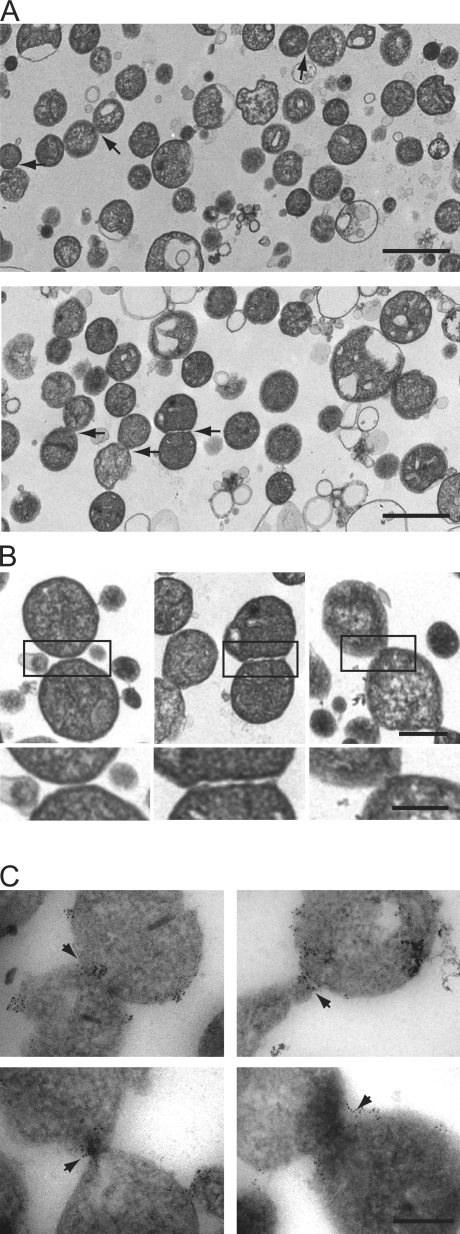
We looked for outer membrane–tethered structures that were functionally relevant for mitochondrial fusion in our yeast in vitro assay under S1 conditions. Under S1 conditions with wild-type mitochondria, EM analysis revealed a population of closely associated mitochondria (22%, n = 350; Fig. 6 A). To discern structures that were engaged from those that were touching indiscriminately, we counted only those with distinct morphological features consistent with membrane-tethered intermediates (4.5%, n = 350; Fig. 6 A). Specifically, in regions of mitochondrial association, we looked for the outer membranes to be reciprocally deformed and evenly spaced apart with an electron-dense region between them (Fig. 6, A [arrows] and B [boxes]). These structures were less abundant under S2 conditions that promote both outer and inner membrane fusion, which is consistent with the interpretation that they are productive intermediates (1.6%, n = 400). To further determine whether these structures are bona fide outer membrane–tethered intermediates, we localized Fzo1 by immuno-EM analysis of wild-type mitochondria under S1 assay conditions. Consistent with its proposed role in outer membrane tethering, we observed that Fzo1 is localized on the outer membrane and is enriched at the interface of adjacent tethered mitochondria (Fig. 6 C, gold particles). In addition, S1 reactions containing fzo1-1 and wild-type mitochondria treated with the nonhydrolyzable GTP analogue GMP-PCP (β,γ-methyleneguanosine 5′-triphosphate) accumulated outer membrane–tethered structures to levels greater than those observed in wild-type S1 reactions, which is consistent with being blocked at a step preceding outer membrane fusion (Fig. 7 A; Meeusen et al., 2004). In contrast, treatment of wild-type mitochondria with the protease trypsin significantly decreased the amount of outer membrane–tethered structures observed (Fig. 7 A). Collectively, these observations indicate that we have identified outer membrane–tethered intermediates in the pathway of fusion and are consistent with the proposed role of Fzo1 in outer membrane tethering.
Figure 6.
Identification of mitochondrial outer membrane fusion intermediates. (A) Analysis of outer membrane tethering. Representative electron micrograph fields of wild-type (top) and ugo1-2 (bottom) mitochondria subjected to in vitro outer membrane fusion at the nonpermissive temperature are shown. Tethered mitochondria are indicated by arrows. (B) Representative images of outer membrane–tethered mitochondria from analysis described in A. The boxed regions are enlarged below. Bars: (top) 0.5 µm; and (bottom) 0.25 µm. (C) Immuno-EM analysis of Fzo1 localization in wild-type mitochondria subjected to S1 in vitro fusion conditions in the presence of the nonhydrolysable GTP analogue GMP-PCP. Arrows indicate the interface at the outer membrane formed by two mitochondria, decorated Fzo1-specific immunogold particles. (A and C) Bars, 0.5 µm.
