Abstract
In an initial epigenetic characterization of diffuse large B-cell lymphoma (DLBCL), we evaluated the DNA methylation levels of over 500 CpG islands. Twelve CpG islands (AR, CDKN1C, DLC1, DRD2, GATA4, GDNF, GRIN2B, MTHFR, MYOD1, NEUROD1, ONECUT2, and TFAP2A) showed significant methylation in over 85% of tumors. Interestingly, the methylation levels of a CpG island proximal to FLJ21062 differed between the activated B-cell-like (ABC-DLBCL) and germinal center B-cell-like (GCB-DLBCL) subtypes. In addition, we compared the methylation and expression status of sixty-seven genes proximal (within 500-bp) to the methylation assays. We frequently observed that hypermethylated CpG islands are proximal to genes that are expressed at low or undetectable levels in tumors. However, many of these same genes were also poorly expressed in DLBCL tumors where their cognate CpG islands were hypomethylated. Nevertheless, the proportional reductions in BNIP3, MGMT, RBP1, GATA4, IGSF4, CRABP1, and FLJ21062 expression with increasing methylation suggests that epigenetic processes strongly influence these genes. Lastly, the moderate expression of several genes proximal to hypermethylated CpG tracts suggests that DNA methylation assays are not always accurate predictors of gene silencing. Overall, further investigation of the highlighted CpG islands as potential clinical biomarkers is warranted.
Keywords: epigenetics, genomics, CpG island, microarray, gene expression
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive malignancy of the mature B-lymphocyte that accounts for approximately one-third of non-Hodgkin lymphoma.1, 2 Extensive analyses of gene expression profiles3-5 and genomic copy number6-9 in DLBCL have provided valuable insights into their cellular origins and the molecular bases for their variable clinical behaviors. For example, gene expression analyses have identified two major DLBCL subtypes, germinal center B-cell-like (GCB-DLBCL) and activated B-cell-like (ABC-DLBCL), that originate from different stages of normal B-cell development.4 Patients with GCB-DLBCL have a substantially longer median overall survival than patients with ABC-DLBCL.3, 4, 10
In contrast to these genetic characterizations, much less is known about epigenetic changes present in DLBCL. Although wide-spread epigenetic analyses could yield insights into disease origins and provide diagnostic and prognostic biomarkers, larger-scale studies have focused on NHL cell lines.11 Here, we have taken the initial steps in the wide-spread epigenetic characterization of DLBCL by comparing the DNA methylation levels of over five hundred gene-associated CpG islands in tumors. Using a two-phase methylation screening strategy, we identified genes that are frequently methylated in all DLBCL tumors and uncovered evidence of epigenetic differences between ABC-DLBCL and GCB-DLBCL tumor subtypes. Furthermore, we identified genes that show proportional reductions in expression in response to increased methylation levels in nearby CpG islands. Overall, we highlight candidate DNA methylation changes associated with DLBCL that also could warrant further investigation as potential clinical biomarkers.
Materials and Methods
Samples and patients
Frozen tissue sample DNA from diagnostic tumor biopsies from ten ABC-DLBCL and seventeen GCB-DLBCL patients acquired prior to anthracycline-based chemotherapy were obtained from the University of Nebraska Medical Center and the Norwegian Radium Hospital. There was consensus central pathology re-review of the specimens to confirm the diagnosis of DLBCL and that the samples had >75% tumor cells. Clinical information has been obtained from all patients according to a protocol approved by the University of Nebraska Medical Center Institutional Review Board.
CpG island microarray assays
CpG island microarrays, consisting of 4,395 PCR products, and nucleic acid targets were prepared as previously described and used for two-color hybridization analyses (Supplementary Figure 1).12, 13 Briefly, tumor DNA was digested with MseI and ligated to double-stranded linkers prior to division into equal test and reference fractions. The test fraction was digested with the methylation-dependent McrBC endonuclease (5′ … Pu mC [N40-3,000] Pu mC … 3′) while the reference fraction was untreated. Afterwards, both fractions were subjected to amplification with linker-specific primers. In theory, only those fragments in the test fraction lacking a methylated McrBC recognition sequence remain intact after digestion and serve as PCR templates. The amplified test and reference fractions were random-prime labeled with Cy3- or Cy5- dUTP and combined in a cocktail prior to hybridization to CpG island microarrays.12, 13 Afterwards, the microarrays were subject to two washes, dried by centrifugation, and imaged using the ScanArray 5000 (GSI Lumonics, Inc.) with ScanArray Express software (Perkin Elmer). Signal intensities and quality metrics for each clone were calculated using ImaGene microarray software (BioDiscovery, Inc.).
Microarray data processing
Log2-transformed background subtracted hybridization signals were obtained for both test and reference targets and imported into Microsoft Excel. We employed multiple filters to ensure that our final analysis was limited to non-repetitive clones yielding the most robust data. First, we eliminated clones that either failed to amplify or gave multiple PCR products. We also disregarded data from clones with poor spot morphology or whose reference signal intensity was less than twice that of the local background. Lastly, we only analyzed clones with a reference signal intensity < 30,000 units in order to reduce the effects of excessive cross-hybridization.
