Abstract
Objective
The degree of liver fibrosis is a determinant for initiation of therapy for HCV. Liver biopsy is invasive, risky and costly, but is required to assess fibrosis. This study intended to identify novel non-invasive markers to accurately assess fibrosis in HIV/HCV co-infection.
Methods
Using 100 biopsies from 68 HIV/HCV co-infected patients, we developed a predictive model consisting of six serum markers along with age and ART experience. DNA microarray analysis of PBMCs associated with a subset of 51 biopsies obtained from 28 patients was performed and incorporated into a second model. Results: The 8-marker model yielded an AUROC of 0.904. Combined analysis of clinical and DNA microarray data in the 51 biopsy subset identified two genes (alanine amino peptidase-N and mitogen-activated protein kinase kinase-3) that predicted fibrosis with high significance. The 4-marker model that included the two genes and two serum markers had an AUROC of 0.852, which did not differ significantly from the 8-marker model on this subset (AUROC = 0.856, p=0.96).
Conclusion
Both models accurately predicted fibrosis with an accuracy of 87.9%, thereby sparing 83% of patients from obtaining a biopsy. DNA microarray analysis can be invaluable in identifying novel biomarkers of liver fibrosis.
Keywords: Genomics, HCV, HIV, liver fibrosis, biopsy
Introduction
Infection by Hepatitis C virus (HCV) is commonly complicated by co-infection with HIV due to shared routes of transmission. Co-infection is found in up to 30% of HIV-infected individuals overall and up to 95% of HIV-infected injection drugs users (1). HCV/HIV co-infection has increased morbidity and mortality relative to infection by either HIV or HCV alone. Compared with HCV infection alone, co-infection with HIV increases HCV levels in plasma and is associated with a 50% accelerated rate of liver fibrosis progression (2).
Given the significant adverse event profile of current HCV therapy, it is important to balance the likelihood of liver disease progression with the risks of these side effects when deciding whether to initiate therapy. The degree of liver fibrosis is a determinant for the initiation of therapy in HIV/HCV co-infected patients (3). Liver biopsy is the gold standard for the assessment of fibrosis, but is invasive, expensive, and associated with complications such as pain and bleeding that may be life-threatening. Additionally, because it samples only 1/10,000–1/50,000 of the liver, it is highly variable and subject to sampling error (4). Liver biopsy is not an ideal test and is poorly suited to repeated follow-up, which may be particularly relevant in the setting of HIV/HCV co-infection, where progression of fibrosis is accelerated and more frequent monitoring of the degree of fibrosis may be required (5).
There has been considerable interest in generating a non-invasive test that would allow simple repeated testing to more easily monitor fibrosis progression, and provide a more global estimate of fibrosis. Several new assays to measure fibrosis have been generated that involve the use of panels of serum markers. These markers include routine clinical tests that measure liver function and inflammation such as AST, ALT, GGT, prothrombin time international normalized ratio (PT-INR), and total bilirubin (6). Less commonly used markers have also been proposed and incorporated into panels. These include components of the extracellular matrix (ECM) such as hyaluronic acid (HA), procollagen I, and collagen IV, enzymes that regulate the ECM such as YKL-40, regulators of matrix metalloproteases, such as TIMP-1, as well as acute-phase proteins such as α2-macroglobulin and haptoglobin (7, 8).
Several of the panels of currently available biomarkers have demonstrated good accuracy with respect to differentiating mild from severe fibrosis. However, the ability to differentiate more finely between different stages of fibrosis is limited (9). Thus, it is important to develop new and more robust biomarkers that increase the accuracy and sensitivity of non-invasive models of fibrosis. The use of DNA microarray may allow the identification of novel biomarkers by comparing differences in gene expression in the peripheral blood mononuclear cells of individuals with different stages of fibrosis. The aims of this study were first, to validate existing markers of fibrosis in a cohort of HIV/HCV co-infected patients and second, to determine whether DNA microarray analysis could be used to discover novel biomarkers that could be added to currently available clinical and biochemical markers to develop a more accurate model of liver fibrosis, which can be validated in larger number of patients.
