Abstract
Nicotinic acetylcholine receptors (nAChR) are cation-selective, ligand-gated ion channels of the cysteine (Cys)-loop gene superfamily. The recent crystal structure of a bacterial homolog from Erwinia chrysanthemi (ELIC) agrees with previous structures of the N-terminal domain of AChBP (acetylcholine-binding protein) and of the electron-microscopy-derived Torpedo nAChR structure. However, the ELIC transmembrane domain is significantly more tightly packed than the corresponding region of the Torpedo nAChR. We investigated the tightness of protein packing surrounding the extracellular end of the M2 transmembrane segment and around the loop connecting the M2 and M3 segments using the substituted cysteine accessibility method. The M2 20′ to 27′ residues were highly water accessible and the variation in reaction rates were consistent with this region being α-helical. At all positions tested, the presence of ACh changed methanethiosulfonate ethylammonium (MTSEA) modification rates by <10-fold. In the presence of ACh, reaction rates for residues in the last extracellular α-helical turn of M2 and in the M2M3 loop increased, whereas rates in the penultimate α-helical turn of M2 decreased. Only three of eight M2M3 loop residues were accessible to MTSEA in both the presence and absence of ACh. We infer that the protein packing around the M2M3 loop is tight, consistent with its location at the interdomain interface where it is involved in the transduction of ligand binding in the extracellular domain to gating in the transmembrane domain. Our data indicate that the Torpedo nAChR transmembrane domain structure is a better model than the ELIC structure for eukaryotic Cys-loop receptors.
Keywords: acetylcholine, nicotine, serotonin, Cys loop, ion channel, gating
Introduction
Functional cysteine (Cys)-loop receptors can assemble from the same subunit to form homopentameric ligand gated ion channels or from different subunits to form heteropentameric receptors. The individual subunits share the same transmembrane topology. The ∼200 amino acid long extracellular N-terminal domain contains the ligand-binding site as well as the eponymous disulfide-linked loop. Four transmembrane segments (M1–M4) form the transmembrane domain. These are linked by two short loops between M1 and M2 on the cytosolic side and between M2 and M3 on the extracellular side. A long loop between M3 and M4 is the major contributor to the intracellular domain that is involved in modulating conductance, trafficking and localization, but that is not an absolute requirement for assembly and function as an ion channel (Jansen et al., 2008). The extracellular domain consists mainly of two antiparallel β-sheets. High-resolution crystal structures are available for AChBP (acetylcholine-binding protein) with different ligands bound (Brejc et al., 2001; Celie et al., 2004; Dutertre and Lewis, 2006), and also for a recombinant mouse α1 nicotinic acetylcholine receptor (nAChR) extracellular domain (Dellisanti et al., 2007). The electron-microscopy-derived structure of almost the complete Torpedo nAChR at 4.0 Å showed that the transmembrane segments are α-helical (Unwin, 2005). The M2 segments from all five subunits line the ion-conducting channel, whereas the other segments shield the M2 segments from the lipids, with M4 having the greatest exposure to membrane lipids (Blanton and Cohen, 1994; Unwin, 2005; Guzmán et al., 2006). The top of M2 and the M2M3 loop segment at the interface between the N-terminal and the transmembrane domain are crucial for the propagation of the conformational change from ligand binding to channel gating (Kash et al., 2003; Bouzat et al., 2004; Lee and Sine, 2005; Mukhtasimova et al., 2005; Reeves et al., 2005). Recently, the crystal structures from prokaryotic Cys-loop receptor homologs, ELIC, from Erwinia chrysanthemi (closed state, 3.3 Å) (Hilf and Dutzler, 2008) and GLIC, from Gloeobacter violaceus (potentially open state, 3.1 and 2.9 Å) were solved (Bocquet et al., 2009; Hilf and Dutzler, 2009). In these structures, the conformation of the N-terminal domain is essentially identical to the one from Torpedo nAChR. However, whereas the nAChR and GLIC structures depict a narrow open pore in the center of the M2 helices, the ELIC structure is entirely closed by several lipophilic side chains in the extracellular third of the channel. Interestingly, the entire transmembrane domain in ELIC is more tightly packed compared with the Torpedo nAChR structure that showed, in agreement with previous experimental data, that the transmembrane segments are surrounded by water filled crevices (Akabas and Karlin, 1995; Williams and Akabas, 1999; Bera et al., 2002; Goren et al., 2004). In the present study we used the substituted cysteine accessibility method (SCAM) (Akabas et al., 1992) to investigate the accessibility of the top of the M2 segment as well as the M2M3 loop and its changes during gating in muscle nAChR with a subunit stoichiometry of (α)2βγδ.
Materials and Methods
Mutagenesis.
M2 segment residues are named using an index numbering system that facilitates comparison between M2 segments of different members of the superfamily. At the cytoplasmic end, the conserved positively charged residue (nAChR αK242, GABAA α1R255, 5-HT3A R278) is defined as the 0′ position (Miller, 1989). More C-terminal residues are assigned consecutively 1′, 2′, 3′… and residues N-terminal to 0′ are assigned −1′, −2′… The 20′ position is the residue aligned with the nAChR extracellular ring of charge, a conserved ring of charged residues in Cys-loop subunits (Imoto et al., 1988).