After data filtration, test and reference fraction hybridization signals were normalized using an interactive linear regression approach based on the signal intensities of mitochondrial clones 12. Since mitochondrial DNA is unmethylated,12, 14 the signal intensities for both Cy3 and Cy5 are expected to be equal. Following normalization, the ratios of test and reference signals were calculated. In order to minimize experimental noise, ratios were truncated to a maximum value of one. These values reflect the fraction of unmethylated alleles in a given tumor sample. Overall, we obtained DNA methylation measurements for 592 CpG islands adjacent to 442 unique annotated genes in a minimum of four GCB-DLBCL and four ABC-DLBCL (Supplementary Table 1 and Supplementary Figure 2).
MethyLight
Using published protocols, tumor DNA was subjected to sodium bisulfite conversion and individual loci were amplified with methylation-specific primers that flank a methylation-specific reporter oligonucleotide (Supplementary Table 2).15 Samples were analyzed on an Opticon DNA Engine Continuous Fluorescence Detector (MJ Research/Bio-Rad). Relative measurements of DNA methylation (reported as PMR, percentage of methylated reference) values were calculated based upon the performance of a normalizing control reaction (Alu: HB-313) in a 1:25 dilution series of in vitro methylated human reference sample.15
Bisulfite sequencing
Bisulfite PCR products representing specific CpG islands from selected DNA samples were subcloned and individual colonies sequenced as described in Supplementary Figure 3.
Gene expression profiling
Total RNA from frozen tumor biopsies was isolated and subjected to analysis on Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA) according to the manufacturer's recommended protocols. We report normalized log2-transformed gene expression data for probe tilings with minimal cross-hybridization potential (i.e. designated by Affymetrix as “_at” tilings) that interrogate NCBI-designated Reference Sequence (RefSeq) transcripts located within 500-bp of MethyLight reactions (Supplementary Table 3).
Results and Discussion
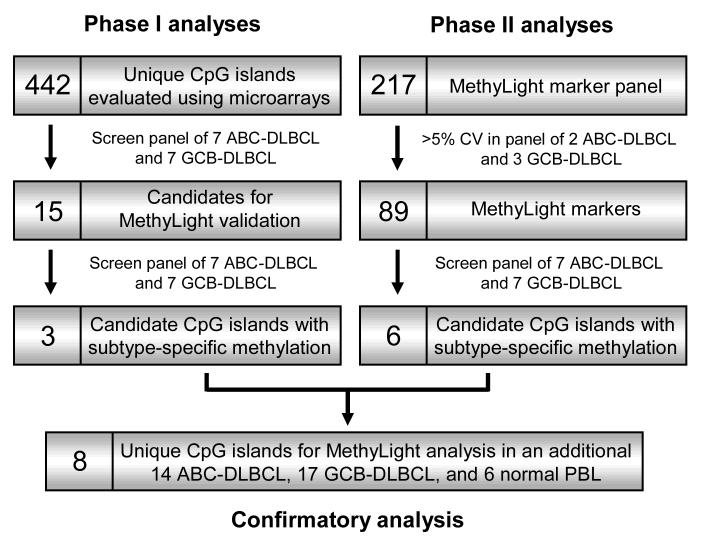
In Phase I of our two-phase strategy to characterize epigenetic phenomena in DLBCL, we measured DNA methylation levels in seven ABC-DLBCL and seven GCB-DLBCL tumors using CpG island microarray assays 12, 16, 17 (Figure 1). This provided a rapid means of evaluating the methylation-dependent cleavage of 442 unique gene-associated CpG islands by McrBC endonuclease (Supplementary Table 1 and Supplementary Figure 2). Due to the semi-quantitative nature of CpG island microarray-based analyses,18 we validated selected results using MethyLight, a quantitative bisulfite PCR platform (Table 1).15, 19
Figure 1.
Flowchart of study design. Phase I and Phase II analyses were conducted independent of one another. The identities and relevant methylation data from all candidate CpG islands with subtype-specific methylation levels are provided in Table 1. CV refers to the coefficient of variation. Note that only seven gene-associated CpG islands are interrogated by both the Phase I CpG island microarray assays and Phase II MethyLight assays.
Table 1.
