Abstract
Exit from mitosis is characterized by a precipitous decline in cyclin-dependent kinase (Cdk) activity, dissolution of mitotic structures, and cytokinesis. In Saccharomyces cerevisiae, mitotic exit is driven by a protein phosphatase, Cdc14, which is in part responsible for counteracting Cdk activity. Throughout interphase, Cdc14 is sequestered in the nucleolus, but successful anaphase activates the mitotic exit network (MEN), which triggers dispersal of Cdc14 throughout the cell by a mechanism that has remained unknown. In this study, we show that a MEN component, protein kinase Dbf2–Mob1, promotes transfer of Cdc14 to the cytoplasm and consequent exit from mitosis by direct phosphorylation of Cdc14 on serine and threonine residues adjacent to a nuclear localization signal (NLS), thereby abrogating its NLS activity. Our results define a mechanism by which the MEN promotes exit from mitosis.
Introduction
Exit from mitosis in the budding yeast Saccharomyces cerevisiae is characterized by a precipitous decline of B-type cyclins, dissolution of the spindle, and completion of cytokinesis. Cdc14 phosphatase plays a key role in initiating this transition by reversing Clb/Cdk phosphorylations while simultaneously promoting the destruction of B-type cyclins via the ubiquitin ligase anaphase-promoting complex (APC)–Cdh1 (Taylor et al., 1997; Jaspersen et al., 1998; Visintin et al., 1998; Zachariae et al., 1998; Schwab et al., 2001). To ensure that the newly duplicated genome is faithfully segregated before mitotic exit and cytokinesis, Cdc14 is sequestered in the RENT (regulator of nucleolar silencing and telophase) complex where it is tightly bound to and inhibited by Net1 (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999). The mechanisms that control dissociation of Cdc14 form Net1 fall under multiple layers of checkpoint and cell cycle control, thereby ensuring an orderly exit from mitosis (Wang et al., 2000, 2003; Bouck and Bloom, 2005; D'Aquino et al., 2005).
Two temporally distinct mechanisms govern Cdc14 release. The 14 early anaphase release network provides a transient burst of Cdc14 phosphatase activity in early anaphase that is primarily restricted to the nucleus and promotes spindle stability, spindle elongation, and ribosomal DNA segregation. In late anaphase, the mitotic exit network (MEN) pathway affects a complete and sustained release of Cdc14 that results in its dispersal throughout the cell and drives exit from mitosis (Jaspersen et al., 1998; Stegmeier et al., 2002; Marston et al., 2003; Azzam et al., 2004; D'Amours and Amon, 2004; Sullivan et al., 2004; Queralt et al., 2006). Notably, Cdc14 substrates that act during late anaphase/telophase (e.g., Cdh1 and Swi5) are held in the cytoplasm until MEN is activated, whereas those that act during mid anaphase (e.g., Sli15, Ask1, and Fin1) are associated with the nucleus (Moll et al., 1991; Jaquenoud et al., 2002; Pereira and Schiebel, 2003; Geymonat et al., 2004; Higuchi and Uhlmann, 2005; Woodbury and Morgan, 2007).
MEN-dependent activation of Cdc14 is controlled by the G protein Tem1 and by protein kinases Cdc15 and Dbf2 (Bardin et al., 2000; Pereira and Schiebel, 2001; Visintin and Amon, 2001). Genetic and biochemical analyses of the MEN components suggest that Dbf2 and its regulatory partner Mob1 comprise the most downstream signaling component of MEN. A recent study indicates that Dbf2–Mob1 enters the nucleus during telophase and colocalizes with Cdc14 at kinetochores, suggesting that Dbf2–Mob1 could directly modify either Cdc14, Net1, or an as yet unidentified member of the nucleolar complex to initiate mitotic exit (Stoepel et al., 2005). Furthermore, a small quantity of Cdc14 appears at the cytoplasmic surface of the spindle pole body in mid anaphase. Collectively, it appears that some fraction of Cdc14 interacts with Dbf2–Mob1 before complete release of Cdc14 from the nucleolus in telophase, allowing for direct control of Cdc14 activation by Dbf2–Mob1.
Evidence that nuclear import and export play a part in restraining Cdc14 activity to telophase has been mounting. Screens designed to identify mutants that bypass the essential requirement for the MEN pathway identified alleles of KAP104 and SRP1 (Shou et al., 2001; Asakawa and Toh-e, 2002; Shou and Deshaies, 2002). More recently, identification of a leucine-rich nuclear export signal (NES) within Cdc14 that contributes to its localization and function hints at a role for nuclear/cytoplasmic shuttling in control of mitotic exit (Bembenek et al., 2005). Furthermore, regulation of the Schizosaccharomyces pombe homologue of Cdc14, Clp1, may also involve cell cycle–dependent nuclear/cytoplasmic shuttling. Although the control of mitotic exit and the biological function of the MEN pathway are surprisingly divergent in S. pombe and S. cerevisiae, the two species may use similar mechanisms to regulate Cdc14/Clp1 (Trautmann and McCollum, 2005).
Results
The C terminus of Cdc14 contains an NLS
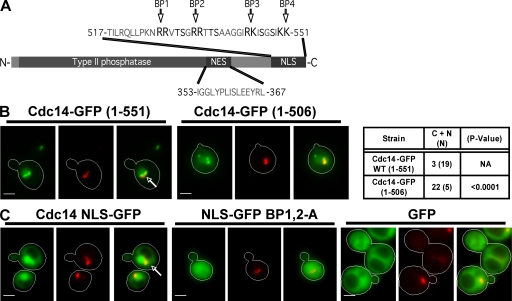
As described in the Introduction, several lines of evidence suggest that regulated nucleocytoplasmic shuttling of Cdc14 may play an important role in exit from mitosis. To explore this possibility, we sought to define an NLS sequence. Interestingly, the extreme C-terminal region of Cdc14 contains four pairs of basic amino acids with no intervening Asp or Glu residues (Fig. 1 A). To test whether this basic patch (BP) mediates nuclear localization of Cdc14, we generated a C-terminal truncation fused to GFP and expressed from the galactose-inducible GAL1,10 promoter. Under the inducing conditions used for this experiment (75 min in 3% galactose at 30°C), Cdc14-GFP was overexpressed relative to endogenous Cdc14 by only twofold or less (Fig. S1, available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1). Deletion of the C-terminal 45 amino acids of Cdc14 resulted in a pattern of localization that differed from full-length Cdc14-GFP (Fig. 1 B). The truncated protein still accumulated in the nucleolus, but a substantial fraction was distributed throughout the nucleus and cytoplasm. A larger deletion that encompasses NES (Fig. 1 A; Bembenek et al., 2005) partially restored the pattern seen with the full-length reporter (not depicted). These observations suggest that there is an NLS within the C-terminal 45 amino acids of Cdc14 and a second NLS within the N-terminal 345 residues. The N-terminal two thirds of the protein does not contain a recognizable NLS motif but binds to substrates, including Net1, which might piggyback Cdc14 to the nucleus (Taylor et al., 1997).
Figure 1.
BPs near the C terminus of Cdc14 comprise an NLS. (A) Diagram of Cdc14 showing the relative positions of the type II phosphatase domain and putative NES and NLS. BP1–4 refer to four pairs of basic amino acids. (B) The C-terminal domain of Cdc14 promotes nuclear localization. Cdc14 containing the indicated number of amino acids (full-length protein is 551 residues) to GFP and expressed from the GAL1,10 promoter. Fusion protein expression was induced for 75 min with 3% galactose, and cells were imaged by fluorescence microscopy. The table shows the number of cells for each genotype in which the GFP signal was present in both the cytoplasm and nucleus (C + N) or restricted to the nucleus (N). The statistical significance of the deviation from wild-type behavior (calculated using a χ2 test) is shown in the third column. (C) The C-terminal region of Cdc14 contains NLS activity. (left) The last 100 amino acids of Cdc14 were fused to the N terminus of GFP and transiently expressed from the GAL1,10 promoter. NLS-GFP accumulated in the nucleus of unbudded and small-budded cells. (middle) Arg 527, 528, 533, and 534 of Cdc14 were substituted for Ala (BP1,2A). (right) GFP alone was present in both the cytoplasm and nucleus. Arrows indicate the colocalization of Cdc14-GFP (green) with nucleolar Nop1-DsRed (red; γ adjusted). Bars, 2 µm.
To determine whether Cdc14’s C-terminal domain is sufficient to drive nuclear localization, we joined the last 100 amino acids of Cdc14 to GFP and expressed the resulting fusion protein from the GAL1,10 promoter. Expression of this construct resulted in concentration of fluorescence in the nucleus in interphase cells (Fig. 1 C). In contrast, unfused GFP was evenly distributed between the nucleus and cytoplasm (Fig. 1 C). Owing to its strong nuclear localization, we refer to this construct hereafter as NLS-GFP. The last 50 amino acids of Cdc14 contain four vicinal pairs of Args and/or Lys (BP 1–4; Fig. 1 A) with no intervening acidic residues. Because these residues appeared to be the best candidates for an NLS, we changed Args 527, 528, 533, and 534 to Ala (BP1,2A) and compared the subcellular distribution of the wild-type and BP1,2A NLS-GFP reporters by live cell fluorescence microscopy (Fig. 1 C). Mutation of BP1 and BP2 effectively eliminated concentration of the fusion protein in the nucleus of interphase cells. Thus, these pairs of Args comprise an essential determinant of the C-terminal NLS.