The exact starts and ends of the α-helical transmembrane segments are not well defined. The PDB-file remarks to Unwin's structure (Unwin, 2005) read: “Users should bear in mind that because of the limited resolution the conformations of the side chains and their atomic coordinates are not individually reliable. Also the exact extents of the α helices and β strands are uncertain by at least one residue.” This is exemplified by the end of the α-helical assignment for the two α-subunits per receptor in Unwin's structure. One of them (strand A) has the α-helical part of M2 end with ELIPSTSSA (αA270 = 28′), whereas the other (strand D) ends several residues earlier with ELIPST (αT267 = 25′). For our study, we define M2 to end with αA270 (28′) and the M2M3 loop to cover residues αV271 to αY277 (29′ to 35′). In the ELIC or GLIC structures, the α-helical content of M2 extends for 25 residues, FSERLQTSFTLMLTVVAYAFYTSNI or YEANVTLVVSTLIAHIAFNILVETN in ELIC or GLIC, respectively. The final, most extracellular residue of M2 corresponds to αL263 (21′) in our alignment or αT267 (25′) in the structure-based alignment by Hilf and Dutzler (2008). The M2M3 loop spans seven residues in ELIC/GLIC and 5–8 in Unwin's structure. The start of the α-helical content of M3 is essentially identical in all structures, corresponding to αG275 or αK276 (33′ or 34′).
Mouse muscle nAChR subunits in the pSP64T plasmid were used (Akabas et al., 1994). Generation of αE262C has been described previously (Akabas et al., 1994). Cys were introduced, one at a time, in the α-subunit by PCR using the appropriate forward and reverse mutational primers, DpnI (all enzymes from New England Biolabs) digest and transformation into competent Escherichia coli. All mutant plasmids were sequenced to confirm the mutations. Plasmids were linearized (α- and γ-subunits with XbaI, β with SacI, and δ with BamHI) before in vitro mRNA transcription with SP6 RNA polymerase (SP6 mMessage mMachine kit, Ambion). mRNA was purified with the mMega Clear kit (Ambion), precipitated with ammonium acetate, dissolved in diethylpyrocarbonate-treated water, and stored at –80°C.
Reagents.
Stock solutions of methanethiosulfonate ethylammonium (MTSEA) and methanethiosulfonate ethylsulfonate (MTSES) (100 mm) (Biotium) were prepared in water on the day of the experiment and kept on ice. Appropriate working solutions were diluted in Ca2+-free frog Ringer buffer (CFFR) [(in mm) 115 NaCl, 2.5 KCl, 1.8 MgCl2, 10 HEPES, pH 7.5, with NaOH] immediately before application.
Expression in X. laevis oocytes.
Oocytes were harvested and defolliculated as described (Jansen and Akabas, 2006). One day after isolation each oocyte was injected with a 2:1:1:1 mixture (α:β:γ:δ) of 10 ng of mRNA. Oocytes were kept in SOS medium (in mm) 82.5 NaCl, 2.5 KCl, 1 MgCl2, 5 HEPES, pH 7.5 with 100 IU/ml penicillin, 100 μg/ml streptomycin, 250 ng/ml amphotericin B (Invitrogen) and 5% horse serum (Sigma) at 16°C. Experiments were conducted 3–5 d after injection.
Two-electrode voltage clamp.
For two-electrode voltage-clamp experiments, oocytes were continuously superfused at 5 ml/min with CFFR at room temperature (RT). Holding potential was −40 mV. The ground electrode was connected by a 3 m KCl/Agar bridge to the bath. Glass microelectrode resistance was <2 MΩ when filled with 3 m KCl. Data were acquired at 200 Hz and analyzed using a TEV-200 amplifier (Dagan Instruments), a Digidata 1322A data interface and pClamp 8 software (Molecular Devices). Currents (IACh) elicited by ACh applications were separated by sufficient CFFR wash to allow for complete recovery from desensitization. Currents were judged to be stable if the variation between consecutive IACh was ≤10%.
Concentration–response analysis.
After obtaining a stable IACh with an approximately EC50 ACh concentration, progressively increasing ACh concentrations were applied to oocytes expressing wild-type or mutant receptors. Currents were normalized to the maximal ACh-induced current (Imax). The ACh concentration–response relationship was determined for wild-type and each mutant by least-squares minimization (GraphPad Prism version 5.01 for Windows, GraphPad Software) of the currents to a logistic equation of the form: I/Imax*100 = 1/(1 + 10^((logEC50 − [ACh])*nH), where nH is the Hill coefficient and EC50 is the ACh concentration that gives rise to 50% of the maximal current. Parameters from several oocytes were averaged to obtain the mean EC50 and Hill coefficient. Data are presented as mean ± SEM.
Modification of engineered Cys with MTS reagents.
MTS reagents H3CSO2-R react with Cys (Cys-SH) to add the moiety R to the Cys sulfur (Cys-SR) with sulfinic acid (H3CSO2H) as the leaving group. The reagents MTSEA (r = SCH2CH2NH3+) and MTSES (r = SCH2CH2SO3−) were applied to wild-type and mutant nAChR-expressing oocytes, and their effect on the ACh-induced current amplitude (I) at the ACh EC30–70 was investigated. After a stable ACh response (IACh) was recorded from two or more consecutive ACh pulses, MTS reagent was applied (MTSEA 2 mm for 2 min, MTSES 5 mm for 2 min), and I was determined again. The effect of MTS modification was calculated as follows: % effect = ((Iafter/Iinitial) − 1)*100, where Iinitial is the ACh-induced current amplitude before and Iafter is the amplitude after MTS reagent application. Significance of effect was determined by one-way ANOVA with Dunnett's posttest with wild type as a control (n = 3–6). MTSEA modification was assayed at all positions, whereas MTSES was only used at those positions where MTSEA did not cause a significant effect, and in addition on αS269C.
