Abstract
Telomeres protect chromosome ends from being viewed as double-strand breaks and from eliciting a DNA damage response. Deprotection of chromosome ends occurs when telomeres become critically short because of replicative attrition or inhibition of TRF2. In this study, we report a novel form of deprotection that occurs exclusively after DNA replication in S/G2 phase of the cell cycle. In cells deficient in the telomeric poly(adenosine diphosphate ribose) polymerase tankyrase 1, sister telomere resolution is blocked. Unexpectedly, cohered sister telomeres become deprotected and are inappropriately fused. In contrast to telomeres rendered dysfunctional by TRF2, which engage in chromatid fusions predominantly between chromatids from different chromosomes (Bailey, S.M., M.N. Cornforth, A. Kurimasa, D.J. Chen, and E.H. Goodwin. 2001. Science. 293:2462–2465; Smogorzewska, A., J. Karlseder, H. Holtgreve-Grez, A. Jauch, and T. de Lange. 2002. Curr. Biol. 12:1635–1644), telomeres rendered dysfunctional by tankyrase 1 engage in chromatid fusions almost exclusively between sister chromatids. We show that cohered sister telomeres are fused by DNA ligase IV–mediated nonhomologous end joining. These results demonstrate that the timely removal of sister telomere cohesion is essential for the formation of a protective structure at chromosome ends after DNA replication in S/G2 phase of the cell cycle.
Introduction
Telomeres protect chromosome ends from being viewed as double-strand breaks and serve as templates for replication by telomerase (for reviews see Verdun and Karlseder, 2007; Palm and de Lange, 2008). Mammalian telomeres consist of tandem arrays of double-stranded TTAGGG repeats that run out to the end of the chromosome as a single-stranded 3′ overhang (Makarov et al., 1997). Telomeres are bound by a six-subunit protein complex termed shelterin (for review see de Lange, 2005) containing two double-stranded DNA-binding proteins: TRF2 (Bilaud et al., 1997; Broccoli et al., 1997), which provides the protective function (van Steensel et al., 1998), and TRF1 (Chong et al., 1995), which regulates telomere length (van Steensel and de Lange, 1997). TRF1 acts in cis to block access of telomerase; inhibition of TRF1 leads to telomere elongation (van Steensel and de Lange, 1997; Ancelin et al., 2002). In contrast, inhibition of TRF2 leads to deprotection of telomeres and end to end fusions (van Steensel et al., 1998). TRF2 promotes a specialized structure at telomeres termed the t loop, in which the single-stranded overhang is buried (and thus protected) in the double-stranded telomere tracts (Griffith et al., 1999). In the absence of TRF2, telomeres become exposed to the nonhomologous end joining (NHEJ) machinery and are inappropriately fused (Smogorzewska et al., 2002; Celli et al., 2006).
Tankyrase 1 is a poly(ADP ribose) polymerase that modifies TRF1, regulating its localization and function at telomeres (Smith et al., 1998; for review see Hsiao and Smith, 2008). Overexpression of tankyrase 1 in nuclei of human cells releases TRF1 (but not TRF2) from telomeres (Smith and de Lange, 2000). Once released from telomeric DNA, TRF1 is ubiquitinated and degraded by the proteasome (Chang et al., 2003), leading to telomere elongation by telomerase (Smith and de Lange, 2000; Cook et al., 2002). Conversely, partial depletion of tankyrase 1 leads to telomere shortening (Donigian and de Lange, 2007), indicating tankyrase 1 as a positive regulator of telomere length.
Tankyrase 1 is also required after DNA replication in S/G2 phase of the cell cycle to resolve sister telomere cohesion before mitosis. Depletion of tankyrase 1 in HeLa cells using siRNA resulted in mitotic arrest (Dynek and Smith, 2004; Chang et al., 2005). In the absence of tankyrase 1, chromosomes aligned normally on the metaphase plate, but sister chromatids were unable to segregate to daughter poles. FISH using chromosome-specific probes revealed that although sister chromatids were separated (appeared as doublets) at their centromeres and along their arms, they remained associated (appeared as singlets) at their telomeres (Dynek and Smith, 2004). Several lines of evidence revealed that this persistent sister telomere association was not the result of covalent ligation but rather the result of protein–protein interactions. Density gradient fractionation of telomeres after BrdU incorporation showed that persistent telomere association was not caused by unreplicated telomeric DNA (Dynek and Smith, 2004). Moreover, persistent sister telomere association did not survive the hypotonic treatment used in metaphase spread preparation (Dynek and Smith, 2004; Canudas et al., 2007). Consistent with this, FISH analysis using chromosome-specific probes showed that telomeric singlets could be resolved into doublets by treatment with hypotonic buffer (Canudas et al., 2007). Together, the data indicate that the persistent sister telomere association in tankyrase 1–deficient cells is caused by protein–protein interactions.
Sister chromatids are held together from the time of their replication in S phase until their separation in mitosis by cohesin, a multisubunit ring-shaped complex comprised of Smc1, Smc3, Scc1, and Scc3 (which exists as two orthologues, SA1 or SA2, in vertebrate cells; for reviews see Losada and Hirano, 2005; Nasmyth and Haering, 2005). Biochemical and genetic analyses indicated that the persistent sister telomere association observed in tankyrase 1–depleted cells was mediated by protein–protein interactions between the SA1 cohesin complex and two shelterin subunits, TRF1 and its associated factor TIN2 (Canudas et al., 2007). Indeed, depletion of the SA1 cohesin subunit or the telomeric proteins TIN2 and TRF1 restored the normal resolution of sister telomeres in mitosis in tankyrase 1–depleted cells (Canudas et al., 2007), indicating that telomeres have a unique form of cohesin that requires tankyrase 1 for removal.
In this study, we describe an unexpected consequence of persistent sister telomere cohesion: sister chromatid end to end fusion. FISH analysis using a telomere repeat probe of metaphase chromosomes from tankyrase 1–depleted cells revealed a dramatic increase in chromatid-type telomere fusions, which occurred almost exclusively between sister chromatids. Sister telomere fusions were dependent on TIN2 and on persistent sister telomere cohesion. In addition, sister telomere fusions required the DNA ligase IV component of the NHEJ machinery; in the absence of DNA ligase IV, sister telomeres remained cohered but were not fused. Our experiments indicate that the inability to remove cohesion from telomeres leads to deprotection of sister chromatids and inappropriate end to end fusion by NHEJ. We hypothesize that the timely removal of sister telomere cohesion is required to assemble a protective structure at telomeres, thus ensuring genomic stability.