Figure 7.
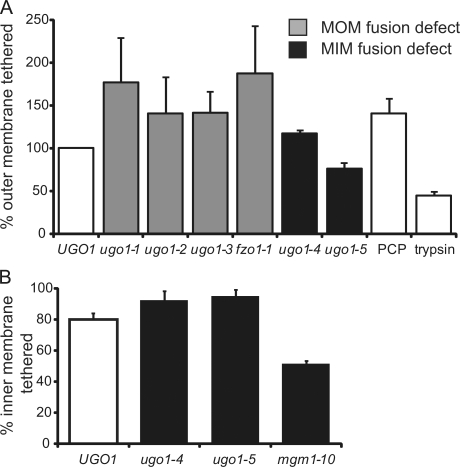
Quantification of mitochondrial outer and inner membrane tethering efficiency. (A) Comparison of the abundance of tethered mitochondria observed in mitochondria isolated from UGO1, ugo1ts, and fzo1-1 strains subjected to S1 in vitro fusion conditions. Wild-type mitochondria were also either treated with the nonhydrolysable GTP derivative PCP or pretreated with trypsin to remove cytosolic portions of outer membrane proteins before S1 in vitro fusion conditions. The absolute values of the mean of at least two independent experiments (percentage of total mitochondria, n > 350) are UGO1, 4.5; ugo1-1, 7.9; ugo1-2, 6.3; ugo1-3, 6.3; ugo1-4, 5.3; ugo1-5, 3.4; fzo1-1, 8.4; PCP, 6.3; and trypsinized, 1.98. (B) Comparison of the abundance of tight inner membrane interfaces in outer membrane–fused structure observed in the in vitro fusion mitochondria isolated from UGO1, ugo1-4, ugo1-5, and mgm1-10 strains. The absolute values of the mean of at least two independent experiments (percentage of total mitochondria, n > 350) are UGO1, 80; ugo1-4, 100; ugo1-5, 100; and mgm1-10, 51. White bars show wild-type control strains. MOM, mitochondrial outer membrane; MIM, mitochondrial inner membrane. Data are represented as mean ± SEM.
To pinpoint the role of Ugo1 in outer and inner membrane fusion events, we quantified populations of outer and inner membrane–tethered structures in reactions containing ugo1ts mitochondria by EM (Fig. 7, A and B). Interestingly, although the amount of outer and inner membrane fusion is significantly affected by Ugo1 function in an allele-specific manner, the levels of outer membrane–tethered structures were similar to that observed for wild-type mitochondria for all ugo1ts reactions analyzed (Fig. 7 A). We also found that inner membrane tethering efficiency in ugo1ts alleles blocked for inner membrane fusion (ugo1-4 and ugo1-5) was comparable with wild type in contrast to mgm1-10, which is a mutant previously shown to have a defect in inner membrane tethering (Fig. 7 B; Meeusen et al., 2006). This indicates that the outer and inner membrane fusion defects observed in the ugo1ts alleles are not caused by gross defects in membrane tethering but instead are likely to be a consequence of a defect in a later step required for membrane mixing and fusion.
Ugo1 is not required on both mitochondrial partners
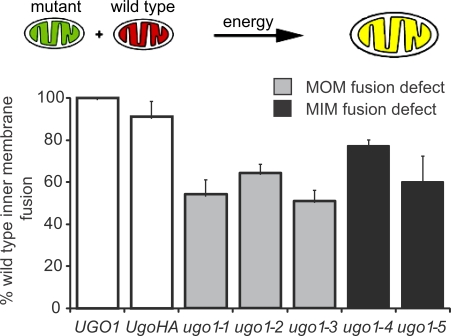
To gain further insight into the role of Ugo1 in fusion, we tested the in vitro fusion efficiency of the mutants in heteroallelic reactions containing equal amounts of mitochondria isolated from UGO1 and temperature-sensitive ugo1 cells. We observed that in these heteroallelic reactions, the fusion activity is increased considerably compared with the levels observed in homoallelic reactions (compare Fig. 3 with Fig. 8). This is in contrast to heteroallelic reactions containing wild-type and either mgm1 or fzo1 mitochondria in which mitochondrial fusion activity remains significantly below the activity observed in comparable reactions containing wild-type mitochondria, which is consistent with the proposal that trans-interactions of fusion DRPs are important for membrane tethering and fusion (Meeusen et al., 2004, 2006). In contrast, our data indicate that Ugo1 function is required only on one mitochondrial partner to fulfill its role in fusion, which is consistent with our observation that Ugo1 is not required for outer or inner membrane tethering events. Our characterization of Ugo1 supports a model in which it functions in cis at both the outer and inner membranes together with the fusion DRPs at a postmembrane tethering step to facilitate lipid mixing of each mitochondrial membrane.