Modification of engineered Cys with MTSEA in the presence of ACh.
For all positions where MTSEA application did not have a significant effect on subsequent ACh-induced currents when applied alone, we tested whether MTSEA (2 mm, 2 min) had an effect when applied together with ACh (EC80–EC100 concentration).
Rates of reaction with MTSEA.
Once a stable initial ACh-induced current (Iinitial) was obtained, MTSEA was applied repeatedly for brief periods. After each reagent application, I was recorded, and current amplitudes were normalized to Iinitial, plotted as a function of the cumulative MTS-reagent application time, and fitted with a monoexponential function of the form: I = (Iinitial − I∞)e−tτ′ + I∞, where Iinitial is the value of the ACh-induced current amplitude before modification, I∞ is the current amplitude at the end of the reaction, t is the cumulative MTS-reagent application time, and τ′ is the pseudo first-order rate constant (s−1). The second-order rate constants, τ, were calculated by dividing the pseudo first-order time constants τ′ by the MTSEA concentration. Second-order rate constants were determined at two different concentrations to verify that the rates were independent of the reagent concentration. In all cases the second-order rate constants were independent of the MTS-reagent concentrations. Data are presented as mean ± SEM.
At all positions where MTSEA application caused a significant effect when applied alone or with ACh, the second-order rate constants were determined both in the absence and presence of ACh (EC80–EC100 concentration) with MTSEA. The experimental procedure to determine MTSEA reaction rates was essentially similar in the absence and presence of ACh.
Homology modeling.
A model of the mouse muscle nAChR was built based on the Torpedo nAChR structure (PDB: 2BG9) with the Swiss-Pdb Viewer 3.7 (http://spdbv.vital-it.ch) (Guex and Peitsch, 1997). Alignment scores for individual subunits between Torpedo and mouse nAChR are high, 82 for α, 62 for β, γ, and δ, facilitating the manual alignment and model building inside the software.
Three additional models of mouse muscle nAChR were constructed based on the recently published structures of prokaryotic Cys-loop receptor homologs, one in the closed state (from Erwinia chrysanthemi, ELIC, PDB: 2VL0) (Hilf and Dutzler, 2008) and two in the potentially open state (from Gloeobacter violaceus, GLIC, PDB: 3EHZ, 3EAM) (Bocquet et al., 2009; Hilf and Dutzler, 2009). The two GLIC models are essentially identical and we therefore will only refer to the one build on 3EHZ. ELIC and GLIC share 18% of identical amino acids. The sequence identity between prokaryotic and eukaryotic homologs is low (16% between ELIC and nAChR-α). To improve the reliability of the alignment, we performed a multiple alignment with >50 members of the Cys-loop superfamily involving a wide variety of Cys-loop receptor families. Sequences were obtained from UniProt (Universal Protein Resource; http://www.ebi.uniprot.org/index.shtml) or RCSB PDB (Research Collaboratory for Structural Bioinformatics; http://www.rcsb.org/pdb/), and aligned using ClustalW (http://www.ebi.ac.uk/clustalw/index.html). The alignment in the investigated M2 M3 area (supplemental Fig. 1, available at www.jneurosci.org as supplemental material) yielded an absolutely conserved proline toward the C-terminal end of M2 (corresponding to nAChR αP265 = 23′). The 9′ position (corresponding to nAChR αL251) is highly conserved, most subunits bear a Leu at 9′, however, in both eukaryotic and prokaryotic members residues different from Leu can be found (Ile and Val). The alignment we obtained is in agreement with the one published with the GLIC structure (Bocquet et al., 2009), but differs from the one published with the ELIC structure (Hilf and Dutzler, 2008). The length of the linker between M2 and M3 varies by up to two amino acids (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
Results
Expression and functional characterization of Cys mutants
We expressed 16 different nAChR α-subunit single Cys mutants from E262C to Y277C covering the extracellular end of M2, the M2M3 loop, and the first three residues of M3 together with wild-type β, γ, and δ-subunits in X. laevis oocytes (Fig. 1). For orientation the investigated stretch is depicted with side chains in CPK colors in stick representation in Figure 6A,B. All constructs trafficked to the plasma membrane and were functional as evidenced by ACh-induced currents in two-electrode voltage-clamp experiments. The ACh EC50s over all mutants varied <200-fold: for wild-type receptors the EC50 was 2.96 ± 0.54 μm, whereas the lowest determined EC50 was 0.18 ± 0.03 μm for αS269Cβγδ and the highest was 34.8 ± 4.5 μm for αI274Cβγδ (Table 1, Fig. 2). At nine positions the log(EC50) was significantly different from wild-type by one-way ANOVA with Dunnett's posttest (Table 1). It is notable that substitution of αP272 with Cys suggested to undergo a cis-to trans isomerization during gating in the 5HT3 receptor (Lummis et al., 2005), had a minimal effect on ACh EC50. The Hill coefficients for all mutants were not significantly different from wild-type values (nH = 1.30 ± 0.11).
Figure 1.