Results
Tankyrase 1 depletion leads to DNA damage and a senescence-like phenotype in both tumor cells and normal cells
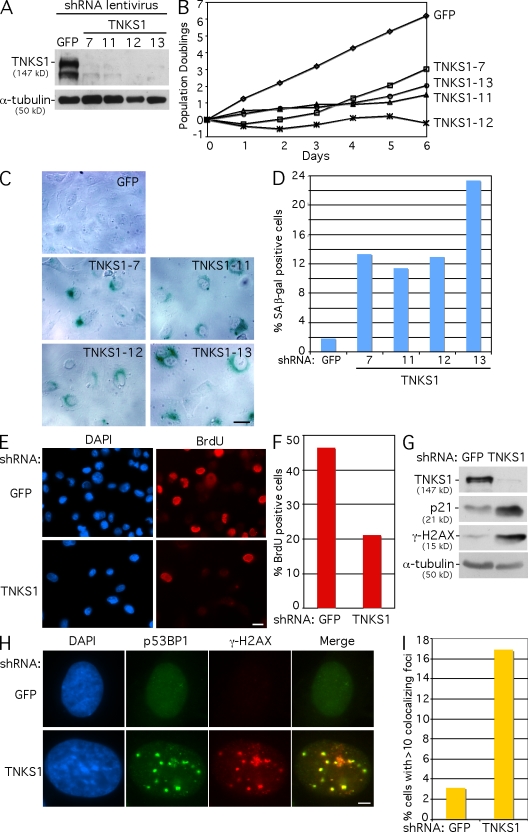
Our previous study showed that depletion of tankyrase 1 using transient transfection with siRNA oligonucleotides resulted in persistent sister telomere associations and mitotic arrest in HeLa cells (Dynek and Smith, 2004). We sought to determine whether tankyrase 1 depletion would lead to a similar phenotype in other human cell lines. Unexpectedly, we found that treatment of a different human tumor cell line, HTC75 (a derivative of HT1080), with tankyrase 1 siRNA oligonucleotides did not lead to a mitotic arrest despite efficient knockdown of tankyrase 1 protein (unpublished data). To determine whether long-term (rather than transient) depletion of tankyrase 1 would influence HTC75 cell growth, we generated stable cell lines lacking tankyrase 1 using lentiviral short hairpin RNAs (shRNAs). HTC75 cells were infected with four different tankyrase 1 shRNAs and a GFP shRNA as a control, and stable lines were selected with puromycin and harvested for analysis 48 h after infection. Immunoblot analysis shows efficient depletion of tankyrase 1 protein (Fig. 1 A). To determine the effect of tankyrase 1 depletion on cell growth, stable cell lines were passaged from day 0 (48 h after infection) to day 6, and the population doublings (PDs) were determined. As predicted from transient siRNA experiments, HTC75 cells lacking tankyrase 1 did not arrest in mitosis. However, they did have a striking phenotype; cell proliferation was inhibited (Fig. 1 B). Moreover, analysis of the cells on day 0 revealed hallmarks of cellular senescence: large vacuolated, multinuclear cells that stained positive for the senescence-associated (SA) β-galactosidase marker that is active at pH 6.0 (Fig. 1, C and D; Dimri et al., 1995). These effects were observed with four distinct lentiviral shRNAs (Fig. 1, A–D). For all subsequent experiments, the TNKS1-13 lentivirus was used, and the analysis was performed at 48 h after infection (day 0). Consistent with a senescence-like G1 arrest, tankyrase 1–depleted cells show a block in BrdU incorporation (Fig. 1, E and F) and an up-regulation of p21 (Fig. 1 G; Brown et al., 1997).
Figure 1.
Tankyrase 1 depletion leads to DNA damage and a senescence-like phenotype in HTC75 tumor cells. (A) Immunoblot analysis of extracts from HTC75 cells harvested 48 h after infection with GFP, TNKS1-7, TNKS1-11, TNKS1-12, or TNKS1-13 lentiviral shRNAs. (B) Graphical representation of the effect of tankyrase 1 depletion on HTC75 cell growth. Growth was monitored from 48 h after infection (day 0) to day 6. Cell growth was plotted as PDs versus days in culture. (C) Phase contrast microscopic images of HTC75 cells stained for SA β-galactosidase activity 48 h after infection with the indicated lentiviral shRNAs. (D) Quantification of SA β-galactosidase–positive cells. Approximately 500 cells were scored for each sample. (E) Immunofluorescence analysis of HTC75 cells infected with GFP or TNKS1 lentiviral shRNA. 48 h after infection, cells were incubated with BrdU for 1 h, ethanol fixed, and probed with anti-BrdU antibody (red); DNA was stained with DAPI (blue). (F) Quantification of BrdU-positive cells. Approximately 1,000 cells were scored for each sample. (G) Immunoblot analysis of extracts from HTC75 cells harvested 48 h after infection with GFP or TNKS1 lentiviral shRNA shows up-regulation of p21 expression and induction of γ-H2AX in tankyrase 1–depleted cells. (H) Immunofluorescence analysis of HTC75 cells infected with GFP or TNKS1 lentiviral shRNA, fixed with paraformaldehyde 48 h after infection, and stained with antibodies against p53BP1 (green) and γ-H2AX (red); DNA was stained with DAPI (blue). (I) Quantification of DNA damage foci. Cells with >10 foci that costained with 53BP1 and γ-H2AX were scored. Approximately 1,000 cells were scored for each sample. Bars: (C and E) 20 µm; (H) 5 µm.
Several studies have shown that a senescence response in certain tumor cell lines (including HT1080) can be induced by DNA damage (for review see Shay and Roninson, 2004). To determine whether this was the case here, we analyzed cells for DNA damage by immunofluorescence for dual staining with the early DNA damage markers γ-H2AX (Rogakou et al., 1999) and 53BP1 (Schultz et al., 2000). HTC75 cells infected with GFP or tankyrase 1 lentiviral shRNAs with >10 colocalizing DNA damage foci were scored (Fig. 1 H). As shown in Fig. 1 I, tankyrase 1 shRNA led to a greater than fivefold increase in cells with DNA damage foci. In addition, immunoblot analysis shows a clear increase in γ-H2AX protein levels (Fig. 1 G). Together, these data indicate that depletion of tankyrase 1 in HTC75 cells leads to a DNA damage response and a senescence-like phenotype.
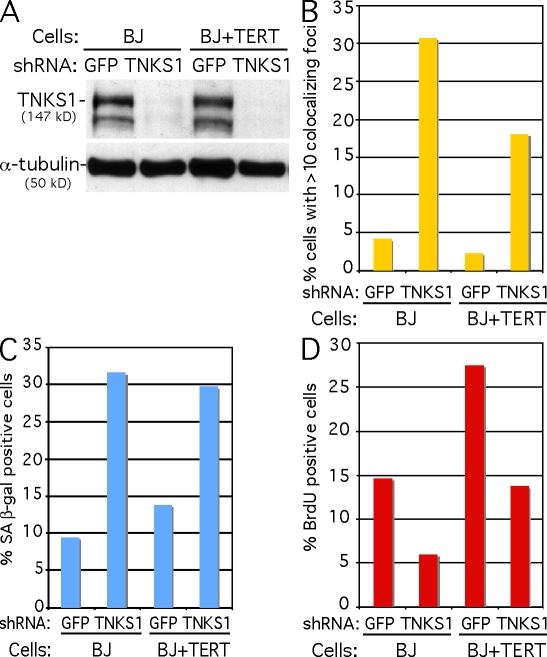
To determine whether tankyrase 1 had a similar effect in normal human cells, primary human BJ fibroblasts or BJ cells immortalized with human telomerase reverse transcriptase (TERT; BJ + TERT) were infected with GFP or tankyrase 1 shRNA lentivirus. Immunoblot analysis shows efficient depletion of tankyrase 1 protein (Fig. 2 A). Depletion of tankyrase 1 in BJ or BJ + TERT cells led to induction of DNA damage foci (Fig. 2 B), an increase in SA β-galactosidase–positive cells (Fig. 2 C), and a concomitant decrease in BrdU incorporation (Fig. 2 D), which are consistent with DNA damage response and senescence-like G1 arrest.
Figure 2.
Tankyrase 1 depletion leads to DNA damage and a senescence-like phenotype in normal human BJ fibroblast cells. (A) Immunoblot analysis of cell extracts from BJ or BJ + TERT cells harvested 48 h after infection with GFP or TNKS1 lentiviral shRNA. (B) Quantification of DNA damage foci. Cells with >10 foci that costained with 53BP1 and γ-H2AX were scored. Approximately 1,000 cells were scored for each sample. (C) Quantification of SA β-galactosidase–positive cells. Approximately 500 cells were scored for each sample. (D) Quantification of BrdU-positive cells. Approximately 1,000 cells were scored for each sample.
