Abstract
Expression of the Keratin 13 (KRT13) gene, which encodes a cytoskeletal protein thought to play important roles in breast cancer growth and metastasis, is differentially regulated by estradiol (E2) and the selective estrogen receptor modulators (SERMs) tamoxifen and raloxifene. While stimulation of KRT13 by tamoxifen is robust and prolonged, stimulation by E2 is more transient and raloxifene has virtually no effect. To investigate the mechanistic basis for the differential ligand regulation of KRT13, we have defined the regulatory regions of KRT13, compared gene expression by E2 and SERMs, and explored the magnitudes and time courses of estrogen receptor (ER) and cofactor recruitment patterns on these regions. Using a ChIP scanning approach and reporter transactivation assays, we identified a 2.5 kb upstream ER-binding regulatory region for KRT13. Directed composite mutations in this region revealed that three estrogen response elements and three Sp1 sites were involved in its ligand dependent regulation. Differential recruitment of ERα and cofactors to the KRT13 regulatory sites paralleled the different time course and magnitude of regulation by these ligands: There was almost no ERα or cofactor recruitment with raloxifene, whereas there was strong, prolonged ER recruitment and histone acetylation with tamoxifen, and an early and more transient recruitment with E2. Taken together, our results suggest that the different ligand regulations of KRT13 are due to ligand-differential recruitment of ER and coactivators, and they provide insight into the mechanisms responsible for the different agonistic activities and differential gene regulation by estradiol and the SERMs tamoxifen and raloxifene.
Keywords: estrogen receptor, estradiol, tamoxifen, SERM, gene expression, breast cancer
Introduction
Estrogens are critical for the development and maintenance of reproductive tissues and have important roles in additional tissues such as bone and cardiovascular, and in nervous system function (Deroo and Korach, 2006). These actions are mediated through estrogen receptors alpha and beta (ERα and ERβ, respectively), which are members of the nuclear receptor superfamily. By binding to cognate ligands or pharmacologic agents such as the selective estrogen receptor modulators (SERMs) tamoxifen and raloxifene, the ER undergoes conformational changes and is able to display protein surfaces which allow the receptor to bind to DNA regulatory sequences in chromatin directly or indirectly through protein intermediaries. Direct ER binding sites include palindromic estrogen response elements (EREs), non-consensus EREs and ERE half sites. When binding to chromatin indirectly, ER regulates gene expression by tethering to other transcription factors, such as AP-1 and Sp1.
Depending on the properties of ligands, availability of coregulators in a given cell, and chromatin contexts, the ER is able to recruit different complexes of coregulators to either stimulate or repress the expression of target genes (Brzozowski et al., 1997, Kumar and Chambon, 1988, Lonard et al., 2007, Murdoch and Gorski, 1991). In the context of breast cancers, estrogens have long been known to promote tumor cell growth, presumably through regulating certain growth-promoting gene subsets such as stimulatory cell cycle-related genes (Frasor et al., 2003, Katzenellenbogen and Frasor, 2004). The SERM tamoxifen is widely utilized in breast cancer therapies (Smith and Good, 2003, Strassmer-Weippl and Goss, 2003), and exerts its effects by binding ER and differentially regulating target genes. By genome-wide microarray gene transcriptional profiling, Frasor et al. (Frasor et al., 2003, Frasor et al., 2004, Frasor et al., 2006) identified genes that were differentially regulated by E2 and SERMs. In most cases, genes were up-regulated or down-regulated by E2, with this regulation being reversed by the SERMs tamoxifen and raloxifene and the antiestrogen ICI 182, 780. However, some genes such as keratin 13 (KRT13) were highly stimulated by the SERM tamoxifen.
Keratins comprise the intermediate filament cytoplasmic cytoskeleton and are the major structural proteins of epithelial cells. Intermediate filaments are made up of heterodimeric polymers composed of both type I and type II keratins which are encoded by more than 50 genes in separate clusters on chromosomes 12 and 17 (Hesse et al., 2001). The specific structural or regulatory functions required for keratin filaments to act in a tissue-specific manner are likely to result from the directed transcriptional regulation of distinct keratin genes and the properties of the proteins they encode (Magin et al., 2007). Keratin 13, a type I keratin (KRT10-KRT-20), often pairs with KRT4, a type II keratin (KRT1-9), and both are expressed in suprabasal layers of non-cornified stratified squamous epithelium (e.g. mucosa). Keratin 13 expression is known to be regulated by calcium and nuclear receptor ligands such as retinoids and 1α,25-dihydroxyvitamin D3, but little is known about KRT13 regulation by estrogens or SERMs (Waseem et al., 1998). Olson et al. showed that keratin 13 was up-regulated in human luminal epithelial cells of secretory phase endometrium, suggesting that KRT13 might play a role in preparation for the implantation process (Olson et al., 2002).
In this study, we have characterized the differential regulation of KRT13 gene expression by E2 and the SERMs tamoxifen and raloxifene in breast cancer cells. We have identified the estrogen-responsive regulatory regions of KRT13 by ChIP assay, and studied the recruitment of ERα and the cofactors p300, SRC-2, and SRC-3 and the acetylation of histones on the KRT13 regulatory regions in response to the different ER ligands over time. Also, to identify specific sites involved in KRT13 gene regulation, we made an extensive series of deletions and mutations in the regulatory region of KRT13 and tested their individual and combined responses to ER ligands. Our studies highlight marked differences between tamoxifen and raloxifene and indicate that the differential regulation of this gene by different ER-ligand complexes is associated with differential recruitment of ER and coactivators and changes in histone acetylation.
Materials and Methods
Cell Culture and RNAi Studies
MCF-7 cells were maintained in Minimum Essential Medium (MEM) plus phenol red supplemented with 5% calf serum in a 5% CO2 incubator. Cells were then cultured for 3 days in MEM (no phenol red) supplemented with 5% calf serum. 2.5 × 105 cells/35-mm-well were plated in MEM plus 5% charcoal-dextran-treated calf serum (CDCS) 1 day before siRNA transfection. Transient transfections were performed using 100 pmol siRNA and 10 μl Lipofectamine 2000 reagent (Invitrogen Corp.). The cells were incubated at 37 °C in a 5% CO2-containing incubator and were treated with ligands at 24 or 44 hrs after siRNA transfection and were harvested 48 hrs after transfection. siRNA duplexes targeting ERα (forward, UCAUCGCAUUCCUUGCAAAdTdT; reverse, UUUGCAAGGAAUGCGAUGAdTdT, and control (GL3 luciferase, #D-001400-01) were obtained from Dharmacon (Lafayette, CO).