Alignment of amino acid sequences spanning the M2 segment, the M2M3 loop, and part of the M3 segment of the mouse muscle nAChR α (ACHa1mumu), β (ACHb1mumu), γ (ACHg1mumu), and δ (ACHd1mumu) subunits, and Torpedo nAChR α (2BG9_A, 2BG9_D), β (2BG9_B), γ (2BG9_E), and δ (2BG9_C) subunits (PDB entry 2BG9). Segments that are α-helical in the Torpedo nAChR structure are underlined. The conserved 9′ leucine and the conserved 23′ proline are boxed. The 16 positions in the α-subunit investigated in this study that were individually mutated to Cys are in bold and also indicated by a bold italic “C” above the residue. Two possible alignments for the prokaryotic Cys-loop receptor homolog from ELIC are shown. The upper alignment was obtained by aligning all known Cys-loop receptor sequences; the lower alignment is a structure-based alignment from Hilf and Dutzler (2009). Numbers at the right of each row indicate the amino acid number of the last residue shown. Symbols below alignment denoting the degree of conservation observed in each column are as follows: an asterisk means that the residues or nucleotides in that column are identical in all sequences in the alignment, a colon means that conserved substitutions have been observed, and a period means that semiconserved substitutions are observed.
Figure 6.
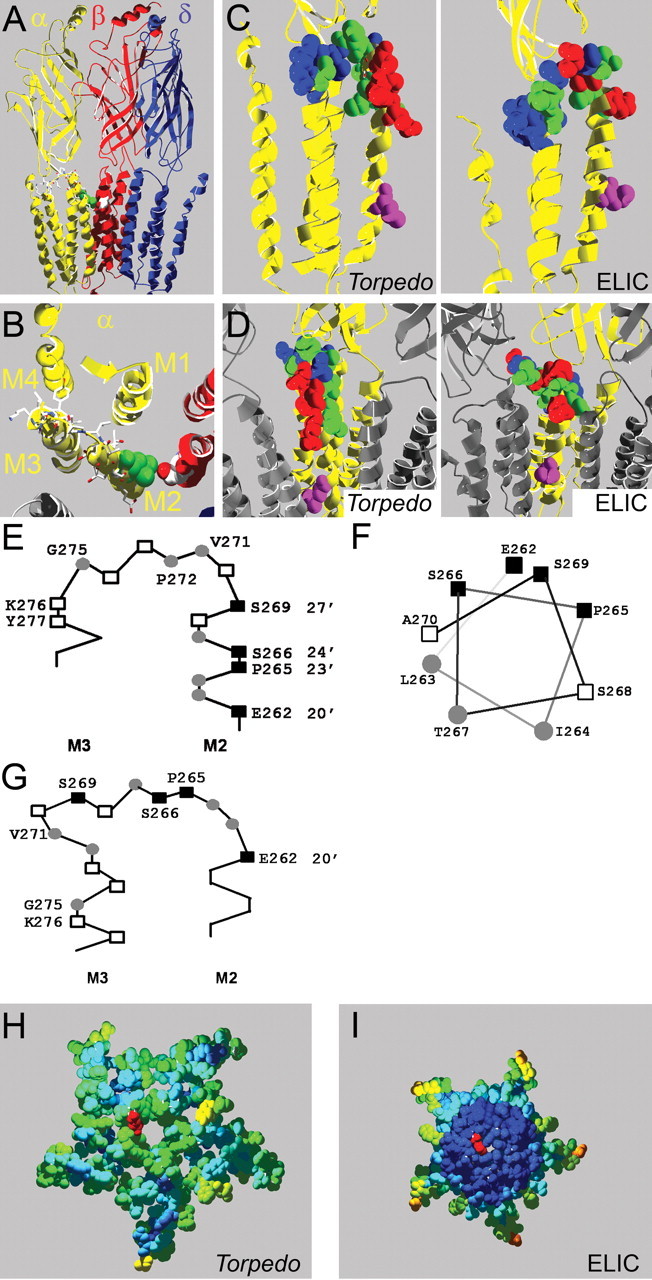
Homology models of the mouse muscle nAChR. A, Side view of homology model based on the Torpedo nAChR structure; one α-subunit and the γ-subunit are omitted for clarity. The intracellular domain, most of which was not resolved in the Torpedo nAChR structure, is not shown. Residue αL263 is in green, other residues mutated to Cys in α-subunits in stick representation and CPK colors, and βD268 is shown in space-filling representation and CPK colors. B, Same model as in A but viewed from the extracellular side, focusing on the α-subunit (yellow). Only the slice of the receptor in the M2M3 loop area that we investigated is shown; the extracellular domain is not shown. C, Rate of MTSEA modification indicated in homology models of the nAChR α-subunit based on the Torpedo (left) and ELIC (right) structures. The rate of MTSEA reaction is color coded: red = fast reacting (>10,000 m−1 s−1), green = slow reacting (<1000 m−1 s−1), blue = nonreactive, pink = 9′ Leu, viewed from the side. D, Same models as in C except viewed from the channel lumen with two neighboring subunits in gray for orientation purposes. E, F, Schematic representation of the residues we studied based on Unwin's structure; positions with fast modification rates (■), slow rates (○), and residues where MTSEA application did not cause an effect (□) viewed from the side (E) and M2 positions studied plotted on an α-helical wheel viewed from the extracellular side (F). G, Same as in E, except based on the ELIC structure. H, I, Computed overall water accessibility of the Torpedo (H) and ELIC (I) structures of a top-view slice of the receptors encompassing the investigated area. One 20′ αE262 residue for orientation is in red. Otherwise, color coding of investigated amino acids is as per accessibility of a 1.4 Å sphere. Scale is red to blue, where red is highly accessible and dark blue is inaccessible. Note that in contrast to the Torpedo structure, the center of the ELIC structure consists entirely of water-inaccessible (dark blue) residues, which in general indicated higher compactness.
Table 1.