Figure 8.
Ugo1 is not required on both mitochondrial partners for efficient mitochondrial fusion. A schematic representation of the in vitro inner membrane fusion assay with heterologous reactions is shown. The graph represents a comparison of in vitro mitochondrial fusion efficiency in UGO1 × UGO1, UGO1 × UGO1-HA, and UGO1 × ugo1ts heterologous reactions at the nonpermissive temperature (37°C). The absolute values of the mean at three independent experiments (percentage of total mitochondria, n > 350) are UGO1, 8.5; UGO1-HA, 7.7; ugo1-1, 4.6; ugo1-2, 5.5; ugo1-3, 4.3; ugo1-4, 6.6; and ugo1-5, 5.1. White bars show wild-type control strains. MOM, mitochondrial outer membrane; MIM, mitochondrial inner membrane. Data are represented as mean ± SEM.
Ugo1 mutations destabilize the Ugo1 dimer but have no apparent effect on Ugo1–DRP interactions
Ugo1 has been shown to interact independently with both the outer and inner membrane fusion DRPs, which has led to the proposal that it may act as a physical adapter and coordinate outer and inner membrane fusion events. Thus, we tested the ability of the temperature-sensitive mutant Ugo1 proteins to interact with Fzo1 and Mgm1. Immunoprecipitation of Ugo1 was performed on chemically cross-linked mitochondria isolated from ugo1ts strains using anti-HA antibodies. After immunoprecipitation, cross-links were reversed with reducing agents and precipitates were analyzed by SDS-PAGE and Western blotting. This analysis revealed that, in each case, Fzo1 and Mgm1 were observed in mutant Ugo1 protein immunoprecipitates (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200809099/DC1). In contrast, the outer membrane protein porin was absent in immunoprecipitates, demonstrating that the interactions were specific. These data show that Ugo1 mutants defective for either outer or inner membrane fusion are capable of interacting with the fusion DRPs. However, analysis of these interactions by immunoprecipitation may not detect changes in their affinity or any interaction-dependent conformational changes in these proteins that are functionally relevant.
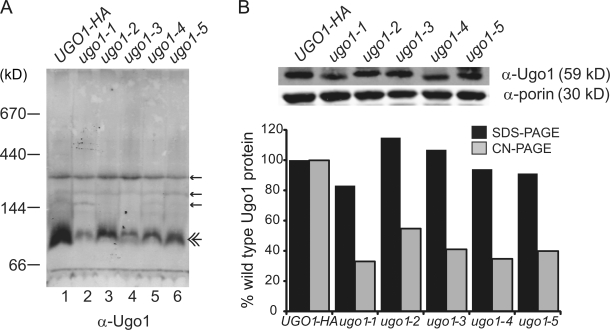
Our structural analysis of Ugo1 suggests that it exists as a dimer that is functionally important. Thus, we examined whether our temperature-sensitive mutations in UGO1 altered the stability of the 115-kD Ugo1 observed by hrCN-PAGE. An analysis of mitochondria isolated from ugo1ts cells on hrCN-PAGE indicated that the abundance of the 115-kD Ugo1 species was decreased in all cases, although most severely in ugo1-1, which is consistent with the fact that the I314T mutation in this allele is located in the critical second ETM (Fig. 9). These data are consistent with a model in which the Ugo1 dimer is fundamentally important for Ugo1's role in both mitochondrial outer and inner membrane fusion.
Figure 9.
