Abstract
The utilization of photopolymerized phospholipids for the preparation of spatially-defined, chemically-functionalized, μm-sized domains within enclosed fluidic channels was recently reported. Fabrication of the phospholipid patterns is achieved via self-assembly of photoreactive phospholipid membranes that are subsequently cross-linked via UV-irradiation through a photomask. In this work, we have characterized the chemical and physical stability of the self-assembled, chemically-functionalized, cross-linked phospholipid patterns, and extended this approach to the preparation of cross-linked phospholipid patterns with multiple chemical functionalities. Poly(bis-SorbPC) patterns were found to withstand a number of chemical and physical challenges, including drying/rehydration, solvent or surfactant rinse, and extended storage without compromising the size or morphology of the cross-linked phospholipid patterns. Non-specific adsorption of proteins was found to be markedly reduced in the presence of UV-photopolymerized poly(bis-SorbPC) compared to bare silica capillaries. The resulting barcode-like patterns were used to prepare protein-functionalized domains via covalent attachment of fluorescent proteins and active enzymes to chemically-functionalized lipid headgroups. We also demonstrate multiple component polymer lipid patterns with adjacent chemically-functionalized polymer lipid regions. The unique combination of stability, biocompatibility, reduced non-specific protein adsorption and the availability of numerous chemically functionalized lipid headgroups suggests the utility of this approach for preparing a widely applicable platform for multi-component, high-throughput chemical sensing and screening applications.
INTRODUCTION
The preparation of spatially-defined chemical patterns on planar surfaces is well established, most notably via the application of microcontact printing or photolithography with chemical etching. Microcontact printing has been extensively used to prepare biocompatible and biologically active chemical patterns using biological components, e.g. proteins1-4 and fluid lipid bilayers;5;6 however, microcontact printing is limited by the stamp feature size and transfer efficiency5;7 and patterns prepared with microcontact printing are susceptible to a number of deleterious effects, including protein denaturation and non-specific binding.7 Photolithographic methods have been used to prepare physical barriers on planar surfaces that: direct photophysical deposition and attachment of biomolecules;8;9 corral fluid lipid bilayers;10-13 and enhance photodegradation of regions within a supported phospholipid bilayer (SPB).14;15 Photolithographic patterning has been used to prepare biomolecular patterns on curved substrates16;17 and inside microfluidic channels,8;18;19 though preparation of photolithographic patterns on non-planar (e.g. curved) and completely enclosed surfaces (e.g. channels) is non-trivial. In many cases, preparation of chemically-patterned microfluidic channels results from chemical functionalization of only one surface in the three-dimensional channel structure. Thus, the application of microcontact printing and photolithography/etching may be less practical for preparing (bio)chemically functionalized patterns on three-dimensional and/or completely enclosed substrates.
SPBs have been utilized for a variety of chemical screening and sensing applications including: ligand-receptor binding assays;20 enzyme kinetics;21 tethering functional lipid vesicles;22;23 and protein resistant coatings for electrophoresis capillaries.24;25 SPBs have received considerable attention for the preparation of (bio)chemical patterns on both planar and curved surfaces where functionalized SPBs present a number of advantages over traditional covalent attachment or adsorption of biological components (e.g. proteins, peptides, etc.).16;18;21;26 SPBs are naturally biocompatible, minimize non-specific protein adsorption, self-assemble via vesicle fusion on many substrate materials, and can be chemically functionalized using a rich variety of headgroup chemistries. In particular, self-assembly of SPBs on surfaces increases their versatility and has been exploited in situations where planar lipid deposition techniques, for example, Langmuir-Blodgett (LB) and Langmuir-Schaefer (LS), can not be used.27 Incorporation of chemically functionalized lipids provides a basis for immobilization of proteins or other biologically active compounds in SPBs through both covalent and non-covalent interactions.26 Moreover, SPBs support the function and integrity of many membrane proteins, allowing investigation of ligand-receptor interactions.28-32
While the chemical properties of SPBs are very promising, a number of inherent structural disadvantages limit widespread use of SPBs for fabricating spatially-defined, chemically-functionalized surfaces. Primarily, lipid bilayers tend to diffuse laterally on solid supports, leading to expansion of the bilayer area, and mixing of patterned domains unless lipid resistant barriers have been fabricated.12;13 In most cases, these barriers do not add chemical functionality to the surface.12;13 Additionally, SPBs are not stable to drying without introducing a protective layer (e.g. protein, trehalose, PEG-linked lipids, etc.).33-36 Preparation of (bio)chemically patterned SPBs that are chemically and environmentally stabilized may help to alleviate these limitations.
A number of approaches have been explored for stabilizing SPBs including: preparation of hybrid bilayers;37 covalent attachment of lipids to the surface;38 and adsorption of a protective protein layer on top of the SPB.34 The most robust lipid structures are prepared through stabilization of SPBs via polymerization of the lipids themselves, increasing the stability of the SPB against physical and chemical challenges, while maintaining the desirable attributes of the SPB. This approach has been used most commonly with diacetylene-functionalized phosphorylcholine (DAPC) lipid.39;40 Preparation of SPB patterns via formation of polymerized lipid boundary regions using DAPC has been accomplished on planar surfaces41 and has proven particularly useful for surface plasmon resonance imaging applications where fluid lipids are corralled within the poly(DAPC) barriers. Though useful, poly(DAPC) barriers are limited to planar surfaces as DAPC requires LB or LS deposition rather than self-assembly.41;42
Sorbyl-functionalized reactive lipids developed by O’Brien and coworkers provide an attractive alternative for developing polymer lipid patterns.43;44 Cross-linked sorbyl lipids have increased mechanical stabilization relative to unpolymerizable lipids, yield less rigid structures following polymerization, can be polymerized via UV irradiation or thermal and/or redox radical initiators, and self-assemble on a variety of solid substrates. For example, bis-SorbPC (1,2-bis[10-(2′,4′-hexadieoyloxy)decanoyl]-sn-glycero-2-phosphocholine), forms smooth, continuous SPBs on planar substrates.43;45 Following polymerization, the resultant films are stable to drying/rehydration and surfactant rinses, and retain the resistance to non-specific protein absorption seen in fluid SPBs.45 Furthermore, at least one transmembrane protein has been demonstrated to retain function in a polymerized bis-SorbPC film.46
We have previously demonstrated the capability to fabricate spatially-defined, chemically functionalized molecular patterns on the interior surfaces of enclosed fused silica capillaries via self-assembly and subsequent photopolymerization of bis-SorbPC.47 Here, we present an expanded characterization of the resulting self-assembled, bis-SorbPC polymer patterns and demonstrate that the resultant cross-linked bis-SorbPC polymer networks are chemically and environmentally stabilized to a number of physical and chemical challenges that typically destroy SPBs, and can be stored for over a year without compromising the patterned structures. We also demonstrate the spatially-defined, functional immobilization of an enzyme and fluorescent protein via covalent linkage chemistry using polymer lipid patterning. Further, we demonstrate the capacity of this patterning approach for preparing multiple component polymer lipid patterns.