Study Subjects
Data from all 100 liver biopsies performed in HIV-HCV co-infected patients at the Warren Grant Magnusson Clinical Research Center during the past seven years for whom biochemical data were available were obtained from 68 patients. Biochemical data was available on all patients at the time of liver biopsy. Hepatic fibrosis was assessed by the modified Ishak fibrosis score as described elsewhere (10). An Ishak fibrosis score greater than or equal to 3 was considered advanced, while a score less than 3 was considered mild. Nine serum markers (aspartate aminotransferase [AST], alanine aminotransferase [ALT], γ-glutamyltransferase [GGT], platelets, alpha-fetoprotein [AFP], total cholesterol, total bilirubin, , α2-macroglobulin [A2M], haptoglobin, and apolipoprotein A1), one cell marker (platelet count), and one plasma marker (prothrombin international normalized ratio [INR]) were assayed from samples collected within 12 weeks of biopsy. Hyaluronic acid (HA) was measured by an enzyme-linked protein-binding assay on a 96-well microplate (Corgenix Inc. Denver, CO, USA). Sex, age at biopsy, and CD4+ count were assessed. Cumulative months of anti-retroviral therapy (ART) and nevirapine (NVP) use were determined via chart review, and ART experience was additionally calculated as a dichotomous (Yes/No) value (Table 1). Patients were not receiving interferon therapy at the time of either biopsy or serum collection. All patients were urged to abstain from alcohol consumption; however, a comprehensive history of alcohol consumption was not collected. The study was reviewed and approved by the Institutional Review Board of the National Institute of Allergy and Infectious Diseases.
Table 1.
Stage of fibrosis of patients at time of liver biopsy. Mild fibrosis is defined as Ishak score <3, while advanced fibrosis is defined as Ishak score ≥3.
| 100 biopsy set | Genomics subset | |||
|---|---|---|---|---|
| Mild | Advanced | Mild | Advanced | |
| N | 65 | 35 | 28 | 23 |
| Ishak score 0: | 9 | 2 | ||
| 1 | 18 | 10 | ||
| 2 | 38 | 16 | ||
| 3 | 20 | 14 | ||
| 4 | 8 | 5 | ||
| 5 | 1 | 1 | ||
| 6 | 6 | 3 | ||
Genomics
Peripheral blood mononuclear cells (PBMC) were collected by Ficoll-Hypaque separation technique for DNA microarray analysis from a subset of 28 HCV/HIV patients at the same time of their liver biopsy (total 51 liver biopsies). This subset was selected based on the availability of DNA microarray and proteomics data, which were performed only in this subset. Otherwise there was no statistically significant difference between the subset and the larger set of patients. Total RNA and labeled cRNA synthesis and hybridization to the Affymetrix U133A human microarray, representing approximately 14,500 genes, were performed according to the manufacturer’s recommended protocol as described elsewhere (11). One way ANOVA analysis was used to identify significantly modulated genes (p<0.05) that additionally had an absolute fold change difference >1.3.These genes were included in modeling.
Model
Logistic regression analysis was performed to develop a predictive model from the 68 subjects with 100 biopsies. To account for repeated diagnostic tests (biopsies) within a subject, a generalized estimating equation [26] was adopted. Backward elimination was used for model selection: we started with the feature set of all factors significantly correlated with fibrosis (p<0.05), and sequentially deleted the factors until the performance of the model began to decline. The performance of the predictive model was evaluated by AUROC, and the positive and negative predictive values. Bootstrap method [27] was used to correct for the overestimation of AUROC caused by the fact that the model was both trained and tested on the same dataset.
Over the subset of 28 subjects with 51 biopsies, two predictive models were generated from logistic regression: one contained only clinical and biochemical markers, the other contained 2 biochemical markers and 2 genes . The two models were compared in terms of AUROC by the method of DeLong et al (12).
Results
Patient characteristics
The characteristics of the patients at time of biopsy and of the genomics subset broken out are shown in Table 1. The patients in the larger set were predominantly male (81.5%) and the majority (65%) had a mild stage (Ishak score <3) of liver fibrosis. Comparison between the groups with mild and advanced fibrosis are shown in Table 2. The two groups differed significantly at all markers except for sex, CD4+ T cell count, Nevirapine use, ALT, AST/ALT ratio, serum cholesterol, and apolipoprotein A1, though the failure of sex to reach statistical significance may have influenced by the low number of females overall. The significant differences observed with respect to platelet counts and hyaluronic acid level might be due to the low platelet and high HA level among the participants with advanced fibrosis in this study.
Table 2.