ACh EC50, ANOVA, and nH for the Cys mutants
| Construct | EC50 (μm) | ANOVAa | nH | n |
|---|---|---|---|---|
| αβγδ | ||||
| wild type | 2.96 ± 0.54 | 1.30 ± 0.11 | 4 | |
| αY277C | 8.42 ± 0.72 | ** | 1.15 ± 0.04 | 3 |
| αK276C | 4.42 ± 0.46 | ns | 1.31 ± 0.02 | 3 |
| αG275C | 0.64 ± 0.03 | *** | 1.18 ± 0.06 | 3 |
| αI274C | 34.8 ± 4.5 | *** | 1.34 ± 0.02 | 3 |
| αL273C | 1.45 ± 0.15 | ns | 1.36 ± 0.03 | 3 |
| αP272C | 1.34 ± 0.07 | ns | 1.42 ± 0.06 | 3 |
| αV271C | 2.74 ± 0.17 | ns | 1.47 ± 0.21 | 3 |
| αA270C | 2.37 ± 0.60 | ns | 1.08 ± 0.14 | 4 |
| αS269C | 0.18 ± 0.03 | *** | 1.41 ± 0.06 | 4 |
| αS268C | 0.79 ± 0.13 | *** | 1.45 ± 0.08 | 5 |
| αT267C | 28.0 ± 5.2 | *** | 1.37 ± 0.07 | 5 |
| αS266C | 17.0 ± 2.9 | *** | 1.26 ± 0.08 | 3 |
| αP265C | 3.25 ± 0.43 | ns | 1.11 ± 0.04 | 3 |
| αI264C | 9.0 ± 1.6 | *** | 1.38 ± 0.05 | 8 |
| αL263C | 0.40 ± 0.06 | *** | 1.41 ± 0.05 | 4 |
| αE262C | 2.52 ± 0.40 | ns | 1.27 ± 0.06 | 3 |
The two mutants that varied most from wild type and for which the dose–response curves are shown in Figure 2 are in bold.
aOne-way ANOVA of log(EC50) with Dunnett's posttest; ns, p > 0.05, not significant; **0.001 < p < 0.01, very significant; ***p < 0.0001, extremely significant.
Figure 2.
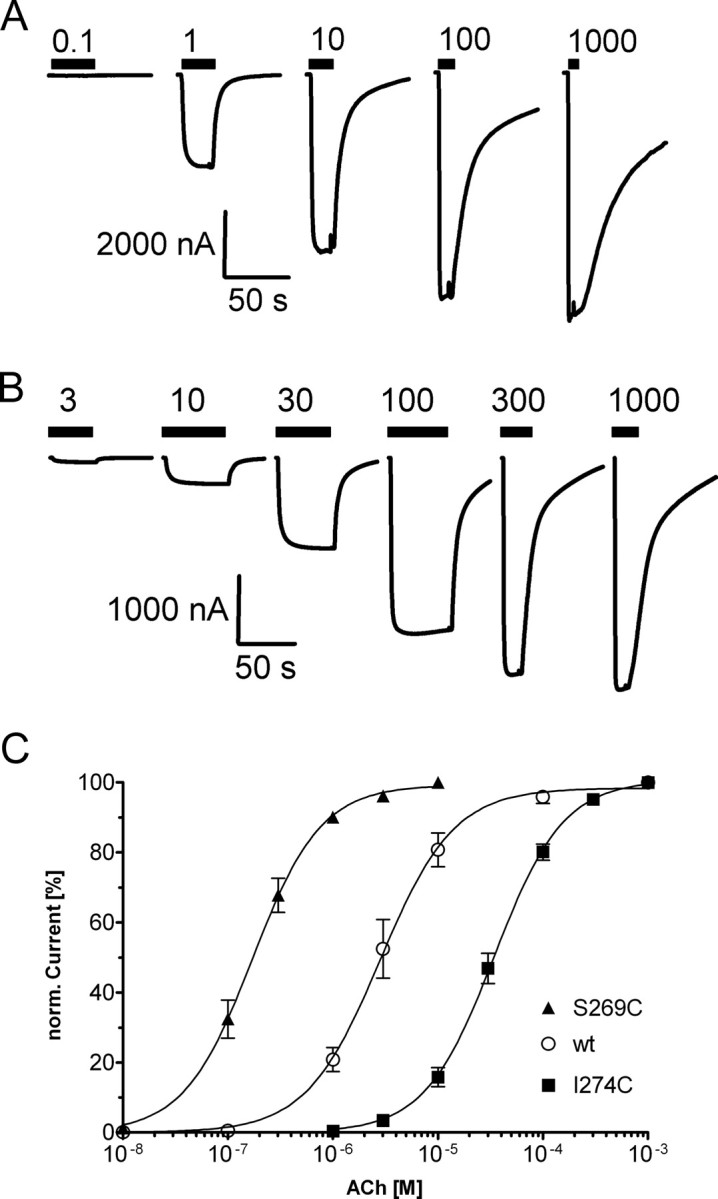
ACh concentration–response curves of wild-type and mutant αβγδ nAChR. A, B, Representative current traces for wild-type (A) and mutant (B) αI274Cβγδ receptors upon increasing ACh concentrations are shown. Application of ACh is indicated by black bars above the current traces, and micromolar ACh concentrations are given above bars. C, ACh concentration–response curves from oocytes expressing αβγδ wild-type (○), αS269Cβγδ (▴), or αI274Cβγδ (■) receptors. Currents were normalized (norm.) to the maximum current for each oocyte. Data points represent the mean ± SEM from three or four independent experiments. Data were fit by nonlinear regression analysis as described in Materials and Methods. ACh EC50 and nH values are reported in Table 1.