Tankyrase 1 depletion leads to persistent telomere cohesion in multiple human cell types
As described in the previous section, depletion of tankyrase 1 in three different human cell lines (HTC75, BJ, and BJ + TERT) led to a DNA damage response and a senescence-like G1 arrest. This cellular response was dramatically different from the mitotic arrest observed upon tankyrase 1 depletion in HeLa cells (Dynek and Smith, 2004; Chang et al., 2005). Therefore, we wondered whether (despite the different phenotypes observed) tankyrase 1 depletion was still causing the same telomere dysfunction in the aforementioned HTC75 and BJ cell lines. To address this question, HTC75 cells were infected with tankyrase 1 or GFP shRNA lentivirus. Cells were isolated 48 h after infection by mitotic shake-off and analyzed by chromosome-specific telomere FISH using a 16pter subtelomere probe. As shown in Fig. 3 A, in control mitotic cells (GFP shRNA), sister telomeres appear as doublets, indicating normal resolution of sister telomere cohesion. In contrast, in tankyrase 1–depleted cells (TNKS1 shRNA), sister telomeres appear as singlets, indicating a block in resolution of sister telomere cohesion. Quantification of this analysis shows a >10-fold increase in unresolved sister telomeres (Fig. 3 B). Thus, tankyrase 1–depleted HTC75 cells, like HeLa cells (Dynek and Smith, 2004; Canudas et al., 2007), show a block in resolution of sister telomere cohesion. Similar results were obtained for BJ (Fig. 3, C and D) and BJ + TERT (Fig. 3, E and F) cells.
Figure 3.
Tankyrase 1 depletion leads to persistent sister telomere cohesion in HTC75, BJ, and BJ + TERT cells. (A–F) Visualization and quantification of chromosome-specific FISH analysis of HTC75 (A and B), BJ (C and D), or BJ + TERT (E and F) cells after infection with GFP or TNKS1 lentiviral shRNA. At 48 h after infection, colcemid was added for 12 h, and cells were collected by mitotic shake-off. Cells were fixed directly with methanol/acetic acid without hypotonic swelling and hybridized to a subtelomeric 16pter probe (green). DNA is stained with DAPI (blue). (B, D, and F) Quantification of mitotic cells with unseparated telomeres. Approximately 100 mitotic cells were scored for each sample. Bar, 2 µm.
Tankyrase 1 depletion leads to sister chromatid fusions in HTC75 cells
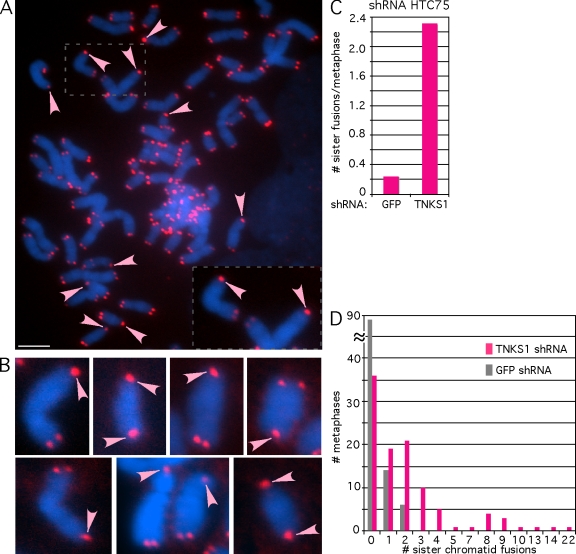
Next, we considered what might give rise to the DNA damage observed in tankyrase 1–depleted HTC75 cells. The observation that these cells (unlike HeLa cells) go through mitosis raised the possibility that covalent telomere fusions (generated during the cell cycle) could give rise to DNA breaks after passage through mitosis. However, the persistent sister telomere cohesion (revealed by the aforementioned chromosome-specific FISH; Fig. 3) would not be predicted to give rise to DNA breaks because (as described in the Introduction) it is caused by protein–protein interactions. Indeed, persistent sister telomere cohesion does not survive the hypotonic treatment used in metaphase spread preparations (Dynek and Smith, 2004; Canudas et al., 2007). Thus, to assay for a low level of covalent telomere fusions that might result in DNA damage after mitosis, we subjected HTC75 cells (48 h after infection with GFP or tankyrase 1 shRNA lentivirus) to hypotonic treatment and metaphase spread analysis, which removes the protein–protein interactions and persistent cohesion but should reveal covalent end to end fusions, and probed telomeres with a peptide nucleic acid (PNA) FISH telomere repeat probe. As shown in Fig. 4 and Table I, we observe a dramatic increase in sister chromatid fusions. The fusion of sister telomeres is made apparent by the single telomere signal that appears between the sisters and is often double the intensity of the normal signal found on each sister at the nonfused end of the chromosome (Fig. 4, A and B). In some cases, both ends of the same chromosome had sister chromatid fusions (Fig. 4 B).
Figure 4.
Tankyrase 1 depletion leads to sister chromatid fusions in HTC75 cells. (A–D) Visualization and quantification of telomeric PNA FISH analysis of metaphase spreads of HTC75 cells after infection with GFP or TNKS1 lentiviral shRNA. At 48 h after infection, colcemid was added for 12 h, and cells were collected by trypsinization, swollen in hypotonic buffer, and fixed in methanol/acetic acid. Telomere repeats were detected by using a Cy3-(CCCTAA)3 PNA probe (red). DNA is stained with DAPI (blue). (A) A metaphase spread from TNKS1 lentiviral shRNA–infected HTC75 cells showing multiple sister chromatid fusions (indicated with arrowheads). The inset is an enlargement of two chromosomes. (B) Examples of sister chromatid fusions (arrowheads) taken from different metaphase spreads from TNKS1 lentiviral shRNA–infected HTC75 cells. Note that in some cases, sister chromatid fusions were present at both ends of the chromosome. (C) Quantification of the frequency of sister chromatid fusions per metaphase in HTC75 cells. (D) Graph showing the distribution of the number of sister chromatid fusions observed in each metaphase. 104 metaphase spreads from two independent experiments were analyzed (Table I, experiments 1 and 2). Bar, 5 µm.
Table I.
Analysis of chromosome and chromatid fusions in tankyrase 1–depleted cells
| Experiment | Cell line | RNAi | Metaphases analyzed | Chromosome fusions | Nonsister chromatid fusions | Sister chromatid fusions | Sister chromatid fusions per metaphase |
| 1 | HTC75 | GFP | 52 | 0 | 0 | 18 | 0.35 |
| 1 | HTC75 | TNKS1 | 52 | 16 | 5 | 160 | 3.08 |
| 2 | HTC75 | GFP | 52 | 0 | 0 | 8 | 0.15 |
| 2 | HTC75 | TNKS1 | 52 | 3 | 5 | 86 | 1.165 |
| 3 | BJ | GFP | 50 | 0 | 0 | 6 | 0.12 |
| 3 | BJ | TNKS1 | 50 | 0 | 0 | 17 | 0.34 |
| 4 | BJ + TERT | GFP | 50 | 0 | 0 | 4 | 0.08 |
| 4 | BJ + TERT | TNKS1 | 50 | 0 | 0 | 12 | 0.24 |
| 5 | HeLaI.2.11 | GFP | 52 | 2 | 2 | 9 | 0.17 |
| 5 | HeLaI.2.11 | TNKS1 | 52 | 1 | 1 | 55 | 1.06 |
| 6 | HTC75 | GFP | 52 | 0 | 0 | 13 | 0.25 |
| 6 | HTC75 | TNKS1 | 52 | 1 | 1 | 86 | 1.65 |
Experiments 1–4 used lentivirus shRNA infection, and experiments 5 and 6 used siRNA transfection.