Characteristics of patients at time of liver biopsy for the set consisting of 100 biopsies. All data are mean +/− SD or proportions [(n(%)]. For continuous variables, t-tests were performed on natural logarithmic transformed values.
| Marker | Mild fibrosis (Ishak <3) | Advanced fibrosis (Ishak≥3) | p value |
|---|---|---|---|
| Age at Biopsy (yrs) | 45.2 ± 6.4 | 49.7 ± 8.4 | 0.008 |
| Sex (# male) (%) | 53 (81.5) | 32 (91.4) | 0.15 |
| CD4+ (cells/uL) | 533.2 ± 244.9 | 505.9 ± 254.5 | 0.6 |
| ART use (mo) | 44.3 ± 38.9 | 70.1 ± 50.5 | 0.01 |
| ART use | 0.80 | 0.94 | 0.03 |
| (naïve=0, experienced=1) | ± 0.40 | ± 0.24 | |
| NVP use (mo) | 9.7 ± 26.8 | 6.6 ± 28.3 | 0.6 |
| AST (IU/L) | 47.7 ± 28.0 | 77.6 ± 91.1 | 0.003 |
| ALT (IU/L) | 57.2 ± 46.0 | 79.2 ± 65.8 | 0.06 |
| AST/ALT ratio | 0.96 ± 0.33 | 1.07 ± 0.43 | 0.17 |
| Platelets (x10^9) | 229.4 ± 71.3 | 161.5 ± 57.1 | <0.0001 |
| GGT (IU/L) | 87.1 ± 100.6 | 115.7 ± 92.5 | 0.005 |
| AFP (IU/L) | 4.2 ± 2.6 | 9.4± 11.5 | 0.004 |
| Cholesterol (mg/dl) | 172.3 ± 35.5 | 172.5 ± 34.7 | 0.99 |
| Bilirubin (umol/L) | 0.73 ± 0.47 | 0.84 ± 0.39 | 0.04 |
| PT-INR | 1.01 ± 0.10 | 1.06 ± 0.09 | 0.007 |
| A2M (g/L) | 238.7 ± 87.9 | 364.4 ± 113.3 | <0.0001 |
| Haptoglobin (g/L) | 87.7 ± 71.6 | 44.8 ± 40.2 | 0.0007 |
| Apo A1 (g/L) | 125.5 ± 31.9 | 124.9 ± 31.3 | 0.95 |
| HA (ng/mL) | 60.1 ± 51.9 | 152.0 ± 241.8 | 0.003 |
Univariate analyses of the association between the clinical and biochemical markers with fibrosis stage were shown in Table 3. Eleven of the seventeen markers evaluated were significantly correlated with fibrosis stage. These were considered for inclusion in the logistic regression for predictive model. Increased A2M, decreased platelets, and increased hyaluronic acid were most closely related to increased stage of fibrosis. The other significant factors, in order of decreasing significance were: AFP, age, haptoglobin, GGT, cumulative ART experience, AST/Platelet ratio, PT-INR, and bilirubin.
Table 3.
Univariate analysis of marker correlation with fibrosis, ranked by significance. Correlation was performed between markers and all stages of fibrosis, rather than mild versus advanced. Dichotomous variables, marked by (*) were analyzed using the Fisher Exact test.
| p value | Correlation coefficient | Marker |
|---|---|---|
| < 0.0001 | 0.55 | Alpha-2-macroglobulin |
| < 0.0001 | −0.39 | Platelets |
| < 0.0001 | 0.38 | Hyaluronic acid |
| 0.001 | 0.33 | AFP |
| 0.001 | 0.32 | Age at biopsy |
| 0.003 | −0.29 | Haptoglobin |
| 0.004 | 0.28 | GGT |
| 0.004 | 0.29 | Cumulative ART experience (mo) |
| 0.016 | 0.24 | Prothrombin INR |
| 0.046 | 0.20 | Total bilirubin |
| 0.056 | 0.19 | ALT |
| 0.078 | * | ART experience (yes/no) |
| 0.086 | 0.17 | AST/ALT ratio |
| 0.146 | 0.15 | AST |
| 0.147 | −0.15 | Total cholesterol |
| 0.247 | * | Male sex |
| 0.321 | −0.10 | Cumulative NVP use (mo) |
| 0.506 | −0.07 | CD4+ |
| 0.794 | −0.03 | Apolipoprotein A1 |
Model Generation
Through model selection by backward elimination, eight out of the 11 markers which were significantly related to fibrosis stage were included in the predictive model. In addition, ART experience (yes/no), instead of the cumulative ART experience (which was continuous), was used in the model, since information about this binary factor was easier to obtain and it also lead to a slightly better performance than the cumulative ART experience. The resultant diagnostic score was: S = 6.281 + 0.370*(ART experience) −3.739*ln(Platelets) + 0.063*ln(GGT) + 0.058*(Age) + 0.054*(AFP) + 0.236*(Total bilirubin) + 2.000*ln(Alpha-2-macroglobulin) − 0.663*ln(Haptoglobin).