EXPERIMENTAL
Materials
Bis-SorbPC was synthesized by a modification of the procedure reported by Lamparski et al.43;48 DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine), maleimide lipid (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[maleimide(polyethylene glycol) 2000] (ammonium salt)), biotin lipid (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(Cap Biotinyl), Rh-DPPE (1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(lissamine rhodamine B sulfonyl) (ammonium salt)) and NBD-DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-(7-nitro-2-1,3-benzoxadiazol-4-yl) (ammonium salt)) were from Avanti Polar Lipids (Alabaster, AL). DOPE-PEG-pNP (p-nitrophenylcarbonyl-PEG-1,2-dioleoyl-sn-glycero-3-phosphoethanolamine) was synthesized and purified following published procedures.49;50 FM 1-43, biotin-DHPE (N-(biotinoyl)-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine, triethylammonium salt), streptavidin-R-phycoerythrin (SA-RPE), streptavidin-fluorescein conjugate and 3-(2-furoyl)quinoline-2-carboxaldehyde (FQ) were from Molecular Probes (Eugene, OR). Ribonuclease A, streptavidin, tris(2-carboxyethyl)phosphine hydrochloride (TCEP), β-galactosidase from E. coli, and fluorescein di-β-D-galactopyranoside were from Sigma (St. Louis, MO). Penta-His biotin conjugate antibody was from Qiagen (Valencia, CA). Fused silica capillaries (360 μm o.d., i.d. as indicated in text) were from Polymicro Technologies (Phoenix, AZ). Six-histidine tagged enhanced green fluorescent (6xHis-EGFP), six-histidine tagged Cerulean (6xHis-Cerulean) and six-histidine tagged DsRed (6xHis-DsRed) protein was purified using Ni2+-NTA metal affinity chromatography from transfected E. coli. A Barnstead (Dubuque, IA) EASYpure UV/UF was used to prepare all H2O used in this work.
Instrumentation
Fluorescent images were acquired on a Nikon Eclipse TE300 Quantum inverted microscope using a 4x/0.13 N.A. objective. Fluorescence images were obtained using the following filter sets: rhodamine (λex = 540/25 nm; λem = 620/60 nm), fluorescein/NBD (λex = 480/30 nm; λem = 535/40 nm) and Cerulean (λex = 425/40 nm; λem = 480/30 nm). Images were collected using a Quantix 57 back illuminated CCD camera (Roper Scientific, Tucson, AZ). MetaVue software (Universal Imaging, Downingtown, PA) was used to capture and analyze all images.
Fluorescence intensities in Figures 4 and 5 were calculated by averaging the intensity over a ca. 2 mm area of a capillary for non-pattered capillaries (Figure 4) or individual patterned features (light or dark, Figure 5) in patterned capillaries (see Supporting Information Figure S1) The mean intensities of each feature of a specific type within patterned capillaries were then averaged to obtain the mean fluorescence intensity for the compiled “light” regions and “dark” regions of the capillary presented the plots. Images are background corrected by subtracting an image taken under the same conditions (light settings, integration times, etc.) without a capillary present and/or with a bare, uncoated capillary.
Figure 4.
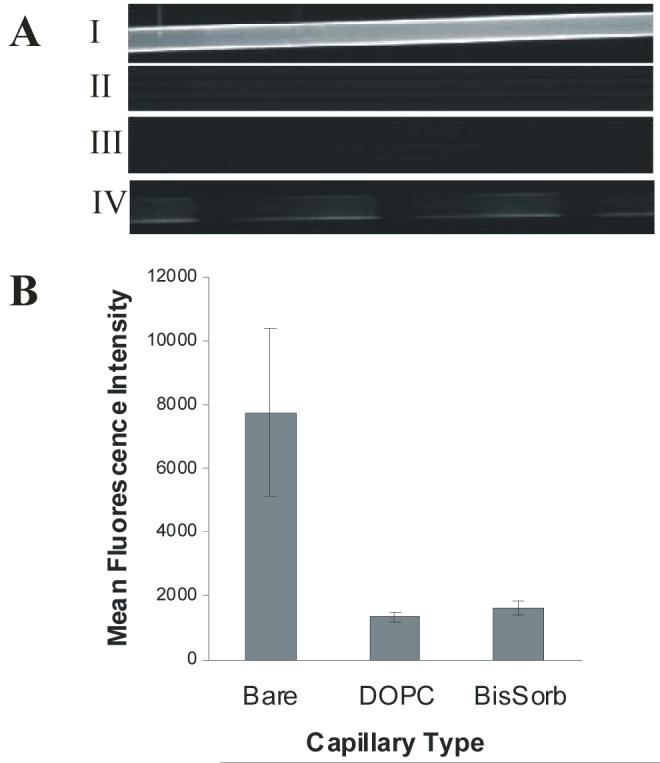
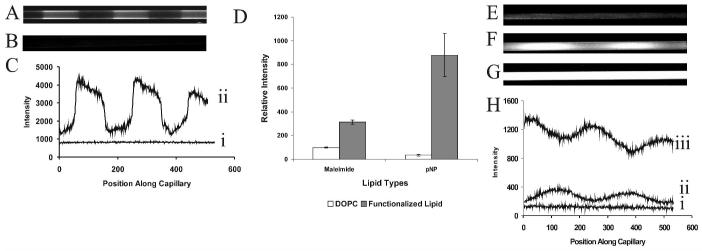
Nonspecific protein staining of silica capillary surfaces. A) SA-RPE was introduced to: I) Bare capillary, II) DOPC coated capillary, III) Poly(bis-SorbPC) patterned capillary in which the void regions were filled with DOPC, IV) Poly(bis-SorbPC) patterned capillary with void regions left bare. N = 3 capillaries of each type. B) Mean fluorescence intensity (± 1 s.d.) vs. wall coating is shown.
Figure 5.
Covalent functionalization of poly(lipid) patterned capillaries. A) Fluorescence image of a poly(bis-SorbPC) patterned capillary in which the void regions were filled with maleimide-doped DOPC SUVs, followed by covalent immobilization of FQ-labeled, reduced ribonuclease A. Scale bar = 60 μm. B) Fluorescence image of poly(bis-SorbPC) patterned capillary with maleimide-doped DOPC, followed by introduction of FQ-labeled unreduced ribonuclease A. C) Linescans showing the fluorescence intensity of capillaries shown in Figure 5A (ii) and Figure 5B (i) as a function of position along the capillary (pixel number). The linescans were taken along the inner capillary wall. D) Relative fluorescence intensity versus functionalization (± 1 s.d.), n=3 for each capillary type. Bare regions of the pattern were functionalized using DOPC (white bars) or DOPC doped with headgroup functionalized lipid (gray bars). E) Fluorescent image of poly(bis-SorbPC) patterned capillary with the void regions filled with DOPC following introduction of β-galactosidase and fluorescein di( β-D) galactopyranoside (FDG) (t = 3 min). F) Fluorescent image of poly(bis-SorbPC) patterned capillary containing β-galactosidase immobilized via 20% maleimide-doped DOPC following introduction of FDG (t = 30 s). G Fluorescent image of poly(bis-SorbPC) patterned capillary containing β-galactosidase immobilized via 20% maleimide-doped DOPC following introduction of FDG (t = 3 min). Images in E, F and G are shown on the same scale. G) Linescans through the center of capillaries shown in Figure 5E (i), Figure 5F (ii), and Figure 5G (iii).