Analysis of 104 metaphases from two independent experiments showed a 10-fold increase in sister telomere fusions in tankyrase 1–depleted cells compared with the GFP control: 2.3 fusions versus 0.25 fusions per metaphase, respectively (Fig. 4 C and Table I, experiments 1 and 2). There was a range in the number of sister chromatid fusions observed in tankyrase 1–depleted cells. The majority of metaphases had one to three sister chromatid fusions, but several metaphases had three or more (Fig. 4, A and D). In contrast, only a small fraction of control metaphases had sister chromatid fusions, and not more than two per metaphase (Fig. 4 D).
Chromatid-type fusions occur after DNA replication in S/G2 phase. They can occur between chromatids on the same chromosome (the aforementioned sister chromatid fusions) or between chromatids of different chromosomes. Previous studies have shown that in cells lacking normal TRF2, the majority of postreplicative chromatid-type fusions occurred between chromatids of different chromosomes (Bailey et al., 2001; Smogorzewska et al., 2002). In contrast, in tankyrase 1–depleted cells, the vast majority of fusions were between chromatids of the same chromosomes (sister chromatids). Thus, for the aforementioned 104 metaphases, we observed a total of 256 chromatid-type fusions. Remarkably, 246 (96%) were between chromatids on the same chromosome (sisters), and only 10 (4%) were between chromatids of different chromosomes (Table I, experiments 1 and 2).
In addition to chromatid-type telomere fusions, we observed a small number of chromosome-type telomere fusions. This type of fusion most likely occurs by fusion of telomeres from two different chromosomes in G1 phase, which is then subsequently replicated in S phase (Smogorzewska et al., 2002). For the aforementioned 104 metaphases, we observed a total of 275 telomere fusions. 256 (93%) were chromatid-type fusions, and only 19 (7%) were chromosome-type fusions (Table I, experiments 1 and 2). All together, these data indicate that telomere fusions in tankyrase 1–depleted cells occur predominantly between sister chromatids after DNA replication in S/G2 phase of the cell cycle.
Finally, we note that although we observe as much as a 10-fold increase in sister chromatid fusions in tankyrase 1–depleted cells, the frequency at which chromosomes fuse is much lower than the frequency at which they remain cohered. Thus, as measured by 16pter hybridization, 30–55% of mitotic cells show persistent telomere cohesion (Fig. 3), and similar results were obtained with telomere-specific probes for other chromosomes (Dynek and Smith, 2004), suggesting that potentially all telomeres are persistently cohered in 30–55% of tankyrase 1–depleted cells. In contrast, sister chromatid fusions, as measured by PNA FISH with a telomere repeat probe, occur at a much lower frequency of approximately two per metaphase (Fig. 4). These observations suggest that only a fraction of cohered telomeres become fused.
Tankyrase 1 depletion leads to sister chromatid fusions in multiple human cell lines
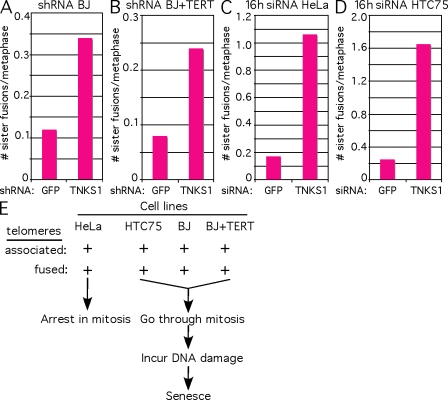
The preponderance of sister chromatid fusions uniquely in tankyrase 1–depleted HTC75 cells raised the possibility that these fusions were the direct result of the persistent sister telomere cohesion observed in these cells. Thus, we asked whether the other human cell lines (BJ, BJ + TERT, and HeLa) that showed persistent sister telomere cohesion also had sister telomere fusions. BJ and BJ + TERT cells were infected with GFP or tankyrase 1 shRNA lentivirus and subjected to metaphase spread analysis with a PNA FISH telomere repeat probe. As shown in Fig. 5 (A and B), we observed a modest but clear increase in sister telomere fusions in tankyrase 1–depleted BJ and BJ + TERT cells (Table I, experiments 3 and 4, respectively).
Figure 5.
Tankyrase 1 depletion leads to sister chromatid fusions in multiple cell lines. (A–D) Quantification of the frequency of sister chromatid fusions in BJ (A) and BJ + TERT (B) cells analyzed 48 h after infection with GFP or TNKS1 lentivirus shRNA and in HeLa (C) and HTC75 (D) cells analyzed after 16-h transient transfection with GFP or TNKS1 siRNA. Colcemid was added for 12 h, and cells were collected by trypsinization, swollen in hypotonic buffer, and fixed in methanol/acetic acid. Metaphase spreads were analyzed by telomeric PNA FISH. Approximately 50 metaphase spreads were analyzed for each sample (Table I, experiments 3–6). (E) Schematic diagram showing the phenotypes of the four different human cell lines in response to tankyrase 1 depletion.
For analysis of HeLa cells, we used transient transfection with tankyrase 1 siRNA. We showed previously that a short (16 h) treatment of HeLa cells with tankyrase 1 siRNA was sufficient to yield a dramatic increase in persistent sister telomere cohesion (Canudas et al., 2007). Thus, we used the same format to probe for sister telomere fusions. HeLa cells were transiently transfected with GFP or tankyrase 1 siRNA for 16 h and subjected to metaphase spread analysis with a PNA FISH telomere repeat probe. As shown in Fig. 5 C and Table I (experiment 5), tankyrase 1 depletion leads to sister chromatid fusions in HeLa cells. The observation here that the telomere fusions occur at the same early time point as persistent telomere cohesion is consistent with the notion that the cohered sister telomeres give rise to sister fusions. Finally, we subjected HTC75 cells to metaphase spread analysis using the same aforementioned conditions for HeLa cells (after 16-h siRNA treatment) and found a similar increase in sister chromatid fusions (Fig. 5 D and Table I, experiment 6).
Together, our data indicate that depletion of tankyrase 1 leads to the same telomere dysfunction (persistent sister telomere cohesion and sister telomere fusions) in four different human cell lines. However, the cells respond differently. HeLa cells arrest in mitosis, whereas HTC75, BJ, and BJ + TERT cells continue through the cell cycle, incurring DNA damage and ultimately arresting with a senescence-like phenotype (Fig. 5 E).