Mutations in Ugo1 destabilize the dimer. (A) The stability of the Ugo1 115-kD species is affected in the ugo1ts alleles. Mitochondria isolated from UGO1-HA and the ugo1ts strains were solubilized in 2% digitonin and analyzed by hrCN-PAGE and Western analysis with Ugo1 polyclonal antiserum. Small amounts of nonspecific bands are visible on the blots (single arrows) in addition to the Ugo1 species (double arrow). (B) To examine protein stability, the digitonin-solubilized mitochondria in A (10 µg) were separated by SDS-PAGE and visualized by Western analysis with α-Ugo1 or α-porin. The graph represents the quantification of digitonin lysate Ugo1 protein normalized to porin as detected by SDS-PAGE analysis versus that detected in the 115-kD Ugo1 species detected by hrCN-PAGE analysis. Quantification was performed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Discussion
This work defines the topology Ugo1 as a three membrane–spanning protein that places the two more highly conserved ETMs at the membrane, which is consistent with Ugo1's classification as a member of the mitochondrial transport protein family. It seems likely that this modified structure, in comparison with typical transport protein family members, evolved to tailor the structure of Ugo1 to its function in mitochondrial fusion. Specifically, at the cost of additional TMDs, Ugo1 contains a large IMS domain at the C terminus that is likely important for its interaction with Mgm1 at the inner membrane and/or its role in membrane fusion.
Analysis of the role of ETMs in Ugo1 structure indicates that they participate in the formation of a Ugo1 115-kD species that is likely a homodimer. We postulate that ETMs participate in dimer formation via an intermolecular salt bridge network. This is in contrast to the mitochondrial transport protein family in which the motifs take part in intramolecular interactions (Pebay-Peyroula et al., 2003). Interestingly, a Ugo1 dimer contains six TMDs, which is a feature common to members of the mitochondrial transport protein family, albeit with a different predicted structural symmetry. Thus, although Ugo1 is a divergent member of this family, the basic function of ETMs may be similar in that they are structurally important for the formation of a six TMD unit. The effect of the charge reversal mutations within the first and second ETMs on Ugo1 function as scored by mitochondrial morphology correlate with dimer stability, but importantly, they differ in severity. Thus, in the simplest model for the dimer formation, the interface would be comprised of trans-interactions between identical motifs in each Ugo1 partner. In conventional mitochondrial transport proteins, the salt bridges lock the TMDs together on the matrix side of the inner membrane, and these must be broken for a substrate to be transported. Thus, it is also possible that Ugo1 dimer formation is dynamic and in this capacity may facilitate postmembrane tethering fusion events.
A role for Ugo1 in the fusion of both outer and inner membranes is supported by our discovery of ugo1 mutations that selectively block each fusion event. This observation raises the question of how an outer membrane protein can affect both outer and inner membrane fusion events. The interaction of Ugo1 with both Mgm1 and Fzo1 suggests a simple model in which Ugo1 functions as an adapter to coordinate fusion. However, given that loss of function does not correlate with loss of the interaction of Ugo1 with Fzo1 and/or Mgm1, our data suggest that Ugo1 plays a role more complex than that of a simple adapter. Although the role of Ugo1 in outer and inner membrane fusion is genetically resolvable, mutations in both phenotypic classes cause the destabilization of the Ugo1 115-kD species, suggesting that the Ugo1 dimer stability is important but not sufficient in each fusion event. However, in this context, it should be noted that hrCN-PAGE analysis is stringent and thus likely underestimates the steady-state level of the Ugo1 115-kD species in vivo.
Our data also indicate that Ugo1 function is required only after membrane tethering has occurred, likely at the lipid-mixing step of both outer and inner membrane fusion. At present, we can only speculate on the exact mechanistic role of Ugo1 in fusion. One rational possibility is that Ugo1 functions by regulating outer and inner membrane fusion DRP assembly and/or disassembly, which are activities critical for mitochondrial fusion. In this manner, Ugo1 may also temporally and spatially couple the fusion of the outer and inner membrane. Although immunoprecipitation data indicate that Ugots proteins are capable of interacting with Fzo1 and Mgm1, higher resolution assays are required to determine whether these mutant proteins are defective in the formation of potentially transient assembled Fzo1 and Mgm1 structures at sites of fusion. A second possibility for Ugo1's function in fusion is suggested by the transporter-like structure of Ugo1. Specifically, Ugo1 could function as more than a dynamic DRP scaffold during fusion and may, by an unknown transport event, facilitate lipid mixing by more directly modulating the lipid and/or chemical environment at the site of fusion.
Although there is no readily identifiable mammalian orthologue to Ugo1 at present, there are ∼50 members of the transport/carrier protein family in humans, raising the possibility that one exists. Indeed, based on immunoprecipitation analysis, the mammalian outer and inner membrane fusion DRPs, Mfn1 or Mfn2 and Opa1, are present in a complex in mammalian mitochondria (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200809099/DC1; Guillery et al., 2008). Like Fzo1, Mfn1 and Mfn2 possess only a very small loop of amino acids in IMS, which is unlikely to be sufficient to directly mediate a stable interaction with OPA1 at the inner membrane. Indeed, in yeast, the Fzo1–Mgm1 interaction is dependent on Ugo1 (Sesaki et al., 2003). Thus, the existence of an Mfn1/2–Opa1 complex suggests that a mammalian functional, if not structural, equivalent of Ugo1 exists and that the fundamental mechanism of mitochondrial fusion is highly conserved.