Cdc14’s C-terminal NLS is regulated by MEN
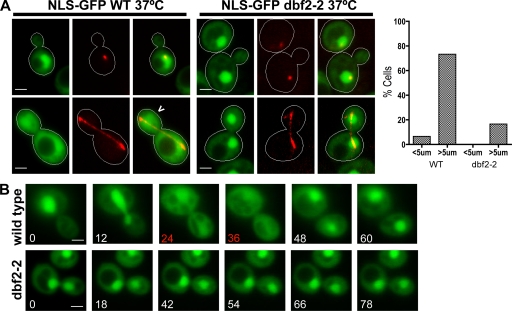
An interesting feature of NLS-GFP is that its localization appeared to change during the cell cycle (unpublished data). An appealing hypothesis is that when MEN switches on in cells undergoing anaphase, it inactivates NLS, causing NLS-GFP to disperse throughout the cell. To test this hypothesis, we quantified the localization of the NLS-GFP reporter in wild-type and temperature-sensitive dbf2-2 strains that also expressed a red fluorescent marker of the mitotic spindle, mCherry-Tub1 (α-tubulin). Transformants were grown at 25°C, induced to express the NLS-GFP reporter (90 min in 3% galactose), and shifted to 37°C for 45 min to inactivate dbf2-2. Cells were resuspended in TE (10 mM Tris, pH 8.0, and 1 mM EDTA) before microscopy to arrest growth and preserve protein localization. The number of cells that exhibited cytoplasmic versus nuclear fluorescence were counted, and spindle length was measured for each cell. NLS-GFP was delocalized to the cytoplasm in 73% (n = 75) of wild-type cells with late anaphase spindles >5 µm in length as opposed to only 16% (n = 50) of dbf2-2 cells with late anaphase spindles (Fig. 2 A), indicating that Dbf2 activity is required for the late anaphase inactivation of the C-terminal NLS. In contrast, analysis of GFP-Crz1 (Boustany and Cyert, 2002) revealed that nuclear export was not globally impeded in dbf2-2 cells arrested in anaphase (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1).
Figure 2.
Cytoplasmic localization of NLS-GFP during late mitosis is dependent on MEN. (A) Localization of NLS-GFP (green) in temperature-sensitive dbf2-2 and wild-type (WT) W303 grown at 37°C (nonpermissive). mCherry-Tub1 was imaged to identify interphase (short spindle) and late anaphase (long spindle) cells. 73% of wild-type (n = 75) cells with spindles >5 µm exhibited pancellular localization of the NLS-GFP reporter (carrot) compared with 16% of dbf2-2 (n = 50) cells, indicating that Dbf2 activity is required for inactivation of Cdc14’s C-terminal NLS during late anaphase. Only 4% of wild-type (n = 50) cells and no dbf2-2 (n = 50) cells with spindles <5 µm exhibited pancellular GFP staining. The graph shows the percentage of cells with released Cdc14 (i.e., both cytoplasmic and nuclear fluorescence) as a function of spindle length (<5 or >5 µm). (B) Wild-type and temperature-sensitive dbf2-2 yeast strains were grown at 37°C for 45 min, resuspended in 2.5% LMP agarose with 1× synthetic complete medium, and placed on slides. Slides were held at 37°C by a temperature-controlled stage for ≥30 min before imaging the cells every 6 min. Representative time-lapse series are shown for wild-type (n = 7) and dbf2-2 (n = 5) cells. The NLS-GFP reporter was released from the nucleus for approximately two time points (red) in all of the wild-type cells but exhibited persistent nuclear localization in all five dbf2-2 cells. Bars, 2 µm.
To more directly observe cell cycle–dependent regulation of NLS-GFP, we mounted wild-type and dbf2-2 cells on agarose pads after growth at 37°C for 30 min and placed them on a temperature-controlled 37°C microscope stage. Fluorescence and differential interference contrast (DIC) images were captured every 6 min to visualize division of the nucleus and its partitioning to daughter cells. In the representative wild-type cell shown (n = 7) in Fig. 2 B, the NLS-GFP reporter was rapidly released into the cytoplasm between 12 and 24 min, which is coincident with partitioning of the nucleus. This is exactly the time in the cell cycle that MEN is switched on (Stoepel et al., 2005). Once nuclear division was completed, the reporter was resequestered in the nucleus between 36 and 48 min. Meanwhile, all five dbf2-2 cells examined failed to divide and exhibited stable nuclear accumulation of NLS-GFP throughout parallel time-lapse experiments (Fig. 2 B). These observations suggest that the C-terminal NLS of Cdc14 is transiently inactivated in late mitosis by a mechanism that depends on MEN.
Dbf2–Mob1 phosphorylates Cdc14 in vitro and in vivo
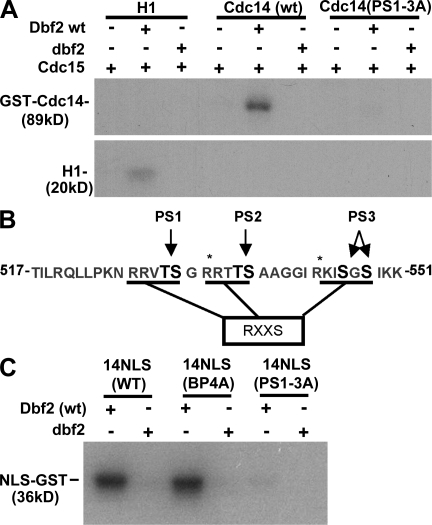
Given that DBF2 regulates the C-terminal NLS of Cdc14 and Dbf2 is known to colocalize with Cdc14 at kinetochores (Stoepel et al., 2005), we sought to determine whether Cdc14 can be phosphorylated by Dbf2–Mob1. His6-tagged Dbf2–Mob1 was purified from insect cells and activated with purified protein kinase Cdc15 as described previously by Mah et al. (2001). When full-length GST-Cdc14 was incubated with activated Dbf2–Mob1 and γ-[32P]ATP for 20 min, we observed incorporation of label into Cdc14 that was much greater than that incorporated into the nonspecific substrate histone H1 (Fig. 3 A). Although wild-type Dbf2 efficiently incorporated label into GST-Cdc14, the kinase-dead version did not, indicating that the Cdc15 used to activate Dbf2–Mob1 did not phosphorylate Cdc14 appreciably.
Figure 3.
Cdc14 is phosphorylated by Dbf2–Mob1. (A) Phosphorylation of Cdc14 by Dbf2–Mob1. Purified GST-Cdc14, GST-Cdc14(PS1–3A), or histone H1 were treated with wild-type (wt) or kinase-dead Dbf2–Mob1 (dbf2) complex. Dbf2–Mob1 was activated in vitro by protein kinase Cdc15. Phosphorylated Cdc14 was detected by SDS-PAGE and autoradiography. PS1–3A refers to a hextuple mutant in which the residues in PA pairs 1–3 (PS1–3; B) were changed to Ala. (B) The C terminus of Cdc14 contains three potential regions of Dbf2–Mob1 phosphorylation. The sites that were altered for this study, PS1–3, are indicated in black font. RXXS refers to the preferred site for phosphorylation by Dbf2 (Mah et al., 2005). (C) Phosphorylation of Cdc14’s NLS region by Dbf2–Mob1 depends on PS1–3. The last 100 amino acids of the C terminus of Cdc14 were fused to GST. Purified GST-NLS was treated with either wild-type Dbf2–Mob1 or kinase-dead Dbf2–Mob1. Dbf2–Mob1 complexes were activated with protein kinase Cdc15. Phosphorylation of wild type, a BP mutant, BP4A (Fig. 1 A), and a six-site Ser/Thr to Ala mutant, PS1–3A, were assayed. Asterisks indicate predicted trypsin cleavage sites.
Previously, our laboratory reported that Dbf2–Mob1, like other kinases of the Akt and PKA class, prefers to phosphorylate the sequence motif RXXS (Mah et al., 2005). Analysis of the primary amino acid sequence of Cdc14 revealed three pairs of Sers and/or Thrs (for a total of six residues) in the extreme C terminus of Cdc14 that are potential sites of Dbf2–Mob1 phosphorylation (Fig. 3 B). For simplicity, we refer to these sites as phosphosite (PS) pairs 1–3 (PS1–3). Interestingly, PS1–3 are immediately adjacent to and interdigitated between the BPs (BP1–4) that govern nuclear localization of NLS-GFP (Fig. 3 B). To determine whether these C-terminal sites are targeted by Dbf2–Mob1, we fused the last 100 amino acids of Cdc14 to GST. The resulting fusion protein purified from Escherichia coli was efficiently phosphorylated by Dbf2–Mob1 (Fig. 3 C). However, a mutant in which the PS1–3 sites were mutated to Ala exhibited greatly diminished 32P incorporation, suggesting that PS1–3 are the major targets of Dbf2–Mob1 within the C-terminal 100 amino acids of Cdc14 (Fig. 3 C). Mutation of two C-terminal Lys (BP4) that do not fall within predicted Dbf2–Mob1 consensus sites did not affect phosphorylation (Fig. 3 C).
The C-terminal 100 amino acids of Cdc14 contain 24 Ser and Thr residues, yet mutation of only six sites nearly abolished 32P incorporation by Dbf2–Mob1. To determine whether other sites of Dbf2–Mob1 phosphorylation were likely to be found within the full-length protein, we mutated the same residues to create a PS1–3A allele of GST-Cdc14. Activated Dbf2–Mob1 failed to incorporate significant 32P into this substrate (Fig. 3 A), suggesting that the six Ser and Thr residues that lie within RXXS motifs in the extreme C-terminal region of Cdc14 are the primary target.
To confirm that the C-terminal residues implicated by mutation are true Dbf2–Mob1 phosphorylation sites, we used mass spectrometry (MS). GST-Cdc14 was treated with activated Dbf2–Mob1 in vitro and subsequently isolated by SDS-PAGE. Gel bands corresponding to Cdc14 were excised, digested with AspN and trypsin, and analyzed for phosphorylation content by phosphopeptide-selective precursor ion-scanning liquid chromatography (LC) MS. This revealed two dominant phosphopeptide species that were identified by tandem LC-MS/MS as TTpSAAGGIR (amino acids 535–543) and KIpSGSIKKEA (amino acids 544–551; Fig. S1). The extra Glu and Ala residues at the end of the second peptide were added during assembly of the recombinant construct. We were unable to identify a peptide corresponding to phosphorylated PS1, most likely because of its extremely small size (five residues).