Chromatin Immunoprecipitation (ChIP)
MCF-7 cells maintained in MEM (plus phenol red) supplemented with 5% calf serum, were then cultured in MEM plus 5% CDCS at 37 °C in a 5% CO2-containing incubator for 3 days prior to treatment with vehicle, 10−8 M estradiol or 10−7 M trans-hydroxytamoxifen (Tam) or 10−7 raloxifene (Ral) for the times indicated. Chromatin was crosslinked using 1% formaldehyde at 37 °C for 10 min and cells were then washed twice with PBS and harvested in ice-cold PBS plus protease inhibitor cocktail (Roche) plus 1 mM DTT. Cell pellets were first resuspended in nuclei isolation buffer (50 mM Tris, pH 8.0. 60 mM KCl, 0.5% NP40, protease inhibitor and 10 mM DTT), centrifuged at 1000 g for 3 min and then resuspended in lysis buffer (0.5% SDS, 10 mM EDTA, 0.5 mM EGTA, 50 mM Tris-HCl, pH 8.0, protease inhibitor and 10 mM DTT) and sonicated (Fisher Scientific, Sonic Dismembrator Model 100) at 80% of maximum power three times for 10 seconds. The cell lysate was centrifuged at 14,000 g. The supernatant was diluted 1:4 with dilution buffer (1% Triton X-100, 2mM EDTA, 150mM NaCl, 20 mM Tris-HCl, pH8.0, protease inhibitor and 10 mM DTT ) and precleared with 15 μl preimmune IgG (Santa Cruz Inc.), 2μg salmon sperm DNA, 50 μl 25% protein A agarose slurry (Santa Cruz Inc.). Complexes were incubated at 4°C overnight with 2–5 μg antibody, then pulled down at 4°C for 1 h with 60 μl of 25% protein A-agarose slurry containing 2 μg salmon sperm DNA. Precipitates were sequentially washed with 1ml washing buffer I (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.0), 150 mM NaCl ), 1 ml washing bufferII (0.1% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris-HCl (pH 8.0), 500 mM NaCl), 1ml washing bufferIII (0.25 mM LiCl, 1% NP40, 1% sodium deoxycholate, 1 mM EDTA, 10 mM Tris-HCl (pH 8.0)) and twice with1 ml TE buffer (1 mM EDTA, 10 mM Tris-HCl (pH 8.0)). Chromatin complexes were incubated at room temperature for 20 min with 100–300 μl elution buffer (1% SDS, 0.1 M NaHCO3). The crosslinking was reversed by incubating at 65°C overnight with 200 mM NaCl and 200 μg/ml proteinase K (Invitrogen Corp.). DNA was purified with QIAquick columns (Qiagen) and quantified by real-time PCR.
Real-time PCR
Real-time PCR was performed to quantify gene expression levels and ChIP samples. Each real-time PCR reaction contained 4 μl of diluted cDNA or ChIP sample, 5 μl of 2X SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA), 0.5 μl of 1.25 μM forward and reverse primers and 0.5 μl of ddH2O. The real time PCR was carried out in an ABI Prism 7900 HT Sequence Detection System (Applied Biosystems) for 40 cycles (95°C for 15 s and 60°C for 1 min) after an initial 10 min incubation at 95°C. The primers used for real-time PCR are listed in Table 1. The fold change in expression of each gene was calculated as described previously, with 36B4 cDNA as an internal control (Frasor et al., 2003). The ChIP results were calculated as percent of input chromatin.
Table 1.
| Table 1A. Primers used for real-time PCR | ||
|---|---|---|
| Forward primer | Reverse primer | |
| 36B4 | 5′-GTGTTCGACAATGGCAGCAT | 5′-GACACCCTCCAGGAAGCGA |
| KRT13 | 5′-GGCAGAGATGAGGGAGCAGTA | 5′-TCTTGGCGTGGAACCATTC |
| SRC-1 | 5′-CTCATGGTGTGGCTCGTTCAT | 5′-TGCTGGCGGTTTATTCTGGT |
| SRC-2 | 5′-AGGACAGCCCACCTCCATG | 5′-TGACTGGGTGGGATTCGAG |
| SRC-3 | 5′-CCATGGGAAGACCAGGAGG | 5′-CGAAGAGGCAATGTGGGAA |
| KRT13-P1 | 5′-CCCTTGCTGAGTTCACTGGG | 5′-AGCCTCAATGCCTTAGGCTCTT |
| KRT13-P2 | 5′-GAACCCAGGAAAACAGTGCAC | 5′-ACCTCGCCCTGTGTATCTCTTC |
| KRT13-P2.5 | 5′-GGCATTACAAGTCCCCGCTAA | 5′-CAATGCTGCAGATCCTCCAA |
| KRT13-P3 | 5′-CTGCTGATACCCCTGCGG | 5′-GGTTCTCCTGGAACCCCCT |
| KRT13-P3.5 | 5′-TGGGTCCTGTGTCACATCCTC | 5′-TCACATCCAGGAAGACGGC |
| KRT13-P4 | 5′-CCCCAAGTTGTGACAACCAAA | 5′-CAATTACCTGACTCCACACCCC |
| KRT13-TSS | 5′-AAGAGCCACAGTCCTCGGC | 5′-TGGCAGAGGAGCTCTGCAG |
| Table 1B. Primers used for Cloning | ||
|---|---|---|
| Forward primer | Reverse primer | |
| KRT13wt | 5′-ATGGTACCGTTGTTGTTGTTGTTTTTGCCA | 5′-TATGATCTCCATAGCCCCCAGCTGATCCC |
| KRT13T1 | 5′-ATTGGTACCGCCCTAAAGCAGTCAGCGTAAT | 5′-TATGATCTCCATAGCCCCCAGCTGATCCC |
| KRT13T2 | 5′-ATTGGTACCGGCAATAGTAGGACCCTCCTCC | 5′-TATGATCTCCATAGCCCCCAGCTGATCCC |
| KRT13T3 | 5′-ATTGGTACCGGCAGGTGATTCTATCGAACCA | 5′-TATGATCTCCATAGCCCCCAGCTGATCCC |
| KRT13T4 | 5′-ATTGGTACCTGTCCCCTCTGAATCATTCTTTTT | 5′-TATGATCTCCATAGCCCCCAGCTGATCCC |
Genomic Cloning, Mutagenesis, and Luciferase Reporter Assays
The KRT13 promoter region was amplified from human genomic DNA (Roche Corp.) and then cloned into the pGL3-promoter reporter plasmid using primers as indicated in Table 1B. Mutagenesis of putative response elements was performed using the QuickChange Site-Directed Mutagenesis protocol. The original sequences and mutagenesis primers are listed in Table 2, with the mutated sequences highlighted. All reporter constructs were sequenced for verification. Briefly, 1000 ng pGL3 reporter vector and 25 ng pRL-SV40 were co-transfected into MCF-7 cells in 24-well plates using Lipofectamine 2000 in OptiMEM as per manufacturer’s instructions (Invitrogen, Carlsbad, CA). Cells were transfected for 6 h, washed, and treated with indicated ligands prior to cell lysis in 1x passive lysis buffer (Promega, Madison, WI) and measurement of luciferase activity in an MLX Microtiter Plate Luminometer (Dynex Technologies, Chantilly, VA).
Table 2.