Depletion of TIN2 rescues persistent sister telomere cohesion and sister telomere fusions
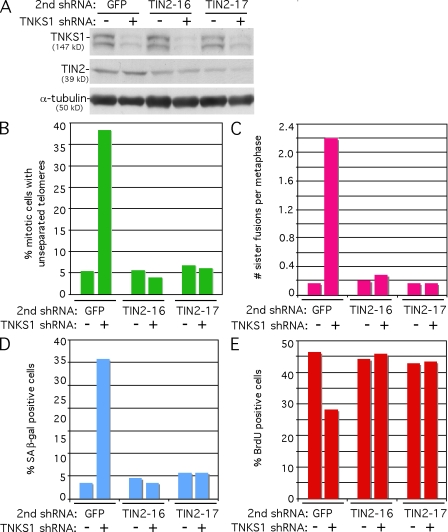
We showed previously that the persistent sister telomere cohesion observed in tankyrase 1–depleted HeLa cells was mediated by binding of the shelterin components TRF1 and TIN2 to the SA1 cohesin complex (Canudas et al., 2007). We found that depletion of TIN2, which leads to concomitant degradation of TRF1, abrogates the requirement for tankyrase 1 in resolution of sister telomere cohesion and in mitotic progression in HeLa cells. Thus, we sought to determine whether TIN2 depletion could rescue the sister telomere fusions and senescence-like G1 arrest in tankyrase 1–depleted HTC75 cells. To address this question, cells were doubly infected with tankyrase 1 shRNA lentivirus and a control (GFP) shRNA or two different TIN2 shRNA lentiviruses. Immunoblot analysis shows depletion of tankyrase 1 and TIN2 (Fig. 6 A). Cells were isolated by mitotic shake-off and analyzed by chromosome-specific telomere FISH. As shown in Fig. 6 B, depletion of TIN2 rescued the persistent sister telomere cohesion in tankyrase 1–depleted cells. To analyze sister telomere fusions, cells were subjected to hypotonic treatment and metaphase spread analysis and probed with a PNA FISH telomere repeat probe. As shown in Fig. 6 C, depletion of TIN2 also rescued the sister telomere fusions in tankyrase 1–depleted cells, which is consistent with the notion that sister telomere fusions arise from persistent telomere cohesion. Finally, we show that depletion of TIN2 also rescues the senescence-like G1 arrest phenotype (Fig. 6, D and E).
Figure 6.
Depletion of TIN2 rescues sister telomere cohesion and sister telomere fusions. (A) Immunoblot analysis of extracts harvested from HTC75 cells 48 h after double infection with lentiviral tankyrase 1 shRNA and GFP shRNA or two different lentiviral TIN2 shRNAs. (B) Quantification of mitotic cells with persistent sister telomere associations as measured by chromosome-specific telomere FISH using a subtelomeric 16pter probe. Approximately 100 mitotic cells were scored for each sample. (C) Quantification of the frequency of sister chromatid fusions as measured by telomere PNA FISH. 25 metaphase spreads were scored for each sample. (D) Quantification of SA β-galactosidase–positive cells. Approximately 500 cells were scored for each sample. (E) Quantification of BrdU-positive cells. Approximately 1,000 cells were scored for each sample.
DNA ligase IV is required for sister telomere fusions but not for persistent sister telomere cohesion
Dysfunctional telomeres can become substrates for the NHEJ machinery (for review see Riha et al., 2006). In mammals, NHEJ is mediated by DNA ligase IV–XRCC4, Ku70/Ku80 heterodimer, and DNA-dependent protein kinase catalytic subunit (DNA-PKcs). Previous studies have shown that telomeres rendered dysfunctional by removal of TRF2 are fused in a DNA ligase IV–dependent manner (Smogorzewska et al., 2002; Celli et al., 2006). Thus, we asked whether a similar mechanism was acting to fuse sister chromatids in tankyrase 1–depleted cells. To address this question, cells were doubly infected with tankyrase 1 shRNA lentivirus and a control (GFP) shRNA or two different DNA ligase IV shRNA lentiviruses. Immunoblot analysis shows depletion of tankyrase 1 and DNA ligase IV (Fig. 7 A). Cells were isolated by mitotic shake-off and analyzed by chromosome-specific telomere FISH. As shown in Fig. 7 B, DNA ligase IV depletion (unlike TIN2 depletion) has no effect on persistent sister telomere cohesion, which is consistent with the noncovalent nature of this association. To analyze sister telomere fusions, cells were subjected to hypotonic treatment and metaphase spread analysis and probed with a PNA FISH telomere repeat probe. As shown in Fig. 7 C, depletion of DNA ligase IV prevented fusion of sister telomeres in tankyrase 1–depleted cells, which is consistent with the idea that sister chromatids are fused by ligase IV–dependent NHEJ. We note that the low background of sister telomere fusions observed in control cells (<0.2 per metaphase) is unaffected by ligase IV depletion, suggesting that these could be caused by another mechanism or by preparation artifacts.
Figure 7.
Ligase IV is required for sister telomere fusions but not for persistent sister telomere cohesion. (A) Immunoblot analysis of extracts harvested from HTC75 cells 48 h after double infection with lentiviral tankyrase 1 shRNA and GFP shRNA or two different lentiviral DNA ligase IV shRNAs. (B) Quantification of mitotic cells with persistent sister telomere associations as measured by chromosome-specific telomere FISH using a subtelomeric 16pter probe. At least 100 mitotic cells were scored for each sample. (C) Quantification of the frequency of sister chromatid fusions as measured by telomere PNA FISH. 25 metaphase spreads were scored for each sample. (D) Schematic diagram showing the fate of sister chromatids in G2. After removal of cohesion, separated sister telomeres can form a protected structure, whereas cohered sister telomeres cannot and thus become susceptible to NHEJ.
Discussion
Our experiments demonstrate that cohered sister telomeres are substrates for ligase IV–mediated fusion after DNA replication in S/G2 phase of the cell cycle. Normally, sister telomeres separate in S/G2 via a tankyrase 1–dependent mechanism. In the absence of tankyrase 1, sister telomere cohesion persists, leading to inappropriate fusion. These experiments reveal a novel form of telomere deprotection and further suggest that resolution of sister telomere associations in S/G2 is essential for telomere integrity.
Tankyrase 1 functions at telomeres in multiple human cell types
Tankyrase 1 depletion leads to persistent sister telomere cohesion and sister chromatid fusions in the four human cell lines that were tested, including tumor cells and normal cells with or without telomerase, suggesting a conserved role at telomeres in human cells. However, despite having the same telomere dysfunction, tankyrase 1–depleted cells show very different growth phenotypes; HeLa cells arrest in mitosis without DNA damage (Dynek and Smith, 2004), whereas HTC75, BJ, and BJ + TERT cells pass through mitosis, incur DNA damage, and arrest with a senescence-like phenotype (this study). The observation that the growth phenotype (whether mitotic arrest or senescence) can be rescued by depletion of TIN2 (Canudas et al., 2007 or Fig. 6, respectively) is consistent with telomere dysfunction as the causative agent.
It is not clear why HeLa cells respond to this dysfunction by arresting in mitosis. Our previous study showed that HeLa cells with very long or short telomeres and GM847 cells (a telomerase-negative tumor cell line that maintains its telomeres through alternative lengthening of telomeres [for review see Cesare and Reddel, 2008]) arrested in mitosis in response to tankyrase 1 depletion (Dynek and Smith, 2004). Thus, the mitotic arrest response is not caused by telomere length or telomerase status. Mitotic progression in some cells may be particularly sensitive to tankyrase 1 depletion and to persistent sister telomere associations or fusions. In contrast, other cell types such as BJ and HTC75 pass through mitosis. However, these cells incur DNA damage, most likely through breakage of fused chromosomes, and ultimately undergo a senescence-like growth arrest.
Previous studies have shown that cells can respond differently to the same telomeric dysfunction. Inhibition of the shelterin subunit TRF2 leads to apoptosis in HeLa cells (Karlseder et al., 1999) but to a senescence-like growth arrest in HTC75 cells (van Steensel et al., 1998) and IMR90 primary cells (Smogorzewska and de Lange, 2002). The mechanism is not known, but different cells may have an innate preference to respond by different pathways. Further work is needed to determine what accounts for these different cellular responses, but we speculate that the difference may involve p53 and/or retinoblastoma (Rb) status. BJ and BJ + TERT are primary cells and have wild-type p53 and Rb, as do HTC75 cells (derived from HT1080 cells with wild-type p53 [Sharma et al., 1993; Tarunina and Jenkins, 1993] and wild-type Rb [Li et al., 1995]). In contrast, the functions of p53 and Rb in HeLa cells and GM847 cells may be altered or abrogated, respectively, by expression of human papilloma virus E6 and E7 proteins or by simian virus 40 T antigen.