Effect of MTS modification
The accessibility of engineered Cys was probed for all 16 positions with the positively charged reagent MTSEA (2 mm, 2 min) (Fig. 3). MTS reagents react 109 times faster with ionized thiolates (-S−) than with protonated thiols (-SH) (Roberts et al., 1986). Only in an aqueous environment is the Cys likely to ionize to the thiolate form. The extent of ionization can also be influenced by local electrostatic factors. Other factors influencing reactivity/accessibility of a certain Cys toward MTS reagents are steric factors (access to the site and steric constraints at the site), electrostatic interactions between charged reagents and charged residues lining the access pathway or the site of reaction. At five positions (αI264C, αS266C, αT267C, αS269C, αV271C) MTSEA application in the absence of ACh significantly increased subsequent ACh-induced current amplitudes by 57 ± 16% to 197 ± 37%. At five other positions (αE262C, αL263C, αP265C, αP272C, αG275C), MTSEA application in the absence of ACh significantly reduced subsequent ACh-induced current amplitudes by 63 ± 1% to 97 ± 0.4% (Fig. 3). For the six positions where there was no effect of MTSEA application (αS268C, αA270C, αL273C, αI274C, αK276C, αY277C) and also αS269C, we tested the effect of the negatively charged MTSES (5 mm, 2 min) (Fig. 3F). MTSES application significantly increased subsequent ACh-induced current amplitudes for αS268C and αS269C by 85 ± 12 and 177 ± 37% compared with 1 ± 8% for wild type. For αS268C, we also tried successive application of MTSEA and afterward MTSES to the same oocyte to discriminate between nonreaction and silent reaction of MTSEA at this position. MTSES applied after MTSEA similarly increased subsequent ACh-induced currents compared with when applied alone, indicating that MTSEA does not react at this position (Fig. 3F).
Figure 3.
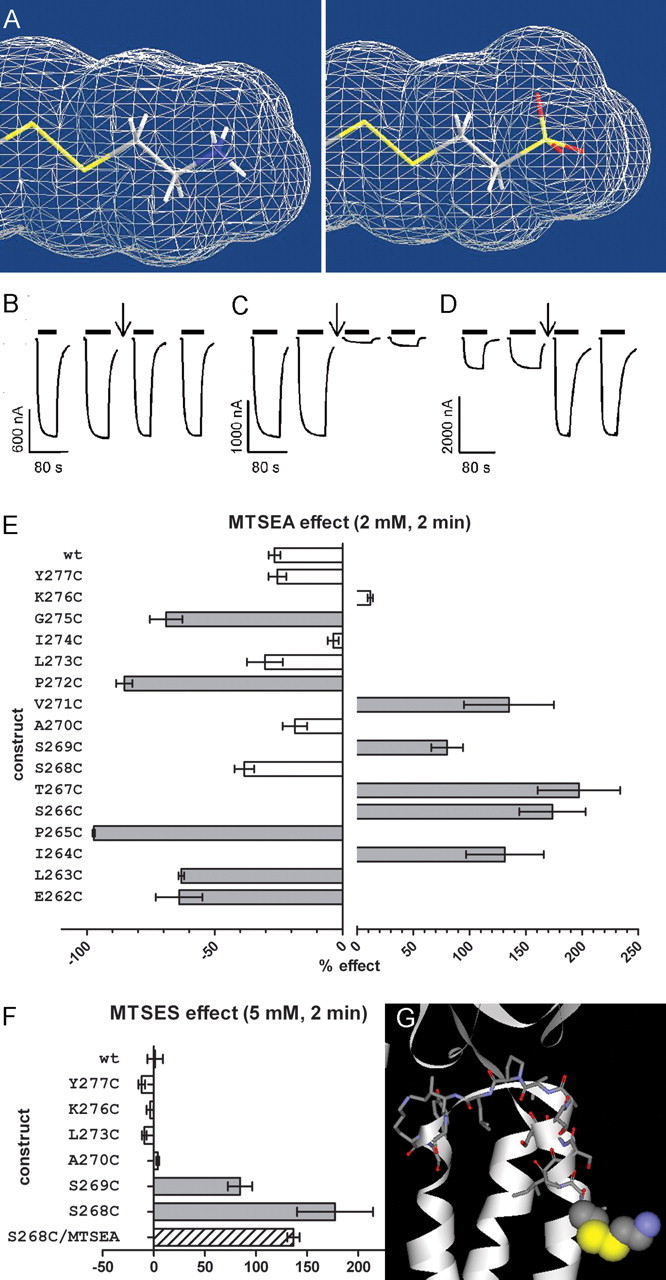
Effect of MTS reagent application on subsequent ACh-induced EC30–70 currents. A, Structure and size of the moiety that is added onto the Cys sulfur by MTSEA (left) and MTSES (right). Solvent-accessible surface shown. MTSES adds a moiety to the Cys sulfur that is slightly larger in volume than the one added by MTSEA modification. The distances between the added sulfur and the furthest atom of the added moiety are 4.8 and 5.3 Å for MTSEA and MTSES modification, respectively (Chem3D Ultra; CambridgeSoft). B–D, Sample current traces showing no effect of MTSEA application (2 mm, 2 min, at downward arrow) on ACh-induced current amplitudes for αI274Cβγδ (B), inhibition for αP272Cβγδ (C), or potentiation for αS266Cβγδ receptors (D). Current traces during reagent application not shown. E, Effects of MTSEA application (2 mm, 2 min) on wild-type and mutant receptors. Percentage effect on ACh-induced current amplitude (IACh) after MTSEA treatment is shown. Note the difference in scales for inhibitory and potentiating effect. Significance of effect was determined by one-way ANOVA with Dunnett's posttest with wild type as a control (n = 3–6, p < 0.05). F, Effect of MTSES application (5 mm, 2 min) on ACh- induced EC30–70 currents. Percentage effect on ACh-induced current amplitude (IACh) after MTSES treatment shown. The striped bar indicates the effect of MTSES application after MTSEA treatment (2 mm, 2 min) where the MTSEA treatment did not have a significant effect. Negative values represent an inhibition of IACh after MTS reaction, whereas positive values represent an increase in IACh. Data are represented by mean ± SEM. Effects that were significant different compared with wild type are indicated in dark gray or striped bars, nonsignificant effects in white bars. G, nAChR model of αE262C modified by MTSEA. αE262Cys-S-S-EA is in spacefill representation and the other 15 investigated positions are in stick representation. Hydrogens are omitted for clarity. Carbon = gray, oxygen = red, sulfur = yellow, nitrogen = blue. The addition of the aminoethanesulfide moiety increases the volume of the side chain of the engineered Cys to a size that is in between the size of a Lys or Arg side chain.
