Abstract
Cryptococcus neoformans is the causative agent of cryptococcal meningoencephalitis, most frequently occurring in immunocompromised individuals. There are three varieties of C. neoformans, var. grubii, var. neoformans, and var. gatti. Worldwide var. grubii is the most prevalent clinical isolate. However, few tools for the study of essential genes in var. grubii exist. Here we describe three endogenous inducible promoters for use in the study of this important opportunistic pathogen. We identified eight potential homologs of S. cerevisiae galactose genes in var. grubii. We found that GAL1, GAL7, and UGE2 were regulated by glucose and galactose and can be used successfully during mating. Our analysis indicated these promoters should prove to be excellent tools for analysis of genes in var. grubii.
Keywords: GAL1, GAL7, GAL10, UGE1, UGE2 mating, ADE2, Serotype A
Introduction
Cryptococcus neoformans is a pathogenic yeast that is a threat to immunocompromised individuals, causing respiratory infection and meningoencephalitis. Kwon-Chung and Bennett found that 75% of C. neoformans clinical isolates were var. grubii, 7% were var. neoformans and the remaining 18% were var. gatti indicating the majority of cryptococcosis infections were due to var. grubii (Kwon-Chung, K.J. and Bennett, J.E. 1978; Lengeler, K.B., Wang, P. et al. 2000). Additionally, the incidence of cryptococcosis has risen dramatically over the past 25 years, corresponding to the increase in HIV infection (Casadevall, A. and Perfect, J.R. 1998). In spite of therapeutic advances, developing nations are still experiencing significant problems with cryptococcosis, which accounts for up to half the cases of neurological disease in HIV patients (Banerjee, U., Datta, K. et al. 2001).
Paralleling the increased incidence of cryptococcosis, study of the biology and pathogenesis of C. neoformans has greatly increased. Varieties neoformans and grubii have been sequenced and annotated ((Loftus, B.J., Fung, E. et al. 2005), www.broad.mit.edu), allowing for greater utilization of available molecular techniques, such as gene disruption or specific mutation by homologous recombination of DNA introduced through biolistic transformation or electroporation (Edman, J.C. and Kwon-Chung, K.J. 1990; Toffaletti, D.L., Rude, T.H. et al. 1993; Lodge, J.K., Jackson-Machelski, E. et al. 1994). Although the above techniques have proven useful they are limited with respect to the study of essential genes.
Current established techniques for genetic analysis in C. neoformans include conditional lethality, anti-sense gene orientation, RNA interference, mating with segregation analysis, and the use of inducible promoters. Mutations that produce conditional lethality may cause auxotrophy or temperature sensitivity. A classic example of using conditional lethality to demonstrate the essentiality of a gene in C. neoformans is the deletion of ADE2. This gene encodes an enzyme, phosphoribosylaminoimidazole carboxylase, which is involved in the production of endogenous adenine biosynthesis. Strains with this gene deleted are conditionally lethal on minimal medium in the absence of adenine (Perfect, J.R., Toffaletti, D.L. et al. 1993). Lodge and colleagues reported the use of temperature sensitive strains for the determination of an essential gene that encodes Myristoyl-CoA:protein N-myristoyltransferase (Nmt). A mutation, NMTG487D, was introduced into the gene and resulted in temperature-sensitive myristic acid auxotrophs at 37°C (Lodge, J.K., Jackson-Machelski, E. et al. 1994; Lodge, J.K., Jackson-Machelski, E. et al. 1998). Both of these methods are limited and are only successful when investigating genes that lead to conditional lethality. The C. neoformans β-(1,3)-glucan synthase, Fks1, has been shown to be essential. Using anti-snse orientation Thompson et al. established this via homologous integrative transformation that relied upon a plasmid that integrated in two orientations, one orientation disrupted the gene function of FKS1 and led to the loss of viability (Thompson, J.R., Douglas, C.M. et al. 1999). RNA interference (RNAi) has been used successfully in var. neoformans. In this technique double-stranded RNA homologous to ADE2 resulted in specific degradation of ADE2 mRNA and thus reduced translation of this gene (Liu, H., Cottrell, T.R. et al. 2002). They found that the ADE2 RNAi strain was phenotypically similar to that of the ade2Δ. Additionally, Liu and colleagues demonstrated that RNAi could be used to target two genes simultaneous through the expression of chimeric double-stranded RNA (Liu, H., Cottrell, T.R. et al. 2002). RNAi techniques for the study of essential genes in var. grubii have not been successfully established. Similar to RNAi, Gorlach et al. used antisense to repress the expression of two genes coding for calcineurin A and laccase 1. The deletions of these genes cause temperature sensitivity at 37°C and loss of melanin pigment, respectively, and both transcripts were successful repressed using this method (Gorlach, J.M., McDade, H.C. et al. 2002). Antisense repression relies on the ectopic integration of the antisense construct. Ectopic integration can lead to the inadvertent disruption of other genes, which makes this method less desirable. To determine synthetic lethality between two gene deletions, mating and segregation analysis is a commonly employed procedure. However this method can be limiting due to several factors such as: First, the deletions may affect the ability of C. neoformans to mate, thus affecting the production of basidiospores. Secondly, if spores are produced, isolating them is a time consuming and tedious procedure, and being able to acquire the number of progeny needed to statically support synthetic lethality can be problematic. Lastly, random spore analysis can be used to produce multiple deletion strains, a faster method than isolation of single spores, and readily provides enough progeny for statistical analysis. Again, this method is only feasible if mating is possible and viable spores are produced. Finally, inducible promoters have also been used in C. neoformans to establish gene essentiality and are described below.