To verify that Ser and Thr residues within the NLS are phosphorylated in vivo, we purified epitope-tagged Cdc14 from yeast cells for analysis by MS. Rapidly growing cells were quenched with TCA, proteins were extracted into buffer containing 8 M urea, and tagged Cdc14 was isolated with nickel resin and separated by SDS-PAGE. Bands corresponding to tagged Cdc14 were excised and treated with both AspN and trypsin. LC-MS/MS analysis indicated that KIpSGSIK and TTpSAAGGIR were also present in vivo (Fig. S2, available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1).
Phosphorylations adjacent to BPs regulate the activity of the C-terminal NLS
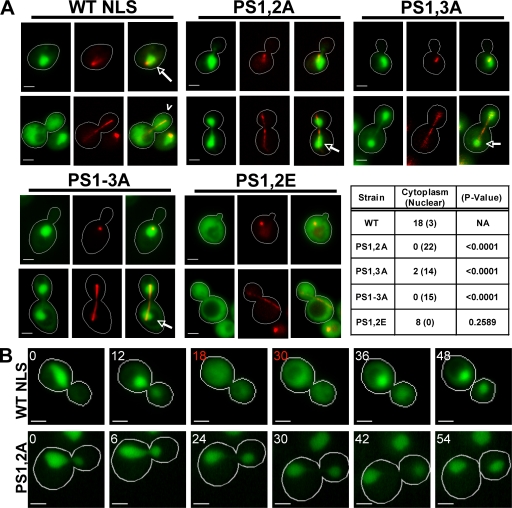
Our data so far indicate that the C-terminal region of Cdc14 contained NLS activity, was relocalized to the cytoplasm in late mitosis in a DBF2-dependent manner, and was directly phosphorylated by Dbf2–Mob1. Collectively, these observations suggested that Dbf2 phosphorylates residues adjacent to NLS in late mitosis, which inactivates it and results in redistribution of NLS-GFP to the cytoplasm. To test this hypothesis, we individually mutated phosphorylation sites within our NLS-GFP reporter and assayed the effects of these mutations on nuclear localization (Fig. 4 A). Wild-type NLS-GFP expressed for 90 min from the GAL1,10 promoter was concentrated in the nucleus in interphase (short spindle) cells. In late mitotic (long spindle) cells, the same construct was largely dispersed throughout the cell. In contrast, the double mutants PS1,2A and PS1,3A, as well as a mutant lacking all six sites (PS1–3A) were tightly restricted to the nucleus in both interphase and late mitotic cells (Fig. 4 A).
Figure 4.
Phosphorylation sites within Cdc14’s NLS are required for its cell cycle regulation. (A) Mutation of Dbf2–Mob1 phosphorylation sites in NLS changes the nuclear/cytoplasmic distribution of NLS-GFP (green) reporter fusions. Wild-type (WT), PS1,2A, PS1,3A, PS1–3A, and PS1,2E NLS-GFP reporters are shown. Red fluorescent mCherry-Tub1 (red; γ adjusted) was used to identify interphase (short spindle) and late anaphase (long spindle) cells. The indicated NLS constructs were transiently expressed (3% galactose for 90 min) from the GAL1,10 promoter. Live cells were imaged. Wild-type NLS-GFP was concentrated in the nucleus of interphase cells (short spindle; arrows) and dispersed into the cytoplasm of late anaphase cells (long spindle; carrot). NLS-GFP alleles PS1,2A, PS1,3A, and PS1–3A do not exhibit cytoplasmic release of the reporter. In the table, the first column shows genotypes, the second column shows the number of cells for which the spindle was >5 µm long and the GFP signal was dispersed into the cytoplasm or concentrated in the nucleus, and the third column shows the statistical significance of the deviation from wild type (χ2 test). In contrast to the nonphosphorylatable mutants, PS1,2E, a putative phosphomimetic mutant, is defective for nuclear accumulation. The residues that comprise PS1–3 are specified in Fig. 3 B. (B) The PS mutant NLS-GFP is not released from the nucleus during late mitosis. Time-lapse microscopy was performed as described in Fig. 2 B. Representative time-lapse series are shown for wild-type and PS1,2A reporters. The wild-type NLS-GFP fusion was dispersed throughout the cell for approximately two time points (indicated by red numbers; n = 5). In contrast, mutant PS1,2A reporters did not redistribute to the cytoplasm for the duration of the time course (n = 5). Bars, 2 µm.
The constitutive nuclear localization of PS mutants suggested that phosphorylation normally impedes the function of NLS. To determine whether phosphorylation of these sites is sufficient to redistribute NLS-GFP, we mutated Ser and Thr residues of PS1 and PS2 to Glu (PS1,2E) to mimic constitutive phosphorylation. The PS1,2E mutant was distributed nearly equally throughout the cytoplasm and the nucleus of interphase and late mitotic cells, indicating that the nuclear localization function had been inhibited (Fig. 4 A).
To further test the idea that nearby phosphorylations during late mitosis inhibit NLS function, we followed the localization of wild-type and mutant reporters by time-lapse fluorescence microscopy. Yeast cells embedded in agarose were observed every 6 min while dividing and rebudding. NLS-GFP accumulated in the nucleus throughout interphase but was released from the nucleus for approximately two time points during the large-budded stage of the cell cycle, which is coincident with the time of nuclear division (Fig. 4 B). In contrast, the PS1,2A mutant NLS-GFP reporter was never released into the cytoplasm in cells undergoing nuclear division (n = 5), indicating that the mutated NLS was no longer subject to cell cycle–dependent regulation.
Localization of full-length Cdc14-GFP fusions is altered by NLS mutations
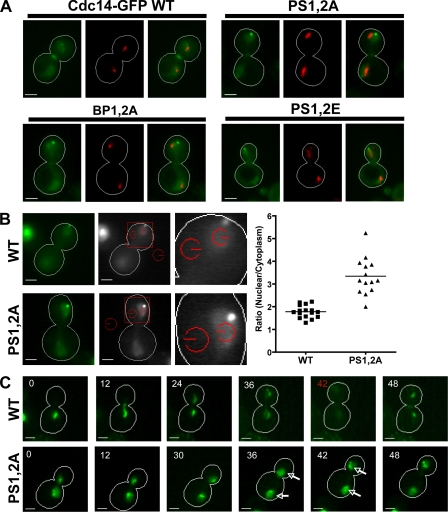
Our results so far provide strong evidence that the extreme C terminus of Cdc14 exhibits NLS activity, and this activity can be controlled during progression through late mitosis by Dbf2-dependent phosphorylation. To address whether this regulatory mechanism normally controls Cdc14, we sought to test a key prediction of the preceding findings shown in Figs. 1–4, which is that a PS1,2A mutant of full-length Cdc14 should not be widely dispersed throughout the cell during late anaphase because the C-terminal NLS cannot be phosphorylated by Dbf2–Mob1 and therefore remains active. We assayed the localization of NLS mutants of full-length Cdc14 fused to GFP expressed from the GAL1,10 promoter. To control the levels of Cdc14-GFP achieved from galactose induction, we created a yeast strain lacking the divergent promoter required for transcription of the GAL1,10 locus (gal1,10Δp), thereby eliminating galactose consumption (Schneider et al., 2004). In this strain, galactose can be used as a gratuitous inducer. At 0.015% galactose, plasmid-borne Cdc14-GFP alleles were expressed at a level that was lower than endogenous Cdc14 (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1). When expressed at physiological levels, full-length Cdc14-GFP mimicked the behavior of the NLS-GFP constructs. Although wild-type Cdc14-GFP was widely dispersed in large-budded cells with a divided nucleolus (i.e., cells in late anaphase; Torres-Rosell et al., 2004), the PS1,2A mutant was more concentrated in the nucleus. In contrast to the nuclear retention of the PS1,2A mutant that has an uninhibitable C-terminal NLS, the mutants in which this element is completely inactivated (BP1,2A and PS1,2E) were readily dispersed throughout the cell in anaphase (Fig. 5 A).
Figure 5.
Basic residues and phosphorylation sites within the C-terminal NLS are required for proper cell cycle–dependent control of Cdc14-GFP localization. (A) Localization of full-length Cdc14-GFP fusions (green) with either wild-type (WT) or mutated NLSs. Wild-type, PS1,2A, BP1,2A, and PS1,2E mutants expressed for 16 h from the GAL1,10 promoter with the addition of galactose to 0.015% are shown. Nop1-DsRed (red; γ adjusted) marks the nucleolus and relative position of the nucleus. Late anaphase cells with segregated nucleoli are shown. (B) Cdc14-GFP reporters were expressed from the GAL1,10 as described in A. Red fluorescent mCherry-Tub1 (red; γ adjusted) was used to select cells in late anaphase (spindles >5 µm). The intensity of Cdc14-GFP fluorescence inside the nucleus (red circle nearest the spindle pole) and inside the cytoplasm (red circle adjacent to the cell wall) was determined. (left) Representative images for wild-type and PS1,2A reporters are shown. (middle) Red squares define the regions of interest shown to the right. Red circles define the areas for which mean pixel intensity was calculated. (right) The ratio of fluorescence intensities (nuclear/cytoplasm) was calculated for multiple cells and plotted for each reporter. The mean ratio ± SD was calculated for wild type (1.8 ± 0.3 SD, n = 16) and PS1,2A (3.3 ± 0.8 SD, n = 14). The difference between the mean ratios (1.5 ± 0.2 SEM) was determined to be significant by an unpaired t test (P < 0.0001). (C) Defective dispersion of a full-length Cdc14-GFP PS mutant during late mitosis. Full-length Cdc14-GFP constructs were transiently expressed from the GAL1,10 promoter by treating cells with 3% galactose for 75 min. Wild-type and PS1,2A Cdc14-GFP reporters were observed through a single cell division by time-lapse fluorescence microscopy as described in Fig. 2 B. Representative time series of wild type (top; n = 5) and PS1,2A (bottom; n = 5) are shown. Wild-type Cdc14-GFP accumulated in the nucleolus and was released during late anaphase. The time point when Cdc14-GFP was maximally released is indicated in red. Note that GFP fluorescence is difficult to see at this time point because of its dispersion throughout the cell. In contrast, mutant Cdc14 (PS1,2A)-GFP accumulated in the nucleolus and nucleus before division and was released from the nucleolus but retained in the nucleus during anaphase (arrows). Bars, 2 µm.