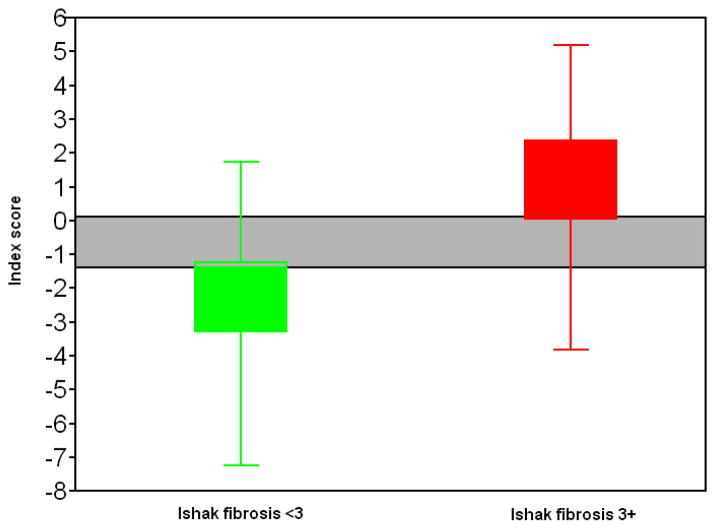
The AUROC for the above diagnostic score was 0.904. To achieve 90% sensitivity, the cutoff for S needs to be no more than −1.3; to achieve 90% specificity, the cut-off needs to be no less than 0.1. At cutoff of −1.3, the sensitivity was 91.4%, with specificity 72.3%, positive predictive value 64%, and negative predictive value 94%. At cutoff of 0.1, the specificity was 90.8%, with sensitivity 74.3%, the positive predictive value 81.2% and negative predictive value 86.8%. 83% of the biopsies fell either above or below the cut-offs and could thus be classified with 90% sensitivity and specificity. Of these 83% of biopsies, 87.9% were correctly identified. The ROC curve was shown in (Figure 1).
Figure 1.
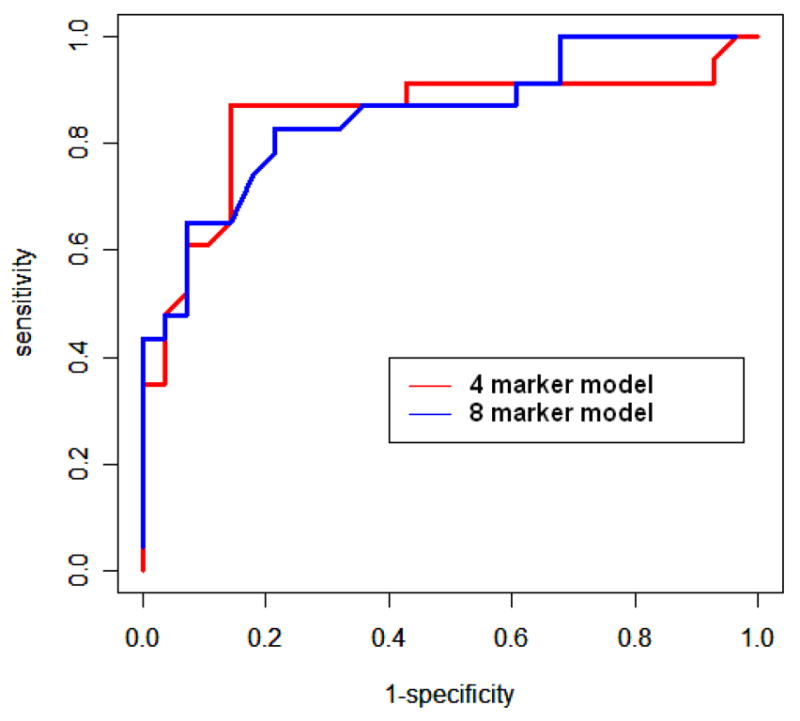
ROC curves using the 51 biopsy dataset for the four marker model containing genomics data (AUROC = 0.852) and the eight marker model containing clinical data (AUROC = 0.856). The area under the two curves is not significantly different.
The above AUROC and the model performance evaluation could be too optimistic, due to the fact that the diagnostic score was both trained and tested on the same data. The validation by bootstrap, which compared favorably with the standard cross-validation techniques especially for sample sizes, was applied to correct the possible overestimation of the diagnostic score performance. The following were results from the validation: the validated AUROC was 0.8154. At the cutoff of −1.3, the sensitivity was 76.2%, the specificity was 69.9%, the PPV was 58.4%, and the NPV was 85.2%; at the cutoff of 0.1, the sensitivity was 64.2%, specificity was 86.9%, PPV was 72.9% and NPV was 82.0%.