Sister telomeres are fused in S/G2 by NHEJ in tankyrase 1–depleted cells
Deprotected telomeres are inappropriately recognized by the cell as double-strand breaks and are fused by the cell’s DNA repair machinery. Telomeres can be rendered dysfunctional by inhibition of TRF2 or by the telomere attrition that occurs in the absence of telomerase. Telomere fusions that arise from TRF2 deficiency are significantly reduced in mouse cells deficient for DNA ligase IV, indicating a role for NHEJ in telomere fusions (Smogorzewska et al., 2002; Celli and de Lange, 2005). For fusions that arise from telomere attrition, one group has shown suppression of fusions lacking telomeric sequence in late generation mice deficient for telomerase and Ku80 or DNA-PKcs (core NHEJ factors; Espejel et al., 2002a,b). However, another group has observed no change in fusions in late generation telomerase- and DNA-PKcs–deficient mice or in mouse cells deficient for telomerase and DNA ligase IV (Maser et al., 2007). Thus, it remains to be determined whether fusion of shortened telomeres occurs by NHEJ or by an alternative pathway. Our work in this study on tankyrase 1–depleted cells shows that the frequency of sister chromatid fusions is suppressed by depletion of DNA ligase IV (Fig. 7), indicating that these fusions occur by NHEJ.
In addition to NHEJ, dysfunctional telomeres can be repaired by homologous recombination. The choice of pathway is thought to be dictated by the cell cycle stage, where NHEJ is the predominant pathway in G1 and homologous recombination is the predominant double-strand break repair pathway in G2 (for review see Riha et al., 2006). Because the sister telomere fusions in tankyrase 1–depleted cells occur postreplicatively in S/G2, it was surprising to observe that they occur via NHEJ. However, recent work has shown that NHEJ can be equally or more active in G2 than in G1 (Guirouilh-Barbat et al., 2008; Mao et al., 2008).
A recent study of telomeres rendered dysfunctional by loss of TRF2 suggests that fusion occurs by NHEJ primarily in G1, but it can occur at a low level postreplicatively in S/G2 (Konishi and de Lange, 2008). Analysis of postreplicative chromatid-type fusions from TRF2-inhibited cells showed that they occur predominantly between chromatids from different chromosomes (Bailey et al., 2001; Smogorzewska et al., 2002). In contrast, the fusions in tankyrase 1–depleted cells occur predominantly between chromatids of the same chromosome (sisters). Thus, these differences (in the timing and the nature of the fusions) indicate different mechanisms for deprotection in TRF2-depleted versus tankyrase 1–depleted cells.
A role for tankyrase 1 and sister telomere cohesion in chromosome end protection
Our observation that suppression of persistent sister telomere cohesion by depletion of TIN2 also leads to the suppression of sister telomere fusions in tankyrase 1–depleted cells (Fig. 6) indicates that persistent cohesion has a role in sister telomere fusions. How might persistent telomere cohesion lead to deprotection? Studies suggest that after DNA replication, telomeres are processed to generate 3′ overhangs before formation of a protective t-loop structure (for review see Verdun and Karlseder, 2007), although the mechanisms and nucleases involved in this process are not well understood. Also, after DNA replication, cohesion must be removed from telomeres by tankyrase 1 (Dynek and Smith, 2004; Canudas et al., 2007). We hypothesize that removal of telomere cohesion may be required for access of proteins that process the 3′ ends. Thus, in this (highly speculative) scenario, persistently cohered telomeres would be unable to form t loops. These deprotected sister telomeres, held in close proximity by persistent cohesion, would then be susceptible to fusion by NHEJ (Fig. 7 D). In this scenario, it is the cohered telomeres that lead directly to deprotection. An alternative hypothesis is that tankyrase 1 acts at telomeres in two steps: first to regulate postreplicative processing (by an unknown mechanism) and, second, to remove cohesion. Insight into the mechanism will require more analysis. However, we consider one reason why it might be advantageous to link 3′ end processing with removal of cohesion. The formation of t loops on sister telomeres requires that each sister undergo cis strand invasion of the 3′ overhang into the double-stranded telomere repeats; trans-invasion of the sister telomere would compromise telomere integrity. Ensuring that the 3′ overhang is formed only after telomere cohesion is removed and sister telomeres are separated could prevent deleterious trans-invasion.
Materials and methods
Plasmids
Lentiviral vectors contained shRNA sequences targeting GFP, tankyrase 1 (TNKS1-7 TRCN0000018346, TNKS1-11 TRCN000040185, TNKS1-12 TRCN0000040186, and TNKS1-13 TRCN0000040187), TIN2 (TIN2-16 TRCN0000039988 and TIN2-17 TRCN0000039989; provided by B. Hahn, Dana-Farber Cancer Institute, Boston, MA), and DNA ligase IV (LigIV-45 TRCN0000009847 and LigIV-47 TRCN0000040003; Open Biosystems).
Lentiviruses and cell lines
Lentiviruses were produced by transfection of 293FT (Invitrogen) packaging cells with a three-plasmid system as described previously (Naldini et al., 1996; Zufferey et al., 1997). 293FT cells were seeded in a 6-cm dish at 1.2 × 106 cells and, 24 h later, were transfected with 1 µg lentiviral vector, 1 µg pCMVΔR.89 packaging plasmid, and 100 ng pMD.G envelope plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Lentiviral supernatants were collected at 24 and 48 h after transfection, filtered with a 0.45-µm filter (Millipore), and either used immediately to infect target cells or frozen at −80°C. 24 h before infection of target cells, HTC75 cells (van Steensel and de Lange, 1997) were seeded at a density of 2.2 × 105, and BJ (at PD 28) and BJ + TERT cells (American Type Culture Collection) were seeded at a density of 3.3 × 105 in 6-cm dishes. Target cells were twice infected overnight with lentiviral supernatants supplemented with 8 µg/ml polybrene (Sigma-Aldrich). After a 24-h recovery period in fresh medium, cells were subcultured 1:2 into medium containing 2 µg/ml puromycin and, upon confluence (usually 24 h later), were harvested and analyzed. This point of harvest, which is ∼48 h after infection, was designated as PD 0 and day 0 on the growth curve in Fig. 1 B. For the growth curve, PDs were calculated using the formula ΔPD = log(Nt/No)/log 2, where No is the number of cells plated and Nt is the number of cells at confluency.
Immunoblotting
Cell extracts prepared as described previously (Cook et al., 2002) were fractionated by SDS-PAGE and transferred to nitrocellulose membrane. Blots were incubated with the following primary antibodies: 1.8 µg/ml rabbit anti–tankyrase 1 762 (Scherthan et al., 2000), 1:500,000 mouse anti–α-tubulin ascites (Sigma-Aldrich), 1:2,000 mouse anti–p21 Waf1/Cip1 (Cell Signaling Technology), 0.2 µg/ml mouse anti–γ-H2AX (Millipore), 0.5 µg/ml rabbit anti–TIN2 701 (Houghtaling et al., 2004), and 1:1,000 rabbit anti–DNA ligase IV serum (GeneTex).
Indirect immunofluorescence
Cells were processed for indirect immunofluorescence as described previously (Cook et al., 2002). In brief, cells were fixed in 2% paraformaldehyde in PBS, permeabilized in 0.5% NP-40/PBS, and blocked in 1% BSA in PBS. Cells were incubated with the following primary antibodies: 0.2 µg/ml mouse anti–γ-H2AX (Millipore) and 0.1 µg/ml rabbit anti-53BP1 (Novus Biologicals) followed by TRITC-conjugated donkey anti–mouse and FITC-conjugated donkey anti–rabbit (Jackson ImmunoResearch Laboratories). DNA was counterstained with 0.2 µg/ml DAPI.
siRNA transfections
siRNA transfections were performed using Oligofectamine (Invitrogen) according to the manufacturer’s instructions. 100 nM GFP Duplex I siRNA (Thermo Fisher Scientific) or tankyrase 1 siRNA oligonucleotide (5′-AACAAUUCACCGUCGUCCUCU-3′; Dynek and Smith, 2004) was transfected into HTC75 or HeLaI.2.11 cells for 16 h.