To verify that the qualitative differences seen in the microscope images represented a statistically significant difference in nuclear localization, we directly measured the fluorescence intensity of wild-type or PS1,2A proteins in the nucleus and the cytoplasm of late anaphase cells. Cdc14-GFP proteins were expressed at subphysiological levels from a GAL1,10 promoter in a gal1,10Δp strain that also expressed mCherry-Tub1. Late anaphase cells (spindles >5 µm) were imaged, and fluorescence of the GFP reporter (mean pixel intensity) was quantified within the nucleus and cytoplasm of multiple cells. The ratio of fluorescence inside the nucleus versus the fluorescence within the cytoplasm for both wild-type and PS1,2A proteins was determined (Fig. 5 B). An unpaired t test of the mean ratios calculated from multiple wild-type (1.8 ± 0.3, n = 16) and PS1,2A (3.3 ± 0.8, n = 14) cells indicated that the difference was significant (P < 0.0001) and that mutation of PSs within a full-length Cdc14-GFP fusion protein impairs its relocalization to the cytoplasm in late anaphase.
To confirm that the aforementioned experiment with asynchronous cells did not miss dispersion of Cdc14 that might occur despite the PS1,2A mutation, we performed time-lapse microscopy on wild-type cells expressing Cdc14-GFP or Cdc14(PS1,2A)-GFP (Fig. 5 C). Although wild-type Cdc14-GFP was sequentially released from the nucleolus and the nucleus (at which point the signal becomes difficult to see because of its diffusion throughout the cells; Fig. 5 C, 42 panel), Cdc14-PS1,2A remained concentrated within the nucleus throughout nuclear division.
Mutation of Dbf2–Mob1 phosphorylation sites in Cdc14 compromises resumption of mitotic exit
To determine whether Dbf2–Mob1-mediated inactivation of Cdc14’s C-terminal NLS influences progression through anaphase, we constructed strains in which native CDC14 was deleted, and mutant alleles of CDC14 driven by natural promoter and termination sequences were integrated at the TRP1 locus. All of the mutants that we tested were viable, and with only one exception (cdc14-BP1,2A), each of the CDC14 alleles grew with approximately wild-type kinetics at 30°C (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1).
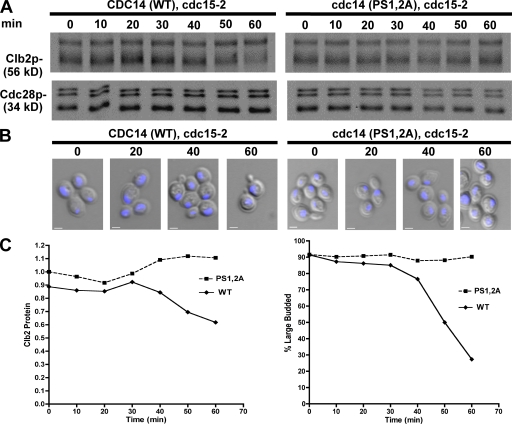
The robust growth of cells sustained by Cdc14-PS1,2A suggested that Dbf2–Mob1 needn’t inactivate the C-terminal NLS region of Cdc14 for cells to exit from mitosis. To reconcile the profound effect these mutations have on NLS function with their ability to complement cdc14Δ, we sought to determine whether these mutants were at least partially impaired by using a more sensitive assay. The temperature-sensitive cdc15-2 allele was introduced into CDC14 and cdc14-PS1,2A strains, and the cells were synchronized in late anaphase by shifting them to the nonpermissive temperature. Once the cultures had arrested at the cdc15-2 block, they were returned to the permissive temperature to restore Cdc15 function, and we tracked two hallmarks of mitotic exit: degradation of mitotic cyclin Clb2 and the completion of cytokinesis. CDC14 cells efficiently escaped mitotic arrest after reversal of the cdc15-2 block. Clb2 protein levels began to drop between 40 and 50 min after shifting the culture to 25°C (Fig. 6, A and C), and cells executed cytokinesis, resulting in a sharp reduction of the number of large-budded cells with segregated nuclei (Fig. 6, B and C). In contrast, cdc14-PS1,2A cells failed to resume cell division after reversal of the cdc15-2 block, resulting in high levels of Clb2 protein for the duration of the experiment and a failure to undergo cytokinesis (Fig. 6, A–C). The cytokinetic defect is consistent with the behavior of a Cdc14 NES mutant that also fails to accumulate in the cytoplasm (Bembenek et al., 2005). Thus, MEN-dependent inactivation of Cdc14’s C-terminal NLS is important for exit from mitosis when the activity of MEN has been compromised by the cdc15-2 mutation.
Figure 6.
Phosphorylation sites within NLS are required for efficient release from a late mitotic block. (A) Mutation of NLS phosphorylation sites impaired mitotic exit after reversal of cell cycle arrest. CDC14 and cdc14-(PS1,2A) cultures were placed at 37°C for 2 h to achieve a late mitotic cdc15-2 arrest and returned to permissive temperature (25°C). Clb2p and Cdc28 levels were followed by immunoblot analysis. (B) Cell aliquots from A were stained with DAPI and evaluated by fluorescence microscopy. Combined DIC and DAPI fluorescence images are shown. (C, left) Clb2 protein detected in A was quantified and normalized to Cdc28 levels. Relative units of fluorescence intensity were plotted for each time point. (right) The frequency of large-budded cells (n > 200) with segregated nuclei (B) was plotted for each time point. WT, wild type. Bars, 2 µm.
Inactivation of NLS promotes bypass of CDC15
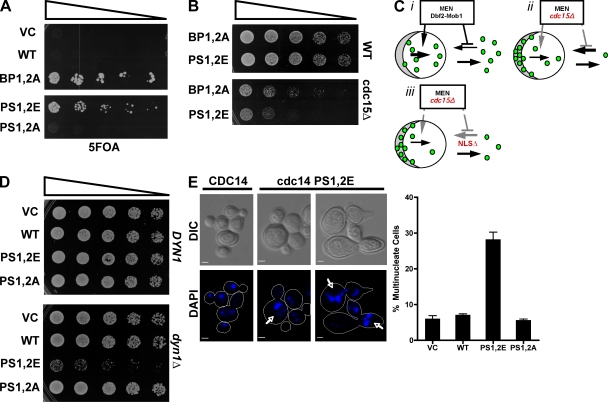
If dispersion of Cdc14 throughout the cell contributes to the exit from mitosis, we reasoned that constitutive inactivation of Cdc14’s C-terminal NLS might reduce the requirement for MEN. To address this question, we transformed a cdc15Δ strain kept alive by a CDC15 URA3 plasmid with CEN plasmid-borne CDC14 alleles and selected for loss of the CDC15 vector by plating cells on 5FOA. BP mutant CDC14-BP1,2A allowed bypass of cdc15Δ, whereas wild-type CDC14 did not (Fig. 7 A). Moreover, the CDC14-PS1,2E phosphomimetic allele also enabled bypass of cdc15Δ, although their growth relative to cells with a fully functional CDC15 gene was compromised (Fig. 7, A and B). This experiment, together with the experiment shown in Fig. 6, provides strong evidence that phosphorylation of Cdc14’s C-terminal NLS by Dbf2–Mob1 is one (but not the only) mechanism by which MEN promotes exit from mitosis.
Figure 7.
Bypass of MEN by inactivation of Cdc14’s C-terminal NLS compromises the checkpoint that monitors spindle position. (A) Inactivation of Cdc14’s C-terminal NLS bypasses the essential function of the MEN kinase Cdc15. Cdc15Δ yeast carrying plasmid-borne copies of the indicated CDC14 alleles and kept alive by a CDC15 URA3 plasmid were serially diluted and plated on 5-fluoroorotic acid to select for transformants that were able to grow without the CDC15 URA3 plasmid. (B) Growth of cdc15Δ and CDC15+ cells harboring the endogenous CDC14 gene and plasmid (pRS315)-borne alleles. Growth was measured by threefold serial dilution of yeast cells that were spotted onto synthetic media lacking leucine and incubated at 30°C. Mutant PS1,2E and BP1,2A alleles bypass cdc15Δ but grow more slowly than wild-type (WT) cells, suggesting that NLS mutations do not fully restore CDC15’s essential functions. (C) Model for MEN regulation of Cdc14. (i) MEN activity promotes release of Cdc14 from nucleolar Net1, and some of this Cdc14 makes its way to the cytoplasm. MEN also prevents reuptake of this cytoplasmic Cdc14 into the nucleus by inactivating the C-terminal NLS, allowing Cdc14 to accumulate in the cytoplasm. (ii) In a MEN mutant (e.g., cdc15Δ), release of Cdc14 from the nucleolus is inhibited. Those few molecules that are released and translocated to the cytoplasm fail to accumulate there because the C-terminal NLS remains active. Consequently, both the nuclear and cytoplasmic pools of Cdc14 are very low. (iii) The same as in ii is shown except that the C-terminal NLS is mutated. Consequently, the small number of Cdc14 molecules that leak through to the cytoplasm can accumulate there, and thereby enable suppression of cdc15Δ. (D) Constitutive inactivation of Cdc14’s C-terminal NLS compromises growth of dyn1Δ cells. Threefold serial dilutions of strains carrying the indicated CDC14 NLS mutant alleles on a low copy centromeric plasmid in either CDC14 dyn1Δ or CDC14 DYN1 backgrounds are shown. (E, left) DIC and DAPI fluorescence images of dyn1Δ cells harboring the indicated CDC14 alleles on a low copy centromeric plasmid. Yeast cultures were grown at 25°C. Cells with multiple nuclei are indicated by arrows. (right) Graph indicating the frequency with which dyn1Δ cells accumulated multiple nuclei in the presence of various CDC14 alleles (error bars indicate SD of three replicate experiments; n > 350 for each strain). VC, vector control. Bars, 2 µm.