Primers used for Site-Directed Mutagenesis
| Original Sequence | Forward primer | Reverse primer | |
|---|---|---|---|
| KRT13mut1 | GGGGCATTGTTACCC | 5′-GGTTTGGGGCATTGTTTAGCCAGGCATGGCAGGTTC | 5′-GAACCTGCCATGCCTGGCTAAACAATGCCCCAAACC |
| KRT13mut2 | GGGCCAGACTGACCC | 5′-CAACCCGAGGGCCAGACTGGAACAGGCATCTCAACCTCAAC | 5′-GTTGAGGTTGAGATGCCTGTTCCAGTCTGGCCCTCGGGTTG |
| KRT13mut3 | CTGTCACTGTGACTT | 5′-GGTCTCTGTCACTGTGGAATTTTCTGATGGCCTCGG | 5′-CCGAGGCCATCAGAAAATTCCACAGTGACAGAGACC |
| KRT13mut4 | GGGCTACGGTGACCT | 5′-GTCGAGGGCTACGGTGGAATTGCAAAGCACAGAG | 5′-CTCTGTGCTTTGCAATTCCACCGTAGCCCTCGAC |
| KRT13mutA | CGGGCAGGGT | 5′-GGGCAGGAGGGCCGATAAGGGTCAGGGGTTAG | 5′-CTAACCCCTGACCCTTATCGGCCCTCCTGCCC |
| KRT13mutB | ACCCAGCCCC | 5′-GTGCCACCCCACCCATATCCTGGCCTGGTATC | 5′-GATACCAGGCCAGGATATGGGTGGGGTGGCAC |
| KRT13mutC | AACCAGCCCC | 5′-GGGAGGAGAGAAGATAACCATATCCTATGGAGGTGTATAAAAG | 5′-CTTTTATACACCTCCATAGGATATGGTTATCTTCTCTCCTCCC |
The mutation sequences are bolded.
Results
Differential up-regulation of KRT13 mRNA level by E2, Tamoxifen and Raloxifene
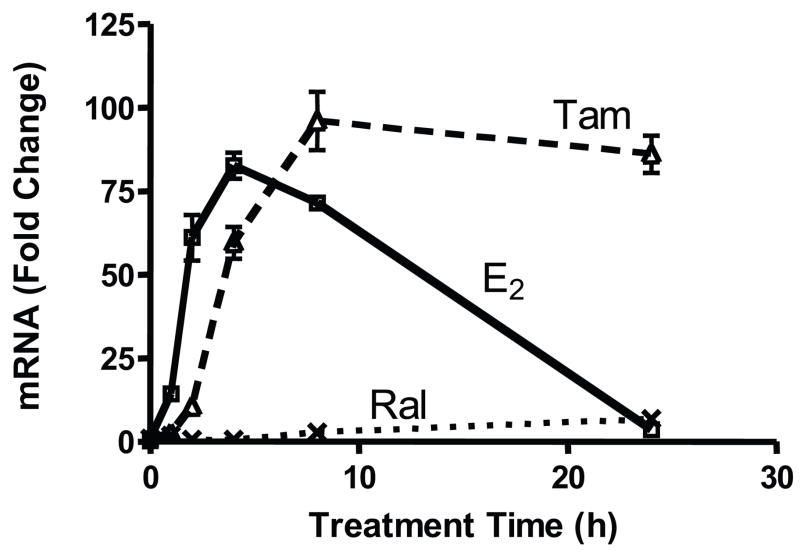
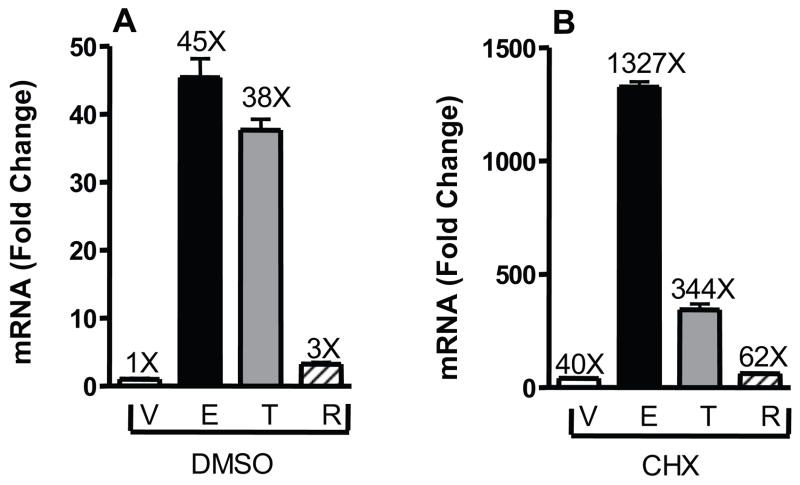
Our previous work identified KRT13 as being differentially regulated by estrogen and SERMs and suggested that KRT13 might serve as a marker of breast tumor responsiveness to tamoxifen therapy (Frasor et al., 2006). To characterize the regulation of KRT13, ligand treatment time course experiments were performed in MCF-7 breast cancer cells treated with estradiol (E2), tamoxifen or raloxifene and KRT13 mRNA levels were quantified by real-time PCR (Fig. 1). KRT13 was differentially regulated by E2, tamoxifen and raloxifene in a time-dependent manner. KRT13 mRNA increased upon 1 h E2 treatment, peaked at 4 h and returned to almost the control basal level at 24 h. Compared to the E2 treatment curve, the tamoxifen treatment curve was shifted right, and showed a slower increase in KRT13 mRNA with a maximum response at 8h that remained high through 24 h. By contrast, raloxifene had only a very mild effect on KRT13 expression at all times. To investigate whether the gene expression stimulations were primary responses to the ligands, cells were treated with DMSO vehicle or the translation inhibitor cycloheximide together with vehicle, E2, tamoxifen or raloxifene for 6 h. Fig. 2 shows that cycloheximide treatment still allowed up-regulation of KRT13 mRNA expression by E2 and SERMs, indicating that KRT13 is likely a primary, direct gene target of the ER. We cannot, however, rule out possible involvement of secondary effects in the tamoxifen stimulations since the fold change with tamoxifen was reduced relative to basal in the cycloheximide vs. no cycloheximide treatment groups.
Figure 1.
Differential up-regulation over time of KRT13 mRNA levels by E2, Tam (trans-hydroxytamoxifen) and Ral. MCF-7 cells were treated with 10 nM E2, 100 nM Tam or 100 nM Ral for 0, 1, 2, 4, 8, and 24 h. RNA was extracted and reverse transcribed, and real time PCR was carried out. Each value represents the mean ± SEM from three separate experiments.
Figure 2.
Examination of the effect of cycloheximide on the up-regulation of KRT13 gene expression by E2, Tam and Ral. MCF-7 cells were pre-treated with DMSO vehicle (A) or 10 μg/mL cycloheximide (B) for 30 min and then treated with vehicle, 10 nM E2, 100 nM Tam (trans-hydroxytamoxifen) or 100 nM Ral with or without 10 μg/mL cycloheximide for 6h. RNA was extracted and reverse transcribed, and real time PCR was carried out. Each value represents the mean ± SEM from three separate experiments.