BrdU labeling and SA β-galactosidase staining
Cells were labeled in 10 µM BrdU (Sigma-Aldrich) for 1 h, fixed in 70% ethanol in 50 mM glycine, pH 2.0, for 20 min at −20°C, washed three times in PBS, and blocked in 1% BSA in PBS. Cells were incubated at 37°C for 30 min with mouse anti-BrdU (Roche), washed five times in 1% BSA in PBS, and incubated for 15 min at room temperature with TRITC-conjugated donkey anti–mouse (Jackson ImmunoResearch Laboratories). DNA was stained with 0.2 µg/ml DAPI for 1 min at room temperature. For the SA β-galactosidase assay (Dimri et al., 1995), cells were fixed in 2% formaldehyde and 0.2% glutaraldehyde in PBS for 5 min, washed three times in PBS, and stained for 6–8 h at 37°C in staining solution (1 mg/ml X-gal, 150 mM NaCl, 2 mM MgCl2, 5 mM K3Fe[CN]6, 5 mM K4Fe[CN]6, and 40 mM NaPi, pH 6.0).
Chromosome-specific FISH
Cells were treated with 0.5 µg/ml colcemid (Invitrogen) for 12 h, collected by mitotic shake-off, fixed, and processed for chromosome-specific FISH as previously described (Dynek and Smith, 2004). In brief, cells were fixed twice in methanol/acetic acid (3:1) for 15 min, cytospun (Shandon Cytospin; Thermo Fisher Scientific) at 2,000 rpm for 2 min onto slides, rehydrated in 2× SSC at 37°C for 2 min, and dehydrated in an ethanol series of 70, 80, and 95% for 2 min each. Cells were denatured at 75°C for 2 min and hybridized overnight at 37°C with a 16pter subtelomeric probe (Cytocell). Cells were washed in 0.4× SSC at 72°C for 2 min and in 2× SSC with 0.05% Tween 20 at room temperature for 30 s. DNA was counterstained with 0.2 µg/ml DAPI.
Metaphase spreads and PNA FISH
Metaphase spreads and PNA FISH were performed as described previously (Dynek and Smith, 2004). In brief, cells were treated with 0.5 µg/ml colcemid for 12 h, hypotonically swollen in 10 mM Tris, pH 7.4, 10 mM NaCl, and 5 mM MgCl2 for 10 min at 37°C, and fixed twice for 15 min in methanol/acetic acid (3:1). Metaphase spreads were prepared by dropping fixed cells on slides. Slides were rehydrated in PBS for 5 min and fixed in 3.7% formaldehyde for 2 min. After three 5-min washes in PBS, slides were incubated with 1 mg/ml pepsin in 10 mM glycine, pH 2.8, at 37°C for 10 min and fixed again in 3.7% formaldehyde for 2 min. Slides were washed three times in PBS, dehydrated in an ethanol series of 70, 95, and 100%, and denatured at 75°C for 10 min in 70% formamide in 2× SSC. Cells were hybridized for 2 h at room temperature with 0.5 µg/ml of a Cy3-conjugated (CCCTAA)3 PNA probe in 10 mM NaHPO4, 10 mM NaCl, 20 mM Tris, pH 7.5, 70% formamide, 0.1 µg/ml tRNA, and 0.1 µg/ml herring sperm DNA. Slides were washed twice for 15 min in 70% formamide, 10 mM Tris, pH 7.5, and 0.1% BSA and were washed three times for 5 min in 0.1 M Tris, pH 7.5, 0.15 M NaCl, and 0.08% Tween 20. After the washes, cells were dehydrated in an ethanol series of 70, 95, and 100%, and DNA was counterstained with 0.5 µg/ml DAPI.
Image acquisition
Images were acquired using a microscope (Axioplan 2; Carl Zeiss, Inc.) with Plan Apochromat 63× NA 1.4, Plan Neofluar 100× NA 1.3, or 40× NA 1.3 oil immersion lenses (Carl Zeiss, Inc.) and a digital camera (C4742-95; Hamamatsu Photonics). Images were acquired and processed using Openlab software (PerkinElmer).
Acknowledgments
We are grateful to Bill Hahn for reagents and advice for lentiviral infection. We thank Tom Meier and members of the Smith laboratory for critical comments on the manuscript and helpful discussions.
This work was supported by grants from the National Institutes of Health (RO1 CA95099 and R01 CA116352).
Footnotes
Abbreviations used in this paper: DNA-PKcs, DNA-dependent protein kinase catalytic subunit; NHEJ, nonhomologous end joining; PD, population doubling; PNA, peptide nucleic acid; Rb, retinoblastoma; SA, senescence associated; shRNA, short hairpin RNA; TERT, telomerase reverse transcriptase.
References
- Ancelin K., Brunori M., Bauwens S., Koering C.E., Brun C., Ricoul M., Pommier J.P., Sabatier L., Gilson E. 2002. Targeting assay to study the cis functions of human telomeric proteins: evidence for inhibition of telomerase by TRF1 and for activation of telomere degradation by TRF2.Mol. Cell. Biol. 22:3474–3487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S.M., Cornforth M.N., Kurimasa A., Chen D.J., Goodwin E.H. 2001. Strand-specific postreplicative processing of mammalian telomeres.Science. 293:2462–2465 [DOI] [PubMed] [Google Scholar]
- Bilaud T., Brun C., Ancelin K., Koering C.E., Laroche T., Gilson E. 1997. Telomeric localization of TRF2, a novel human telobox protein.Nat. Genet. 17:236–239 [DOI] [PubMed] [Google Scholar]
- Broccoli D., Smogorzewska A., Chong L., de Lange T. 1997. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2.Nat. Genet. 17:231–235 [DOI] [PubMed] [Google Scholar]
- Brown J.P., Wei W., Sedivy J.M. 1997. Bypass of senescence after disruption of p21CIP1/WAF1 gene in normal diploid human fibroblasts.Science. 277:831–834 [DOI] [PubMed] [Google Scholar]
- Canudas S., Houghtaling B.R., Kim J.Y., Dynek J.N., Chang W.G., Smith S. 2007. Protein requirements for sister telomere association in human cells.EMBO J. 26:4867–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celli G.B., de Lange T. 2005. DNA processing is not required for ATM-mediated telomere damage response after TRF2 deletion.Nat. Cell Biol. 7:712–718 [DOI] [PubMed] [Google Scholar]
- Celli G.B., Denchi E.L., de Lange T. 2006. Ku70 stimulates fusion of dysfunctional telomeres yet protects chromosome ends from homologous recombination.Nat. Cell Biol. 8:885–890 [DOI] [PubMed] [Google Scholar]
- Cesare A.J., Reddel R.R. 2008. Telomere uncapping and alternative lengthening of telomeres.Mech. Ageing Dev. 129:99–108 [DOI] [PubMed] [Google Scholar]
- Chang P., Coughlin M., Mitchison T.J. 2005. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function.Nat. Cell Biol. 7:1133–1139 [DOI] [PubMed] [Google Scholar]
- Chang W., Dynek J.N., Smith S. 2003. TRF1 is degraded by ubiquitin-mediated proteolysis after release from telomeres.Genes Dev. 17:1328–1333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong L., van Steensel B., Broccoli D., Erdjument-Bromage H., Hanish J., Tempst P., de Lange T. 1995. A human telomeric protein.Science. 270:1663–1667 [DOI] [PubMed] [Google Scholar]