Preparation of polymerized bis-SorbPC patterns
Polymerized bis-SorbPC coated capillaries were prepared similarly to the method briefly described in previous work.47 Detailed preparation protocols were as follows. Patterned regions of ca. 1.5 cm in length were prepared in the center of 5 cm capillary segments. Illumination windows were formed by removing the polyimide capillary coating using a butane lighter. Capillaries were first washed with 70% piranha solution (70% H2SO4, 30% H2O2) and rinsed thoroughly with H2O. Following overnight vacuum removal of solvent (benzene), a solution of 1 mg/mL bis-SorbPC was prepared in H2O. The resulting aqueous lipid solution was sonicated to form small unilamellar vesicles (SUVs) ca. 50-70 nm in diameter. The vesicle solution was then introduced into the capillary using a modified vial that allowed gravity-driven flow of the vesicle solution into the capillary. Upon introduction to the capillary, vesicles fuse along the capillary wall leading to a solid supported bilayer membrane.24 UV-induced polymerization was initiated by irradiation from a low-pressure mercury pen lamp with a rated intensity of 4500 μW/cm2 (UVP Inc, Upland, CA) at 254 nm for 10 min through a photomask (Photo Etch Technologies, Lowell, MA) (Figure 1).51 Following irradiation, unpolymerized lipid was removed from the capillary by exposure to alternating rinses with 1 % Triton X-100 and H2O. Capillaries were rinsed at 25 μL/min for 10 seconds, followed by a 7 min equilibration before the next rinse cycle. The resulting lipid patterned capillaries were allowed to dry at room temperature and stored until further use. Prior to use, polymerized lipid structures were hydrated with H2O or 10 mM phosphate buffer (pH 7.4).
Figure 1.
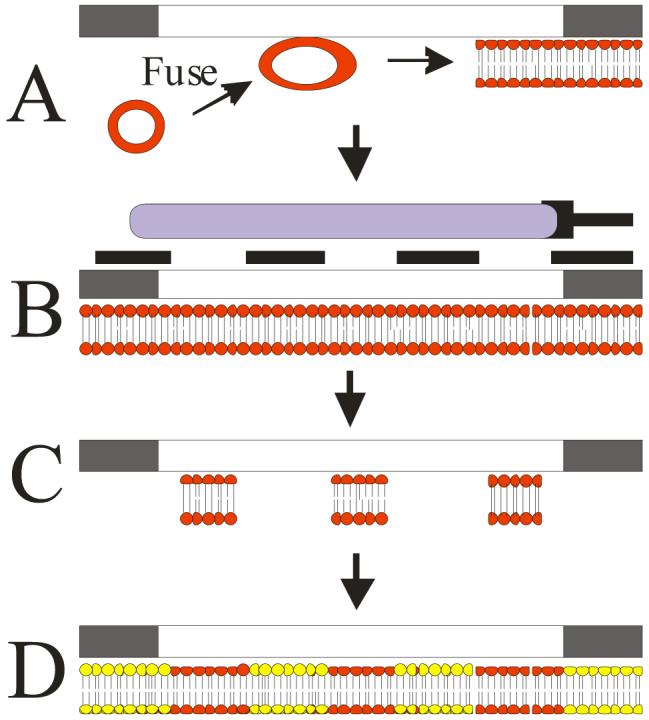
Schematic representation of polymerized phospholipid patterning process. A) SUVs prepared using doped or undoped bis-SorbPC are fused to the capillary surface in situ to create a uniform SPB within the capillary. B) The coating is polymerized via UV irradiation through a photomask placed over the capillary. C) Unpolymerized lipid is removed from the capillary to yield patterned regions containing the stabilized SPB. D) SUVs comprised of other lipid compositions are then deposited into the bare silica regions between poly(bis-SorbPC) structures, generating chemically-functionalized spatial patterns.
Control capillaries coated with DOPC were prepared following a previously published procedure.24
Chemical functionalization of phospholipid patterns
Chemical functionalization of phospholipid patterns was performed by deposition of doped SUVs into the bare silica regions of the pattern. SUVs were prepared from DOPC doped with 0.5 - 20 % (w/w) functionalized lipid and allowed to self-assemble onto the capillary walls. Excess lipid was removed prior to use via a series of H2O washes. Polymerized bis-sorbPC patterned capillaries were stored dry until use, at which point they were rehydrated and functionalized fluid lipids were subsequently deposited.Capillaries patterned with poly(bis-SorbPC) can be reused for additional experiments containing the same or different functionalities by removing fluid lipid regions via surfactant/air/water rinses followed by deposition of functionalized SUVs into the now resulting silica regions. Data presented within this paper was obtained during the first functionalization of individual capillaries, with the exception of the long-term storage experiments wherein capillaries had been functionalized and utilized multiple times.
For multifunctionalized phospholipid patterns, doped bis-SorbPC SUVs were deposited onto the capillary surface. Doped bis-SorbPC SUVs were prepared by vacuum drying bis-SorbPC with either 2% Rh-DPPE or 10% biotin-DHPE. The resulting lipid mixture was hydrated to 1 mg/mL total lipid concentration with 20 mM MOPS buffer, pH 7.4, and sonicated to clarity. Patterns were prepared by UV-irradiation of functionalized bis-SorbPC regions followed by removal of unpolymerized lipid and subsequent deposition of functionalized SUVs prepared using DOPC and/or bis-SorbPC as the primary lipid component.
Poly(lipid) pattern stability studies
Feature sizes, visualized via FM 1-43 staining, of poly(bis-SorbPC) patterned domains were measured before and after addition of surfactant, drying/rehydration cycles or chloroform. In each case, the same features were measured before and after the addition of the chemical or physical challenge. Triton X-100 or chloroform was introduced to the patterned capillary at 25 μL/min, followed by R 10 min equilibration time. Following exposure, the capillary was rinsed with H2O followed by addition of FM 1-43. For drying/rehydration, the capillary was dried via air purges for > 2 min. FM 1-43 was introduced upon rehydration and the feature size was measured following each of 10 drying/rehydration cycles. A minimum of three capillaries was used for each series of conditions.
Covalent attachment of proteins
Polymerized bis-SorbPC chemical patterns were prepared as described. Following surfactant wash and buffer rinse, the void regions in the polymer lipid chemical patterns were back filled with binary mixtures of DOPC containing either 20% w/w maleimide functionalized lipid for covalent linkage through thiol groups or 5% w/w DOPE-PEG-pNP lipid for attachment via primary amine coupling. For covalent attachment via free thiol coupling, a solution of ribonuclease A was first labeled with the amine-reactive fluorogenic dye FQ to allow fluorescence visualization. For this, a solution containing 200 nmol FQ in methanol was dried of solvent then reacted at 40°C with 18 μL 0.40 mM ribonuclease A and 2 μL 25 mM KCN for 20 minutes. After the reaction was complete, 100 μL TCEP was added to a final concentration of 28 mM to reduce disulfide bonds. Reduced FQ-ribonuclease A-SH was injected into capillaries and allowed to react for 30 min at room temperature.52 For amine linkage, a solution of 1 μM 6xHis-EGFP in 10 mM borate buffer, pH 10, was introduced to the DOPE-PEG-pNP functionalized patterned capillary and reacted for 30 min at room temperature.49;50 Excess 6xHis-EGFP was removed via rinsing prior to use.