Genomic analysis
22 genes were identified as significantly modulated between biopsies with mild and advanced fibrosis. The two most significantly modulated genes were the cell surface marker alanine aminopeptidase N (AAN) (p<0.005), also known as CD13, and mitogen-activated protein kinase kinase 3 (MKK3) (p<0.006).
Generation and Comparison of Models in the Genomics Subset
The eight markers identified as predictive in the larger set were included in logistic regression modeling in the 51 biopsy subset. The resulting eight-marker diagnostic score was S = 2.66 + 0.41*(ART experience) + 0.04*(Age) + 0.11*(AFP) −1.11*(Bilirubin) − 2.83*ln(Platelets) −0.16*ln(GGT) + 2.16*log(A2M) −0.40*log(Haptoglobin). The AUROC for the diagnostic score was 0.856.
Microarray data from the 22 genes previously identified by ANOVA analysis were then included with the eight clinical markers and logistic regression modeling was again performed to yield a second model. Four markers (A2M, haptoglobin, AAN, MKK3) were retained as significantly predictive in this model. The resulting 4-marker diagnostic score was: S = −20.6 + 0.79*AAN +1.03*MKK3 +1.65*ln(A2M) −0.36*ln(Haptoglobin). The AUROC was 0.852.
Though the 8-parameter model seemed to have slightly higher AUROC than the 4-parameter model, the difference between the two was not statistically significant (p=0.96). It was thus reasonable to expect that the 4-parameter model should have similar performance as the 8-parameter model if validated over the full data of 100 biopsies. In fact, over the 51 biopsies, the validated AUROC for the 4-parameter model was 0.796.
Discussion
In this study, we describe a novel set of four markers (A2M, haptoglobin, MAP kinase kinase 3 gene and alanine aminopeptidase N gene), which accurately predicted the stage of liver fibrosis in HIV-infected individuals. This set of four markers includes two novel genetic markers expressed in PBMCs functioning as surrogate markers of hepatic fibrosis. Our findings suggest that the novel 4 biomarker set had the same accuracy in predicting the degree of liver fibrosis when compared to that of a more elaborate 8 biomarker set. Our study also illustrates the validity of utilizing novel approaches such as whole genome wide analysis of transcriptional profiling in PBMC to identify novel biomarkers.
Determination of the stage of liver fibrosis is necessary to assess the stage of liver disease, monitor disease progression and also as a determinant for treatment of HCV and for follow-up of response to therapy in HIV/HCV co-infected patients. Liver biopsy is the gold standard. However, it is invasive, associated with significant morbidity, and is highly variable due to the small sample size relative to the entire liver. Several non-invasive models of liver fibrosis have thus been proposed, most using various combinations of serum markers representing inflammation and extracellular matrix breakdown and formation. While these models have generally performed well with regards to the differentiation of fibrosis, it remains important to identify novel biomarkers that may prove more robust. Furthermore, many of the existing biomarkers were developed retrospectively using laboratory data routinely collected for monitoring liver disease. Therefore, identification of novel biomarkers will not only enable us to differentiate degrees of liver fibrosis, but also to understand the systemic pathophysiology of liver fibrosis in these patients.
The use of “-omics” technology has not been widely applied to the creation of noninvasive models of liver fibrosis. Utsonomiya et al found that a model based on hepatocyte gene-expression signatures accurately predicted fibrosis, though this technique was invasive (13). Poon et al combined serum proteomics and clinical data to create a noninvasive model of fibrosis in chronic Hepatitis B (14). However, no study to our knowledge has thus far used genomic analysis to discover new serum biomarkers of liver fibrosis.
In our study, we first created a model based on clinical and biochemical markers. We found that a model consisting of six serum markers (A2M, platelets, haptoglobin, GGT, AFP, bilirubin) and two clinical markers (age, ART experience) accurately predicted fibrosis, with an AUROC of 0.904 (bootstrapped AUROC = 0.8154), which compared favorably with previous models. Using cut-offs set to 90% sensitivity and specificity, 83% of the biopsies were classifiable and the model correctly identified 87.9% of these biopsies. Based on our model, 83% of all liver biopsies would thus have been avoidable with high accuracy. The accuracy of this model may approach the limit of accuracy when compared to biopsy. Mathematical modeling by Afdhal et al has indicated that assuming an 80–90% accuracy of biopsy largely secondary to sampling error, non-invasive models cannot achieve an AUROC better than 0.9 (15, 16).