Materials and methods
Antibody production and purification
Antiserum was raised against His6-tagged fusion proteins comprised of full-length mouse dihydrofolate reductase and the N terminus of Ugo1 (residues 1–125) or an internal region of Fzo1 (residues 422–516; Covance). Fusion proteins were purified on nickel nitrilotriacetic acid columns (QIAGEN) in 8 M urea according to the manufacturer's instructions except that the proteins were eluted in 0.1% SDS and 10 mM Tris-Cl, pH 7.4.
Generation of UGO1 internal HA tags and point mutants
To construct the Ugo1 internal HA tags, Ugo1 was amplified in two fragments with XbaI and XhoI sites at the 5′ and 3′ ends, respectively. The 5′ PCR fragment included ∼500 base pairs of promoter sequence and the coding sequence up to the internal tag site (amino acid 393 or 438). The 3′ fragment included ugo1 coding sequence 3′ of the internal tag site and ∼500 base pairs of terminator sequence. The internal primers also each encode one half of the triple HA sequence with homologous sticky ends for ligation. These fragments were simultaneously ligated into pRS315 digested with XbaI and XhoI, and plasmid products with both fragments were sequenced to confirm correct insertion of the triple HA tag sequence.
The Ugo1 point mutations (Ugo1D134R, Ugo1R137D, Ugo1D312R, or Ugo1R315D) were created by site-directed mutagenesis of a genomic copy of Ugo1 (cloned into pRS315 XbaI and XhoI sites) using a complementary primer PCR method with primers of 20–30 nucleotides on either side of the altered nucleotides. Whole plasmid amplification was performed, and PCR products were digested with DpnI for 2 h at 37°C to remove template DNA. The amplified plasmid DNA was transformed into DH5-α cells. Plasmid DNA was isolated from selected colonies and sequenced to confirm mutations.
Protease protection analysis
Mitochondria (∼50 µg of total mitochondrial protein) were resuspended in 500 µl of NMIB buffer (0.6 M sorbitol, 5 mM MgCl2, 50 mM KCl, 0.1 M KOAc, and 20 mM Hepes-KOH, pH 7.4), mitoplast buffer (20 mM Hepes-KOH, pH 7.4), or solubilizing buffer (0.6 M sorbitol, 5 mM MgCl2, 50 mM KCl, 0.1 M KOAc, 20 mM Hepes-KOH, pH 7.4, and 1% Triton X-100). After a 15-min incubation on ice, outer mitochondrial membranes in the mitoplast samples were disrupted by gently pipetting up and down 15 times. Trypsin was added to the indicated samples to a final concentration of 100 µg/ml, and samples were incubated on ice for 15 min. The reaction was stopped by adding 2 mM PMSF and incubating on ice for 5 min. Mitochondria from the supernatant were collected by centrifugation at 16,000 g for 10 min at 4°C. The pellets of the intact mitochondria were resuspended in NMIB containing 2 mM PMSF, and all protein, including solubilized samples and mitoplast supernatants, were subjected to precipitation by the addition of 12.5% TCA. The proteins were pelleted by centrifugation at 16,000 g for 10 min at 4°C, washed with acetone, dried, and resuspended in SDS sample buffer. The samples were analyzed by SDS-PAGE and Western blotting. The following antibodies were used: mouse α-HA (1:1,000; Covance), rabbit α-Ugo1 (1:250; Covance), rabbit α-Fzo1 (1:2,000; Covance), α-Tim23 and α-cyt b2 (1:10,000; provided by C. Koehler), and α-Abf2 (1:1,000; Meeusen et al., 1999). For all Western blots, α–rabbit or α–mouse secondary antibodies were used at 1:10,000.
Western blot analysis and quantification
Proteins transferred to nitrocellulose or PVDF were detected using primary rabbit or mouse antibodies and visualized with the appropriate secondary antibodies conjugated to either HRP (SouthernBiotech) or IRDye (800CW; LI-COR Biosciences). Quantification was performed using the Odyssey Infrared Imaging System (LI-COR Biosciences).