Summary of Phase I and Phase II analyses to identify DLBCL subtype-specific methylation
| Phase I CpG island microarray screen |
Phase I MethyLight confirmation |
|||||||
|---|---|---|---|---|---|---|---|---|
| Gene | ABCa | GCBa | Pb | ABCc | GCBc | Pb | ML ID | |
| CEBPG | 0.991 | 0.947 | 0.0099 | 0.0 | 0.0 | 1.000 | HB-480 | |
| CENPH | 0.944 | 0.913 | 0.0104 | 0.0 | 0.0 | 0.320 | HB-428 | |
| CPVL | 0.948 | 0.843 | 0.0163 | 61.3 | 60.5 | 0.340 | HB-427d | |
| FLJ21062 | 0.967 | 0.897 | 0.0446 | 0.0 | 23.4 | 0.009 | HB-442d | |
| GNMT | 0.934 | 0.794 | 0.0062 | 0.0 | 55.6 | 0.005 | HB-426d | |
| HOXC9 | 0.953 | 0.892 | 0.0105 | 69.1 | 80.4 | 0.650 | HB-440d | |
| HTRA4 | 0.892 | 0.787 | 0.0472 | 0.1 | 0.30 | 0.064 | HB-484 | |
| KLHL14 | 0.973 | 0.938 | 0.0209 | 2.2 | 3.6 | 0.220 | HB-481 | |
| NOS1AP | 0.900 | 0.985 | 0.0472 | 0.0 | 0.0 | 0.940 | HB-485 | |
| ONECUT2 | 0.826 | 0.974 | 0.0143 | 97.4 | 50.5 | 0.018 | HB-242d | |
| ONECUT2 | 0.826 | 0.974 | 0.0143 | 82.0 | 36.7 | 0.025 | HB-446d | |
| PFDN5 | 0.919 | 0.812 | 0.0074 | 0.0 | 0.0 | 0.940 | HB-424d | |
| PFDN5 | 0.919 | 0.812 | 0.0074 | 0.0 | 0.0 | 0.320 | HB-425 | |
| PHC2 | 0.900 | 0.841 | 0.0424 | 0.0 | 0.0 | 0.320 | HB-483 | |
| PRIMA1 | 0.875 | 0.802 | 0.0321 | 31.5 | 64.6 | 0.140 | HB-482 | |
| TP53I11 | 0.863 | 0.797 | 0.0285 | 0.0 | 0.0 | 1.000 | HB-443 | |
| ZNF615 | 0.939 | 0.795 | 0.0223 | 0.0 | 0.0 | 0.950 | HB-431d | |
| Phase II Independent MethyLight Screen | ||||||||
| Gene | ABCa | GCBa | Pb | ML ID | ||||
| CYP27B1 | 4.5 | 56.9 | 0.0127 | HB-233 | ||||
| ONECUT2 | 55.9 | 36.6 | 0.0127 | HB-243 | ||||
| NEUROG1 | 29.3 | 11.7 | 0.0348 | HB-261 | ||||
| KL | 6.3 | 25.8 | 0.0467 | HB-175 | ||||
| MINT2 | 24.7 | 0.0 | 0.0467 | HB-187 | ||||
| DRD1 | 43.6 | 28.5 | 0.0476 | HB-252 | ||||
Median fraction of unmethylated alleles in a subtype, as provided by CpG island microarray analyses.
Uncorrected Wilcoxon t-test. Data are bolded if P < 0.05.
Median MethyLight PMR (Percent of Methylated Reference) value that reflects the fraction of fully methylated alleles in a subtype.
Based on one or more replicate reactions.
Based on our Phase I CpG island microarray analyses, fifteen candidate CpG islands revealed differences in methylation between subtypes of DLBCL (uncorrected Wilcoxon P < 0.05 and greater than 5% difference in median methylation levels) (Table 1 and Supplementary Table 1). We used modest criteria to identify differential methylation since cross-hybridization of CpG island sequences can artificially compress methylation estimates. In our Phase I confirmatory analyses, we developed novel MethyLight assays for each of these fifteen candidate CpG islands and interrogated their methylation levels in the same group of fourteen DLBCL (Table 1 and Supplementary Table 4). The methylation levels of six (CPVL, FLJ21062, GNMT, HOXC9, ONECUT2, and PRIMA1) of these fifteen CpG islands showed PMR (percentage of methylated reference) values greater than ten in more than one tumor. Interestingly, based on an uncorrected Wilcoxon signed-rank test, MethyLight assays for FLJ21062 (HB-442, P = 0.009), GNMT (HB-426, P = 0.005), and ONECUT2 (HB-242, P = 0.018; HB-446, P = 0.025) showed differences between ABC-DLBCL and GCB-DLBCL (Table 1). The results from all replicate experiments were in excellent agreement (Supplementary Table 4).
The fact that nine (CEBPG, CENPH, HTRA4, KHL14, NOSAP1, PFDN5, PHC2, TP53I11, and ZNF615) of fifteen CpG islands demonstrate methylation in DLBCL by CpG island microarray assays, but not by MethyLight, partially reflects fundamental differences in these platforms. Based on the McrBC endonuclease recognition sequence, our microarray assays can score a CpG island as being methylated if it contains at least two 5-methylcytosines that are between forty bases and thirty kilobases apart. In contrast, our MethyLight assays are designed to only score CpG islands as being methylated if they contain an average of eight closely spaced 5-methylcytosines. This highlights the complex nature of methylation within individual CpG islands and the advantages of using complimentary technologies to evaluate their status. Importantly, the CpG island microarrays succeeded in rapidly identifying viable candidates for confirmation using more quantitative methods.
In our Phase II studies, we analyzed the DNA methylation status of a focused group of CpG islands proximal to genes whose methylation status is either known or suspected to be associated with the development and progression of cancer (Figure 1). We first surveyed the methylation levels of 217 unique CpG island sequences in two ABC-DLBCL and three GCB-DLBCL using pre-existing MethyLight assays. We identified 80 unique CpG islands with at least moderate differences in methylation levels among these five randomly selected DLBCL (i.e. coefficient of variation greater than 5%) (Supplementary Table 5). Next, we analyzed the DNA methylation levels of these 80 CpG islands in the fourteen DLBCL from the Phase I study. Twelve CpG islands (AR, CDKN1C, DLC1, DRD2, GATA4, GDNF, GRIN2B, MTHFR, MYOD1, NEUROD1, ONECUT2, and TFAP2A) showed substantial methylation (PMR>20) in over 85% of the samples (Supplementary Table 6). Furthermore, seven CpG islands previously reported to be methylated in DLBCL (AR,20 CDKN2B (aka TP15),21 CDKN2A (aka p16INK4),21, 22 CYP27B1,11 DLC1,11 MGMT,23-26 and RARB (aka RARβ2)11) showed substantial methylation (PMR>20) in at least one tumor. However, note that the methylation status of AR could be influenced by gender since it is located on the X chromosome and subject to inactivation in females.