Interestingly, the inclusion of ART experience improved the prediction of fibrosis in this study, while cumulative ART use was not sufficiently independently predictive to be included in the model. There are conflicting reports regarding the role of ART in liver fibrosis in HIV/HCV co-infected patients. Some studies have found that ART therapy slows the progression of liver fibrosis, while other have found that ART use, accelerates fibrosis (17). The stronger predictive performance for the dichotomous versus continuous variable with fibrosis in our study is difficult to interpret, but may indicate an initial hepatotoxic response followed by a long-term protective response. A second interpretation may be that initiation of ART was the result of HIV disease progression, and associated with HCV progression, though CD4+ count was not associated with fibrosis in our study. Nevirapine, in particular, has been associated with liver fibrosis (18); however our study did not find an association, though this may be due to the small number of patients on nevirapine.
Two models were then created using data from a subset of biopsies that had associated genomics data. One model contained the same eight clinical and biochemical markers as the first model. The second additionally contained data from genomic analysis of PBMCs and resulted in a model combining two biochemical markers and two genomic markers. This four-marker model performed equivalently to the eight-marker model, with AUROCs of 0.852 and 0.856 respectively, a difference that was statistically insignificant. Given the smaller number of parameters, this model is more likely to be robust compared to the eight-marker model when applied to a validation set of HIV/HCV co-infected patients, due to a smaller risk of “over-fitting.” Additionally, because it lacks an ART parameter, the four-marker model is more likely to be applicable to predicting fibrosis in a more diverse array of chronic liver disease, if this biomarker set is validated in a larger clinical cohort of HIV/HCV co-infected patients. The approach of using DNA microarrays is to generate a model, which could then be validated by using less expensive methods such as multiplex bDNA amplification or real time PCR along with the two clinical parameters. This methodology is much more cost effective and adaptable in many centers.
The two genes identified to be most significantly positively associated with fibrosis and to best predict fibrosis in this analysis of PBMCs were mitogen-associated protein kinase kinase 3 (MKK3) and alanine aminopeptidase N (AAN), which has been shown to be identical to CD13 (19). These genes are both biologically plausible with regards to the pathogenesis of liver fibrosis and are thus likely to be robust markers. AAN is a membrane-associated metalloproteinase expressed by a range of cells, including macrophages, neutrophils, fibroblasts and epithelial cells. It has been shown to both be involved in the chemotaxis of T lymphocytes (20), as well in the degeneration of the extra cellular matrix (ECM), particularly in the setting of tumor angiogenesis (21). While it has not been previously implicated in liver fibrosis, increased expression of AAN has been shown to be associated with poor outcome in biliary atresia, also an inflammatory process of the liver (22).
MKK3 is an upstream component of the p38 mitogen-associated protein kinase (MAPK) signal transduction pathway, which is activated predominantly by cytokines and by environmental stresses such as osmotic changes and inflammation. Transforming growth factor-β1 (TGF-β1) has been shown to potently induce the p38 MAPK pathway, inducing ECM synthesis and fibrotic change in renal mesangial cells and hepatic stellate cells (HSC) (23). Genomic analysis of HSCs in response to the fibrogenic agent CCl4 has indicated that up-regulation of the p38 MAPK pathway is strongly associated with fibrosis progression (24). Lastly, a selective inhibitor of the p38 MAPK pathway has been shown to improve cirrhosis in a rat model (25).
In conclusion, our model, incorporating eight markers (six biochemical and two clinical), accurately predicts fibrosis in HIV/HCV co-infected patients and correctly classifies biopsies as containing either mild or advanced fibrosis. Thus, this model could spare biopsy for 83% of these patients by accurately predicting their status of fibrosis. In a subset of biopsies with genomic data, a four-marker model containing two novel candidate genetic markers, AAN and MKK3, performed equivalently compared to the eight-marker model in predicting fibrosis. While AAN and MKK3 are biologically plausible markers of fibrosis, this is the first study that has linked their up-regulation in PBMCs to liver fibrosis. The association between these genes and the role of PBMCs in fibrogenesis are thus deserving of further investigation. It is highly plausible that the gene expression profile in PBMCs serves as a surrogate for the inflammatory response in the liver. It will also be important to validate our models in larger clinical trials and to compare how they perform prospectively and in relation to previously published models. It is also necessary to test the validity of this model and novel biomarkers in other subsets of patients that include HIV/HBV co-infected, HIV, HCV, and HBV mono-infected individuals. Such studies will have to be performed prospectively given the lack of availability of stored samples that is needed to quantify levels of expression of AAN and MKK3. These studies will help us determine whether the clinical utility of our model is restricted to a subset of patients co-infected with HCV and HIV or a global biomarker for liver fibrosis irrespective of the causality of chronic liver disease. Nevertheless, this study indicates that use of DNA microarray technology is a viable approach to the development of biomarkers for non-invasive modeling of liver fibrosis.