Clear native gel electrophoresis
For analyzing the oligomeric state of Ugo1, 100 µg mitochondria was lysed in digitonin buffer (2% [wt/vol] digitonin, 50 mM NaCl, 50 mM imidizole-HCl, pH 7.0, 2 mM 6-aminohexanoic acid, 1 mM EDTA, and 1 mM PMSF) for 15 min on ice and centrifuged at 16,000 g for 30 min at 4°C. hrCN-PAGE sample buffer (0.1% Ponceau S and 50% glycerol) was added, and the extract was separated on a 4–13% or 4–15% polyacrylamide gradient gel at 4°C. The molecular mass standards for thyroglobulin (670 kD), apoferritin (440 kD), alcohol dehydrogenase (144 kD), and bovine serum albumin (66 kD) were used to generate a standard curve and estimate the size of native Ugo1.
Sucrose gradient sedimentation
For analyzing the hydrodynamic state of Ugo1, 250 µg mitochondria was lysed in digitonin buffer (1% [wt/vol] digitonin, 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 1 mM EDTA, and 1 mM PMSF) for 15 min on ice and centrifuged at 30,000 g for 30 min (TLA55; Sorvall) at 4°C. The supernatant was layered on top of a 5–20% sucrose gradient followed by centrifugation at 200,000 g at 4°C (SW55 Ti; Sorvall) for 4.5 h. Molecular mass markers with known sedimentation coefficients (GE Healthcare) that were simultaneously run over separate sucrose gradients were used to construct standard curves of fraction number versus sedimentation coefficient and to estimate the Svedberg coefficients of Ugo1 and Ugo1D312R/R315D.
Generation and characterization of ugo1 temperature-sensitive alleles
The ugo1 temperature-sensitive strains with C-terminal HA epitope tags were created by a PCR-mediated technique (Longtine et al., 1998). The mutant alleles were generated by mutagenic PCR of the entire coding region, and these were amplified with an HA tag and kanamycin resistance cassette from pFA6-3HA-kanMX6 by splicing by overlap extension PCR (Horton et al., 1990; Longtine et al., 1998). The resultant PCR products were transformed into wild-type W303 haploids by the lithium acetate method, and transformants were plated on YPD (yeast extract, peptone, and dextrose) + 300 µg/ml Geneticin (Invitrogen) to select for the homologous recombination event, resulting in the replacement of the endogenous UGO1 locus. Temperature-sensitive strains were screened for a growth defect on nonfermentable YPEG (yeast extract, peptone, ethanol, and glycerol) media at elevated temperatures (37°C) compared with the permissive temperature (23°C). Based on this growth assay, five strains were selected for further analysis. These strains were backcrossed to wild type twice, and segregation of the temperature-sensitive phenotype was confirmed to be 2:2. To identify the mutations within the ugo1ts alleles, the UGO1 locus was amplified by PCR from ugo1ts genomic DNA, and the resulting PCR products were sequenced (College of Biological Sciences Sequencing Facility, University of California, Davis, Davis, CA).
Preparation of fusion-competent mitochondria and in vitro mitochondrial fusion
Enriched mitochondrial fractions were prepared from wild-type and temperature-sensitive strains as in Meeusen et al. (2004) with the following exceptions: cell growth and subfractionation were conducted at 23°C, and spheroplasting was performed with 4 mg/ml Zymolyase 20T (MP Biomedicals) for 60 min in 1.2 M sorbitol. Inner and outer membrane fusion assays were conducted as described in Meeusen et al. (2004) except for the following changes: (a) The energetic components of the S2 reaction mix include 1.2 mg/ml creatine phosphokinase, 40 mM creatine phosphate, and 1.5 mM GTP; (b) matrix-targeted BFP was replaced with the enhanced BFP variant azurite (Mena et al., 2006); and (c) S1 or S2 reactions were performed at either 23 or 37°C, as indicated in Figs. 4–8. Each experiment was performed three times, and >350 mitochondria were analyzed for each experimental condition.
Analysis of fusion by light microscopy
Mitochondria were mounted in 3% low melt agarose in NMIB and viewed at room temperature with a microscope (IX70 Deltavision; Olympus) using a 60× 1.4 NA objective (Olympus) and a 100-W mercury lamp (Applied Precision, LLC). Two- and three-dimensional light microscopy data were collected using an integrated, cooled charge-coupled device–based camera (MicroMax; Princeton) equipped with an interline chip (Sony). Three-dimensional datasets were processed using DeltaVision's iterative, constrained three-dimensional deconvolution method to remove out of focus light. Deconvolved images were analyzed in SoftWorx (Applied Precision, LLC).