Typically inducible promoters have been the best method to confirm the essentiality of genes. However, inducible promoters for use in var. grubii are limited. There are three well-characterized inducible promoters in C. neoformans. The first is the promoter of GAL7 from var. neoformans, which is repressed by glucose and induced by galactose (Wickes, B.L. and Edman, J.C. 1995). Ory and colleagues found var. neoformans’ PGAL7 was endogenously induced up to 83 fold; however, when this same promoter was expressed heterogenously in var. grubii they observed only three fold induction, thus limiting the usefulness of var. neoformans’ PGAL7 in var. grubii (Ory, J.J., Griffith, C.L. et al. 2004). Second is the promoter of MFα1 from var. neoformans, which is induced on V8 medium (del Poeta, M., Toffaletti, D.L. et al. 1999). Although this promoter drives expression well in both varieties, the V8 medium causes slow growth and induces sporulation; as such this specific growth requirement restricts PMFα1’s general use. Finally, the PCTR4 is a copper repressible promoter that is induced by copper chelation (Ory, J.J., Griffith, C.L. et al. 2004). It is the best available option for investigators of var. grubii: first it is tightly regulated and secondly it exhibits higher levels of induction than the other available promoters. However, it requires the use of copper-free medium, which may not be suitable for all experiments. Therefore, the need exists for additional reliable, endogenous, and easily regulated inducible promoters for use in var. grubii.
The Leloir pathway has been well established in Saccharomyces cerevisiae and components of this pathway have been exploited for genetic analysis (reviewed in, Johnston, M. 1987). The enzymes in this pathway that convert galactose to glucose-6-phosphate are Gal1p (a kinase), Gal7p (a transferase), Gal10p (an epimerase). Other Gal enzymes are regulators of these three key enzymes, however, they are not directly part of the Leloir pathway, yet they retain the Gal designation. One exception is an α Galactosidase that is designated Mel1p. Three of the Gal proteins, Gal3p, Gal4p/Gal81p, and Gal80p function in the regulation of Mel1p, Gal1p (a kinase), Gal7p (a transferase), Gal10p (an epimerase), Gal2p (a galactose permease) (Johnston, M. 1987), and Gal5p (a phosphoglucomutase) (Oh, D. and Hopper, J.E. 1990). In S. cerevisiae GAL1, GAL7, and GAL10 are clustered together on chromosome two, but are transcribed from separate promoters. GAL2 and MEL1 lie on chromosome number twelve (Leloir, L.F. 1951; Torchia, T.E., Hamilton, R.W. et al. 1984; Torchia, T.E. and Hopper, J.E. 1986; Tschopp, J.F., Emr, S.D. et al. 1986). GAL4 encodes a protein that activates transcription of these five genes. Gal80p negatively regulates Gal4p while Gal3p, which is induced by galactose, activates Gal4p indirectly by negatively regulating Gal80p (Johnston, M. 1987).
Therefore, to find new endogenous promoters that could be regulated in vitro in var. grubii, we focused our search on the Leloir pathway and those enzymes known to be involved in galactose metabolism. We investigated homologs to nine S. cerevisiae GAL genes to determine which were repressed by glucose and induced by galactose in var. grubii. Here we report and characterize three endogenous galactose inducible promoters PGAL1, PGAL7, and PUGE2 for use in var. grubii.
Material and Methods
Fungal strains and media
The strain KN99α of C. neoformans var. grubii (Nielsen, K., Cox, G.M. et al. 2003), was used as the wild type strain, and all promoter swap constructs were introduced into it. An adeΔ strain (in the H99 background) var. grubii was used as the positive control (Perfect et al., 1993). Strains were grown on rich media at 30 C, YPD or YPG (1% yeast extract, 2% bacto-peptone, and 2% dextrose or 2% galactose, respectively) and on minimal media SD-ade (.67% YNB with NH4SO4 without amino acids and dextrose,.008% CSM-ade, 2% bacto agar and 2% dextrose or galactose with 0.2% dextrose). Solid media contained 2% bacto-agar. Selective YPD or YPG media contained 100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany) and/or 200 μg/ml gentamycin (Invitrogen, Carlsbad, CA).
Identification of putative galactose inducible promoters
Amino acid sequences were retrieved from Saccharomyces Genome Database for well-known genes dependent on galactose for expression in S. cerevisiae (GAL1, GAL2, GAL3, GAL4, GAL5, GAL7, GAL10, GAL80, MEL1) (www.yeastgenome.org). The amino acid sequences were BLASTed against Broad H99 var. grubii predicted proteins (www.broad.mit.edu/annotation/genome/cryptococcus_neoformans). The protein sequences were retrieved and used as queries to locate the genomic DNA and the genomic sequences were used in a BLAST search to find the transcripts. These coding sequences were used to design probes for Northern blots. Gene nomenclature was assigned as described by Gerik et al. (Gerik, K.J., Donlin, M.J. et al. 2005) with the following exceptions: For clarity lower case letters were used to designate multiple homologues to S. cerevisiae GAL genes instead of numbers. The UXS1, UGE1, and UGE2 designations were used instead of GAL10 or GAL10c because these have previously been described in the literature (Bar-Peled, M., Griffith, C.L. et al. 2001; Moyrand, F., Fontaine, T. et al. 2007). Additionally, GAL10b was not used because it has previously been deposited into Genbank (AAR84604) as UGE2.
RNA extraction for northern blot analysis
C. neoformans strain KN99 was grown for 24 hours at 30°C in liquid YPD or YPG broth. For RNA extraction and poly-A RNA purification cells were collected by centrifugation at 1,800 × g for five minutes, washed once with distilled water, and lyophilized overnight. The lyophilized pellet was then vortexed with 3 ml glass beads (1 mm; Biospec, Inc., Bartlesville, OK) and re-suspended in 4 ml TRIzol Reagent (Invitrogen, Carlsbad, CA). After sitting at room temperature for five minutes, 800 μl of chloroform was added and the mixture was shaken for 30 sec. This cell lysate was then centrifuged at 4,000 rpm for ten minutes and the supernatant was transferred to a new tube. Two ml isopropanol was added, incubated for 10 min at room temperature, and centrifuged at 4,000 rpm for ten minutes. After washing the pellet with 75% EtOH, it was resuspended in water and incubated with DNase I at 37°C for 1 hr. The RNA was extracted again with TRIzol and chloroform and precipitated with isopropanol as above. The dried pellet was resuspended in 300 μl RNase-free water (Invitrogen, Carlsbad, CA). Poly-A RNA was purified from the total RNA sample using an oligo-Tex RNA purification kit (Qiagen, Valencia, CA) following the manufacturer’s specifications.