When MTSEA was applied in the presence of ACh one additional position showed a significant effect on subsequent ACh-induced current amplitudes (αI274C).
Rate constants for modification of Cys with MTSEA
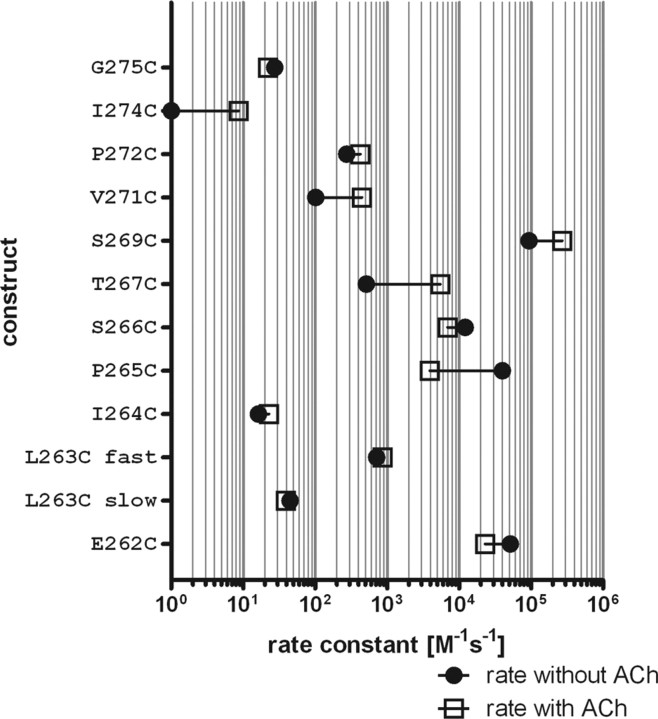
For the 10 mutants where MTSEA application alone caused a significant change in ACh-induced currents we determined the second-order rate constants (Fig. 4). Second-order rate constants covered five orders of magnitude, varying between 16.3 ± 2.1 m−1 s−1 for αI264C and 93,800 ± 9100 m−1 s−1 for αS269C (Fig. 5, Table 2). For four of these mutants (αE262C, αP265C, αS266C, and αS269C) the rate constants were fast (>10,000 m−1 s−1) and comparable with the order of magnitude of the rate constant in free solution (76,000 m−1 s−1) (Karlin and Akabas, 1998). The rate constants for the remaining six positions (αL263C, αI264C, αT267C, αV271C, αP272C, αG275C) were slow (<1000 m−1 s−1). When the residues αE262C to αS269C are plotted on an α-helical wheel the positions with a fast second-order rate constant are on one side of the α-helix whereas the slow reacting ones are on the other side (Fig. 6C,D). The α-helical part of M2 extends for two more helical turns past αE262C above the membrane confirming the Torpedo nAChR structure. In this α-helical stretch of eight positions that we investigated, five reacted at a fast reaction rate with MTSEA, two at a slow rate, and one position only reacted with MTSES. In the proximal M2M3 loop, of eight residues that we investigated, three reacted with a slow rate, and five did not react with either MTSEA or MTSES. We therefore infer that the 20′ to 27′ extracellular α-helical M2 segment is more accessible than the M2M3 loop. The lack of reactivity at five of eight positions in the M2M3 loop implies that the M2M3 loop is more tightly packed with other regions of the protein than the M2 segment region that we studied.
Figure 4.