Evaluation of β-galactosidase activity
Poly(bis-SorbPC) patterned capillaries were prepared as described with the void regions filled with either DOPC or 20% w/w maleimide functionalized lipid. All capillaries were rinsed with a blocking buffer of 100 mM Tris, 150 mM NaCl, and 2% w/v BSA, pH 7.4. For covalent attachment, β-galactosidase (0.33 μM in 50 mM phosphate buffer, pH 7.4 with 2 mM MgCl2) was introduced to the capillary and allowed to react for 30 minutes. Excess enzyme was rinsed from the capillary with H2O. A 200 nM solution of fluorescein di-β-D-galactopyranoside was introduced the capillary immediately prior to imaging.
RESULTS AND DISCUSSION
Preparation and evaluation of cross-linked phospholipid bilayer patterns
Patterned poly(bis-SorbPC) capillaries were prepared as previously described47 and depicted in Figure 1. SUVs prepared from bis-SorbPC were introduced into the capillary at low flow rates and allowed to fuse to the capillary surface to yield a bis-SorbPC bilayer on the capillary walls (Figure 1A). After 30 min, excess lipid was removed by a series of rinses and the resulting bis-SorbPC coated capillary was polymerized via UV-irradiation through a photomask (Figure 1B). Following irradiation, unpolymerized lipid, residing in the non-illuminated regions of the pattern was removed using two rinsing cycles of surfactant and H2O, leaving the polymerized lipid patterns (Figure 1C). Following removal of the unpolymerized lipid, the void regions between poly(lipid) barriers were back-filled by vesicle fusion using chemically-functionalized SUVs (Figure 1D) comprised of either DOPC or bis-SorbPC as the primary lipid constituent.
A potential application for this chemical patterning approach is the fabrication of high density, low-volume functionalized arrays for chemical assays. In small i.d. channels, the large surface area to volume ratios provide an efficient environment for enhancing contact between surface bound species and subsequent ligands residing in free solution. As the volume of the patterned feature increases with the square of the channel radius, it is desirable to prepare patterns in small i.d. channels. For example, when phospholipid patterns of 150 μm feature length are prepared in 150 μm i.d. fused silica capillaries, the volume is ca. 2.65 nL, whereas this volume can be reduced to ca. 12 pL in 10 μm i.d. capillaries.
To investigate the dependence of capillary size on pattern formation, a series of fused silica capillaries ranging from 10-150 μm i.d. were used as substrates for lipid assembly and pattern formation (Figure 2). Following polymerization of the lipid pattern and removal of unpolymerized lipid, the resulting poly(bis-SorbPC) patterns were developed to determine pattern quality. Figure 2A and 2B show fluorescent images collected using FM 1-43, a fluorogenic membrane probe that experiences a marked increase in quantum yield upon insertion into the outer leaflet of the lipid bilayer. Linescans of the capillaries in Figure 2 are provided in Supporting Information Figure S2. Based on FM 1-43 development, stable poly(bis-SorbPC) films with well-defined, spatially-discrete patterns were obtained in 95% of patterned capillaries, making this a robust, reproducible method for preparing chemical patterns in situ in enclosed environments. In the cases where poorly patterned capillaries (ca. 5% of patterned capillaries) were obtained, non-uniform staining was observed. Additionally, staining of uncoated capillaries resulted in no detectable FM 1-43 fluorescence, while staining of unpatterned bis-SorbPC or DOPC-coated capillaries resulted in uniformly high fluorescence throughout the length of the capillary.53
Figure 2.

Dependence of channel size on poly(bis-SorbPC) pattern formation. Fluorescent images were collected for: A) 150 μm i.d. capillary stained with FM 1-43. B) 75 μm i.d. capillary stained with FM 1-43. C) 50 μm i.d. capillary with 2% Rh-DPPE/98% DOPC fused into void regions between poly(bis-SorbPC) regions. D) 10 μm i.d. capillary with 2% Rh-DPPE/98% DOPC fused into void regions between poly(bis-SorbPC) regions.
Figures 2C and 2D show fluorescence images of poly(bis-SorbPC) patterned capillaries developed via introduction and fusion of Rh-DPPE. SUVs prepared from DOPC containing 2% Rh-DPPE were deposited in regions devoid of polymerized lipid. Interestingly, there is little incorporation of functionalized lipid into the preformed poly(bis-SorbPC) regions, suggesting that the poly(SPB) presents few defects or that the defects are small enough to inhibit fusion of functionalized vesicles. In most poorly patterned capillaries (< 5 % of poly(bis-SorbPC) patterned capillaries), Rh-DPPE fluorescence throughout the capillary is uniform, further supporting the lack of a uniform poly(bis-SorbPC) structure and providing a basis for quality control of the resulting capillary patterns. Rarely, it was observed that Rh-DPPE deposits inconsistently on the surface, creating a mottled pattern, most likely a result of incomplete rinsing of unpolymerized lipids and indicative of large micron scale defects on the surface (< 1 % of total pattern capillaries). This ability to deposit lipids into defined regions within the pattern provides the basis for preparing multi-functionalized chemical patterns.
Stability of poly(bis-SorbPC) patterns in fused silica capillaries
Implementation of fluid SPBs for biosensing and chemical patterning may be limited by the general fragility of non-covalent lipid structures which affects long-term storage or use in applications requiring surfacants or repeated rinsing and washing steps.33;43;44 Polymerization markedly stabilizes the SPB through the formation of a covalent bond network within the bilayer membrane.43;54 To determine if poly(bis-SorbPC) regions of the chemical patterns are suitably robust, we evaluated the physical and chemical stability of a series of poly(bis-SorbPC) patterns upon exposure to common chemical and physical insults, including drying/rehydration, exposure to organic solvents and surfactants and long-term storage, each of which may be expected to be endured by stable and reusable biosensing devices.
Figure 3 summarizes the stability of a series of poly(bis-SorbPC) patterned capillaries which have been subjected to various chemical and physical challenges. The capability to prepare and store large batches of patterned capillaries is an important consideration for wide scale application of the technique. The long-term stability of poly(bis-SorbPC) patterned capillaries was evaluated to determine the potential for preparing stable, spatially-defined polymer lipid regions. Figure 3A shows a fluorescence image of a poly(bis-SorbPC) patterned capillary which had been prepared and stored dry in excess of one year. A linescan of this capillary is provided in Supporting Information Figure S3. Upon rehydration of the dried lipid film and development with FM 1-43, an image representative of the original pattern is observed. Similar images were obtained for 4 of 4 capillaries stored under the same conditions.
Figure 3.
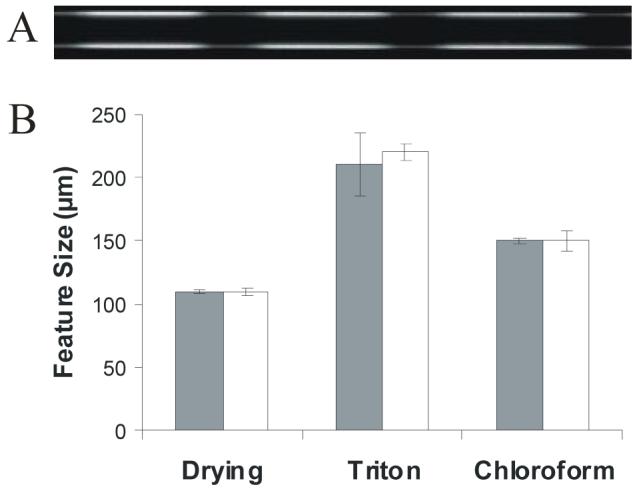
Chemical and physical stability of patterned poly(bis-SorbPC) capillaries. A) Fluorescence image of poly(bis-SorbPC) patterned capillary (50 μm i.d.) following one year of dry storage. B) The stability of poly(bis-SorbPC) patterns was evaluated following exposure to 10 drying/rehydration cycles, 1% Triton X-100 or chloroform by measuring the pattern feature size before (gray bar) and after (white bar) exposure. Patterns were developed using FM 1-43.