The ABC-DLBCL and GCB-DLBCL subtypes could not be discerned based on hierarchical clustering analysis of our Phase II MethyLight data (Figure 2). However, this was not unexpected given that these subtypes also could not be discerned based on hierarchical clustering analysis of gene expression data for these same CpG islands (Figure 2). Nevertheless, we identified six (CYP27B1, DRD1, KL, MINT2, NEUROG1, and ONECUT2) CpG islands in these Phase II analyses that showed subtype-specific differences in methylation levels (uncorrected Wilcoxon signed-rank test P<0.05 and >10 unit difference in median PMR) (Table 1).
Figure 2.

Hierarchical clustering of gene expression and DNA methylation analyses for CpG islands in DLBCL. We performed hierarchical clustering analysis on 89 MethyLight markers (a) generated in our Phase II analyses of seven ABC-DLBCL and seven GCB-DLBCL and gene expression data from proximal genes (b) listed in Supplementary Table 2. Clustering analyses was performed on log-transformed data using Euclidean distance and average linkage. The ABC-DLBCL and GCB-DLBCL subtypes could not be discriminated from one another by either the expression (a) or the methylation analyses (b) conducted on this group of CpG islands.
In preliminary confirmatory studies, we conducted MethyLight analyses of eight candidate CpG islands (FLJ21062, GNMT, ONECUT2, CYP27B1, DRD1, KL, MINT2, and NEUROG1) identified by our Phase I and/or Phase II analyses as having subtype-specific methylation levels on a new test group of fourteen ABC-DLBCL, seventeen GCB-DLBCL, and six normal peripheral blood lymphocytes (PBLs) (Figure 1, Supplementary Table 7). When we pooled all MethyLight data conducted on these eight CpG islands (i.e. data from twenty-one ABC-DLBCL and twenty-four GCB-DLBCL cases), only the FLJ21062 (HB-442, ABC-DLBCL median PMR= 0, GCB-DLBCL median PMR = 27.6, P = 0.001) and ONECUT2 (HB-446, median ABC-DLBCL PMR = 67.7, median GCB-DLBCL PMR = 46.8, P = 0.012) MethyLight reactions showed differences between the DLBCL subtypes (Supplementary Table 7). When considering MethyLight data only from the new test group of fourteen ABC-DLBCL and seventeen GCB-DLBCL, only FLJ21062 (HB-442, ABC-DLBCL median PMR= 4.6, GCB-DLBCL median PMR = 28.0, P = 0.025) showed a difference between the DLCBL subtypes (Supplementary Table 7). Thus, the CpG island proximal to FLJ21062 is a more promising candidate for having subtype-specific methylation levels than the CpG island proximal to ONECUT2. Larger-scale validation studies of ONECUT2 and FLJ21062 CpG island methylation levels in DLBCL are warranted in order to rigorously address questions concerning their subtype-specificity. Lastly, the MethyLight assays for ONECUT2 and FLJ21062 displayed little DNA methylation in the six normal PBLs (i.e. PMR<5 in all cases).
To further elucidate the nature of DNA methylation in the ONECUT2 and FLJ21062 CpG islands, we performed bisulfite sequencing analysis of these CpG islands in four ABC-DLBCL, four GCB-DLBCL, and two normal PBL (Table 2, Figure 3, and Supplementary Figure 3). Overall, bisulfite sequencing of ONECUT2 and FLJ21062 CpG island subclones (average twenty-five per case) yielded results that are in excellent agreement with the MethyLight PMR values (Table 2). While the methylation status of CpG dinucleotides centered within these two islands reflected an all-or-none phenomenon, the status of CpG dinucleotides on their edges was less reflective of the status of the island as a whole (Figure 3 and Supplementary Figure 3).
Table 2.
Confirmatory bisulfite sequencing analyses
| Tumora | CpG Island | # clones sequenced | %b | PMRc |
|---|---|---|---|---|
| ABC - 6 | FLJ21062 | 23 | 1 | 0 |
| ABC - 7 | FLJ21062 | 19 | 2 | 0 |
| ABC - 8 | FLJ21062 | 22 | 19 | 0 |
| ABC - 4 | FLJ21062 | 14 | 16 | 0 |
| GCB - 3 | FLJ21062 | 19 | 4 | 0 |
| GCB - 15 | FLJ21062 | 26 | 0 | 8 |
| GCB - 7 | FLJ21062 | 19 | 45 | 72 |
| GCB - 10 | FLJ21062 | 37 | 73 | 104 |
| PBL - 1 | FLJ21062 | 27 | 0 | 0 |
| PBL - 4 | FLJ21062 | 22 | 2 | 0 |
| ABC - 2 | ONECUT2 | 27 | 71 | 64 |
| ABC - 6 | ONECUT2 | 24 | 78 | 69 |
| ABC - 9 | ONECUT2 | 41 | 70 | 111 |
| ABC - 7 | ONECUT2 | 24 | 65 | 137 |
| GCB - 5 | ONECUT2 | 17 | 4 | 0 |
| GCB - 4 | ONECUT2 | 28 | 53 | 51 |
| GCB - 7 | ONECUT2 | 31 | 69 | 82 |
| GCB - 10 | ONECUT2 | 33 | 93 | 135 |
| PBL - 1 | ONECUT2 | 20 | 2 | 0 |
| PBL - 4 | ONECUT2 | 17 | 7 | 1 |
The type (ABC-DLBCL or GCB-DLBCL) and sample number for each tumor are provided. PBL refers to peripheral blood lymphocyte.