Involvement of ERα in regulation of KRT13 gene expression
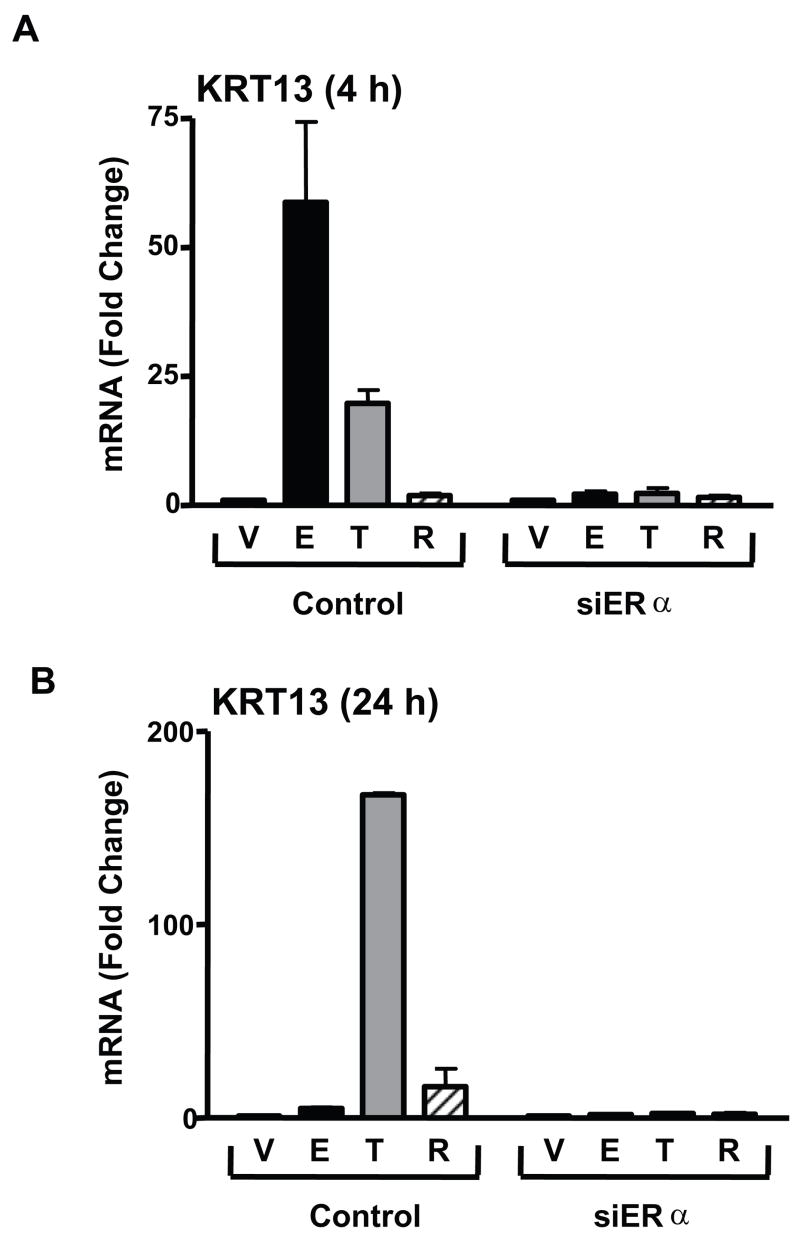
To verify the involvement of ERα in KRT13 regulation, ERα was knocked down by siRNA transfection in MCF-7 cells and KRT13 mRNA levels were then determined after ligand treatments. As shown in Fig. 3 panels A and B, siERα treatment (which reduced ERα protein > 90%) eliminated KRT13 mRNA induction by either E2 or tamoxifen monitored at 4 h and 24 h of treatment. These results imply that ERα is required for the KRT13 mRNA induction by E2 and SERMs. We also obtained no stimulation of KRT13 mRNA by E2 or the SERMs in ER-negative 231 human breast cancer cells (data not shown), further suggesting the importance of ER in the KRT13 response to ligands.
Figure 3.
ERα mediates ligand-induced KRT13 gene expression. MCF-7 cells were transfected with siRNA for control GL3 luciferase, or with siRNA for ERα and were treated with vehicle, 10nM E2, 100nM trans-hydroxytamoxifen or 100 nM Ral for 4 h (panel A) or 24 h (panel B). RNA was extracted and reverse transcribed, and real time PCR was carried out. Each value represents the mean ± SEM from three separate experiments.
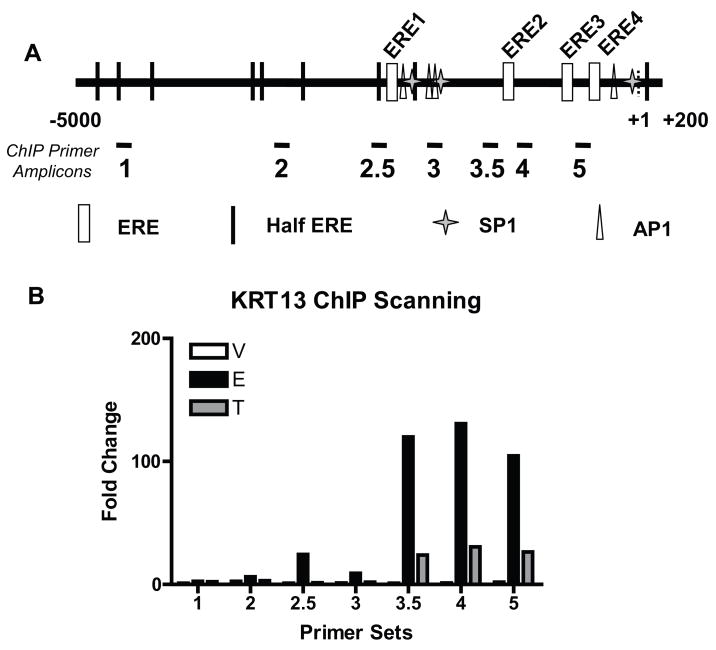
Ligand-dependent ERα recruitment on the KRT13 promoter
Utilizing a bioinformatic sequence analysis of the KRT13 promoter (−5000 to +200 bp of the transcription start site), we found four putative ERE sites, four AP-1 sites, three Sp1 sites and nine half-ERE sites (Fig. 4A). In order to identify the ER binding regions in the KRT13 promoter, ChIP scanning (Wang et al., 2004) was carried out (Fig. 4B). First, cells were treated with vehicle, E2 or tamoxifen for 45 min and a standard ChIP assay was performed using sonicated chromatin with a median size of 500 bp, and the chromatin fragments were immunoprecipitated by antibody against ERα. Second, the precipitated DNA was analyzed by qPCR with 7 primer sets, each targeting a putative regulatory element in the KRT13 promoter (Fig. 4A). As shown in Fig. 4B, regions 3.5, 4 and 5 showed strong ERα recruitment when cells were treated with E2 or tamoxifen, and recruitment at this 45 min time was always greater with E2 vs. Tam treatment. Region 2.5 recruited ERα following E2 treatment but not tamoxifen treatment, suggesting that region 2.5 might be a differential ligand-dependent binding site for ERα.
Figure 4.