Figure 2.
Performance of an eight-marker model showing accuracy of prediction. Grayed area indicates values for which the model is less than 90% sensitive and specific. 83% of biopsies fell outside the grey area and could thus be confidently classified. 87.9% of these biopsies were correctly identified as containing mild or advanced fibrosis.
Acknowledgments
This research was supported in whole by the Intramural Research Program of the NIH, [National Institute of Allergy and Infectious Diseases]. DS conducted the study, MM was the study co-ordinator, ZH performed statistical analysis, DK was the hepatopathologist, BW performed liver biopsies, MV and RL performed assays, AS, MAP,and HM designed the study and SK was the principal investigator.
Footnotes
Presented at the 14th Conference on Retroviruses and Opportunistic infections at Los Angeles, CA Feb 25-28, 2007
Publisher's Disclaimer: Disclaimer
The content of this publication does not necessarily reflect the views of policies of the Department of Health and Human Services, nor does mention of trade names, commercial products or organization imply endorsement by the U.S. Government.
Conflict of Interest Statement
None of the other authors have any conflicts of interest to report.
References
- 1.Alter MJ. Epidemiology of viral hepatitis and HIV co-infection. J Hepatol. 2006;44:S6–9. doi: 10.1016/j.jhep.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Benhamou Y, Bochet M, Di Martino V, Charlotte F, Azria F, Coutellier A, Vidaud M, et al. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. The Multivirc Group Hepatology. 1999;30:1054–1058. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- 3.Tossing G. Management of chronic hepatitis C in HIV-co-infected patients--results from the First International Workshop on HIV and Hepatitis Co-infection, 2nd-4th December 2004, Amsterdam, Netherlands. Eur J Med Res. 2005;10:43–45. [PubMed] [Google Scholar]
- 4.Reiss G, Keeffe EB. Role of liver biopsy in the management of chronic liver disease: selective rather than routine. Rev Gastroenterol Disord. 2005;5:195–205. [PubMed] [Google Scholar]
- 5.Soriano V, Sulkowski M, Bergin C, Hatzakis A, Cacoub P, Katlama C, Cargnel A, et al. Care of patients with chronic hepatitis C and HIV co-infection: recommendations from the HIV-HCV International Panel. Aids. 2002;16:813–828. doi: 10.1097/00002030-200204120-00001. [DOI] [PubMed] [Google Scholar]
- 6.Sterling RK, Lissen E, Clumeck N, Sola R, Correa MC, Montaner J, M SS, et al. Development of a simple noninvasive index to predict significant fibrosis in patients with HIV/HCV coinfection. Hepatology. 2006;43:1317–1325. doi: 10.1002/hep.21178. [DOI] [PubMed] [Google Scholar]
- 7.Rosenberg WM, Voelker M, Thiel R, Becka M, Burt A, Schuppan D, Hubscher S, et al. Serum markers detect the presence of liver fibrosis: a cohort study. Gastroenterology. 2004;127:1704–1713. doi: 10.1053/j.gastro.2004.08.052. [DOI] [PubMed] [Google Scholar]
- 8.Imbert-Bismut F, Ratziu V, Pieroni L, Charlotte F, Benhamou Y, Poynard T. Biochemical markers of liver fibrosis in patients with hepatitis C virus infection: a prospective study. Lancet. 2001;357:1069–1075. doi: 10.1016/S0140-6736(00)04258-6. [DOI] [PubMed] [Google Scholar]
- 9.Rockey DC, Bissell DM. Noninvasive measures of liver fibrosis. Hepatology. 2006;43:S113–120. doi: 10.1002/hep.21046. [DOI] [PubMed] [Google Scholar]
- 10.Ishak K, Baptista A, Bianchi L, Callea F, De Groote J, Gudat F, Denk H, et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22:696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 11.Lempicki RA, Polis MA, Yang J, McLaughlin M, Koratich C, Huang DW, Fullmer B, et al. Gene expression profiles in hepatitis C virus (HCV) and HIV coinfection: class prediction analyses before treatment predict the outcome of anti-HCV therapy among HIV-coinfected persons. J Infect Dis. 2006;193:1172–1177. doi: 10.1086/501365. [DOI] [PubMed] [Google Scholar]