Hypermorphic alleles of Cdc14 exacerbate defects in chromosome segregation
Cells that fail to properly distribute a set of chromosomes to both the mother and daughter cell during anaphase become arrested in late anaphase by a mechanism known as the spindle-positioning checkpoint (Burke, 2000). This checkpoint blocks activation of MEN and thereby delays cytokinesis to ensure that both the mother and daughter cell each receive a set of chromosomes. If our model for how phosphorylation of Cdc14’s NLS regulates mitotic exit is correct, nuclear localization-defective alleles of CDC14 that bypassed cdc15Δ should uncouple partitioning of the nucleus into the bud from mitotic exit and cytokinesis. To test this prediction, we created dynein-deficient dyn1Δ strains sustained by plasmid-borne alleles of CDC14. In dyn1Δ mutants, spindle orientation is compromised, and anaphase often occurs entirely within the mother cell (Li et al., 1993; Saunders et al., 1995). As a consequence, MEN is not activated, and exit from mitosis is delayed (Bardin et al., 2000).
The effect of various CDC14 mutations on the checkpoint-dependent arrest of dyn1Δ cells was assayed in two ways: by growth on synthetic medium at 25°C and by measuring the frequency with which cells became multinucleate. The CDC14-PS1,2E NLS mutant, which bypassed cdc15Δ (Fig. 7 A), grew normally in a DYN1 background but exhibited a synthetic growth defect with dyn1Δ (Fig. 7 D). To directly determine the rate at which spindle defects led to errors in DNA segregation, we grew cells overnight at 25°C and counted the frequency with which individual cell bodies contained multiple nuclei. Many cdc14-PS1,2E dyn1Δ cells had not completed cell separation, although they had clearly undergone one or more nuclear divisions within the mother cell. DAPI staining of fixed cells confirmed that 7% of CDC14 dyn1Δ cells contained multiple nuclei, whereas this value rose to 28% in cdc14-PS1,2E cells (Fig. 7 E).
Discussion
Prior observations coupled with those reported in this study lead us to propose the following sequence of events in exit from mitosis. Accumulation of GTP-bound Tem1 triggered by division of the nucleus along the mother–bud axis leads to sequential activation of the protein kinases Cdc15 and Dbf2–Mob1 (Visintin and Amon, 2001). The activated Dbf2–Mob1 translocates to the nucleus where it comes into contact with protein phosphatase Cdc14 (Stoepel et al., 2005). Dbf2–Mob1 phosphorylates an unknown protein to dislodge Cdc14 from Net1, resulting in diffusion of Cdc14 throughout the nucleus. Dbf2–Mob1 also phosphorylates Cdc14 on several sites that flank C-terminal NLS sequences, thereby inhibiting the NLS. Because of this, phosphorylated Cdc14 molecules that escape to the cytoplasm cannot efficiently return to the nucleus and thus linger in the cytoplasm where they can dephosphorylate substrates such as Cdh1 and Swi5. Although phosphorylated Cdh1 and Swi5 are retained in the cytoplasm (Jans et al., 1995; Knapp et al., 1996; Jaquenoud et al., 2002), the molecules dephosphorylated by cytoplasmic Cdc14 can now gain access to the nucleus, where they activate APC and expression of Sic1, respectively (Visintin et al., 1998; Jaspersen et al., 1999). Cdh1-APC and Sic1 extinguish cyclin B–Cdk activity, thereby enabling a return to G1 phase.
Several key pieces of evidence reported in this study support this model. First, Cdc14 contains a C-terminal NLS that is sufficient to direct localization of a GFP reporter (Fig. 1 C). Second, the activity of this NLS is transiently inhibited in late mitosis in a DBF2-dependent manner (Fig. 2, A and B). Third, the NLS sequences of Cdc14 are flanked by multiple consensus phosphorylation sites for protein kinase Dbf2–Mob1 (Fig. 1 A and Fig. 3 B). We can confirm by direct MS analysis that at least some of these sites are phosphorylated in vivo (Fig. S3) and are phosphorylated by purified Dbf2–Mob1 in vitro (Fig. 3, A–C; and Fig. S1). Fourth, mutation (to Ala) of consensus and validated Dbf2 phosphorylation sites flanking the NLS leads to a loss of DBF2-dependent regulation, such that NLS activity is no longer inhibited during late mitosis (Fig. 4, A and B). Fifth, this key regulatory control is not confined to the overexpressed NLS reporter, as it can also be observed with full-length Cdc14 expressed at subendogenous levels. Although wild-type Cdc14 is dispersed throughout the cell during late anaphase in a MEN-dependent manner (Shou et al., 1999; Visintin et al., 1999), a mutant with a nonphosphorylatable C-terminal NLS is not (Fig. 5, A–C). The role of regulated nuclear transport in control of mitotic exit described in this study adds to a long list of processes that are known to be controlled by phosphorylation-dependent inhibition of NLS activity, dating back to the observation that phosphorylation of consensus Cdk phosphorylation sites adjacent to NLS of Swi5 blocks its uptake into the nucleus during mitosis (Moll et al., 1991).
The behavior of Cdc14 NLS mutants suggests that MEN promotes exit from mitosis by at least two distinct mechanisms. This interpretation is based on the observation that cells can be sustained by an allele of Cdc14 in which the Dbf2–Mob1 phosphorylation sites have been changed to Ala (Fig. S4). Further support is provided by the observation that a phosphomimetic mutant (CDC14-PS1,2E) can bypass cdc15Δ (Fig. 7 A) when expressed from a CEN plasmid. If Cdc14 is the only target (or the only essential target) of Dbf2–Mob1, a nonphosphorylatable allele of Cdc14 should mimic a dbf2 loss of function mutant. This is clearly not the case, which suggests the existence of a second activity for Dbf2–Mob1 that can promote exit from mitosis even if Dbf2–Mob1 cannot phosphorylate the C-terminal NLS of Cdc14 and is required for exit from mitosis even if the C-terminal NLS is inactivated by mutation. However, it is clear that phosphorylation of Cdc14’s NLS contributes to mitotic exit because a mutant lacking these phosphorylation sites is unable to recover efficiently from a cdc15ts block and resume exit from mitosis (Fig. 6, A–C).
The work presented in this study represents a significant step forward in our understanding of exit from mitosis, in which we identify Cdc14 as the first known substrate of Dbf2–Mob1. Our findings thereby yield the first mechanistic insights into how this protein kinase acts directly to trigger mitotic exit, which is a topic that has remained an enigma since it was first discovered nearly a decade ago that Dbf2–Mob1 underlies the release of Cdc14 from the nucleolus during late anaphase (Shou et al., 1999; Visintin et al., 1999). However, a complete description of the molecular basis for the exit from mitosis awaits identification of the additional mechanisms mobilized by Dbf2–Mob1. What is the best candidate for this mechanism? Our data indicate that even though Cdc14 that lacks Dbf2–Mob1 phosphorylation sites is not efficiently localized to the cytoplasm during late mitosis, it is nevertheless released from the nucleolar tether protein Net1 (Fig. 5 A). This implies that the other way in which Dbf2–Mob1 acts is to disrupt the Cdc14–Net1 complex. A Cdc14 mutant lacking the C-terminal phosphorylation sites is essentially not a substrate for Dbf2–Mob1 in vitro (Fig. 3, A and C), but the corresponding phosphomimetic mutant still localizes primarily to the nucleolus in interphase cells (not depicted), suggesting that these phosphorylations do not disrupt the Cdc14–Net1 complex. If Dbf2–Mob1 activates parallel mechanisms to secure mitotic exit, then its second target need not be essential. Multiple proteins implicated in regulator of nucleolar silencing and telophase formation or function are plausible targets, including RNA Pol I, Sir2, Spo12, and Net1. Indeed, Net1 was identified in an in vitro proteomic screen for Dbf2–Mob1 substrates (Mah et al., 2005). Evaluation of this possibility will require analysis of Net1 phosphorylation during exit from mitosis and phenotypic characterization of PS mutants.
A question posed by our results that remains unresolved is why is cytoplasmic retention of Cdc14 not essential for exit from mitosis? We consider two potential explanations. First, the CDC14-PS1,2A mutant may not be a complete PS null. We have attempted to address this by eliminating all three PS clusters, but mutations in PS3 have the paradoxical effect of suppressing cdc15ts (unpublished data), pointing to either a nonspecific effect of the mutations or further complexity in the regulation of Cdc14. Our second explanation relies on the notion that Cdc14 is constantly being exported to the cytoplasm during mitotic exit. Even though Cdc14-PS1,2A molecules are quickly resequestered in the nucleus as a result of action of the uninhibitable mutant C-terminal NLS, they may nonetheless be capable of exerting activity on their substrates during their brief sojourn in the cytoplasm. According to this viewpoint, the chief function of the regulatory circuit we have uncovered in this study is to delay the return of Cdc14 to the nucleus, thereby allowing it to achieve a higher concentration in the cytoplasm. Although this increase would not be as robust in cdc14-PS1,2A cells, it could be sufficient to exceed a threshold level of cytoplasmic Cdc14 activity to enable exit from mitosis unless the other activities of the Cdc15–Dbf2–Mob1 module have been circumscribed.