Mapping ERα-binding regions in the KRT13 promoter by ChIP scanning. (A) Schematic diagram of the −5000 to +200 region of the KRT13 gene. The putative elements and PCR-amplified fragments indicated are: ERE1 (−2129 to −2117), ERE2 (−1289 to −1277), ERE3 (−587 to −575), ERE4 (−394 to −382), amplicon 1 (−4613 to −4511), amplicon 2 (−3314 to −3214), amplicon 2.5 (−2296 to −2171), amplicon 3 (−1888 to −1762), amplicon 3.5 (−1448 to −1334), amplicon 4 (−1157 to −1010), amplicon 5 (−567 to −428). (B) ChIP analysis of E2-induced and tamoxifen-induced ERα-binding to the KRT13 promoter region. MCF-7 cells were treated with vehicle, 10nM E2 or 100nM Tam (trans-hydroxytamoxifen) for 45 min. ChIP assays were performed with antibody against ERα and the ChIP samples were analyzed by real time PCR. Each value represents the mean from two separate experiments.
ERα and factor recruitment on the KRT13 promoter proximal region
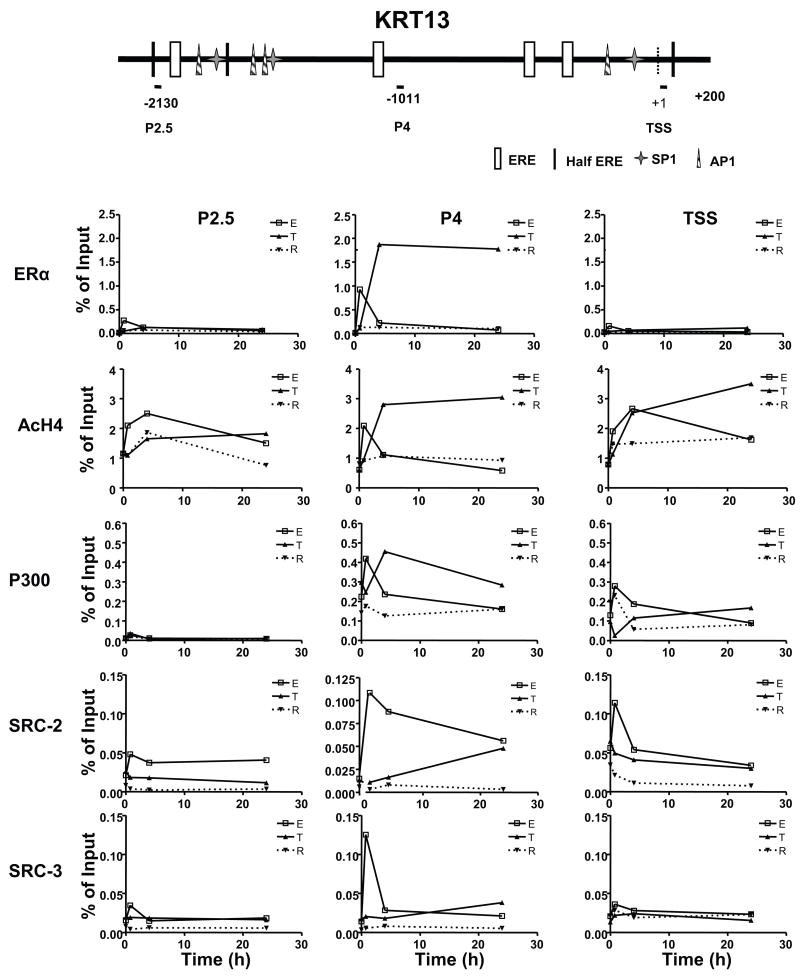
From the findings in Fig. 4, it is clear that estradiol and tamoxifen bound ERα was recruited to specific regions within the KRT13 proximal promoter. To investigate whether other potential regulatory factors are recruited to the same regions as well, cells were treated with estradiol, tamoxifen or raloxifene for times up to 24 h, and factor recruitment on the KRT13 promoter region P2.5, region P4, and the transcription start site (TSS) was analyzed with ChIP and qPCR. In all cases, recruitment was greatest at region P4 (Fig. 5).
Figure 5.
Recruitment of ERα, changes in histone H4 acetylation (AcH4), and recruitment of p300, SRC-2, and SRC-3 on the KRT13 promoter over time in response to E2, Tam or Ral. MCF-7 cells were treated with vehicle, 10nM E2, 100nM Tam or 100nM Ral for 0, 45 min, 4 h and 24 h. ChIP assays were performed with antibody against ERα, AcH4, p300, SRC-2 or SRC-3 and the ChIP samples were analyzed by real time PCR. Recruitments on the KRT13 promoter regions P2.5, P4 and the TSS (transcription start site) are shown. Each value represents the mean from three to five separate experiments.
On the KRT13 P4 region (Fig. 5 middle panels), the recruitment of ERα was markedly increased with estradiol or tamoxifen treatment, but not with raloxifene treatment. With estradiol or tamoxifen, acetylation of histone H4 (AcH4) and recruitment of p300, SRC-2 and SRC-3 were increased and showed different patterns. In estradiol treated cells, recruitment of ERα, p300, SRC-2, SRC-3 and histone acetylation was highest at 45 min, then decreased by 4 h, and further decreased to the basal levels at 24 h (Fig. 5 middle panels). Following tamoxifen treatment, ERα and p300 recruitment and histone H4 acetylation were low at 45 min, rose to high levels at 4 h and remained high at 24 h, whereas SRC-2 and SRC-3 recruitment kept increasing and reached the highest level at 24 h (Fig. 5 middle panels). Of interest, the level of ERα recruitment with tamoxifen was two fold higher than that with E2 at any time monitored and the ERα level was high over a prolonged period of time. By contrast, with raloxifene treatment, recruitment of ERα, cofactors and acetylation of histone H4 changed very little over time, consistent with our observations that this ligand had almost no ability to increase KRT13 gene expression. Thus, of most note was the very different time course of ERα and cofactor recruitment and acetylation of histone H4 at the P4 region by the three ligands, being transiently increased very early after E2 treatment and then declining, whereas the increases with tamoxifen were slower and usually higher and more sustained over time, and no increases were observed with raloxifene.
On the KRT13 P2.5 region (Fig. 5 left panels), and on the KRT13 transcription start site (Fig. 5 right panels, TSS), ERα was not recruited with any ligands, and with estradiol treatment, only weak increases in SRC-2, p300 and AcH4 were seen. In tamoxifen and raloxifene treated cells, there was no increase in recruitment of any cofactors. Hence, the greatest changes in ERα and cofactor recruitments were seen at region P4 (Fig. 5 middle panels).