Stability of poly(bis-SorbPC) patterns to drying/rehydration is a key requirement for preparing multifunctionalized patterns and long-term storage since multiple rinse/wash/dry cycles are required for addition of each different chemical functionality. The stability of poly(bis-SorbPC) patterns was evaluated by measuring the size of the features patterned on the surface (Figure 3B), as the features in poorly polymerized poly(bis-SorbPC) regions rapidly decrease in size upon exposure to common chemical and physical challenges. Using FM 1-43 fluorescence to visualize the polymerized lipid membrane, no measurable change in the length of the poly(bis-SorbPC) pattern features was observed following ten drying/rehydration cycles within a single capillary (110 ± 2 μm before vs. 110 ± 3 μm after, n = 8 regions per capillary over 10 cycles). Poorly polymerized poly(bis-SorbPC) patterns are readily identified, as the poly(bis-SorbPC) regions desorb from the surface beginning with the first drying cycle, resulting in significantly smaller features and poorly defined domain boundaries. Additionally, poly(bis-SorbPC) patterned capillaries were subjected to commonly used membrane disrupting conditions that prove destructive to unpolymerized SPBs, including exposure to chloroform and 1% Triton X-100, respectively. Fluid lipid bilayers are unstable in the presence of organic solvents and readily desorb from the surface; however, poly(bis-SorbPC) is stable in the presence of most common laboratory solvents.43 Additionally, stability against zwitterionic surfactants is important as these compounds are commonly used to integrate hydrophobic analytes and membrane proteins into SPBs.55 The pattern boundaries are maintained and no significant change in poly(bis-SorbPC) feature length is observed upon one exposure to organic solvent (150 ± 3 μm before vs. 150 ± 8 μm after exposure to chloroform, n = 10 regions/capillary; n = 3 capillaries) or surfactant (210 ± 24 μm before vs. 220 ± 7 μm after exposure to 1% Triton X-100, n = 13 regions/capillary; n = 3 capillaries). Thus, the enhanced chemical and physical stability of the poly(SPB) allows for the capillaries to be reused for a variety of assays, whereas unpolymerized, chemically functionalized fluid bilayers can be destroyed and rinsed from the capillary without harming the poly(bis-SorbPC) barriers.
Evaluation of protein adsorption
A key difficulty in using chemically patterned surfaces for biological and chemical sensing and screening is the non-specific adsorption of proteins and other contaminants to the substrate and patterned materials. SPBs terminated with phosphorylcholine have been utilized extensively to minimize non-specific adsorption of proteins and other analytes on planar surfaces,18;21;45 as well as within enclosed silica capillaries and channels.24 Though some polymerized lipid films may minimize non-specific adsorption of proteins (e.g. DAPC), planar SPBs prepared using redox initated poly(bis-SorbPC) reduce protein adsorption to levels comparable to native phospholipids (e.g. DOPC).45 Further, we have shown that redox initiated poly(bis-SorbPC) coated capillaries, in which the coating was assembled and polymerized in situ, reduce non-specific adsorption of protein at levels equivalent to DOPC.53 While reduction of non-specific protein adsorption using redox initiated 45;53 less is known regarding UV-initiated poly(bis-polymerization of bis-SorbPC is established, SorbPC.
To evaluate the feasibility utilizing UV-initiated poly(bis-SorbPC) patterned capillaries for monitoring protein-ligand and protein-protein interactions, we evaluated the degree of non-specific protein adsorption in a series of poly(bis-SorbPC) patterned capillaries. Fused silica capillaries with differing lipid coatings were examined following 30 min exposure to 0.04 mg/mL SA-RPE, a fluorescent protein conjugate (Figure 4). Prior to imaging, SA-RPE was removed by thorough rinsing with phosphate buffer. In bare silica (uncoated) capillaries (Figure 4A, I), uniform fluorescence was observed throughout the length of the capillary due to non-specific adsorption of SA-RPE onto the capillary walls. When a DOPC coated capillary was exposed to SA-RPE (Figure 4A, II), the observed fluorescence was reduced by 83% (± 2%) n = 3 capillaries (Figure 4B). These results are in agreement with those obtained using both planar silica surfaces and fused silica capillary surfaces.24;45 We next evaluated the potential of UV-initiated poly(bis-SorbPC) for preparing protein resistant SPBs in fused silica channels. For poly(bis-SorbPC) patterned capillaries, in which the void regions were filled with DOPC prior to exposure to SA-RPE, the fluorescence is minimized throughout the capillary (Figure 4A, III), to a level where regions containing DOPC and poly(bis-SorbPC) are indistinguishable. Following exposure of a poly(bis-SorbPC) patterned capillary in which the void regions were left bare to SA-RPE and subsequent removal of excess protein, a series of dark and bright regions corresponding to low and high non-specific adsorption of the fluorescent protein were observed (Figure 4A, IV), where the dark regions correspond to poly(bis-SorbPC) domains and the bright regions correspond to bare silica domains resulting from removal of unpolymerized lipids. The nature of the reduced non-specific adsorption to bare silica regions observed in Figure 4A(IV) is unclear, but it uniformly occurs in patterned capillaries. It is not likely that this is due to incomplete removal of the bis-SorbPC, as fluorophore-doped poly(bis-SorbPC) capillaries show no fluorescence after removal of the excess lipid with surfactant in these regions. Overall, a 79% (± 3%), n = 3 capillaries, reduction in fluorescence was observed for poly(bis-SorbPC) coated capillaries compared to bare silica capillaries (Figure 4B), suggesting that non-specific adsorption of proteins can be reduced using poly(bis-SorbPC) to levels comparable to DOPC, though with the added benefit of markedly enhanced chemical and physical stability of the phospholipid pattern.