Percentage of CpG dinucleotides methylated across all clones.
PMR value obtained by MethyLight analysis.
Figure 3.
Bisulfite sequencing of CpG islands proximal to FLJ21062 and ONECUT2. The regions encompassing MethyLight reactions HB-442 (FLJ21062) and HB-446 (ONECUT2) were subject to bisulfite sequencing analysis in four ABC-DLBCL, four GCB-DLBCL, and two PBL samples, as summarized in Table 2 and provided in Supplementary Table 3. Here, we depict representative bisulfite sequencing analyses of FLJ21062 in samples (a) GCB-DLBCL 7 and (b) GCB-DLBCL 10. Likewise, representative bisulfite sequencing analyses of ONECUT2 in samples (c) GCB-DLBCL 7 and (d) ABC-DLBCL 2 are shown. Light gray and blackened circles denote methylated and unmethylated CpG dinucleotides, respectively. The percentage of methylated CpGs in all clones and total number of clones sequenced are provided above each panel along with the corresponding MethyLight PMR value. In addition, the percentage of methylated CpGs in a given clone is provided to the right of each clone. The ONECUT2 amplicon spans nucleotide positions 53256301- 53256419 of chromosome 18 and the FLJ21062 amplicon spans nucleotide positions 89519154- 89519280 of chromosome 7, based on the May 2004 human genome assembly provided at http://genome.ucsc.edu/.
Next, we investigated the relationships between DNA methylation and expression levels of ONECUT2, FLJ21062, and other genes in DLBCL. We were able to compare MethyLight PMR values and oligonucleotide microarray-based gene expression values for 67 genes in thirteen DLBCL (see Figure 4, Supplementary Table 3, and Supplementary Figure 4, where one hundred thirty-four plots are provided that reflect multiple gene expression probe tilings and/or MethyLight reactions for some genes). A total of 39 CpG islands proximal (i.e. within 500-bp in either direction) to the transcription start site of genes showed sufficient variation in PMR values among our DLBCL to justify comment (i.e. greater than twenty unit difference in the second lowest and second highest PMR). For 32 of 39 (82%) of these CpG islands (including ONECUT2), increasing levels of DNA methylation did not result in proportional decreases in gene expression. This was influenced by the fact that genes proximal to CpG islands often showed weak or modest expression (i.e. every log2 expression score was below seven units) regardless of methylation level (e.g. CALCA, CDX1, DRD1, GABRA2, GATA3, GNMT, KL, LDLR, MTHFR, NEUROD1, NOS1AP, ONECUT2, TFPI2, and TWIST1) (Supplemental Figure 4). It is possible that such genes are strongly expressed in normal precursor cells, but are silenced in all DLCBL via genetic and/or epigenetic mechanisms. Alternatively, such genes could be weakly or modestly expressed in normal precursor cells prior to methylation incurred during the development of cancer. The latter possibility would be consistent with a study showing that 69% (118/170) of genes that are methylated in colon tumor samples are expressed at low levels in normal colon as well as in colorectal adenocarcinomas.27 Overall, we favor the interpretation that DNA methylation is not frequently involved in initiating the silencing of highly expressed genes.
Figure 4.
Relationships among DNA methylation and gene expression status. MethyLight (ML) PMR values (X-axis) were plotted against expression value (Y-axis) for all genes showing statistically significant trends (Benjamini-Hochberg corrected P<0.05) for decreasing expression with increasing levels of methylation. The location of each ML reaction relative to the start of the corresponding RefSeq is provided in parentheses. Panel (a) BNIP3 (32-bp upstream of RefSeq NM_004052), (b) MGMT (exon 48-bp downstream of RefSeq NM_002412), (c) RBP1 (exon 43-bp downstream of RefSeq NM_002899), (d) GATA4 (intron 394-bp downstream of RefSeq NM_002052), (e) IGSF4 (exon 37-bp downstream of RefSeq NM_014333), (f) CRABP1 (exon 37-bp downstream of RefSeq NM_004378), and (g) FLJ21062 (21-bp upstream of RefSeq NM_001039706). The ML reaction ID, Affymetrix probe tiling ID, equation for a linear fit of the data, and r-squared value are provided.