Northern Hybridizations
The procedure we used was adapted from Sambrook and Russell (Sambrook, J. and Russell, D. 2001). Approximately 1 μg of poly-A RNA from each strain was mixed with 2μL of DEPC (Sigma, St. Louis, MO) -treated 5X formaldehyde gel running buffer (0.1mM MOPS (Fisher) pH 7.0, 40mM sodium acetate (Sigma, St. Louis, MO), 5mM EDTA pH 8.0), 3.5μL of 37% formaldehyde (Sigma, St. Louis, MO), and 10μL of formamide (Sigma, St. Louis, MO). Samples were then incubated at 65°C for 15 minutes and placed directly on ice. 2μL DEPC – treated formaldehyde gel loading buffer (50% glycerol (Fisher), 1mM EDTA (Fisher), 0.25% bromophenol blue (Sigma, St. Louis, MO)) was added and samples were then loaded on a 1.5% agarose (Roche) gel containing formaldehyde (DEPC treated, 1X formaldehyde gel running buffer, 6.6% formaldehyde) submersed in 1X formaldehyde gel running buffer. An RNA size marker (Promega, Cat. Num. 3191A, Madison, WI) sample was prepared in the same way, however ~1μg ethidium bromide (Sigma, St. Louis, MO) was added before the sample was loaded onto the agarose gel. A positive binding control ladder, containing 100pg of each gene specific DNA of interest, was also loaded on the agarose gel. The primers used to amplify the gene specific DNA fragments for the positive binding control ladder are shown in Supplemental Table S1. Gels were pre-run in 1X formaldehyde gel running buffer for 5 minutes at approximately 5V/cm before samples were loaded, and run at approximately 58V/cm for ~4 hours, with constant recirculation of running buffer, until the bromophenol blue had traveled 8–9 cm. Gels were rinsed in several volumes DEPC-treated water, soaked in several volumes DEPC-treated 50mM NaOH (Fisher) for 20 minutes, and soaked in several volumes 20X SSC for 45 minutes. Fragments were then transferred to charged nylon membranes using a Turbo-Blot apparatus with 20X SSC as transfer buffer. Membranes were then UV cross-linked. Probes for Northern analysis were prepared by random priming (random priming kit; Roche) using 50 μCi [α-32P]dCTP, according to the manufacturer’s instructions. The membranes were incubated in 20ml of ULTRAHyb buffer (Ambion, Austin, TX) for 3 hour at 55°C, then probe was added to this solution, and the membranes were hybridized at 55°C overnight. The membranes were washed one time in ~50 mL of a solution of 1X SSC, 0.1% SDS at 25°C for 10 minutes. Membranes were then washed three times in ~50mL of a solution of 0.5X SSC, 0.1% SDS at 68°C for 10 minutes. The membranes were stripped of probe twice by incubating 2X in ~50mL of 10mM Tris-Cl pH 7.4, 0.2% SDS, heated to boiling for 1.5 hours, and subsequently re-probed following instructions above. Phosphoimager screens were used to detect radiolabels and analyzed using ImageQuant version 5.1.
Generation of promoter swap constructs
Overlap PCR gene deletion technology (Davidson, R., Blankenship, J. et al. 2002) was used to generate promoter-specific exchange cassettes of ADE2, which all included a nourseothricin cassette (McDade, H. and Cox, G. 2001). Primers used to disrupt the ADE2 promoter are shown in Supplemental Table S2. For the ADE2 promoter swap constructs, 500bp preceding the gene were deleted and replaced with the putative galactose inducible promoters. In the promoter swap construct 978bp, 1022bp and1007bp were used for PGAL1, PGAL7 and PUGE2, respectively.
Transformation of C. neoformans
KN99α was transformed using biolistic techniques (Toffaletti, D.L., Rude, T.H. et al. 1993; Hua, J., Meyer, J. et al. 2000). Cells were grown in YPD to late log-phase, concentrated, and plated onto YPD agar for transformation. The cells were bombarded with 0.6 μm gold beads (Bio-Rad, Richmond, CA) that were coated with DNA of the target construct according to the manufacturer’s recommendations. Following the transformation, the cells were incubated at 30°C for 4 hours on nonselective media to allow for recovery and then transferred with 0.8 ml sterile PBS to the nourseothricin selective media. Transformants were observed in 3–5 days.
Analysis of transformants
To isolate stable transformants, they were all passaged five times on nonselective YPD medium and then tested for resistance to the appropriate selective marker. Only those transformants that grew equally well on selective and nonselective media were considered to be stable transformants. A three-primer PCR screen was used to verify homologous integration at both the 5′ and 3′ ends of the deletion cassette. In this manner, homologous recombinants can be distinguished from wild type. A PCR screen using primers outside the transformation construct was used to amplify the entire integration region, demonstrating that a single copy of the transforming DNA had been inserted at the desired locus. Southern blots were performed to screen for single integration in the genome. Single bands were observed on all Southern blots when hybridized with a selectable marker-specific probe. All deletion strains generated for this work had a single deletion construct homologously integrated at the appropriate locus and no other insertions in the genome (data not shown). At least five independent isolates for each mutant were obtained and two were further characterized.
Genomic DNA preparation
Genomic DNA was prepared by a modification of the glass bead DNA extraction protocol described (Fujimura, H. and Sakuma, Y. 1993). Briefly, C. neoformans cells were suspended in a microfuge tube in 500 μl lysis buffer (50 mM Tris-HCl, pH 7.5, 20 mM EDTA, 1% SDS), with 400 mg glass beads (425–600 μm; Sigma G-9268, St Louis, MO). Cells were disrupted by vortexing 10 minutes, followed by 10 min incubation at 70°C. After brief vortexing, 200 μl 5M KOAc and 150 μl 5 M NaCl were added. The tubes were placed on ice for 20 min and centrifuged at 14,000 rpm for 20 min. The supernatant was mixed with 500 μl phenol/chloroform and spun for 5 min at 14,000 rpm. The aqueous phase was then mixed with 450 μl chloroform and spun for 5 min at 14,000 rpm. The DNA was then precipitated by addition of 200 μl ethanol, washed with 70% ethanol, dried, and resuspended in 50 μl deionized water.