Covalent functionalization of poly(lipid) patterned capillaries
Fabrication of chemically functionalized, spatially-defined molecular patterns provides the potential for multi-analyte biomolecular sensing and screening platforms. A key requirement for realizing this potential is the capability to immobilize chemically and biologically active molecules, while retaining function. Proteins and other biologically active compounds are most commonly immobilized via covalent linkages. A series of non-covalent immobilization chemistries have emerged that provide enhanced activity for a number of proteins and enzymes.8;47;56 Due to the availability of a wide range of chemically-functionalized phospholipids, both covalent and non-covalent immobilization chemistries can be readily incorporated into lipid based chemical patterns. In our previous work, we prepared a series of poly(bis-SorbPC) patterned capillaries functionalized with commonly used non-covalent immobilization chemistries including Ni2+-NTA and biotin-streptavidin, where the immobilized protein maintained the corresponding biological function, thereby demonstrating the power of this approach.47
In addition to these non-covalent immobilization chemistries, we have explored the potential for covalent attachment of proteins. In this approach, chemical patterns functionalized with amine-or thiol-reactive lipid headgroups were fabricated using poly(bis-SorbPC) patterned capillaries followed by vesicle fusion and self-assembly of doped SUVs. The data presented in Figure 5 illustrates the capability of attaching proteins to functionalized regions of the molecular pattern. Functionalization was performed via introduction of 20% maleimide-doped DOPC or 5% w/w DOPE-PEG-pNP-doped DOPC SUVs that were used to fill the void regions between poly(bis-SorbPC) regions. FQ-labeled ribonuclease A, which had been reduced to expose free thiol linkages (Figure 5A) or FQ-labeled ribonuclease A that was unreduced (Figure 5B) was introduced into the maleimide functionalized capillary. Linescans taken along the length of the capillary at the wall are shown in (Figure 5C). These data illustrate that the presence of free thiols is required in order to generate a pattern, thereby supporting the specific, covalent nature of the attachment. In addition, 6xHis-EGFP fluorescent protein was immobilized via covalent primary amine linkage with the pNP moiety. Fluorescence images of the resulting protein functionalized patterns were obtained following incubation and removal of excess protein and the fluorescence intensity of the corresponding regions measured. As a comparison, SUVs comprised of DOPC with no dopant lipid were fused into the bare regions of the poly(bis-SorbPC) pattern. Figure 5D summarizes the fluorescence intensity of the protein functionalized regions of the pattern. In both cases, the fluorescence in the DOPC containing regions (white bars) is significantly lower than that seen in the maleimide or DOPE-PEG-pNP functionalized regions (gray bars).
To evaluate whether covalently immobilized proteins may retain function following immobilization, β-galactosidase from E. coli was covalently immobilized to the lipid pattern. β-galactosidase from E.coli presents a high cysteine content that is useful for immobilization.57 β-galactosidase was immobilized into poly(bis-SorbPC) patterned capillaries filled with 20% maleimide-doped DOPC or DOPC in the void regions. Following immobilization, fluorescein di-β-D-galactopyranoside (FDG) was introduced and enzyme activity was visualized via fluorescence imaging. Upon hydrolysis by the enzyme, FDG yields a fluorescent product. The results are shown in Figure 5. When no maleimide functionality was present, introduction of FDG yielded weak fluorescence as seen in Figure 5E and Figure 5H (i). Introduction of FDG to capillaries containing maleimide functionalities, and thus β-galactosidase results in rapid, time-dependent increases in fluorescence. Images collected at t = 30 s and t = 3 min are shown in Figure 5F and 5G, respectively. Linescans obtained along the length of the capillaries are shown in Figure 5H. In the capillary lacking maleimide functionalization little fluorescence is seen (i), while a clear pattern is observed in the maleimide-functionalized capillary at 30 s (ii). The pattern is less obvious at 3 min (iii), due primarily to diffusion of the fluorescent substrate.
Poly(lipid) patterns with multiple binding functionalities
A key potential advantage of this platform is the capacity to prepare poly(lipid) capillary patterns containing multiple, spatially-discrete binding functionalities, thereby opening the door to multicomponent sensor arrays, assays, etc. To realize this potential, chemical functionalization of the poly(bis-SorbPC) regions is required. Successful polymerization of bis-SorbPC with other non-polymerizable lipids has been achieved in poly(bis-SorbPC) vesicles58 and poly(bis-SorbPC) SPBs.43 To evaluate the feasibility of functionalizing poly(bis-SorbPC) regions, biotin lipid was dried together with the bis-SorbPC to form doped-SUVs, which were used for pattern formation. The functionality of the biotin within the patterns was verified via biotin-streptavidin binding (Figure 6A). A poly(bis-SorbPC) capillary in which the void regions were filled with 10% biotin lipid-doped DOPC in the non-polymer regions was also evaluated as a control (Figure 6B). Following pattern formation, streptavidin was introduced into the capillaries to bind available biotin sites. Following removal of excess streptavidin, biotinylated anti-pentaHis antibody was introduced to the capillary and allowed to fuse to the streptavidin. After removing excess antibody, 6xHis-EGFP was introduced allowing the regions containing biotin to be readily identified using fluorescence microscopy (Figure 6A and 6B). As expected, orthogonal fluorescence regions are observed for functionalized poly(bis-SorbPC) (Figure 6A) and functionalized DOPC (Figure 6B) regions, which are readily identified by feature size resulting from the photomask. Further, copolymerization of biotin lipid with bis-SorbPC markedly stabilizes the biotin lipid allowing exposure to surfactant and drying, as these capillaries were rinsed with Triton X-100 and H2O following polymerization, and stored dry until use.
Figure 6.
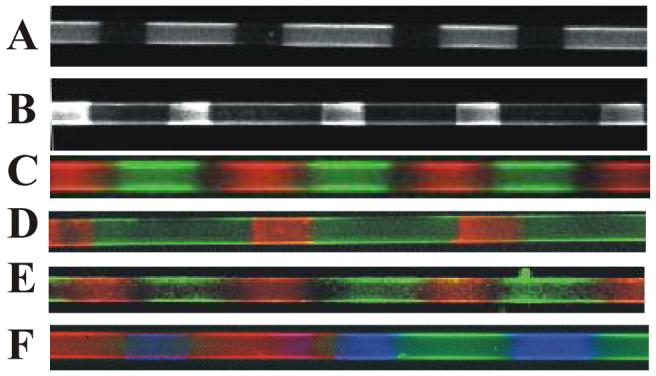
Introduction of multiple functionalities via polymerization of doped bis-SorbPC. A) Poly(biotin lipid/bis-SorbPC) with DOPC fused into the void regions. B) Poly(bis-SorbPC) patterned capillary with 10% biotin lipid/90% DOPC lipid fused into the void regions. For A) and B), patterns were developed via consecutive introduction of streptavidin, biotin-anti-6xHis, and 6xHis-EGFP. C) Poly(Rh-DPPE/bis-SorbPC) (red) with NBD-DOPE-doped DOPC fused into void regions (green). D) Poly(biotin-DHPE/bis-SorbPC) with NTA-doped DOPC fused into the void regions. Streptavidin-fluorecein conjugate and 6xHis-DsRed are introduced into the capillary to develop the regions containing biotin (green) and Ni2+-NTA (red). E) Poly(Rh-DPPE/bis-SorbPC) (red) and poly(NBD-DOPE/bis-SorbPC) (green). F) Poly(Rh-DPPE/bis-SorbPC) (red), poly(NBD-DOPE/bis-SorbPC) (green), and NTA-Ni-6xHis-Cerulean (blue). All capillaries are 50 μm i.d.
To further explore multiple functionalization, patterns were prepared using doped SUVs containing 2% Rh-DPPE in bis-SorbPC. The void regions of the resulting pattern were then filled using 5% NBD-DOPE-doped DOPC SUVs. As seen in Figure 6C, the poly(Rh-DPPE/bis-SorbPC) regions (red) are orthogonal to the NBD-DOPE/DOPC regions (green), demonstrating the capability of introducing two spatially adjacent functionalities.
The capacity to localize binding functionalities, whether they be covalent or non-covalent, in adjacent features within a patterned capillary was explored using biotin/streptavidin chemistry coupled with Ni2+-NTA/6xHis chemistry, wherein the biotin was colocalized within the poly(bis-SorbPC regions. To these capillaries, 10% DOGS-Ni2+-NTA-doped DOPC SUVs were used to fill the void regions within a poly(biotin-DHPE/bis-SorbPC) pattern. A mixture, containing the complementary ligands for each headgroup functionalized lipid, was introduced to the capillary. Following removal of excess ligand, fluorescence images were collected using fluorescein and rhodamine filter cubes, corresponding to the detection of FITC-streptavidin and 6xHis-DsRed, respectively. As seen in Figure 6D, fluorescence from 6xHis-DsRed (red) and FITC-streptavidin (green) are localized in adjacent regions of the pattern, with no significant overlap.