- 12.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837–845. [PubMed] [Google Scholar]
- 13.Utsunomiya T, Okamoto M, Wakiyama S, Hashimoto M, Fukuzawa K, Ezaki T, Aishima S, et al. A specific gene-expression signature quantifies the degree of hepatic fibrosis in patients with chronic liver disease. World J Gastroenterol. 2007;13:383–390. doi: 10.3748/wjg.v13.i3.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Poon TC, Hui AY, Chan HL, Ang IL, Chow SM, Wong N, Sung JJ. Prediction of liver fibrosis and cirrhosis in chronic hepatitis B infection by serum proteomic fingerprinting: a pilot study. Clin Chem. 2005;51:328–335. doi: 10.1373/clinchem.2004.041764. [DOI] [PubMed] [Google Scholar]
- 15.The French METAVIR Cooperative Study Group. Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. Hepatology. 1994;20:15–20. [PubMed] [Google Scholar]
- 16.Afdhal NH, Curry M. Technology evaluation: A critical step in the clinical utilization of novel diagnostic tests for liver fibrosis. J Hepatol. 2007;46:543–545. doi: 10.1016/j.jhep.2007.01.008. [DOI] [PubMed] [Google Scholar]
- 17.Al-Mohri H, Murphy T, Lu Y, Lalonde RG, Klein MB. Evaluating liver fibrosis progression and the impact of antiretroviral therapy in HIV and hepatitis C coinfection using a noninvasive marker. J Acquir Immune Defic Syndr. 2007;44:463–469. doi: 10.1097/QAI.0b013e318030ff8e. [DOI] [PubMed] [Google Scholar]
- 18.Macias J, Castellano V, Merchante N, Palacios RB, Mira JA, Saez C, Garcia-Garcia JA, et al. Effect of antiretroviral drugs on liver fibrosis in HIV-infected patients with chronic hepatitis C: harmful impact of nevirapine. Aids. 2004;18:767–774. doi: 10.1097/00002030-200403260-00007. [DOI] [PubMed] [Google Scholar]
- 19.Look AT, Ashmun RA, Shapiro LH, Peiper SC. Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J Clin Invest. 1989;83:1299–1307. doi: 10.1172/JCI114015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tani K, Ogushi F, Huang L, Kawano T, Tada H, Hariguchi N, Sone S. CD13/aminopeptidase N, a novel chemoattractant for T lymphocytes in pulmonary sarcoidosis. Am J Respir Crit Care Med. 2000;161:1636–1642. doi: 10.1164/ajrccm.161.5.9902008. [DOI] [PubMed] [Google Scholar]
- 21.Fukasawa K, Fujii H, Saitoh Y, Koizumi K, Aozuka Y, Sekine K, Yamada M, et al. Aminopeptidase N (APN/CD13) is selectively expressed in vascular endothelial cells and plays multiple roles in angiogenesis. Cancer Lett. 2006;243:135–143. doi: 10.1016/j.canlet.2005.11.051. [DOI] [PubMed] [Google Scholar]
- 22.Liu C, Chiu JH, Chin T, Wang LS, Tai CH, Li AF, Wei C. Expression of aminopeptidase N in bile canaliculi: a predictor of clinical outcome in biliary atresia and a potential tool to implicate the mechanism of biliary atresia. J Surg Res. 2001;100:76–83. doi: 10.1006/jsre.2001.6205. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, Kwak JH, Kim SI, He Y, Choi ME. Transforming growth factor-beta1 stimulates vascular endothelial growth factor 164 via mitogen-activated protein kinase kinase 3-p38alpha and p38delta mitogen-activated protein kinase-dependent pathway in murine mesangial cells. J Biol Chem. 2004;279:33213–33219. doi: 10.1074/jbc.M403758200. [DOI] [PubMed] [Google Scholar]
- 24.Qiang H, Lin Y, Zhang X, Zeng X, Shi J, Chen YX, Yang MF, et al. Differential expression genes analyzed by cDNA array in the regulation of rat hepatic fibrogenesis. Liver Int. 2006;26:1126–1137. doi: 10.1111/j.1478-3231.2006.01353.x. [DOI] [PubMed] [Google Scholar]
- 25.Hattori S, Dhar DK, Hara N, Tonomoto Y, Onoda T, Ono T, Yamanoi A, et al. FR-167653, a selective p38 MAPK inhibitor, exerts salutary effect on liver cirrhosis through downregulation of Runx2. Lab Invest. 2007 doi: 10.1038/labinvest.3700539. [DOI] [PubMed] [Google Scholar]
- 26.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 27.Efron B, Tishirani R. Improvements on cross-validation: The” 632+ bootstrap method. JASA. 1997;92:548–560. [Google Scholar]