While this manuscript was in revision, a paper appeared from Chen et al. (2008) on the phosphorylation of Clp1 by Sid2. Clp1 is the S. pombe homologue of Cdc14, and Sid2 is a homologue of Dbf2. Although S. pombe contains a pathway very similar to MEN, in fission yeast, this pathway controls the initiation of septation and not exit from mitosis. Chen et al. (2008) show that Sid2 phosphorylates Clp1 on a cluster of sites in vitro and in vivo. The 14-3-3 protein Rad24 binds to phospho-Clp1, which retains Clp1 in the cytoplasm throughout cytokinesis. A nonphosphorylatable mutant, Clp1, returns to the nucleus prematurely during cytokinesis, and this renders the cells sensitive to perturbations of the cytokinetic machinery. There are some interesting differences in the phosphorylation sites of Cdc14 and Clp1. Although the target region of Clp1 spans ∼125 residues and has a significant content of basic (10 Lys and Arg) and acidic (11 Glu and Asp) residues, the target region of Cdc14 is compact, with all sites clustered within a stretch of 25 amino acids (lacking Asp or Glu) and tightly interdigitated with four pairs of Lys or Arg that have NLS activity. NLS for Clp1 has not been mapped. It could be that both yeasts use different strategies to retain Cdc14/Clp1 in the cytoplasm during exit from mitosis/cytokinesis or that a combination of phosphorylation and binding of 14-3-3 protein inactivates a C-terminal NLS in both proteins.
Materials and methods
Yeast strains and plasmids
All strains were made in the W303 background (can1-100, leu2-3, his3-11, trp1-1, ura3-1, and ade2-1). Strain genotypes including plasmids are described in Table S1 (available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1). The wild-type CDC14 gene was cloned with upstream promoter and downstream terminator sequences into pBluescript II SK+ (Agilent Technologies) with flanking primers that introduce XbaI and XhoI restriction endonuclease cleavage sites. CDC14 alleles were generated by site-directed PCR mutagenesis using high fidelity polymerase (LA Taq; Takara) with primers described in Table S2. Subsequent genetic analysis of pRS315 (LEU2)-borne alleles was performed in various mutant backgrounds. CDC14 alleles were also integrated into the chromosome at the TRP1 locus by cloning the CDC14 gene with upstream promoter and downstream terminator sequences into pRS304 and cutting the plasmid with SnaBI before transformation into W303 strains deleted for the endogenous CDC14. When necessary, a complementing copy of wild-type CDC14 in pRS316 (URA3) was evicted by counter selection on synthetic defined medium with 0.1% 5FOA and 50 µg/ml uracil. To measure the growth defects of each allele, 10 µl of threefold serial dilutions starting at 3 × 103 cells/µl were spotted onto selective media.
The full-length CDC14 coding sequence and various truncations were cloned by PCR and inserted into a high copy vector using unique BamHI and HindIII sites that were created during amplification (Table S3, available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1). Mutations within either full-length CDC14 or NLS sequences were created by site-directed PCR mutagenesis as described in this study. In-frame C-terminal GFP(S65T) fusions of full-length and Cdc14 truncations were cloned into pTS508 (provided by T. Stearns, Stanford University, Palo Alto, CA) using BamHI and HindIII sites downstream of the GAL1,10 promoter. The same CDC14 clones were also inserted into a GST fusion vector pGEX-6P-2 that had been modified by site-directed mutagenesis to include an additional translation stop codon. Expression of all N-terminal GST fusions also resulted in the addition of two amino acids, glutamate followed by Ala, at the C terminus. Constructs that expressed GFP-Crz1 from the GAL1,10 promoter were provided by M. Cyert (Stanford University, Palo Alto, CA).
To more tightly control expression of GFP-tagged fusion proteins, we created yeast strains lacking ∼300 base pairs of DNA encompassing the divergent promoter required for transcription of the endogenous GAL1 and the GAL10 genes. A deletion at the GAL1,10 locus was created by double crossover recombination of a hygromycin B resistance cassette. Primers for PCR of the cassette are described in Table S3. Strains harboring pHIS3::mCherry-Tub1 at the TRP1 locus of S. cerevisiae were generated by integrating a pRS304-derived plasmid, pAK010 (provided by E. Schiebel, Center for Molecular Biology, Heidelberg, Germany), after digestion with SnaBI. Strains expressing DsRed-tagged Nop1 were generated by integrating pFA6a-KanMX4(DsRed-Nop1) at the S. cerevisiae NOP1 locus.
Microscopy and imaging
All images were captured with a microscope (Axiovert 200M; Carl Zeiss, Inc.) equipped with a 100× Plan-Apochromat 1.40 NA DIC objective (Carl Zeiss, Inc.) and a charge-coupled device camera (Orca 2; Hamamatsu Photonics). Data were collected and analyzed with Axiovision (version 4.6; Carl Zeiss, Inc.). Cropping of images and adjustments to brightness were made within Axiovision. To visualize Nop1-DsRed and mCherry-Tub1 with GFP reporters, high contrast (γ adjusted) images of the red channel were created within Axiovision. Time-lapse series were captured after mounting live cells in synthetic complete medium with 2.5% low melting point (LMP) agarose (Invitrogen) onto glass slides (Gold Seal) and covered with glass coverslips (VWR). Cells were maintained at the indicated temperatures with a temperature-controlled stage and objective collar. Expression of NLS-GFP (90 min) and Cdc14-GFP (75 min) was induced by the addition of galactose (3% final concentration) to log phase cultures. More modest levels of Cdc14-GFP expression were achieved by adding 0.015% galactose to synthetic medium lacking uracil and supplemented with 3% raffinose for ∼10–16 h at 25°C. Expression of full-length Cdc14-GFP reporter fusions was estimated by immunoblot analysis of yeast cultures induced with either low (0.015%) or high (3%) concentrations of galactose. SDS samples were prepared from log phase cultures after adding 20% TCA (Azzam et al., 2004). Polyclonal anti-Cdc14 and HRP-conjugated goat anti–rabbit IgG (Bio-Rad Laboratories) antibodies were used to detect Cdc14 and Cdc14-GFP in extracts.
To assay localization of NLS-GFP in a conditional dbf2-2 mutant, expression of the NLS-GFP reporter was induced with galactose (3% final concentration) for 90 min. Dextrose (2% final concentration) was added to stop induction, and cells were allowed to grow at 37°C for 45 min. To assay release of NLS-GFP to the cytoplasm, cell metabolism was quenched by washing cultures in 1× TE for 10 min before resuspension in 2.5% LMP agarose in TE. Both NLS-GFP and mCherry-Tub1 (spindle) were imaged. Spindle length was measured with Axiovision. Localization of the wild-type NLS-GFP reporter was determined for wild type (n = 75) and dbf2-2 (n = 50).
A χ2 contingency test of NLS-GFP alleles was performed using Prism 4.0 (GraphPad Software, Inc.). The number of cells with either released (cytoplasm and nuclear) or nuclear localized GFP fluorescence was determined for cells with spindles >5 µm long. Results for each PS allele of NLS-GFP were directly compared with wild type with a two-sided χ2 test. P-values are reported.
To determine the number of nuclei per cell (Fig. 7 C), yeast cultures were grown overnight to late log phase. Before analysis, 350 µl 37% formaldehyde (Mallinckrodt) was added directly to a 5-ml culture and allowed to incubate for 15 min at room temperature. Fixed cells were washed with PBS and dried onto polylysine-treated slides (MP Biomedicals). Mounted yeast cells were briefly dipped in 0.1 µg/ml DAPI (Sigma-Aldrich), rinsed with distilled H2O, and covered with 50% glycerol and a glass coverslip. The number of DAPI-stained nuclei per cell body was assayed. More than one nucleus per compartment was counted as multiple nuclei. Large-budded (undivided) cells with two nuclei in the mother cell were ignored. Values shown are the mean of three independent experiments with >350 cells counted for each strain. Error bars are SD of the three experiments.
Quantification of fluorescence intensity was performed using Axiovision software with quantification modules. Mean pixel intensities within the nucleus or cytoplasm were determined by selecting a single circle of interest within each compartment (Fig. 5 B). Background light was measured by selecting a large circle of interest outside of each cell. Mean pixel intensities (arbitrary units) were calculated by subtracting background from nuclear and cytoplasmic values. The ratio of nuclear versus cytoplasmic fluorescence was determined for each cell. Values reported are the mean of multiple experiments (wild type, n = 16; PS1,2A, n = 14). An unpaired t test (with Welch's correction) comparing the mean ratio of cytoplasmic versus nuclear fluorescence intensity was performed (P < 0.0001).
Cdc15-2 block and release
cdc15-2,CDC14 and cdc15-2,cdc14(PS1,2A) cultures were grown overnight at 25°C in YP (yeast extract with peptone) medium to mid-log phase (OD 0.4). Cultures were transferred to 37°C for 2 h (late anaphase arrest) and shifted back to 25°C. To determine Clb2p levels, 10-ml aliquots were removed at each time point, and growth was stopped with the addition of TCA (20% final concentration) for 20 min on ice. Cells were washed three times to remove TCA. For each wash, cells were pelleted and then resuspended in 10 ml of 100 mM Tris, pH 8.0. After the final wash, cells were pelleted and resuspended in SDS sample buffer with 8 M urea before glass bead disruption. Clb2 and Cdc28 were separated by SDS-PAGE for immunoblot analysis with polyclonal anti-Clb2p and PSTAIRE antibodies (polyclonal anti-Cdc2; Santa Cruz Biotechnology, Inc.). Fluorescently labeled secondary antibodies (IRDye 800–conjugated anti–rabbit IgG; Rockland Biochemicals) were used before imaging (Odyssey infrared imaging system; LI-COR Biosciences). Bands corresponding to Clb2 and Cdc28 were quantified with Odyssey software (version 2.0). Odyssey-captured images were cropped with Photoshop (CS; Adobe). Relative levels of Clb2 protein were plotted in Fig. 6 C after normalizing Clb2 to Cdc28 levels for each time point. To determine the frequency of large-budded cells with segregated nuclei (Fig. 6, B and C), 1-ml aliquots were removed every 20 min after reversal of the cdc15-2 block, and metabolism was quenched by washing pellets into TE before drying onto polylysine-treated slides (MP Biomedicals). Mounted yeast cells were briefly dipped in 0.1 µg/ml DAPI (Sigma-Aldrich), rinsed with distilled H2O, and covered with 50% glycerol and a glass slip before imaging. More than 200 cells were counted for each time point.