Analysis of regulatory elements in the KRT13 promoter
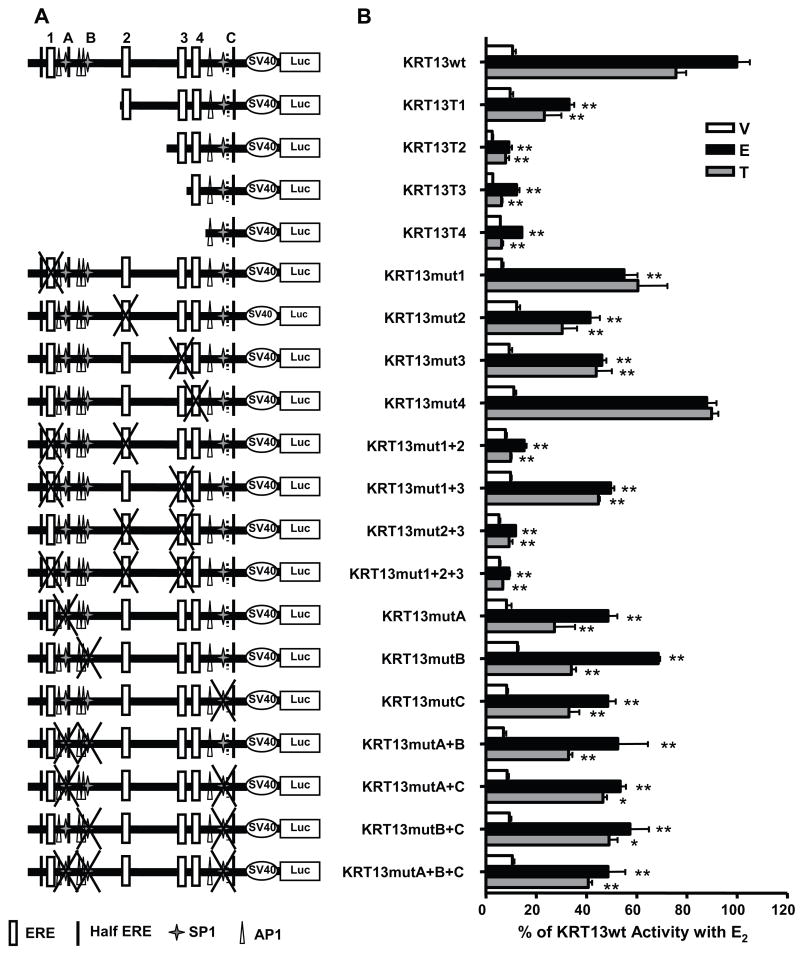
Bioinformatic analysis of the KRT13 promoter region revealed multiple potential EREs, Sp1, AP-1 and half ERE sites. The ChIP assay results in Figs. 4 and 5 showed binding of ER and cofactors to this promoter region. To explore the regulatory activity of the KRT13 promoter in greater detail, the −2500 to +200 bp region flanking the transcription start site of KRT13 was cloned into the pGL3-luciferase reporter plasmid and its transcriptional activity was monitored in MCF-7 cells (Fig. 6A). This full length region showed a 9-fold stimulation with estradiol treatment and a 7-fold stimulation with tamoxifen treatment relative to the vehicle treatment (Fig. 6B).
Figure 6.
Analysis of the E2 and tamoxifen-induced transcriptional activity of the wild type (wt) KRT13 promoter or the KRT13 promoter containing the indicated truncations or mutations. ERE positions are: ERE1 (−2129 to −2117), ERE2 (−1289 to −1277), ERE3 (−587 to −575), ERE4 (−394 to −382). (A) Schematic diagrams of the wild type and mutant constructs. (B) E2 and tamoxifen-induced transcriptional activities of the wild type and KRT13 promoter mutants were evaluated in a luciferase reporter system in MCF-7 cells. Values are the mean ± SEM from three to six separate experiments. Dunnett’s test was performed to compare each mutant with the wild type promoter for each ligand treatment. * p < 0.05, and ** p < 0.01.
In order to evaluate the cis-activation function of different regions in the KRT13 promoter, serial truncated constructs were generated and cloned and their transcriptional activities were analyzed (Fig. 6A). Relative to the full length KRT13 promoter the first truncated construct, denoted KRT13T1 (−1329 - +200 bp), which deleted the most distal ERE (named ERE1, see Fig. 4) and several Sp1 and AP1 sites and a half-ERE, retained approximately 40% of the E2-stimulated activity and 35% of the tamoxifen-stimulated activity seen with the full length promoter. Further truncation to remove also ERE 2, KRT13T2 (−784 - +200 bp), or to remove ERE 3, KRT13T3 (−525 - +200 bp), or to remove all four EREs, KRT13T4 (−204 - +200 bp), yielded constructs that had very low transcription activity with either E2 or tamoxifen (Fig. 6B). These results suggest that the majority of the E2 and tamoxifen inducible activity within the KRT13 promoter is derived from the regulatory elements in the −2500 to −784 region.
To evaluate further the contribution of the ERE and Sp1 sites to the transcription activity of the KRT13 promoter, each site was mutated individually or in various combinations by site directed mutagenesis and the E2 or Tam inducible activities were assessed (Fig. 6). As shown in Fig. 6A and B, individual mutation of ERE 1, 2, or 3 (mut 1, mut2 or mut 3) diminished, but did not abolish, E2 or Tam responses. Mutation of ERE 4 had no effect, consistent with our findings from the promoter truncations also. It is interesting that mutation of ERE 1 reduced only the E2 response, but not the Tam response. Among all four individual ERE mutations, mutation of ERE 2 (mut2) had the greatest impact on both E2 and Tam transcriptional activity. Double ERE mutations of EREs 1–2 and 2–3 and the triple mutation of EREs 1-2-3 further reduced E2/Tam responses almost to the basal level, whereas with the double ERE mutations at sites 1–2 or 2–3 about half of the E2/Tam responses of the wild type promoter were retained. The findings with the ERE mutations demonstrate that EREs 2 and 3 are involved in both E2 and Tam regulation of KRT13, that ERE 1 may be mainly involved in the E2 response, and that ERE 4 does not appear to be involved in regulation of KRT13.
Because truncation T1 more greatly reduced ligand stimulation than was observed with just mutation of ERE 1 (mut1), we examined the possible roles of the two Sp1 sites present near ERE 1 as well as the possible involvement of the Sp1 site nearest to the promoter since even truncation T4 showed some low ligand regulation. In examining the involvement of Sp1 sites, we found that all single, double and triple mutations of the three Sp1 sites gave similarly reduced E2 and Tam responses compared to the wild type KRT13 construct, suggesting that the Sp1 sites are involved in the full activity of the KRT13 promoter. The Sp1 triple mutation still evoked about half of the full E2/Tam responses, this remaining transcriptional activity probably being due to the EREs. These deletion and mutation analyses implicate ERE 2, along with EREs 1 and 3, as being most central in the regulation of KRT13.
Discussion
Our findings reveal differential time dependent up-regulation of keratin 13 gene expression by estradiol and two SERMs. KRT13 was up-regulated by estradiol at early time points, and was up-regulated by tamoxifen at later time points. Thus, for the regulation of KRT13, estradiol was a short-term agonist and tamoxifen was a more long-term agonist, being considerably more effective than E2 in sustaining up-regulation of the mRNA for KRT13 over time. The SERM, raloxifene, by contrast, was quite ineffective in up-regulating KRT13 gene expression.