The potential for creating high density, multiple functionality poly(lipid) patterned capillaries requires the ability to dope individual functionalities into discrete poly(lipid) regions with high feature density. This step requires copolymerization of adjacent regions containing varying functionality yielding a surface comprised primarily of doped-poly(bis-SorbPC) regions. To evaluate this process, a two component pattern was prepared with alternating regions of poly(Rh-DPPE/bis-SorbPC) and poly(NBD-DOPE/bis-SorbPC) using multiple fusion and polymerization steps. Fluorescence images were collected of the resultant two component patterned capillaries using rhodamine and fluorescein filter cubes. As seen in Figure 6E, the poly(Rh-DPPE/bis-SorbPC) regions (red) are readily distinguished from the poly(NBD-DOPE/bis-SorbPC) regions (green), with only slight overlap at the boundary regions.
To demonstrate higher density patterning, capillaries containing three distinct regions were prepared using sequential deposition and polymerization cycles (Figure 6F). In the first cycle, poly(Rh-DPPE/.bis-SorbPC) regions were fabricated on the capillary wall. After removal of excess unpolymerized lipid, the pattern was remasked and NBD-DOPE/bis-SorbPC was deposited and polymerized. Subsequent rinsing steps do not alter the fluorescence intensities of copolymerized lipids, indicating these lipids can be considered as rugged as poly(bis-SorbPC) patterns. After removal of excess unpolymerized lipid, the bare capillary regions between poly(NBD-DOPE/bis-SorbPC) and poly(Rh-DPPE/bis-SorbPC) regions were backfilled with 5% NTA-Ni/DOPC. The resulting 3 component pattern was visualized via fluorescence imaging (Figure 6F). Fluorescence images were obtained individually then combined to produce a three color image. As seen in Figure 6F, an alternating pattern of poly(Rh-DPPE/bis-SorbPC) (red), poly(NBD-DOPE, bis-SorbPC) (green) and NTA-DOPC/6xHis-Cerulean (blue) can be seen within the same capillary. Some spatial overlap of the individual fluorophores is observed in Figure 6F, though this likely results from imprecise registration of the photomask due to the lack of a high precision mask aligner, and not from mixing poly(lipid) regions. The mask registration leads to incomplete polymerization of the region in which fluid lipid can incorporate into these poorly polymerized regions. Thus, preparation of more complex, higher density patterns can be readily envisioned via multiple deposition and polymerization steps using suitably registered photomasks.
CONCLUSION
We have prepared and characterized functional, self-assembled, chemically-functionalized patterns inside completely enclosed fused silica capillaries using polymerized phospholipid bilayers. The patterned polymerized lipid features demonstrate resistance to non-specific protein adsorption and stability to physical and chemical challenges, such as surfactant and organic solvent rinses, drying/rehydration and long-term storage, while retaining chemical function. The resulting chemical pattern can be functionalized using fluid lipids or polymerized lipids to monitor biological binding interactions through non-covalent or covalent attachment of molecules, which retain their activity after surface immobilization. We have further demonstrated the capacity of this patterning approach for preparing multiple component polymer lipid patterns with adjacent chemically-functionalized polymer lipid regions, thereby providing the potential for preparing high density patterns. The unique combination of stability, biocompatibility, reduction in non-specific protein adsorption and the availability of numerous chemically functionalized headgroups suggests the utility of this approach for preparing a universal platform for high-throughput chemical sensing and screening applications.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by funds from NIH (GM074522). We would like to express our gratitude to David L. Roberts for synthesis of DOPE-PEG-pNP lipid and Cliff I. Stains and Indraneel Ghosh for donating the 6xHis-Cerulean.
REFERENCES AND FOOTNOTES
- (1).Bernard A, Delamarche E, Schmid H, Michel B, Bosshard HR, Biebuyck H. Langmuir. 1998;14:2225–2229. [Google Scholar]
- (2).Kam L, Boxer SG. J.Biomed.Mater.Res. 2001;55:487–495. doi: 10.1002/1097-4636(20010615)55:4<487::aid-jbm1041>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- (3).James CD, Davis RC, Kam L, Craighead HG, Isaacson M, Turner JN, Shain W. Langmuir. 1998;14:741–744. [Google Scholar]
- (4).Bernard A, Renault JP, Michel B, Bosshard HR, Delamarche E. Adv.Mater. 2000;12:1067–1070. [Google Scholar]
- (5).Hovis JS, Boxer SG. Langmuir. 2001;17:3400–3405. [Google Scholar]
- (6).Hovis JS, Boxer SG. Langmuir. 2000;16:894–897. [Google Scholar]
- (7).Orth RN, Kameoka J, Zipfel WR, Ilic B, Webb WW, Clark TG, Craighead HG. Biophys.J. 2003;85:3066–3073. doi: 10.1016/S0006-3495(03)74725-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Holden MA, Jung S-Y, Cremer PS. Anal.Chem. 2004;76:1838–1843. doi: 10.1021/ac035234q. [DOI] [PubMed] [Google Scholar]
- (9).Holden MA, Cremer PS. J.Am.Chem.Soc. 2003;125:8074–8075. doi: 10.1021/ja035390e. [DOI] [PubMed] [Google Scholar]
- (10).Groves JT, Ulman N, Boxer SG. Science. 1997;275:651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- (11).Groves JT, Boxer SG. Acc.Chem.Res. 2002;35:149–157. doi: 10.1021/ar950039m. [DOI] [PubMed] [Google Scholar]
- (12).Groves JT, Ulman N, Cremer PS, Boxer SG. Langmuir. 1998;14:3347–3350. [Google Scholar]
- (13).Cremer PS, Boxer SG. J.Phys.Chem.B. 1999;103:2554–2559. [Google Scholar]
- (14).Yee CK, Amweg ML, Parikh AN. Adv.Mater. 2004;16:1184–1189. [Google Scholar]
- (15).Yee CK, Amweg ML, Parikh AN. J.Am.Chem.Soc. 2004;126:13962–13972. doi: 10.1021/ja047714k. [DOI] [PubMed] [Google Scholar]