It should also be noted that specific probe tilings for DLC1, GATA4, NKD2, and RARRES1 indicated at least modest expression levels (i.e. log2 expression score above eight units) even when CpG islands located within 500-bp (in either direction) of their transcription start sites had PMR values greater than 80 units. There are many possible explanations for these observations. For example, we may not be interrogating CpG islands or CpG dinucleotides relevant to the transcriptional regulation of the transcripts pertaining to these probe tilings. Alternatively, if copies of these genes proximal to the residual unmethylated CpG islands were highly expressed, the gene silencing signature associated with methylated CpG islands could be masked. More intriguingly, it is formally possible that these methylated CpG islands are not attracting the appropriate cadre of factors responsible for methylation-associated gene silencing. This could be meaningful given that the relationship of the various nucleic and protein components (e.g. histone modifications28) involved in epigenetic gene silencing have still not been fully defined.
Nevertheless, BNIP3, MGMT, RBP1, GATA4, IGSF4, CRABP1, and FLJ21062 showed significant (Benjamini-Hochberg corrected P<0.05; see Supplementary Table 8) trends for decreasing gene expression with increasing levels of DNA methylation (Figure 4). These represent candidate genes for which the DNA methylation levels of a proximal CpG island is associated with gene silencing in DLBCL. However, some observations (e.g. FLJ21062) could be influenced by experimental noise associated with measuring the abundance of rare transcripts. Nevertheless studies involving the demethylating agent 5-aza-2′-deoxycytidine demonstrate that BNIP3,29 MGMT,30 RBP1 (aka CRBP1),31 GATA4,32 IGSF4 (aka TSLC1),33 and CRABP134 expression are dependent upon CpG island methylation status in various cancer cell culture models.
Next, we examined the expression of the seven (BNIP3, MGMT, RBP1, GATA4, IGSF4, CRABP1, and FLJ21062) candidates as well as the twelve frequently methylated CpG islands (AR, CDKN1C, DLC1, DRD2, GATA4, GDNF, GRIN2B, MTHFR, MYOD1, NEUROD1, ONECUT2, and TFAP2A) in two tonsils and two peripheral blood CD19+ B-cell preparations (Supplementary Table 9). This data derives from published gene expression analyses (http://wombat.gnf.org/index.html).35 Four candidate genes (i.e. BNIP3, MGMT, RBP1, IGSF4) showing decreased expression with increasing methylation in tumors were expressed at ≥0.5% of tonsillar β-actin transcript levels. In addition, two of the seven candidate genes (MGMT and IGSF4) were expressed at ≥0.5% of β-actin transcript levels in CD19+ B-cells. Meanwhile, five of the frequently methylated genes (i.e. AR, CDKN1C, DRD2, GRIN2B, and TFAP2A) were expressed at ≥0.5% of tonsillar β-actin transcript levels. However, only one (DRD2) of the frequently methylated genes met that same criteria in CD19+ B-cells. None of the eighteen unique genes discussed above were expressed at >1.2% of β-actin levels in tonsils or CD19+ B-cells. Although microarray-based comparisons of transcript levels within a single sample should be viewed with caution, we conclude that both the candidate genes showing decreased expression with increasing methylation and the frequently methylated genes in DLBCL are already expressed at low to modest levels in normal B-cells.
Lastly, we compared ONECUT2 expression levels in GCB-DLCBL and ABC-DLCBL with those from normal PBLs and liver using quantitative PCR (qPCR) (Supplementary Table 10). In agreement with our microarray-based gene expression analyses (Supplementary Table 3), ONECUT2 was expressed at low levels in four ABC-DLCBL and two GCB-DLBCL samples. However, ONECUT2 expression was not detected in the four normal PBLs. This suggests that hypermethylation of this CpG island would not affect ONECUT2 expression in a normal lymphocyte sample.
Overall, our epigenetic and genetic analyses have uncovered candidate genes that could warrant further investigation into their functional roles in the development of DLBCL and potential as biomarkers for early detection of disease recurrence. Interesting, the frequently methylated CpG islands we uncovered in DLBCL such as CDKN1C, DLC1, DRD2, GATA4, GDNF, GRIN2B, MTHFR, MYOD1, NEUROD1, ONECUT2, and TFAP2A have been reported to be hypermethylated in cancers outside of DLBCL. This could reflect their status as known or suspected tumor suppressor genes that affect pathways common to multiple cancers. However, it is also possible these genes are not functionally relevant to DLBCL, but are simply located in hypermethylated chromosomal blocks that could contain one or more tumor suppressor genes directly relevant to DLBCL. Recently, hypermethylated chromosomal blocks have been detected in colorectal cancer36 and acute lymphoblastic leukemia (ALL)37. The continued development of technologies for genome-wide DNA methylation analyses is needed to address fundamental questions concerning nature and relevance of hypermethylated chromosome blocks in the development and progression of cancer.
Lastly, the DNA methylation levels observed for specific CpG islands suggest there is considerable epigenetic heterogeneity within the tumors analyzed. This could be related to histological heterogeneity (e.g. levels of tumor-infiltrating immune cells38) or the heterogeneity of cancer cell populations comprising these tumors. Regardless of its origin, epigenetic heterogeneity could confound comparisons of gene expression and methylation profiles. This highlights the value of focusing on individual or limited numbers of cells in cancer genome and epigenome projects. The development of high-throughput DNA methylation profiling technologies that require limited starting materials would facilitate the identification of clinical biomarkers and accelerate studies aimed at defining the roles epigenetic phenomena play in the etiology of different cancers.