To understand the basis for the differential ligand regulation of this gene, we used a ChIP scanning approach to identify the estrogen receptor binding and potential ligand regulated region of KRT13, and we cloned and further characterized this region by site directed mutagenesis and serial truncations. By ChIP scanning, we identified a regulatory region (−2500 bp to +200 bp) that has ligand-dependent ERα binding. The region we identified overlaps with the −2572 bp to −1035 bp region identified by ChIP-on-chip studies of Carroll et al. (Carroll et al., 2006) as an ER binding region close to the KRT13 gene. Within the 2.7-kb region we studied, we found four putative ERE sites and three Sp1 sites using sequence analysis. This 2.7-kb region was further dissected by mutagenesis, and we found that three EREs and three Sp1 sites are involved in the KRT13 hormonal regulation we have examined.
Upon estrogen treatment, ER and other cofactors are known to be recruited to gene regulatory regions and to modulate gene expression (Metivier et al., 2003, Shang et al., 2000). Depending on the promoter context and cellular background, tamoxifen and raloxifene can regulate gene expression in a manner similar to or different from that of estradiol (Frasor et al., 2004, Rivera et al., 2005, Shang and Brown, 2002). In our study, tamoxifen, raloxifene and estradiol regulated KRT13 differentially. Our ChIP time course studies revealed that the time-dependent differential regulation effects of these ligands on KRT13 gene stimulation reflected a differential pattern of recruitment of ER and coregulators to these regulatory sites. Beyond the differential recruitment of coactivators such as SRC-2, SRC-3 and p300, and changes in histone acetylation that we examined, recent work has also highlighted the involvement of FoxA1 and related factors that affect chromatin remodeling that accompanies the changes in gene expression by different ligand-receptor complexes (Carroll et al., 2005, Carroll et al., 2006, Green and Carroll, 2007, Leu et al., 2004, Lupien et al., 2008).
There are several possible explanations for our observations. First, the structures of the ER-tamoxifen and ER-raloxifene complexes are not identical and show some important differences (Brzozowski et al., 1997, Shiau et al., 1998, Wijayaratne et al., 1999) that may contribute to differences in their patterns of gene regulation, and to our observations that tamoxifen was more stimulatory than raloxifene on the KRT13 gene. Second, estradiol-bound ER is reported to be degraded through a ubiquitin proteasome pathway, whereas SERM-bound ER is relatively more stable (Fan et al., 2004, Lonard et al., 2000, Wijayaratne and McDonnell, 2001). Through gene expression profiling, we have observed that tamoxifen and raloxifene can have very different agonistic (stimulatory) activities on different genes, and although the mechanistic basis for these differences have not been elucidated, differences in ligand-dependent degradation of ER may, in part, account for the differential regulation of gene expression in our study. Since the tamoxifen-bound ER appears to be more stable and less susceptible to proteasome-mediated degradation and down-regulation than is the E2-ER complex (Shao et al., 2004, Wijayaratne and McDonnell, 2001), the up-regulation of KRT13 might remain sustained for a longer time by the ER-tamoxifen complex. Finally, it has been shown that 2–24 hour treatments with tamoxifen and raloxifene increased p160 coactivator levels in HeLa and MCF-7 cells (Frasor et al., 2003, Lonard et al., 2004). The elevated levels of coactivators may boost the agonistic effects of tamoxifen on KRT13, especially when the coactivator levels are high, a situation similar to the late time point in our experiments.
Several studies have highlighted marked cell-specific differences in the hormonal regulation of specific genes. For example, whereas GREB1 has been shown to be up-regulated by estrogen in several estrogen receptor-positive breast cancer cell lines, other genes were regulated by estrogen in MCF-7 but not in other estrogen receptor-positive breast cancer cells (Rae et al., 2005, Stender et al., 2007). In this regard, it is of interest that we also observed cell-specific differences in hormonal regulation of KRT13. Although KRT13 was up-regulated by tamoxifen and by estradiol but not by raloxifene in MCF-7 cells, we observed no up-regulation of KRT13 by either of the three ligands over a 48 h time period in 231 breast cancer cells stably expressing ER (at 30% the level of MCF-7 cells), and we observed only weak (2-fold) up-regulation by estradiol at 1 and 4 h and negligible regulation monitored over 48 h by tamoxifen or raloxifene in ZR75 cells, which contained 8% the ER level of the MCF-7 cells. What accounts for these cell-specific differences is unclear, but might include alterations in chromatin modifications in or near the KRT13 gene, differences in ER levels, presence or absence of different coregulators, and differences in additional transcription factors such as FOXA1 in the different ER-positive cells (Eeckhoute et al., 2006, Katzenellenbogen and Katzenellenbogen, 2002, Lupien et al., 2008, Shang and Brown, 2002).
Our results provide insight into the mechanisms responsible for differential gene regulation by estrogen and different SERMs. Although we have reported that another keratin, keratin 19, is regulated by estradiol in breast cancer cells (Choi et al., 2000) and others have shown keratin involvement in breast cancer migration (Chu et al., 1993, Chu et al., 1996, Hendrix et al., 1996, Hendrix et al., 1997), this is the first report on the hormonal regulation of KRT13. Our findings indicate important differences in the regulation of KRT13 by estradiol and different SERMs that might impact on the activity of KRT13 in breast cancer cell migration and metastasis.
Acknowledgments
This work was supported by grants from the NIH (CA18119, BSK, and T32 HD07028 and T32 ES07326, DHB) and The Breast Cancer Research Foundation (BSK).