Southern hybridizations
Approximately 10 μg of genomic DNA from each strain was digested with various restriction endonucleases according to the manufacturer’s recommendations. Restriction fragments were separated on a 1% agarose gel and transferred to nylon membranes using a Turbo-Blot apparatus (Schleicher & Schuell) and 10X SSC as transfer buffer. Probes for Southern analysis were prepared by random priming (random priming kit; Roche) using 50 μCi [α-32P] dCTP (GE Bio-Sciences AA0005, Piscataway, NJ) according to the manufacturer’s instructions. The blots were incubated in 10 ml of buffer (1X phosphate buffer, 7% SDS) solution for 1 hr at 65°C, then probe was added to this solution, and the blots were hybridized at 65°C overnight. The blots were washed twice in 2X SSC, 0.1% SDS at room temperature for 10 min and once for 10 min in 0.2X SSC, 0.1% SDS that had been prewarmed to 65°C. A phosphoimager screen was used to detect the radiolabel and analyzed using ImageQuant version 5.1.
RNA extraction for qPCR
Promoter swap strains were grown for 24 hours at 30°C in liquid YPD or YPG broth, cells were collected by centrifugation at 1,800 rpm for 3 minutes and washed three times with 1 X PBS. The pelleted cells were then lyophilized for 2 hours and rRNA was then purified from the samples using an RNA isolation kit (Agilent, Santa Clara, CA) with modifications to the manufacturer’s protocol for yeast. The samples were homogenized with glass beads for 5 minutes; 600 μL of lysis buffer was added to the samples; vortexed for 5 minutes and collected by centrifugation at 14,000 rpm for 3 minutes. Then 450 μL of lysate was passed through a mini-prefilitration column. The remainder of the manufacturer’s recommendations was followed.
Quantative PCR
One microgram of total rRNA was used for first-strand cDNA synthesis using the First Strand cDNA Synthesis Kit for RT-PCR (AMV) (Roche, Germany) as per manufactures instructions. The resulting cDNA was used as a template in quantitative PCR using SYBR Green PCR reagents (Clontech, Mountain View, CA) according to the manufacturer’s recommendations. Primers used in qPCR are listed in Supplemental Table S3. The DNA Engine Opticon (MJ Research, Inc.) was used as the fluorescence detector with the following protocol for the PCR: 35 s at 94°C, 50 s at 54°C, and 50 s at 72°C, and a plate reading was repeated for a total of 40 cycles after a hot start of 10 min at 94°C. A melting curve was calculated at the end of the reaction to confirm a single product. The data were normalized to actin cDNA expression amplified in the set of qPCRs.
Mating slides
A cell suspension of strains of equal amounts of appropriate opposite mating types (a and α) was mixed together in 500μL 1 X PBS. Approximately 100μL of this suspension was applied to slides covered in V8 agar supplemented with or without 2% glucose or 2% galactose in a mating chamber (100 × 15mm petri dish), then 4mLs of V8 medium (minus agar) was added to aid in maintaining nutrients and chamber humidity. The mating chambers were placed in dark at 25°C for four to five days. Brightfield images were taken on an Olympus Vanox AHBT3 microscope at designated magnifications and are representative of three or more independent experiments.
Mating Plates
Compatible mating types PGAL7:ADE2α (nourseothricin R) and MPK1-FLAGa (gentamycin R) (Gerik, K.J., Bhimireddy, S.R. et al. 2008) were streaked consecutively in patches on V8 mating plates supplemented without or with 2% glucose or 2% galactose. MPK1-FLAGa was chosen because it is a marked strain with no discernible mating defects. The plates were incubated as described for mating slides with the exception of the incubation period being extended to 16 days. Progeny were collected by agar plug at the periphery of the mating patches. Recombinant progeny were selected based on drug resistance to both nourseothricin and gentamycin (100 μg/ml nourseothricin (Werner BioAgents, Jena-Cospeda, Germany) and 200 μg/ml gentamycin (Invitrogen, Carlsbad, CA)).
Results
Identification of galactose genes in var. grubii
To identify putative GAL inducible promoters for var. grubii we first retrieved amino acid sequences from the Saccharomyces Genome Database for nine GAL genes in S. cerevisiae (GAL1, GAL2, GAL3, GAL4, GAL5, GAL7, GAL10, GAL80, MEL1) (www.yeastgenome.org). The amino acid sequences were used in a tBLASTn query of var. grubii, H99, predicted proteins (genome.slu.edu/blast.html). The retrieved protein sequences were used to query the H99 genomic database. Of the nine proteins five, Gal2p, Gal3p, Gal4p, Gal80p, and Mel1p, had no hits indicating that these proteins may not be highly conserved between S.cerevisiae and var. grubii. However, the search yielded a total of eight homologous genes with Gal1p, Gal7p and Gal10p each having multiple putative homologs, two, two, and three, respectively, and Gal5p having one homolog (Table 1).
Table 1.
Galactose Gene Homology
| S. cerevisiae Gene | C. neoformans Protein | C. neoformans Coding Sequence and Designation |
|---|---|---|
| GAL1 | CNAG_06051.1 | CNAT_06051 (GAL1) |
| CNAG_03946.1 | CNAT_03946 (GAL1b) | |
| GAL2 | No Hits | N/A |
| GAL3 | No Hits | N/A |
| GAL4 | No Hits | N/A |
| GAL5 | CNAG_06313.1 | CNAT_06313 (GAL5) |
| GAL7 | CNAG_06052.1 | CNAT_06052 (GAL7) |
| CNAG_03875.1 | CNAT_03875 (GAL7b) | |
| GAL10 | CNAG_00697.1 | CNAT_00697 (UGE1)* |
| CNAG_06050.1 | CNAT_06050 (UGE2)* | |
| CNAG_03322.1 | CNAT_03322 (UXS1)* | |
| GAL80 | No Hits | N/A |
| MEL1 | No Hits | N/A |
UXS1 & UGE1 (Bar-Peled, M., Griffith, C.L. et al. 2001; Moyrand, F., Fontaine, T. et al. 2007); UGE2 Genbank #AAR84604
Three GAL genes are induced by galactose in var. grubii
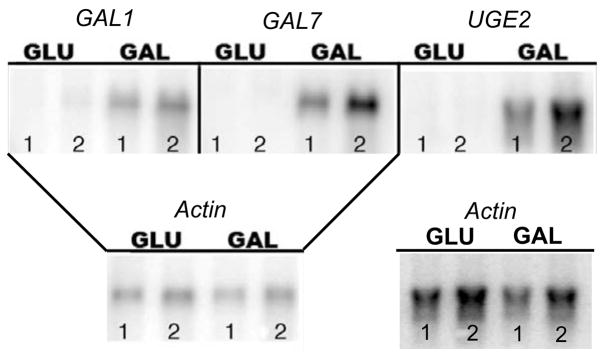
To ascertain which of the eight endogenous promoters were regulated by galactose in var. grubii we determined if their corresponding genes were differentially transcribed in the KN99α wild type strain. Northern blot analysis indicated that GAL1, GAL7 and UGE2 were repressed by glucose and induced by galactose medium (Fig. 1). GAL1b, GAL7b and UGE1 showed no expression under either condition, while GAL5 and UXS1 had equal induction under both conditions (data not shown). This data indicated that the promoters of GAL1, GAL7 and UGE2 were good candidates for further analysis.