Supplementary Material
Acknowledgment of Funding Support
We thank Drs. Larry Brody (NIH), Darren Magda (Pharmacyclics, Inc.) for valuable discussion, and Lou Staudt and Sandeep Dave (NCI) for access to the Affymetrix gene expression data. This study was funded by NIH Grants P50-HG002790 and P30-CA014089, and United States Public Health Service grants CA36727 and CA84967 awarded by the National Cancer Institute, Department of Health and Human Services. TCG is a grantee of the Mantle Cell Lymphoma Research Program of the Lymphoma Research Foundation. This research was supported in part by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health. This investigation was conducted in a facility constructed with support from Research Facilities Improvement Program Grant Number C06 (RR10600-01, CA62528-01, RR14514-01) from the National Center for Research Resources, National Institutes of Health.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu).
REFERENCES
- 1.Lossos IS, Morgensztern D. Prognostic biomarkers in diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:995–1007. doi: 10.1200/JCO.2005.02.4786. [DOI] [PubMed] [Google Scholar]
- 2.Lossos IS. Molecular pathogenesis of diffuse large B-cell lymphoma. J Clin Oncol. 2005;23:6351–6357. doi: 10.1200/JCO.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 5.Wright G, Tan B, Rosenwald A, Hurt EH, Wiestner A, Staudt LM. A gene expression-based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci U S A. 2003;100:9991–9996. doi: 10.1073/pnas.1732008100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bea S, Colomo L, Lopez-Guillermo A, Salaverria I, Puig X, Pinyol M, et al. Clinicopathologic significance and prognostic value of chromosomal imbalances in diffuse large B-cell lymphomas. J Clin Oncol. 2004;22:3498–3506. doi: 10.1200/JCO.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 7.Bea S, Zettl A, Wright G, Salaverria I, Jehn P, Moreno V, et al. Diffuse large B-cell lymphoma subgroups have distinct genetic profiles that influence tumor biology and improve gene-expression-based survival prediction. Blood. 2005;106:3183–3190. doi: 10.1182/blood-2005-04-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tagawa H, Suguro M, Tsuzuki S, Matsuo K, Karnan S, Ohshima K, et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B-cell lymphoma. Blood. 2005;106:1770–1777. doi: 10.1182/blood-2005-02-0542. [DOI] [PubMed] [Google Scholar]
- 9.Chen W, Houldsworth J, Olshen AB, Nanjangud G, Chaganti S, Venkatraman ES, et al. Array comparative genomic hybridization reveals genomic copy number changes associated with outcome in diffuse large B-cell lymphomas. Blood. 2006;107:2477–2485. doi: 10.1182/blood-2005-07-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 11.Shi H, Guo J, Duff DJ, Rahmatpanah F, Chitima-Matsiga R, Al-Kuhlani M, et al. Discovery of novel epigenetic markers in non-Hodgkin's lymphoma. Carcinogenesis. 2007;28:60–70. doi: 10.1093/carcin/bgl092. [DOI] [PubMed] [Google Scholar]
- 12.Nouzova M, Holtan N, Oshiro MM, Isett RB, Munoz-Rodriguez JL, List AF, et al. Epigenomic changes during leukemia cell differentiation: analysis of histone acetylation and cytosine methylation using CpG island microarrays. J Pharmacol Exp Ther. 2004;311:968–981. doi: 10.1124/jpet.104.072488. [DOI] [PubMed] [Google Scholar]
- 13.Pike BL, Groshen S, Hsu YH, Shai RM, Wang X, Holtan N, et al. Comparisons of PCR-based genome amplification systems using CpG island microarrays. Hum Mutat. 2006;27:589–596. doi: 10.1002/humu.20329. [DOI] [PubMed] [Google Scholar]
- 14.Maekawa M, Taniguchi T, Higashi H, Sugimura H, Sugano K, Kanno T. Methylation of mitochondrial DNA is not a useful marker for cancer detection. Clin Chem. 2004;50:1480–1481. doi: 10.1373/clinchem.2004.035139. [DOI] [PubMed] [Google Scholar]
- 15.Weisenberger DJ, Campan M, Long TI, Kim M, Woods C, Fiala E, et al. Analysis of repetitive element DNA methylation by MethyLight. Nucleic Acids Res. 2005;33:6823–6836. doi: 10.1093/nar/gki987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Novak P, Jensen T, Oshiro MM, Wozniak RJ, Nouzova M, Watts GS, et al. Epigenetic inactivation of the HOXA gene cluster in breast cancer. Cancer Res. 2006;66:10664–10670. doi: 10.1158/0008-5472.CAN-06-2761. [DOI] [PubMed] [Google Scholar]
- 17.Ibrahim AE, Thorne NP, Baird K, Barbosa-Morais NL, Tavare S, Collins VP, et al. MMASS: an optimized array-based method for assessing CpG island methylation. Nucleic Acids Res. 2006;34:e136. doi: 10.1093/nar/gkl551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Huang TH. Applications of CpG island microarrays for high-throughput analysis of DNA methylation. J Nutr. 2002;132:2430S–2434S. doi: 10.1093/jn/132.8.2430S. [DOI] [PubMed] [Google Scholar]