Abbreviations
- ChIP
chromatin immunoprecipitation
- E2
17β-estradiol
- ER
estrogen receptor
- ERE
estrogen response element
- KRT13
keratin 13
- Ral
raloxifene
- SERM
selective estrogen receptor modulator
- Tam
tamoxifen
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Choi I, Gudas LJ, Katzenellenbogen BS. Regulation of keratin 19 gene expression by estrogen in human breast cancer cells and identification of the estrogen responsive gene region. Mol Cell Endocrinol. 2000;164:225–37. doi: 10.1016/s0303-7207(00)00197-0. [DOI] [PubMed] [Google Scholar]
- Chu YW, Runyan RB, Oshima RG, Hendrix MJ. Expression of complete keratin filaments in mouse L cells augments cell migration and invasion. Proc Natl Acad Sci U S A. 1993;90:4261–5. doi: 10.1073/pnas.90.9.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu YW, Seftor EA, Romer LH, Hendrix MJ. Experimental coexpression of vimentin and keratin intermediate filaments in human melanoma cells augments motility. Am J Pathol. 1996;148:63–9. [PMC free article] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–70. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–26. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M, Nakshatri H, Nephew KP. Inhibiting proteasomal proteolysis sustains estrogen receptor-alpha activation. Mol Endocrinol. 2004;18:2603–15. doi: 10.1210/me.2004-0164. [DOI] [PubMed] [Google Scholar]
- Frasor J, Chang EC, Komm B, Lin CY, Vega VB, Liu ET, Miller LD, Smeds J, Bergh J, Katzenellenbogen BS. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 2006;66:7334–40. doi: 10.1158/0008-5472.CAN-05-4269. [DOI] [PubMed] [Google Scholar]
- Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology. 2003;144:4562–4574. doi: 10.1210/en.2003-0567. [DOI] [PubMed] [Google Scholar]
- Frasor J, Stossi F, Danes JM, Komm B, Lyttle CR, Katzenellenbogen BS. Selective estrogen receptor modulators: discrimination of agonistic versus antagonistic activities by gene expression profiling in breast cancer cells. Cancer Res. 2004;64:1522–33. doi: 10.1158/0008-5472.can-03-3326. [DOI] [PubMed] [Google Scholar]
- Green KA, Carroll JS. Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer. 2007;7:713–722. doi: 10.1038/nrc2211. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Chu YW, Trevor KT, Seftor RE. Role of intermediate filaments in migration, invasion and metastasis. Cancer Metastasis Rev. 1996;15:507–25. doi: 10.1007/BF00054016. [DOI] [PubMed] [Google Scholar]
- Hendrix MJ, Seftor EA, Seftor RE, Trevor KT. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am J Pathol. 1997;150:483–95. [PMC free article] [PubMed] [Google Scholar]
- Hesse M, Magin TM, Weber K. Genes for intermediate filament proteins and the draft sequence of the human genome: novel keratin genes and a surprisingly high number of pseudogenes related to keratin genes 8 and 18. J Cell Sci. 2001;114:2569–2575. doi: 10.1242/jcs.114.14.2569. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Frasor J. Therapeutic targeting in the estrogen receptor hormonal pathway. Semin Oncol. 2004;31:28–38. doi: 10.1053/j.seminoncol.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Katzenellenbogen BS, Katzenellenbogen JA. Biomedicine. Defining the “S” in SERMs. Science. 2002;295:2380–1. doi: 10.1126/science.1070442. [DOI] [PubMed] [Google Scholar]
- Kumar V, Chambon P. The estrogen receptor binds tightly to its responsive element as a ligand-induced homodimer. Cell. 1988;55:145–56. doi: 10.1016/0092-8674(88)90017-7. [DOI] [PubMed] [Google Scholar]
- Leu YW, Yan PS, Fan M, Jin VX, Liu JC, Curran EM, Welshons WV, Wei SH, Davuluri RV, Plass C, Nephew KP, Huang THM. Loss of estrogen receptor signaling triggers epigenetic silencing of downstream targets in breast cancer. Cancer Res. 2004;64:8184–8192. doi: 10.1158/0008-5472.CAN-04-2045. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Lanz RB, O’malley BW. Nuclear receptor coregulators and human disease. Endocr Rev. 2007;28:575–87. doi: 10.1210/er.2007-0012. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Nawaz Z, Smith CL, O’malley BW. The 26S proteasome is required for estrogen receptor-alpha and coactivator turnover and for efficient estrogen receptor-alpha transactivation. Mol Cell. 2000;5:939–48. doi: 10.1016/s1097-2765(00)80259-2. [DOI] [PubMed] [Google Scholar]
- Lonard DM, Tsai SY, O’malley BW. Selective estrogen receptor modulators 4-hydroxytamoxifen and raloxifene impact the stability and function of SRC-1 and SRC-3 coactivator proteins. Mol Cell Biol. 2004;24:14–24. doi: 10.1128/MCB.24.1.14-24.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magin TM, Vijayaraj P, Leube RE. Structural and regulatory functions of keratins. Exp Cell Res. 2007;313:2021–32. doi: 10.1016/j.yexcr.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–63. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Murdoch FE, Gorski J. The role of ligand in estrogen receptor regulation of gene expression. Mol Cell Endocrinol. 1991;78:C103–8. doi: 10.1016/0303-7207(91)90114-8. [DOI] [PubMed] [Google Scholar]
- Olson GE, Winfrey VP, Blaeuer GL, Palisano JR, Nagdas SK. Stage-specific expression of the intermediate filament protein cytokeratin 13 in luminal epithelial cells of secretory phase human endometrium and peri-implantation stage rabbit endometrium. Biol Reprod. 2002;66:1006–15. doi: 10.1095/biolreprod66.4.1006. [DOI] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME. GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat. 2005;92:141–9. doi: 10.1007/s10549-005-1483-4. [DOI] [PubMed] [Google Scholar]
- Rivera JA, Alturaihi H, Kumar U. Differential regulation of somatostatin receptors 1 and 2 mRNA and protein expression by tamoxifen and estradiol in breast cancer cells. J Carcinog. 2005;4:10. doi: 10.1186/1477-3163-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Brown M. Molecular determinants for the tissue specificity of SERMs. Science. 2002;295:2465–8. doi: 10.1126/science.1068537. [DOI] [PubMed] [Google Scholar]
- Shang Y, Hu X, Direnzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–52. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shao W, Keeton EK, Mcdonnell DP, Brown M. Coactivator AIB1 links estrogen receptor transcriptional activity and stability. Proc Natl Acad Sci U S A. 2004;101:11599–604. doi: 10.1073/pnas.0402997101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–37. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- Smith RE, Good BC. Chemoprevention of breast cancer and the trials of the National Surgical Adjuvant Breast and Bowel Project and others. Endocr Relat Cancer. 2003;10:347–57. doi: 10.1677/erc.0.0100347. [DOI] [PubMed] [Google Scholar]
- Stender JD, Frasor J, Komm B, Chang KC, Kraus WL, Katzenellenbogen BS. Estrogen-regulated gene networks in human breast cancer cells: involvement of E2F1 in the regulation of cell proliferation. Mol Endocrinol. 2007;21:2112–23. doi: 10.1210/me.2006-0474. [DOI] [PubMed] [Google Scholar]
- Strassmer-Weippl K, Goss PE. Prevention of breast cancer using SERMs and aromatase inhibitors. J Mammary Gland Biol Neoplasia. 2003;8:5–18. doi: 10.1023/a:1025727103811. [DOI] [PubMed] [Google Scholar]
- Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq C, Yamamoto KR. Chromatin immunoprecipitation (ChIP) scanning identifies primary glucocorticoid receptor target genes. Proc Natl Acad Sci U S A. 2004;101:15603–8. doi: 10.1073/pnas.0407008101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waseem A, Alam Y, Dogan B, White KN, Leigh IM, Waseem NH. Isolation, sequence and expression of the gene encoding human keratin 13. Gene. 1998;215:269–79. doi: 10.1016/s0378-1119(98)00297-2. [DOI] [PubMed] [Google Scholar]
- Wijayaratne AL, Mcdonnell DP. The human estrogen receptor-alpha is a ubiquitinated protein whose stability is affected differentially by agonists, antagonists, and selective estrogen receptor modulators. J Biol Chem. 2001;276:35684–92. doi: 10.1074/jbc.M101097200. [DOI] [PubMed] [Google Scholar]
- Wijayaratne AL, Nagel SC, Paige LA, Christensen DJ, Norris JD, Fowlkes DM, Mcdonnell DP. Comparative analyses of mechanistic differences among antiestrogens. Endocrinology. 1999;140:5828–40. doi: 10.1210/endo.140.12.7164. [DOI] [PubMed] [Google Scholar]