Protein expression and purification
Full-length GST-Cdc14 and GST-NLS fusion proteins were expressed in E. coli (BL21) and purified in one step over glutathione Sepharose. In brief, 1-L cultures were grown to OD 0.5 at 30°C and induced with IPTG (Sigma-Aldrich) at a final concentration of 1 mM for 3 h. Cells were harvested and resuspended in buffer containing 50 mM Tris, pH 7.5, 100 mM NaCl, 0.2% Triton X-100, 1 mM DTT, and 2 mM EDTA. Cells were disrupted at 25,000 lbs/in.2 using a One Shot (Constant Systems Ltd), and protease inhibitors were added (1 mM PMSF; aprotinin, chymostatin, leupeptin, and pepstatin A all at 5 µg/ml in 90% DMSO) to the crude extract before centrifugation at 35,000 rpm in a Ti60 rotor for 30 min (L8-5M Ultracentrifuge; Beckman Coulter). GST fusion proteins were bound to glutathione Sepharose resin, washed four times with lysis buffer, and eluted with buffer containing 50 mM Tris, pH 7.5, 100 mM NaCl, 0.2% Triton X-100, 1 mM DTT, and 20 mM glutathione. Both GST-Cdc14 and GST-NLS were dialyzed into buffer containing 50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM DTT, and 10% glycerol.
Flag-6His-HA-Dbf2 proteins were expressed either alone or together with 6His-Mob1 in Sf9 insect cells and purified by nickel affinity with MagneHis particles (Promega). Expression of tagged proteins was performed as previously described (Mah et al., 2001). Sf9 cells were resuspended in buffer containing 10 mM Hepes-KOH, pH 7.5, 150 mM NaCl, 20 mM β-glycerophosphate, 5 mM EGTA, and 5 mM β-mercaptoethanol with 1% CHAPS and disrupted by sonication. Extracts were cleared by centrifugation at 15,000 rpm for 15 min (SS-34 rotor; Sorvall), and Dbf2–Mob1 complexes were bound to MagneHis particles and washed four times with buffer containing 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 0.2% Triton X-100, 1 mM DTT, and 20 mM imidazole. Proteins were eluted from resin with 50 mM Tris-Cl, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 2 mM EDTA, 0.2% Triton X-100, 1 mM DTT, and 200 mM imidazole. Purified Dbf2–Mob1 proteins were dialyzed into 50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT, and 10% glycerol. 6His-Cdc15 was expressed in Sf9 cells and purified as previously described (Mah et al., 2001).
Kinase reactions
1 µg Dbf2–Mob1 was activated by 0.2 µg Cdc15 kinase for 15 min at 25°C in Cdc15 kinase buffer (50 mM Hepes-KOH, pH 7.5, 5 mM MgCl2, 2.5 mM MnCl2, 5 mM β-glycerophosphate, 1 mM DTT, and 20 µM ATP). Activated Dbf2–Mob1 was diluted twofold in Dbf2 kinase buffer (50 mM Tris, pH 7.4, 60 mM potassium acetate, 10 mM magnesium chloride, 1 mM DTT, and 10 µM ATP) with 3 µCi [32P]ATP and either 15 µg calf thymus H1 histone (Sigma-Aldrich), 5 µg full-length GST-Cdc14, or 5 µg GST-NLS. Reactions were incubated at 25°C for 20 min and stopped with the addition of 2× SDS sample buffer. Phosphorylated proteins were separated by SDS-PAGE and detected by autoradiography (Biomax MR; Kodak). Gel images were captured with Scan Jet (8270; HP) and Scan Jet software (HP). Images were cropped and adjusted for brightness with Photoshop.
Determination of Dbf2-dependent phosphorylation
To identify sites phosphorylated by Dbf2, GST-Cdc14 was kinased by Cdc15-activated Dbf2–Mob1 as described for in vitro kinase reactions but without the addition of radioactive [32P]ATP. Phosphorylated proteins were separated by SDS-PAGE and stained with SimplyBlue (Invitrogen) to excise gel slices corresponding to GST-Cdc14. Gel slices were equilibrated in 100 mM ammonium bicarbonate, reduced with DTT, and alkylated with iodoacetamide as previously described (Joyal et al., 1997). After incubation overnight at 37°C with 400 ng AspN (Boehringer Mannheim), 50 ng modified trypsin (Promega) was added, and the incubation was continued for either 1 or 4 h at 37°C. Peptides were extracted with acetonitrile and lyophilized.
Phosphopeptide analysis of Cdc14 was accomplished by LC electrospray MS on a hybrid ion trap-triple quadrupole mass spectrometer (4000 QTRAP; Applied Biosystems). Operating as a triple quadrupole mass spectrometer, the instrument selectively detects phosphopeptides as they elute from the LC column using a precursor ion scan for the phosphopeptide marker ion PO3− (mass/charge of 79) in the negative ion mode (Zappacosta et al., 2002). Each precursor scan is followed by a short, narrow mass range scan in the ion trap to determine the monoisotopic mass and charge state for the three most abundant ions detected in the precursor scan. These two scan functions are operating continuously throughout the LC gradient. Select phosphopeptides are sequenced by targeted LC-MS/MS in a separate injection.
In vivo PS mapping
6His-HA-protease3C-ZZ-Cdc14 protein was expressed in wild-type yeast cells (RJD863) by growing the cells in raffinose and inducing the GAL1 promoter of BG1805 (clone YFR028C; Open Biosystems) for 150 min by addition of 3% galactose. In brief, 100-ml cultures expressing Cdc14 were flash killed with the addition of TCA to a final concentration of 20%. Cell pellets were washed five times with 100 mM Tris, pH 8.0, and resuspended in 250 µl of lysis buffer containing 100 mM Hepes, pH 8.0, 8 M urea, 0.1% SDS, and 10 mM imidazole before glass bead agitation (Fast Prep; BIO 101, Inc.). Extracts were allowed to sit at 42°C for 10 min after the addition of protease inhibitors (aprotinin, chymostatin, leupeptin, and pepstatin A at 5 µg/ml in 90% DMSO). Supernatants were cleared by centrifugation at 15,000 rpm in a microcentrifuge (541D; Eppendorf) for 10 min. Half of the extract was added to MagneHis resin and incubated at room temperature for 1 h. Bound proteins and MagneHis resin were washed four times with lysis buffer and eluted with 2× SDS sample buffer containing 8 M urea and 0.5 M imidazole. Proteins were separated by SDS-PAGE and visualized with SimplyBlue stain. Bands corresponding to Cdc14 were cut out and flash frozen in liquid N2 before analysis.
To detect Cdc14 phosphorylation in vivo, we used multiple reaction monitoring MS on the previously described 4000 QTRAP. The multiple reaction monitoring experiment detects the fragmentation reaction of a selected precursor ion undergoing collision-induced dissociation to produce a specific fragment ion. In this study, the production of a select set of fragment ions (Fig. S3) unique to the Cdc14 phosphopeptides KIpSGSIK and TTpSAAGGIR was monitored continuously throughout an LC-MS analysis of tryptically digested Cdc14 from RJD863. Cdc14 digests were prepared as described above (Determination of Dbf2-dependent…) except trypsin incubation was performed for 4 h.
Online supplemental material
Fig. S1 shows expression of full-length Cdc14-GFP with low and high concentrations of galactose. Fig. S2 shows tandem MS mapping of Dbf2–Mob1 phosphorylation sites on GST-Cdc14. Fig. S3 shows the detection of Cdc14 phosphorylation at PS2 and PS3 motifs in vivo by MS. Fig. S4 shows growth of strains sustained by CDC14 alleles integrated at the TRP1 locus. Fig. S5 shows the localization of GFP-Crz1 in temperature-sensitive dbf2-2 mutants. Table S1 shows yeast strains relevant to the preparation of this study. Table S2 shows the primers used to create various CDC14 alleles. Table S3 shows the plasmids used to express various CDC14 alleles in S. cerevisiae. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812022/DC1.
Acknowledgments
We thank the Caltech Protein Expression Facility for technical assistance, T. Stearns, M. Cyert, and E. Schiebel for plasmids, and members of the Deshaies Laboratory, especially W. Shou, R. Azzam, and A. Mah, for their valuable assistance.
R.J. Deshaies is a Howard Hughes Medical Institute investigator. This work was supported by a National Institutes of Health grant (GM059940) to R.J. Deshaies. D.A. Mohl was supported in part by a postdoctoral fellowship from the American Cancer Society.
Footnotes
Abbreviations used in this paper: APC, anaphase-promoting complex; BP, basic patch; DIC, differential interference contrast; LC, liquid chromatography; LMP, low melting point; MEN, mitotic exit network; MS, mass spectrometry; NES, nuclear export signal; PS, phosphosite.