Figure 1. Northern Blots of GAL1, GAL7 and UGE2.
GAL1, GAL7 and UGE2 gene expression is induced by growth in galactose. Lanes 1 and 2 are biological replicates grown under the same conditions. Blots were probed with Actin to show equal loading of mRNA. The same blot was used for both GAL1 and GAL7. Actin blot for UGE2 is separate.
Galactose induces expression of ADE2 under the control of the GAL1, GAL7, and UGE2 promoters
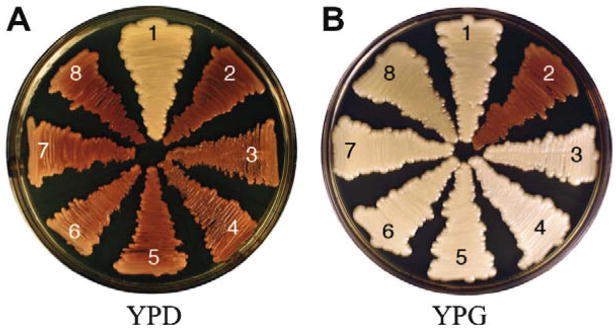
Next, we wanted to establish if the endogenous var. grubii GAL1, GAL7 and UGE2 promoters could be used to induce heterologous gene expression. The Ade2 enzyme, phosphoribosylaminoimidazole carboxylase, is a good candidate for developing tools such as inducible promoters. An ADE2 deletion is conditionally lethal on minimal medium in the absence of adenine, but is viable when grown on yeast peptone dextrose (YPD) where adenine biosynthetic intermediates accumulate which result in pink colonies (Perfect, J.R., Toffaletti, D.L. et al. 1993). We replaced 500 base pairs of the endogenous ADE2 promoter with the predicted promoters of GAL1, GAL7, and UGE2. Each of these promoters was fused to the predicted ADE2 start codon. At least five independent transformants were confirmed for each promoter swap, with two being chosen for further analysis (Table 2). The promoter swap strains were grown on yeast peptone dextrose (YPD) and yeast peptone galactose (YPG) media supplemented with 2% glucose or galactose, respectively. After five days on YPD and YPG, all strains with a PGALX:ADE2 (X = 1, 7 or 10b) cassette were pink or white, respectively (Fig. 2). These data indicated that the promoters could effectively induce ADE2 in the presence of galactose.
Table 2.
Strains Generated for Study
| Strain Name | Strain Number | Marker |
|---|---|---|
| PGAL1:ADE2-1 | JLCN 578 | nourseothricin |
| PGAL1: ADE2-2 | JLCN 584 | nourseothricin |
| PGAL7: ADE2-1 | JLCN 587 | nourseothricin |
| PGAL7: ADE2-2 | JLCN 589 | nourseothricin |
| PUGE2: ADE2-1 | JLCN 594 | nourseothricin |
| PUGE2: ADE2-2 | JLCN 596 | nourseothricin |
Figure 2. PGALX:ADE2 strains grown on YPD and YPG at 30°C for 5 days.
A representative of three biological replicates is shown. 1. KN99α 2. H99 (ade2Δ), 3&4. Independent isolates of PUGE2:ADE2, 5&6. Independent isolates of PGAL7:ADE2, 7&8. Independent isolates of PGAL1:ADE2. On YPD, the PGALX:ADE2 strains are red indicating glucose repression of ADE2 expression, resulting in accumulation of adenine biosynthetic intermediates. On YPG, the PGALX:ADE2 strains are white indicating galactose induction of ADE2 expression, which resulted in wild type phenotype.
To quantitate the level of galactose induction in gene expression using the endogenous var. grubii GAL/UGE promoters, the magnitude of transciption differences in glucose (repression) versus galactose (induction) of the ADE2 transcript was assayed by quantitative PCR (qPCR). All three promoters repressed ADE2 expression in response to growth on glucose and induced ADE2 expression in galactose compared to wild type. For PGAL1, PGAL7, and PUGE2, ADE2 was repressed by 4.8, 6.1 and 5.3 and induced by 15.5, 21.6 and 28.1, respectively, in LOG2 ratio (Table 3). Furthermore, galactose induction was 20.5, 27.8 and 33.5 in LOG2 ratio for PGAL1, PGAL7, and PUGE2, respectively. Galactose did not affect native ADE2 expression levels (data not shown). The data clearly suggested the var. grubii promoters of GAL1, GAL7 and UGE2 were repressed by glucose and induced by galactose.
Table 3.