Analysis of fusion intermediates by EM
S1 fusion reactions were analyzed using conventional EM as described previously (Meeusen et al., 2004), and >300 mitochondria were analyzed for each experimental condition. Analysis of Fzo1 localization in S1 fusion intermediates was conducted with Fzo1 polyclonal antibody generated to amino acids 422–516 (Covance).
Yeast mitochondrial cross-linking and immunoprecipitation
Mitochondria were isolated from ugo1ts allele strains or the Ugo1-HA/Flag strain and resuspended in NMIB (Meeusen et al., 2004) + protease inhibitor cocktail (EMD). 150 µg mitochondria was subjected to chemical cross-linking with 1 mM dithiobis (succinimidyl propionate; Thermo Fisher Scientific) for 1 h on ice. Cross-linker was quenched by the addition of 100 mM glycine, pH 8.0, for 10 min on ice followed by protein precipitation with 10% TCA. Proteins were denatured and solubilized in 100 µl MURB (100 mM MES, pH 7, 1% SDS, and 3 M urea) followed by the addition of 900 µl TWIP (50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5% Tween 20, and 0.1% EDTA). Insoluble proteins were removed by centrifugation at 16,000 g at 4°C. Lysate was preadsorbed with 75 µl TWIP-equilibriated protein A agarose beads (Santa Cruz Biotechnology, Inc.) for 30 min at 4°C and incubated with 8 µl monoclonal HA antibody (Covance) for 1.5 h at 4°C followed by incubation with 75 µl TWIP-equilibriated protein A agarose beads for 45 min at 4°C. Agarose beads were washed three times with 500 µl TWIP, and protein was eluted with MURB + 10% β-mercaptoethanol to reduce cross-links. Analysis by SDS-PAGE and Western blotting was performed on the total fraction and the eluate fractions with the following antibodies: rabbit α-Ugo1 (1:250; Covance), rabbit α-Mgm1 (1:2,000; Covance), rabbit α-Fzo1 (1:2,000; Covance), and α-porin (1:1,000; Invitrogen). For all Western blots, α–rabbit or α–mouse secondary antibodies were used at 1:10,000.
Mammalian mitochondrial isolation and coimmunoprecipitation
Mouse embryonic fibroblasts were plated in a 150-mm dish (25 ml) 24 h before transfection, and 50 µg of total DNA was transfected per plate. To isolate mitochondria, cells were collected 48 h after transfection by cell lifter, washed once with NMIB (0.6 M sorbitol, 5 mM MgCl2, 50 mM KCl, 100 mM KOAc, and 20 mM Hepes, pH 7.4), and homogenized with 100 strokes in a glass-glass dounce (Wheaton) until lysis was ∼60% (approximated by phase-contrast microscopy). Cell debris was separated to the pellet with an 800 g 5-min centrifuge spin, and mitochondria were isolated from the supernatant by centrifugation at 10,000 g for 15 min at 4°C and were subjected to chemical cross-linking with 1 mM dithiobis (succinimidyl propionate; Thermo Fisher Scientific) for 1 h on ice. Cross-linker was quenched by the addition of 100 mM glycine, pH 8.0, for 10 min on ice. Mitochondria were lysed in IPS1 (0.2% SDS, 20 mM Tris, pH 7.52, 150 mM NaCl, and 1% Triton X-100) at 0°C for 30 min, and nonsoluble material was removed by ultracentrifugation at 100,000 g for 30 min at 4°C. Soluble mitochondrial proteins were immunoprecipitated with Flag affinity gel according to the manufacturer's instructions (ANTI-FLAG M2; Sigma-Aldrich), and immunoprecipitated proteins were eluted in 80 µl MURB (100 mM MES, pH 7, 1% SDS, 3 M urea, and 5% β-mercaptoethanol). Analysis by SDS-PAGE and Western blotting was performed on the total fraction and the eluate fractions with the following antibodies: mouse monoclonal α-OPA1 (BD), mouse monoclonal α-Flag antibody (Sigma-Aldrich), and rabbit polyclonal α–voltage-dependent anion channel (porin; Abcam).
Online supplemental material
Fig. S1 shows the hydrodynamic properties of Ugo1 and Ugo1D312R/R315D as determined by sucrose gradient sedimentation. Fig. S2 shows coimmunoprecipitation of Fzo1 and Mgm1 with Ugo1 and ugots alleles. Fig. S3 shows coimmunoprecipitation of OPA1 with Mfn1 and Mfn2. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200809099/DC1.