- (16).Yu C, Parikh AN, Groves JT. Adv.Mater. 2005;17:1477–1480. [Google Scholar]
- (17).Balakirev MY, Porte S, Vernaz-Gris M, Berger M, Arié J-P, Fouqué B, Chatelain F. Anal.Chem. 2005;77:5474–5479. doi: 10.1021/ac0504619. [DOI] [PubMed] [Google Scholar]
- (18).Moran-Mirabal JM, Edel JB, Meyer GD, Throckmorton D, Singh AK, Craighead HG. Biophys.J. 2005;89:296–305. doi: 10.1529/biophysj.104.054346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hird GS, McIntosh TJ, Grinstaff MW. J.Am.Chem.Soc. 2000;122:8097–8098. [Google Scholar]
- (20).Yang T, Simanek EE, Cremer PS. Anal.Chem. 2000;72:2587–2589. doi: 10.1021/ac000131i. [DOI] [PubMed] [Google Scholar]
- (21).Mao H, Yang T, Cremer PS. Anal.Chem. 2002;74:379–385. doi: 10.1021/ac010822u. [DOI] [PubMed] [Google Scholar]
- (22).Boukobza E, Sonnenfeld A, Haran G. J.Phys.Chem.B. 2001;105:12165–12170. [Google Scholar]
- (23).Ajo-Franklin CM, Yoshina-Ishii C, Boxer SG. Langmuir. 2005;21:4976–4983. doi: 10.1021/la0468388. [DOI] [PubMed] [Google Scholar]
- (24).Cunliffe JM, Baryla NE, Lucy CA. Anal.Chem. 2002;74:776–783. doi: 10.1021/ac015627u. [DOI] [PubMed] [Google Scholar]
- (25).Wang C, Lucy CA. Anal.Chem. 2005;77:2015–2021. doi: 10.1021/ac0489622. [DOI] [PubMed] [Google Scholar]
- (26).Cremer PS, Yang T. J.Am.Chem.Soc. 1999;121:8130–8131. [Google Scholar]
- (27).Yang T, Jung S-Y, Mao H, Cremer PS. Anal.Chem. 2001;73:165–169. doi: 10.1021/ac000997o. [DOI] [PubMed] [Google Scholar]
- (28).Elie-Caille C, Fliniauz O, Pantigny J, Mazière J-C, Bourdillon C. Langmuir. 2005;21:4661–4668. doi: 10.1021/la046973k. [DOI] [PubMed] [Google Scholar]
- (29).Martinez KL, Meyer BH, Hovius R, Lundstrom K, Vogel H. Langmuir. 2003;19:10925–10929. [Google Scholar]
- (30).Giess F, Friedrich MG, Heberle J, Naumann RL, Knoll W. Biophys.J. 2004;87:3213–3220. doi: 10.1529/biophysj.104.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Kang X, Cheley S, Rice-Ficht AC, Bayley H. J.Am.Chem.Soc. 2007;129:4701–4705. doi: 10.1021/ja068654g. [DOI] [PubMed] [Google Scholar]
- (32).Slade A, Luh J, Ho S, Yip CM. J.Struct.Biol. 2002;137:283–291. doi: 10.1016/s1047-8477(02)00011-4. [DOI] [PubMed] [Google Scholar]
- (33).Albertario F, Diaz AJ, Yang T, Chapa VA, Kataoka S, Castellana ET, Cremer PS. Langmuir. 2005;21:7476–7482. doi: 10.1021/la050871s. [DOI] [PubMed] [Google Scholar]
- (34).Holden MA, Jung S-Y, Yang T, Castellana ET, Cremer PS. J.Am.Chem.Soc. 2004;126:6512–6513. doi: 10.1021/ja048504a. [DOI] [PubMed] [Google Scholar]
- (35).Crowe JH, Crowe LM, Carpenter JF, Aurell Wistrom C. Biochem.J. 1987;242:10. doi: 10.1042/bj2420001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Crowe LM, Reid DS, Crowe JH. Biophys.J. 1996;71:2087–2093. doi: 10.1016/S0006-3495(96)79407-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Plant AL. Langmuir. 1999;15:5128–5135. [Google Scholar]
- (38).Yang Z, Yu H. Langmuir. 1999;15:1731–1737. [Google Scholar]
- (39).Morigaki K, Baumgart T, Offenhäusser A, Knoll W. Angew.Chem.Int.Ed. 2001;40:172–174. doi: 10.1002/1521-3773(20010105)40:1<172::AID-ANIE172>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- (40).Morigaki K, Kiyosue K, Taguchi T. Langmuir. 2004;20:7729–7735. doi: 10.1021/la049340e. [DOI] [PubMed] [Google Scholar]
- (41).Morigaki K, Schonherr H, Frank CW, Knoll W. Langmuir. 2003;19:6994–7002. [Google Scholar]
- (42).Britt DW, Hofmann UG, Möbius D, Hell SW. Langmuir. 2001;17:3757–3765. [Google Scholar]
- (43).Ross EE, Rozanski LJ, Spratt T, Liu S, O’Brien DF, Saavedra SS. Langmuir. 2003;19:1752–1765. [Google Scholar]
- (44).Ross EE, Bondurant B, Spratt T, Conboy JC, O’Brien DF, Saavedra SS. Langmuir. 2001;17:2305–2307. [Google Scholar]
- (45).Ross EE, Spratt T, Liu S, Rozanski LJ, O’Brien DF, Saavedra SS. Langmuir. 2003;19:1766–1774. [Google Scholar]
- (46).Subramaniam V, Alves ID, Salgado GFJ, Lau P-W, Wysocki RJ, Salamon Z, Tollin G, Hruby VJ, Brown MF, Saavedra SS. J.Am.Chem.Soc. 2005;127:5320–5321. doi: 10.1021/ja0423973. [DOI] [PubMed] [Google Scholar]
- (47).Ross EE, Mansfield E, Aspinwall CA. J.Am.Chem.Soc. 2005;127:16756–16757. doi: 10.1021/ja0539995. [DOI] [PubMed] [Google Scholar]
- (48).Lamparski H, Liman U, Barry JA, Frankel DA, Ramaswami V, Brown MF, O’Brien DF. Biochemistry. 1992;31:685–694. doi: 10.1021/bi00118a008. [DOI] [PubMed] [Google Scholar]
- (49).Torchilin VP, Levchenko TS, Lukyanov AN, Khaw BA, Klibanov AL, Rammohan R, Samokhin GP, Whiteman KR. Biochim.Biophys.Acta. 2001;1511:397–411. doi: 10.1016/s0005-2728(01)00165-7. [DOI] [PubMed] [Google Scholar]
- (50).Torchilin VP, Rammohan R, Weissig V, Levchenko TS. Proc.Natl.Acad.Sci.U.S.A. 2001;98:8786–8791. doi: 10.1073/pnas.151247498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Multiple photomasks have been used and the images within this paper vary in feature size and spacing as a result of the different masks.
- (52).Stoyanov AV, Ahmadzadeh H, Krylov SN. J.Chromatogr., B.Anal.Technol.Biomed.Life Sci. 2002;780:283–287. doi: 10.1016/s1570-0232(02)00535-4. [DOI] [PubMed] [Google Scholar]
- (53).Mansfield E, Ross EE, Aspinwall CA. Anal.Chem. 2007;79:3135–3141. doi: 10.1021/ac0618829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Sisson TM, Lamparski H, Kõlchens S, Elayadi A, O’Brien DF. Macromolecules. 1996;29:8321–8329. [Google Scholar]
- (55).Privé GG. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- (56).Wang C-Y, Hitz S, Andrade JD, Stewart RJ. Anal.Biochem. 1997;246:133–139. doi: 10.1006/abio.1997.2006. [DOI] [PubMed] [Google Scholar]
- (57).Grazú V, Abian O, Mateo C, Batista-Viera F, Fernández-Lafuente R, Guisán JM. Biomacromolecules. 2003;3:1495–1501. doi: 10.1021/bm034262f. [DOI] [PubMed] [Google Scholar]
- (58).Cheng Z, D’Ambruoso GD, Aspinwall CA. Langmuir. 2006;22:9507–9511. doi: 10.1021/la061542i. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.