Average LOG2 Ratio of Repression and Induction of ADE2
| Strain Name | Average Glucose Repression in LOG2 Ratio | Standard Deviation +/− | Average Galactose Induction in LOG2 Ratio | Standard Deviation +/− |
|---|---|---|---|---|
| PGAL1:ADE2-1 | −4.87 | 2.41 | 15.48 | 5.56 |
| PGAL7:ADE2-1 | −6.08 | 3.47 | 21.59 | 9.59 |
| PUGE2:ADE2-1 | −5.31 | 2.09 | 28.06 | 8.79 |
ADE2 driven under the control of the GAL1, GAL7, and UGE2 promoters rescues conditional lethality
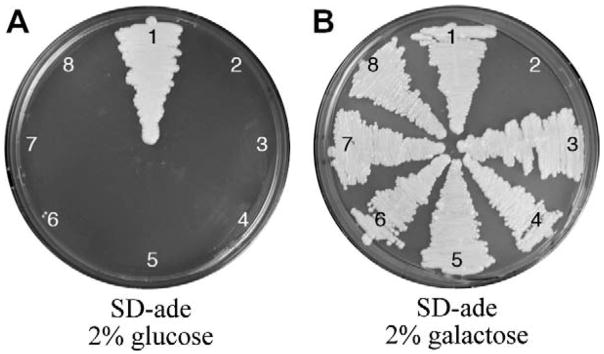
C. neoformans strains without Ade2 do not survive on minimal medium without the addition of adenine in the growth medium. Therefore, we determined if the GAL/UGE promoters could rescue this conditional lethal phenotype. Mutant strains were grown on SD-ade medium, a minimal medium that lacks adenine, supplemented with 2% glucose or galactose for five days at 30°C. As predicted, the PGALX:ADE2 strains died on SD-ade with glucose medium and grew on SD-ade with galactose medium (Fig. 3). These data indicated the GAL promoters were sufficiently repressed on glucose to reveal a conditional lethal phenotype and might prove useful in determining the lethality of other conditional or lethal genes.
Figure 3. PGALX:ADE2 strains grown on SD-ade with glucose and galactose at 30°C for 5 days.
A representative of three biological replicates are shown. The numbering corresponds to the same strains used in Fig. 2. On SD-ade with glucose, the PGALX:ADE2 strains die because ADE2 expression is repressed, creating a conditional lethality. On SD-ade with galactose, the PGALX:ADE2 strains grow because ADE2 is expressed.
The GAL promoters can be used during mating
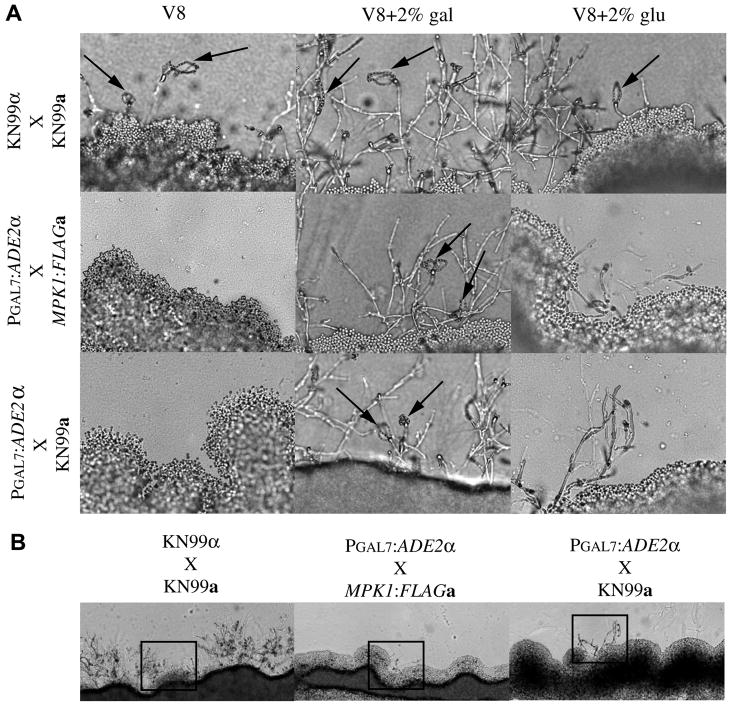
Mating is another tool in C. neoformans that is increasingly utilized to produce multiple deletions or mutations in the same strain background. However, mating of C. neoformans requires V8 agar, a nutrient poor medium (Hull, C.M. and Heitman, J. 2002). To determine if the PGAL promoters could be either repressed or induced during mating we first ascertained if the addition of 2% glucose or galactose to the V8 medium adversely affected wild type mating. Mating slides of compatible wild type crosses, KN99α X KN99a, produced spores on all three media (Fig. 4). However, in addition to sporulation, abundant filamentation was also observed on both of the V8+2% glucose and V8+2% galactose media. These data suggested that the addition of these sugars to the mating medium did not adversely effect sporulation, but did affect the ratio of spore to hyphae.
Figure 4. PGAL7:ADE2 compatible mating crosses.
A. Mating slides of V8 or V8 supplemented with 2% galactose or glucose were inoculated with equal amounts of compatible mating partners α and a, incubated in the dark at 25°C for 4 days. Photos are 40X magnification. Mating strains are indicated to the left of the panels and medium used is indicated on top. B. 10X magnification of mating on V8 supplemented with 2% glucose. Boxed area corresponds to 40X magnification in A. Matings are indicated top of panels. All images are representative of at least three independent experiments.
Having established that the addition of the sugars did not preclude the development of spores, we next assessed the feasibility of using the GAL promoters in mating assays to produce a recombinant strain. To do this we crossed the PGAL7:ADE2α (nourseothricin marker) to another strain MPK1:FLAGa (gentamycin marker) or KN99a. The MPK1:FLAGa strain was chosen for our mating experiments because it has a different marker than the PGAL7:ADE2α strains, has no discernable mating defects, and recombinant progeny could easily be screened on double drug selection. The PGAL7:ADE2α X MPK1:FLAGa or KN99a mating crosses only produced spores when grown on V8+2% galactose slides (Fig. 4). This indicated the V8 medium components are unable to induce the GAL promoters during this short time frame. However, given a longer incubation period that is used with mating on V8 plates, sixteen days instead of 4–5 days on slides, viable progeny from the PGAL7:ADE2α X MPK1:FLAGa cross were recovered from V8 only, V8+2% glucose, and V8+2% galactose media numbering 24, 9, and 156, respectively. These data indicate that the PGAL promoters can be used successfully during mating.