Acknowledgments
We thank the members of the Nunnari laboratory for helpful discussions and comments on the manuscript.
This work was supported by a National Institutes of Health grant (GM62942) to J. Nunnari.
Footnotes
Abbreviations used in this paper: DRP, dynamin-related protein; ETM, energy transfer motif; hrCN-PAGE, high resolution clear native PAGE; IMS, intermembrane space; TMD, transmembrane domain.
References
- Belenkiy R., Haefele A., Eisen M.B., Wohlrab H. 2000. The yeast mitochondrial transport proteins: new sequences and consensus residues, lack of direct relation between consensus residues and transmembrane helices, expression patterns of the transport protein genes, and protein-protein interactions with other proteins.Biochim. Biophys. Acta. 1467:207–218 [DOI] [PubMed] [Google Scholar]
- Coonrod E.M., Karren M.A., Shaw J.M. 2007. Ugo1p is a multipass transmembrane protein with a single carrier domain required for mitochondrial fusion.Traffic. 8:500–511 [DOI] [PubMed] [Google Scholar]
- Griffin E.E., Chan D.C. 2006. Domain interactions within Fzo1 oligomers are essential for mitochondrial fusion.J. Biol. Chem. 281:16599–16606 [DOI] [PubMed] [Google Scholar]
- Guillery O., Malka F., Landes T., Guillou E., Blackstone C., Lombes A., Belenguer P., Arnoult D., Rojo M. 2008. Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential.Biol. Cell. 100:315–325 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria.Annu. Rev. Biochem. 76:751–780 [DOI] [PubMed] [Google Scholar]
- Horton R.M., Cai Z.L., Ho S.N., Pease L.R. 1990. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction.Biotechniques. 8:528–535 [PubMed] [Google Scholar]
- Ishihara N., Eura Y., Mihara K. 2004. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity.J. Cell Sci. 117:6535–6546 [DOI] [PubMed] [Google Scholar]
- Koshiba T., Detmer S., Kaiser J., Chen H., McCaffery J., Chan D. 2004. Structural basis of mitochondrial tethering by mitofusin complexes acting in trans.Science. 305:858–862 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., Demarini D., Shah N., Wach A., Brachat A., Philippsen P., Pringle J. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae.Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Meeusen S., Tieu Q., Wong E., Weiss E., Schieltz D., Yates J.R., Nunnari J. 1999. Mgm101p is a novel component of the mitochondrial nucleoid that binds DNA and is required for the repair of oxidatively damaged mitochondrial DNA.J. Cell Biol. 145:291–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeusen S., McCaffery J.M., Nunnari J. 2004. Mitochondrial fusion intermediates revealed in vitro.Science. 305:1747–1752 [DOI] [PubMed] [Google Scholar]
- Meeusen S., Devay R., Block J., Cassidy-Stone A., Wayson S., McCaffery J.M., Nunnari J. 2006. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1.Cell. 127:383–395 [DOI] [PubMed] [Google Scholar]
- Mena M.A., Treynor T.P., Mayo S.L., Daugherty P.S. 2006. Blue fluorescent proteins with enhanced brightness and photostability from a structurally targeted library.Nat. Biotechnol. 24:1569–1571 [DOI] [PubMed] [Google Scholar]
- Nury H., Dahout-Gonzalez C., Trezeguet V., Lauquin G.J.M., Brandolin G., Pebay-Peyroula E. 2006. Relations between structure and function of the mitochondrial ADP/ATP carrier.Annu. Rev. Biochem. 75:713–741 [DOI] [PubMed] [Google Scholar]
- Pebay-Peyroula E., Dahout-Gonzalez C., Kahn R., Trezeguet V., Lauquin G.J., Brandolin G. 2003. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside.Nature. 426:39–44 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. 2001. UGO1 encodes an outer membrane protein required for mitochondrial fusion.J. Cell Biol. 152:1123–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H., Jensen R.E. 2004. Ugo1p links the Fzo1p and Mgm1p GTPases for mitochondrial fusion.J. Biol. Chem. 279:28298–28303 [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S.M., Yaffe M.P., Jensen R.E. 2003. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane.Mol. Biol. Cell. 14:2342–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittig I., Karas M., Schagger H. 2007. High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes.Mol. Cell. Proteomics. 6:1215–1225 [DOI] [PubMed] [Google Scholar]
- Wong E.D., Wagner J.A., Scott S.V., Okreglak V., Holewinske T.J., Cassidy-Stone A., Nunnari J. 2003. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion.J. Cell Biol. 160:303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]