- Cook B.D., Dynek J.N., Chang W., Shostak G., Smith S. 2002. Role for the related poly(ADP-Ribose) polymerases tankyrase 1 and 2 at human telomeres.Mol. Cell. Biol. 22:332–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Lange T. 2005. Shelterin: the protein complex that shapes and safeguards human telomeres.Genes Dev. 19:2100–2110 [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo.Proc. Natl. Acad. Sci. USA. 92:9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donigian J.R., de Lange T. 2007. The role of the poly(ADP-ribose) polymerase tankyrase1 in telomere length control by the TRF1 component of the shelterin complex.J. Biol. Chem. 282:22662–22667 [DOI] [PubMed] [Google Scholar]
- Dynek J.N., Smith S. 2004. Resolution of sister telomere association is required for progression through mitosis.Science. 304:97–100 [DOI] [PubMed] [Google Scholar]
- Espejel S., Franco S., Rodriguez-Perales S., Bouffler S.D., Cigudosa J.C., Blasco M.A. 2002a. Mammalian Ku86 mediates chromosomal fusions and apoptosis caused by critically short telomeres.EMBO J. 21:2207–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espejel S., Franco S., Sgura A., Gae D., Bailey S.M., Taccioli G.E., Blasco M.A. 2002b. Functional interaction between DNA-PKcs and telomerase in telomere length maintenance.EMBO J. 21:6275–6287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith J.D., Comeau L., Rosenfield S., Stansel R.M., Bianchi A., Moss H., de Lange T. 1999. Mammalian telomeres end in a large duplex loop.Cell. 97:503–514 [DOI] [PubMed] [Google Scholar]
- Guirouilh-Barbat J., Huck S., Lopez B.S. 2008. S-phase progression stimulates both the mutagenic KU-independent pathway and mutagenic processing of KU-dependent intermediates, for nonhomologous end joining.Oncogene. 27:1726–1736 [DOI] [PubMed] [Google Scholar]
- Houghtaling B.R., Cuttonaro L., Chang W., Smith S. 2004. A dynamic molecular link between the telomere length regulator TRF1 and the chromosome end protector TRF2.Curr. Biol. 14:1621–1631 [DOI] [PubMed] [Google Scholar]
- Hsiao S.J., Smith S. 2008. Tankyrase function at telomeres, spindle poles, and beyond.Biochimie. 90:83–92 [DOI] [PubMed] [Google Scholar]
- Karlseder J., Broccoli D., Dai Y., Hardy S., de Lange T. 1999. p53- and ATM-dependent apoptosis induced by telomeres lacking TRF2.Science. 283:1321–1325 [DOI] [PubMed] [Google Scholar]
- Konishi A., de Lange T. 2008. Cell cycle control of telomere protection and NHEJ revealed by a ts mutation in the DNA-binding domain of TRF2.Genes Dev. 22:1221–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Fan J., Hochhauser D., Banerjee D., Zielinski Z., Almasan A., Yin Y., Kelly R., Wahl G.M., Bertino J.R. 1995. Lack of functional retinoblastoma protein mediates increased resistance to antimetabolites in human sarcoma cell lines.Proc. Natl. Acad. Sci. USA. 92:10436–10440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losada A., Hirano T. 2005. Dynamic molecular linkers of the genome: the first decade of SMC proteins.Genes Dev. 19:1269–1287 [DOI] [PubMed] [Google Scholar]
- Makarov V.L., Hirose Y., Langmore J.P. 1997. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening.Cell. 88:657–666 [DOI] [PubMed] [Google Scholar]
- Mao Z., Bozzella M., Seluanov A., Gorbunova V. 2008. DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells.Cell Cycle. 7:2902–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser R.S., Wong K.K., Sahin E., Xia H., Naylor M., Hedberg H.M., Artandi S.E., DePinho R.A. 2007. DNA-dependent protein kinase catalytic subunit is not required for dysfunctional telomere fusion and checkpoint response in the telomerase-deficient mouse.Mol. Cell. Biol. 27:2253–2265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naldini L., Blomer U., Gallay P., Ory D., Mulligan R., Gage F.H., Verma I.M., Trono D. 1996. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector.Science. 272:263–267 [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. 2005. The structure and function of SMC and kleisin complexes.Annu. Rev. Biochem. 74:595–648 [DOI] [PubMed] [Google Scholar]
- Palm W., de Lange T. 2008. How shelterin protects mammalian telomeres.Annu. Rev. Genet. 42:301–334 [DOI] [PubMed] [Google Scholar]
- Riha K., Heacock M.L., Shippen D.E. 2006. The role of the nonhomologous end-joining DNA double-strand break repair pathway in telomere biology.Annu. Rev. Genet. 40:237–277 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Boon C., Redon C., Bonner W.M. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo.J. Cell Biol. 146:905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherthan H., Jerratsch M., Li B., Smith S., Hulten M., Lock T., de Lange T. 2000. Mammalian meiotic telomeres: protein composition and redistribution in relation to nuclear pores.Mol. Biol. Cell. 11:4189–4203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz L.B., Chehab N.H., Malikzay A., Halazonetis T.D. 2000. p53 binding protein 1 (53BP1) is an early participant in the cellular response to DNA double-strand breaks.J. Cell Biol. 151:1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Schwarte-Waldhoff I., Oberhuber H., Schafer R. 1993. Functional interaction of wild-type and mutant p53 transfected into human tumor cell lines carrying activated ras genes.Cell Growth Differ. 4:861–869 [PubMed] [Google Scholar]
- Shay J.W., Roninson I.B. 2004. Hallmarks of senescence in carcinogenesis and cancer therapy.Oncogene. 23:2919–2933 [DOI] [PubMed] [Google Scholar]
- Smith S., de Lange T. 2000. Tankyrase promotes telomere elongation in human cells.Curr. Biol. 10:1299–1302 [DOI] [PubMed] [Google Scholar]
- Smith S., Giriat I., Schmitt A., de Lange T. 1998. Tankyrase, a poly(ADP-ribose) polymerase at human telomeres.Science. 282:1484–1487 [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T. 2002. Different telomere damage signaling pathways in human and mouse cells.EMBO J. 21:4338–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., Karlseder J., Holtgreve-Grez H., Jauch A., de Lange T. 2002. DNA ligase IV-dependent NHEJ of deprotected mammalian telomeres in G1 and G2.Curr. Biol. 12:1635–1644 [DOI] [PubMed] [Google Scholar]
- Tarunina M., Jenkins J.R. 1993. Human p53 binds DNA as a protein homodimer but monomeric variants retain full transcription transactivation activity.Oncogene. 8:3165–3173 [PubMed] [Google Scholar]
- van Steensel B., de Lange T. 1997. Control of telomere length by the human telomeric protein TRF1.Nature. 385:740–743 [DOI] [PubMed] [Google Scholar]
- van Steensel B., Smogorzewska A., de Lange T. 1998. TRF2 protects human telomeres from end-to-end fusions.Cell. 92:401–413 [DOI] [PubMed] [Google Scholar]
- Verdun R.E., Karlseder J. 2007. Replication and protection of telomeres.Nature. 447:924–931 [DOI] [PubMed] [Google Scholar]
- Zufferey R., Nagy D., Mandel R.J., Naldini L., Trono D. 1997. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo.Nat. Biotechnol. 15:871–875 [DOI] [PubMed] [Google Scholar]