Discussion
This is the first study to characterize native inducible promoters of var. grubii. To identify inducible promoters we focused our search on the Leloir pathway (Leloir, L.F. 1951). We found homologs to four GAL genes in S. cerevisiae (GAL1, GAL5, GAL7, and GAL10). In S. cerevisiae the galactose inducible genes GAL1, GAL7, and GAL10 cluster together on chromosome two, and GAL5, on chromosome 13, is unregulated by glucose or galactose (Johnston, M. 1987). A similar situation occurs in C. neoformans, where the genes that are regulated by galactose, GAL1, GAL7, and UGE2 were clustered together in the genome as indicated by their Broad Institute CNAT transcript numbers 06050 (UGE2), 06051(GAL1), and 06052 (GAL7) (Table 1, www.broad.mit.edu/annotation/genome/cryptococcus_neoformans). This suggests that this functional clustering has been conserved through the divergence of the Basidomycetes and Ascomycetes. However, the orientation of the genes within the cluster has not been conserved. In S. cerevisiae the gene order is GAL7, GAL10, and GAL1 (Johnston, M. 1987) compared to UGE2, GAL1, and GAL7. It is interesting that the C. neoformans protein Uge1 (CNAG_00697.1), a UDP-glucose epimerase that reversibly cayalyzes UDP-glucose to UDP-galactose and is part of the Leloir pathway, is most homologous to Gal10 in S. cerevisiae, but was not found in the gene cluster in C. neoformans. Moyrand and colleagues found Uge1 to be necessary for the production of galactoxylomannan (GalXM), one of the two capsular polysacchrides,. GalXM is a minor component of the capsule being approximately 7% of the capsule mass (Moyrand, F., Fontaine, T. et al. 2007), while glucuronoxylomannan (GXM), the other capsular polysaccharide, makes up about 90% of the capsule mass (Bose, I., Reese, A.J. et al. 2003). As Uge2 participates in producing part of the capsule we would hypothesize that it’s expression would increase under capsule inducing conditions, such as growth in low iron medium or in the presence of high CO2. However, our data, under non-capsular conditions, indicated that expression of UGE1 was not regulated by either glucose or galactose (data not shown) and a direct comparison was not performed.
Similar to what is reported in S. cerevisiae, we determined that GAL5 in C. neoformans was not regulated by glucose or galactose, as its expression remained constant under both growth conditions (data not shown). Through our homology based searches of the var. grubii genome we did not find any of the regulatory components of the Leloir pathway GAL2, GAL3, and GAL80 (Leloir, L.F. 1951; Torchia, T.E., Hamilton, R.W. et al. 1984; Torchia, T.E. and Hopper, J.E. 1986; Tschopp, J.F., Emr, S.D. et al. 1986). This may suggest that these regulatory components are not well conserved, or that the GAL/UGE genes in C. neoformans are regulated in a manner different than S. cerevisiae.
We have demonstrated that use of the endogenous inducible promoters in var. grubii is more feasible than using promoters from var. neoformans. In contrast to the three fold induction reported by Ory and colleagues when using the heterogenous promoter from var. neoformans in var. grubii (Ory, J.J., Griffith, C.L. et al. 2004) we found galactose to induce the PGAL7 promoter of var. grubii greater than 1000-fold as compared to glucose suppression. These differences in expression levels argue for using the native promoters in var. grubii.
We have established that these inducible promoters can be used successfully to rescue the conditional lethal phenotype of Ade2 suppression, and they can be used during mating for the creation of recombinant strains. They may even prove to be useful in assaying genes involved in mating. In addition, these promoters should prove to be valuable for investigation into essential genes, as well as the over expression of genes, and other novel purposes. We suspect that PGAL1, PGAL7 and PUGE2 will become indispensable tools for investigators of this variety of C. neoformans.
Supplementary Material
Acknowledgments
We would like Maureen Donlin for bioinformatic input and Leona Campbell, Nicole Gilbert, and Woei Lam for critical reading of this manuscript. This work was supported by an NIH-NIAID grant RO1-AI50184 to JKL and RO1-AI072195.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Banerjee U, Datta K, Majumdar T, Gupta K. Cryptococcosis in India: the awakening of a giant? Med Mycol. 2001;(39):51–67. doi: 10.1080/mmy.39.1.51.67. [DOI] [PubMed] [Google Scholar]
- Bar-Peled M, Griffith CL, Doering TL. Functional cloning and characterization of a UDP- glucuronic acid decarboxylase: The pathogenic fungus Cryptococcus neoformans elucidates UDP-xylose synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(21):12003–12008. doi: 10.1073/pnas.211229198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot Cell. 2003;2(4):655–663. doi: 10.1128/EC.2.4.655-663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Perfect JR. Cryptococcus neoformans. Americian Society for Microbiology; Washington, D.C.: 1998. [Google Scholar]
- Davidson R, Blankenship J, Kraus P, Berrios M, Hull C, D’Souza C, Wang P, Heitman J. A PCR-based strategy to generate integrative targeting alleles with large regions of homology. Microbiology. 2002;138(8):2607–2615. doi: 10.1099/00221287-148-8-2607. [DOI] [PubMed] [Google Scholar]
- del Poeta M, Toffaletti DL, Rude TH, Sparks SD, Heitman J, Perfect JR. Cryptococcus neoformans differential gene expression detected in vitro and in vivo with green fluorescent protein. Infect Immun. 1999;67(4):1812–1820. doi: 10.1128/iai.67.4.1812-1820.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edman JC, Kwon-Chung KJ. Isolation of the URA5 gene from Cryptococcus neoformans var. neoformans and its use as a selective marker for transformation. Mol Cell Biol. 1990;10:4538–4544. doi: 10.1128/mcb.10.9.4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura H, Sakuma Y. Simplified isolation of chromosomal and plasmid DNA from yeasts. Biotechniques. 1993;14(4):538–540. [PubMed] [Google Scholar]