- 19.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Chen CM, Yan P, Huang TH, Shi H, Burger M, et al. The androgen receptor gene is preferentially hypermethylated in follicular non-Hodgkin's lymphomas. Clin Cancer Res. 2003;9:4034–4042. [PubMed] [Google Scholar]
- 21.Garcia MJ, Martinez-Delgado B, Cebrian A, Martinez A, Benitez J, Rivas C. Different incidence and pattern of p15INK4b and p16INK4a promoter region hypermethylation in Hodgkin's and CD30-Positive non-Hodgkin's lymphomas. Am J Pathol. 2002;161:1007–1013. doi: 10.1016/S0002-9440(10)64261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shiozawa E, Takimoto M, Makino R, Adachi D, Saito B, Yamochi-Onizuka T, et al. Hypermethylation of CpG islands in p16 as a prognostic factor for diffuse large B-cell lymphoma in a high-risk group. Leuk Res. 2006;30:859–867. doi: 10.1016/j.leukres.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M, Gaidano G, Goodman SN, Zagonel V, Capello D, Botto B, et al. Hypermethylation of the DNA repair gene O(6)-methylguanine DNA methyltransferase and survival of patients with diffuse large B-cell lymphoma. J Natl Cancer Inst. 2002;94:26–32. doi: 10.1093/jnci/94.1.26. [DOI] [PubMed] [Google Scholar]
- 24.Rossi D, Capello D, Gloghini A, Franceschetti S, Paulli M, Bhatia K, et al. Aberrant promoter methylation of multiple genes throughout the clinico-pathologic spectrum of B-cell neoplasia. Haematologica. 2004;89:154–164. [PubMed] [Google Scholar]
- 25.Al-Kuraya KS, Siraj AK, Al-Dayel FA, Ezzat AA, Al-Jommah NA, Atizado VL, et al. Epigenetic changes and their clinical relevance in Saudi diffuse large B-cell lymphoma. A molecular and tissue microarray analysis of 100 cases. Saudi Med J. 2005;26:1099–1103. [PubMed] [Google Scholar]
- 26.Hiraga J, Kinoshita T, Ohno T, Mori N, Ohashi H, Fukami S, et al. Promoter hypermethylation of the DNA-repair gene O6-methylguanine-DNA methyltransferase and p53 mutation in diffuse large B-cell lymphoma. Int J Hematol. 2006;84:248–255. doi: 10.1532/IJH97.06087. [DOI] [PubMed] [Google Scholar]
- 27.Keshet I, Schlesinger Y, Farkash S, Rand E, Hecht M, Segal E, et al. Evidence for an instructive mechanism of de novo methylation in cancer cells. Nat Genet. 2006;38:149–153. doi: 10.1038/ng1719. [DOI] [PubMed] [Google Scholar]
- 28.Irvine RA, Lin IG, Hsieh CL. DNA methylation has a local effect on transcription and histone acetylation. Mol Cell Biol. 2002;22:6689–6696. doi: 10.1128/MCB.22.19.6689-6696.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murai M, Toyota M, Suzuki H, Satoh A, Sasaki Y, Akino K, et al. Aberrant methylation and silencing of the BNIP3 gene in colorectal and gastric cancer. Clin Cancer Res. 2005;11:1021–1027. [PubMed] [Google Scholar]
- 30.Danam RP, Howell SR, Brent TP, Harris LC. Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histone acetylation and methyl-CpG binding proteins. Mol Cancer Ther. 2005;4:61–69. [PubMed] [Google Scholar]
- 31.Esteller M, Guo M, Moreno V, Peinado MA, Capella G, Galm O, et al. Hypermethylation-associated Inactivation of the Cellular Retinol-Binding-Protein 1 Gene in Human Cancer. Cancer Res. 2002;62:5902–5905. [PubMed] [Google Scholar]
- 32.Guo M, House MG, Akiyama Y, Qi Y, Capagna D, Harmon J, et al. Hypermethylation of the GATA gene family in esophageal cancer. Int J Cancer. 2006;119:2078–2083. doi: 10.1002/ijc.22092. [DOI] [PubMed] [Google Scholar]
- 33.Heller G, Fong KM, Girard L, Seidl S, End-Pfutzenreuter A, Lang G, et al. Expression and methylation pattern of TSLC1 cascade genes in lung carcinomas. Oncogene. 2006;25:959–968. doi: 10.1038/sj.onc.1209115. [DOI] [PubMed] [Google Scholar]
- 34.Lind GE, Kleivi K, Meling GI, Teixeira MR, Thiis-Evensen E, Rognum TO, et al. ADAMTS1, CRABP1, and NR3C1 identified as epigenetically deregulated genes in colorectal tumorigenesis. Cell Oncol. 2006;28:259–272. doi: 10.1155/2006/949506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 37.Taylor KH, Pena-Hernandez KE, Davis JW, Arthur GL, Duff DJ, Shi H, et al. Large-scale CpG methylation analysis identifies novel candidate genes and reveals methylation hotspots in acute lymphoblastic leukemia. Cancer Res. 2007;67:2617–2625. doi: 10.1158/0008-5472.CAN-06-3993. [DOI] [PubMed] [Google Scholar]
- 38.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.