References
- Asakawa K., Toh-e A. 2002. A defect of Kap104 alleviates the requirement of mitotic exit network gene functions in Saccharomyces cerevisiae.Genetics. 162:1545–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam R., Chen S.L., Shou W., Mah A.S., Alexandru G., Nasmyth K., Annan R.S., Carr S.A., Deshaies R.J. 2004. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus.Science. 305:516–519 [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Visintin R., Amon A. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus.Cell. 102:21–31 [DOI] [PubMed] [Google Scholar]
- Bembenek J., Kang J., Kurischko C., Li B., Raab J.R., Belanger K.D., Luca F.C., Yu H. 2005. Crm1-mediated nuclear export of Cdc14 is required for the completion of cytokinesis in budding yeast.Cell Cycle. 4:961–971 [DOI] [PubMed] [Google Scholar]
- Bouck D.C., Bloom K.S. 2005. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast.Proc. Natl. Acad. Sci. USA. 102:5408–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boustany L.M., Cyert M.S. 2002. Calcineurin-dependent regulation of Crz1p nuclear export requires Msn5p and a conserved calcineurin docking site.Genes Dev. 16:608–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke D.J. 2000. Complexity in the spindle checkpoint.Curr. Opin. Genet. Dev. 10:26–31 [DOI] [PubMed] [Google Scholar]
- Chen C.T., Feoktistova A., Chen J.S., Shim Y.S., Clifford D.M., Gould K.L., McCollum D. 2008. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1.Curr. Biol. 18:1594–1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D., Amon A. 2004. At the interface between signaling and executing anaphase–Cdc14 and the FEAR network.Genes Dev. 18:2581–2595 [DOI] [PubMed] [Google Scholar]
- D'Aquino K.E., Monje-Casas F., Paulson J., Reiser V., Charles G.M., Lai L., Shokat K.M., Amon A. 2005. The protein kinase Kin4 inhibits exit from mitosis in response to spindle position defects.Mol. Cell. 19:223–234 [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Wells G.P., Smerdon S.J., Sedgwick S.G. 2004. Clb6/Cdc28 and Cdc14 regulate phosphorylation status and cellular localization of Swi6.Mol. Cell. Biol. 24:2277–2285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. 2005. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation.Nature. 433:171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jans D.A., Moll T., Nasmyth K., Jans P. 1995. Cyclin-dependent kinase site-regulated signal-dependent nuclear localization of the SW15 yeast transcription factor in mammalian cells.J. Biol. Chem. 270:17064–17067 [DOI] [PubMed] [Google Scholar]
- Jaquenoud M., van Drogen F., Peter M. 2002. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1).EMBO J. 21:6515–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., Morgan D.O. 1999. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14.Curr. Biol. 9:227–236 [DOI] [PubMed] [Google Scholar]
- Jaspersen S.L., Charles J.F., Tinker-Kulberg R.L., Morgan D.O. 1998. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae.Mol. Biol. Cell. 9:2803–2817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyal J.L., Annan R.S., Ho Y.D., Huddleston M.E., Carr S.A., Hart M.J., Sacks D.B. 1997. Calmodulin modulates the interaction between IQGAP1 and Cdc42. Identification of IQGAP1 by nanoelectrospray tandem mass spectrometry.J. Biol. Chem. 272:15419–15425 [DOI] [PubMed] [Google Scholar]
- Knapp D., Bhoite L., Stillman D.J., Nasmyth K. 1996. The transcription factor Swi5 regulates expression of the cyclin kinase inhibitor p40SIC1.Mol. Cell. Biol. 16:5701–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Y., Yeh E., Hays T., Bloom K. 1993. Disruption of mitotic spindle orientation in a yeast dynein mutant.Proc. Natl. Acad. Sci. USA. 90:10096–10100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A.S., Elia A.E., Devgan G., Ptacek J., Schutkowski M., Snyder M., Yaffe M.B., Deshaies R.J. 2005. Substrate specificity analysis of protein kinase complex Dbf2-Mob1 by peptide library and proteome array screening.BMC Biochem. 6:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mah A.S., Jang J., Deshaies R.J. 2001. Protein kinase Cdc15 activates the Dbf2-Mob1 kinase complex.Proc. Natl. Acad. Sci. USA. 98:7325–7330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston A.L., Lee B.H., Amon A. 2003. The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation.Dev. Cell. 4:711–726 [DOI] [PubMed] [Google Scholar]
- Moll T., Tebb G., Surana U., Robitsch H., Nasmyth K. 1991. The role of phosphorylation and the CDC28 protein kinase in cell cycle-regulated nuclear import of the S. cerevisiae transcription factor SWI5.Cell. 66:743–758 [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2001. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events.Curr. Opin. Cell Biol. 13:762–769 [DOI] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. 2003. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14.Science. 302:2120–2124 [DOI] [PubMed] [Google Scholar]
- Queralt E., Lehane C., Novak B., Uhlmann F. 2006. Downregulation of PP2A(Cdc55) phosphatase by separase initiates mitotic exit in budding yeast.Cell. 125:719–732 [DOI] [PubMed] [Google Scholar]
- Saunders W.S., Koshland D., Eshel D., Gibbons I.R., Hoyt M.A. 1995. Saccharomyces cerevisiae kinesin- and dynein-related proteins required for anaphase chromosome segregation.J. Cell Biol. 128:617–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider B.L., Zhang J., Markwardt J., Tokiwa G., Volpe T., Honey S., Futcher B. 2004. Growth rate and cell size modulate the synthesis of, and requirement for, G1-phase cyclins at start.Mol. Cell. Biol. 24:10802–10813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M., Neutzner M., Mocker D., Seufert W. 2001. Yeast Hct1 recognizes the mitotic cyclin Clb2 and other substrates of the ubiquitin ligase APC.EMBO J. 20:5165–5175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., Deshaies R.J. 2002. Multiple telophase arrest bypassed (tab) mutants alleviate the essential requirement for Cdc15 in exit from mitosis in S. cerevisiae.BMC Genet. 3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W., Sakamoto K.M., Keener J., Morimoto K.W., Traverso E.E., Azzam R., Hoppe G.J., Feldman R.M., DeModena J., Moazed D., et al. 2001. Net1 stimulates RNA polymerase I transcription and regulates nucleolar structure independently of controlling mitotic exit.Mol. Cell. 8:45–55 [DOI] [PubMed] [Google Scholar]
- Shou W., Seol J.H., Shevchenko A., Baskerville C., Moazed D., Chen Z.W., Jang J., Shevchenko A., Charbonneau H., Deshaies R.J. 1999. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex.Cell. 97:233–244 [DOI] [PubMed] [Google Scholar]
- Stegmeier F., Visintin R., Amon A. 2002. Separase, polo kinase, the kinetochore protein Slk19, and Spo12 function in a network that controls Cdc14 localization during early anaphase.Cell. 108:207–220 [DOI] [PubMed] [Google Scholar]
- Stoepel J., Ottey M.A., Kurischko C., Hieter P., Luca F.C. 2005. The mitotic exit network Mob1p-Dbf2p kinase complex localizes to the nucleus and regulates passenger protein localization.Mol. Biol. Cell. 16:5465–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight A.F., Shou W., Dowd G.J., Turck C.W., Deshaies R.J., Johnson A.D., Moazed D. 1999. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity.Cell. 97:245–256 [DOI] [PubMed] [Google Scholar]
- Sullivan M., Higuchi T., Katis V.L., Uhlmann F. 2004. Cdc14 phosphatase induces rDNA condensation and resolves cohesin-independent cohesion during budding yeast anaphase.Cell. 117:471–482 [DOI] [PubMed] [Google Scholar]
- Taylor G.S., Liu Y., Baskerville C., Charbonneau H. 1997. The activity of Cdc14p, an oligomeric dual specificity protein phosphatase from Saccharomyces cerevisiae, is required for cell cycle progression.J. Biol. Chem. 272:24054–24063 [DOI] [PubMed] [Google Scholar]
- Torres-Rosell J., Machin F., Jarmuz A., Aragon L. 2004. Nucleolar segregation lags behind the rest of the genome and requires Cdc14p activation by the FEAR network.Cell Cycle. 3:496–502 [PubMed] [Google Scholar]
- Trautmann S., McCollum D. 2005. Distinct nuclear and cytoplasmic functions of the S. pombe Cdc14-like phosphatase Clp1p/Flp1p and a role for nuclear shuttling in its regulation.Curr. Biol. 15:1384–1389 [DOI] [PubMed] [Google Scholar]
- Visintin R., Amon A. 2001. Regulation of the mitotic exit protein kinases Cdc15 and Dbf2.Mol. Biol. Cell. 12:2961–2974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R., Craig K., Hwang E.S., Prinz S., Tyers M., Amon A. 1998. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation.Mol. Cell. 2:709–718 [DOI] [PubMed] [Google Scholar]
- Visintin R., Hwang E.S., Amon A. 1999. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus.Nature. 398:818–823 [DOI] [PubMed] [Google Scholar]
- Wang Y., Hu F., Elledge S.J. 2000. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit.Curr. Biol. 10:1379–1382 [DOI] [PubMed] [Google Scholar]
- Wang Y., Shirogane T., Liu D., Harper J.W., Elledge S.J. 2003. Exit from exit: resetting the cell cycle through Amn1 inhibition of G protein signaling.Cell. 112:697–709 [DOI] [PubMed] [Google Scholar]
- Woodbury E.L., Morgan D.O. 2007. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase.Nat. Cell Biol. 9:106–112 [DOI] [PubMed] [Google Scholar]
- Zachariae W., Schwab M., Nasmyth K., Seufert W. 1998. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex.Science. 282:1721–1724 [DOI] [PubMed] [Google Scholar]
- Zappacosta F., Huddleston M.J., Karcher R.L., Gelfand V.I., Carr S.A., Annan R.S. 2002. Improved sensitivity for phosphopeptide mapping using capillary column HPLC and microionspray mass spectrometry: comparative phosphorylation site mapping from gel-derived proteins.Anal. Chem. 74:3221–3231 [DOI] [PubMed] [Google Scholar]