- Gerik KJ, Bhimireddy SR, Ryerse JS, Specht CA, Lodge JK. Protein Kinase C1 (PKC1) is essential for protection against both oxidative and nitrosative stress, cell integrity, and normal manifestation of virulence factors in the pathogenic fungus Cryptococcus neoformans. Eukaryotic Cell. 2008 doi: 10.1128/EC.00146-08. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerik KJ, Donlin MJ, Soto CE, Banks AM, Banks IR, Maligie MA, Selitrennikoff CP, Lodge JK. Cell wall integrity is dependent on the PKC1 signal transduction pathway in Cryptococcus neoformans. Molecular Microbiology. 2005;58(2):393–408. doi: 10.1111/j.1365-2958.2005.04843.x. [DOI] [PubMed] [Google Scholar]
- Gorlach JM, McDade HC, Perfect JR, Cox GM. Antisense repression in Cryptococcus neoformans as a laboratory tool and potential antifungal strategy. Microbiology. 2002;148(1):213–219. doi: 10.1099/00221287-148-1-213. [DOI] [PubMed] [Google Scholar]
- Hua J, Meyer J, Lodge J. Development of positive selectable markers for the fungal pathogen Cryptococcus neoformans. Clin Diagn Lab Immunol. 2000;7(1):125–128. doi: 10.1128/cdli.7.1.125-128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull CM, Heitman J. Genetics of Cryptococcus neoformans. Annual Review of Genetics. 2002;(36):557–615. doi: 10.1146/annurev.genet.36.052402.152652. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1987;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE. Distrubution of α and a mating types of Cryptococcus neoformans among natural and clinical isolates. American Journal of Epidemiology. 1978;108(4):337–340. doi: 10.1093/oxfordjournals.aje.a112628. [DOI] [PubMed] [Google Scholar]
- Leloir LF. Enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch Biochem. 1951;33:186–190. doi: 10.1016/0003-9861(51)90096-3. [DOI] [PubMed] [Google Scholar]
- Lengeler KB, Wang P, Cox GM, Perfect JR, Heitman J. Identification of the MATa mating-type locus of Cryptococcus neoformans reveals a serotype A MATa strain thought to have been extinct. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(26):14455–14460. doi: 10.1073/pnas.97.26.14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Cottrell TR, Pierini LM, Goldman WE, Doering TL. RNA Interference in the pathogenic fungus Cryptococcus neoformans. Genetics. 2002;160(2):463–470. doi: 10.1093/genetics/160.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge JK, Jackson-Machelski E, Higgins M, McWherter CA, Sikorski JA, Devadas B, Gordon JI. Genetic and biochemical studies establish that the fungicidal effect of a fully depeptidized inhibitor of Cryptococcus neoformans myristoyl-CoA:protein N-Myristoyltransferase (Nmt) Is Nmt-dependent. J Biol Chem. 1998;273(20):12482–12491. doi: 10.1074/jbc.273.20.12482. [DOI] [PubMed] [Google Scholar]
- Lodge JK, Jackson-Machelski E, Toffaletti DL, Perfect JR, Gordon JI. Targeted gene replacement demonstrates that myristoyl-CoA: protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. PNAS. 1994;91(25):12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, Vamathevan J, Miranda M, Anderson IJ, Fraser JA, Allen Jonathan E, Bosdet IE, Brent MR, Chiu R, Doering TL, Donlin MJ, D’Souza CA, Fox DS, Grinberg V, Fu J, Fukushima M, Haas BJ, Huang JC, Janbon G, Jones SJM, Koo HL, Krzywinski MI, Kwon-Chung JK, Lengeler Klaus B, Maiti R, Marra MA, Marra RE, Mathewson CA, Mitchell TG, Pertea M, Riggs FR, Salzberg SL, Schein JE, Shvartsbeyn A, Shin H, Shumway M, Specht CA, Suh BB, Tenney A, Utterback TR, Wickes BL, Wortman JR, Wye NH, Kronstad JW, Lodge JK, Heitman J, Davis RW, Fraser CM, Hyman RW. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307(5713):1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDade H, Cox G. A new dominant selectable marker for use in Cryptococcus neoformans. Med Mycol. 2001;39(1):151–154. doi: 10.1080/mmy.39.1.151.154. [DOI] [PubMed] [Google Scholar]
- Moyrand F, Fontaine T, Janbon G. Systematic capsule gene disruption reveals the central role of galactose metabolism on Cryptococcus neoformans virulence. Molecular Microbiology. 2007;64(3):771–781. doi: 10.1111/j.1365-2958.2007.05695.x. [DOI] [PubMed] [Google Scholar]
- Nielsen K, Cox GM, Wang P, Toffaletti DL, Perfect JR, Heitman J. Sexual cycle of Cryptococcus neoformans var. grubii and virulence of congenic a and alpha isolates. Infect Immun. 2003;71(9):4831–4841. doi: 10.1128/IAI.71.9.4831-4841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh D, Hopper JE. Transcription of a yeast phosphoglucomutase isozyme gene is galactose inducible and glucose repressible. Mol Cell Biol. 1990;10(4):1415–1422. doi: 10.1128/mcb.10.4.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ory JJ, Griffith CL, Doering TL. An efficiently regulated promoter system for Cryptococcus neoformans utilizing the CTR4 promoter. Yeast. 2004;21(11):919–926. doi: 10.1002/yea.1139. [DOI] [PubMed] [Google Scholar]
- Perfect JR, Toffaletti DL, Rude TH. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61(10):4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, N.Y: 2001. [Google Scholar]
- Thompson JR, Douglas CM, Li W, Jue CK, Pramanik B, Yuan X, Rude TH, Toffaletti DL, Perfect JR, Kurtz M. A glucan synthase FKS1 homolog in Cryptococcus neoformans is single copy and encodes an essential function. J Bacteriol. 1999;181(2):444–453. doi: 10.1128/jb.181.2.444-453.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformans by use of biolistic delivery of DNA. J Bacteriol. 1993;175(5):1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia TE, Hamilton RW, Cano CL, Hopper JE. Disruption of regulatory gene GAL80 in Saccharomyces cerevisiae: effects on carbon-controlled regulation of the galactose/melibiose pathway genes. Mol Cell Biol. 1984;4(8):1521–1527. doi: 10.1128/mcb.4.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia TE, Hopper JE. Genetic and molecular analysis of the GAL3 gene in the expression of the galactose/melibiose regulon of Saccharomyces cerevisiae. Genetics. 1986;113(2):229–246. doi: 10.1093/genetics/113.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschopp JF, Emr SD, Field C, Schekman R. GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J Bacteriol. 1986;166(1):313–318. doi: 10.1128/jb.166.1.313-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Edman JC. The Cryptococcus neoformans GAL7 gene and its use as an inducible promoter. Mol Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.