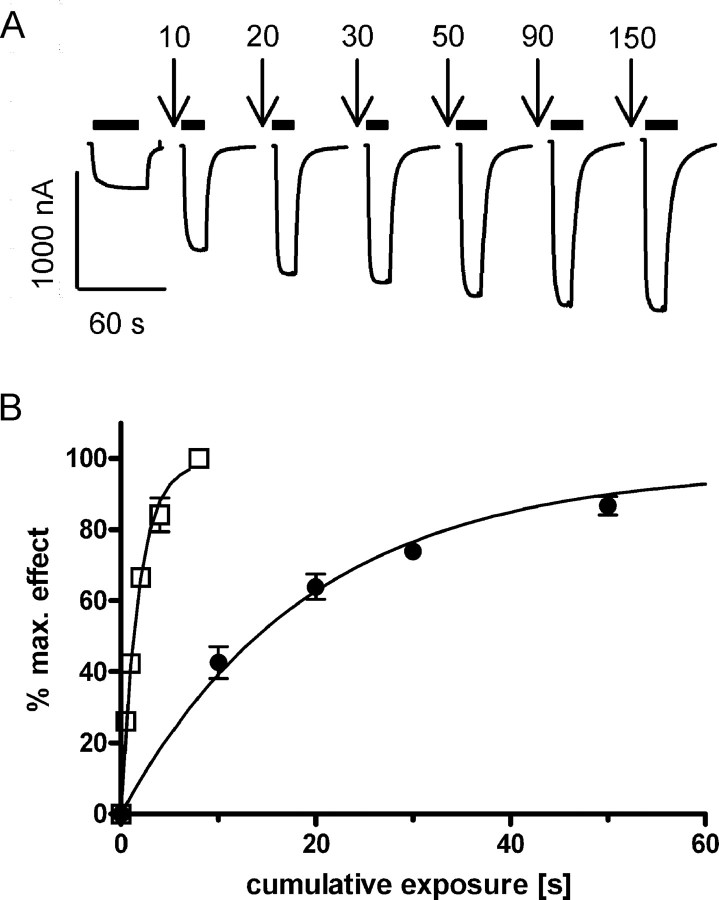
Rate constants for MTSEA modification. A, Representative ACh-induced current traces recorded before and after successive MTSEA applications (100 μm, indicated by downward arrows, cumulative application time in seconds) for αT267Cβγδ receptors. Time of ACh test pulse application is indicated by black bars. B, Fractional effect for αT267Cβγδ was normalized to the maximum effect for each individual oocyte and plotted versus the cumulative exposure time of MTSEA alone (●), and of MTSEA coapplied with ACh (□). Data points represent the mean ± SEM from three or four independent experiments. Note that for some data points the error bars are smaller than the symbol and therefore not visible. Data were fit by nonlinear regression analysis as described in Materials and Methods. Second-order rate constants are reported in Table 2.
Figure 5.
Rate constants for modification by MTSEA applied in the absence (●) and presence (□) of ACh. Horizontal lines connect the mean rate constants for each condition. Each symbol is the mean of at least three independent experiments.
Table 2.
Second-order reaction rate constants for MTSEA applied in the absence or presence of ACh with the Cys mutants
| Construct | τ in the absence of ACh (m−1 s−1) | n | τ in the presence of ACh (m−1 s−1) | n |
|---|---|---|---|---|
| αG275Cβγδ | 27.5 ± 2.4 | 3 | 22.2 ± 3.3 | 3 |
| αI274Cβγδ | nt | 8.7 ± 2.6*** | 3 | |
| αP272Cβγδ | 274 ± 15 | 4 | 426 ± 66* | 3 |
| αV271Cβγδ | 102 ± 10 | 4 | 398 ± 73*** | 3 |
| αS269Cβγδ | 93,800 ± 9100 | 3 | 273,100 ± 31,000** | 3 |
| αT267Cβγδ | 510 ± 54 | 4 | 5,450 ± 86*** | 3 |
| αS266Cβγδ | 12,200 ± 2200 | 4 | 6,970 ± 1800 | 3 |
| αP265Cβγδ | 40,100 ± 3700 | 5 | 3940 ± 620*** | 3 |
| αI264Cβγδ | 16.3 ± 2.1 | 4 | 22.8 ± 6.5 | 4 |
| αL263Cβγδ | 45.0 ± 13.0 | 3 | 39.3 ± 7.4 | 3 |
| 717 ± 105 | 870 ± 360 | |||
| αE262Cβγδ | 51,300 ± 5500 | 4 | 23,000 ± 2100** | 3 |
| Free solution | 76,000 ± 4000a |
nt, Not tested. The rate constant for modification of position αL263C could be better fit with a double exponential equation.
*0.01 < p < 0.05, significant;
**0.001 < p < 0.01, very significant;
***p < 0.001, extremely significant.
αE262C was previously reported to react with a second-order rate constant of 14 m−1 s−1 (Pascual and Karlin, 1998). The rate reported here is 4000 times faster. We verified the nACh α1E262C mRNA by RT PCR and subsequent DNA sequencing to confirm the mutation. We do not know why the rate constant reported in this study is different from the one reported previously.
The rate constant for modification of position αL263C could be better fit with a double exponential equation whereas all others were fit best with a mono exponential function. Position αL263C is facing the intersubunit interface, one αL263C faces the β-subunit and the other the γ-subunit. On examination of the nAChR structure we noted that the charge at one neighboring position that is directly apposed to the αL263C SH is not conserved between the β and γ-subunit. At the M2 21′ position there is an aspartate, βD268, in β and a lysine, γK270, in γ (Fig. 6A,B). To test whether the biexponential fit of the MSTEA reaction rate with αL263C was due to different rates of reaction with the two engineered Cys due to the differing charges on the neighboring residues, we made the mutants βD268K and γK270D. When αL263C was expressed with the mutant βD268K and wild-type γ and δ, MTSEA application significantly decreased subsequent ACh-induced currents (data not shown). However, when αL263C was expressed with γK270D and wild-type β and δ, MTSEA application did not cause any effect (data not shown). MTSES application did not cause an effect in either case (data not shown). Sequential application of MTSES and then MTSEA for αL263C βD268K γδ showed that MTSES did not react silently, since MTSEA still induced its effect (data not shown). We infer that βD268K and γK270D are indeed facing αL263C. For the double mutant αL263CβD268Kγδ where both αL263C Cys face a positive charge (βD268K and γK268), the pKa of the αL263C Cys will be decreased, thus leading to an increase in deprotonation rate which in turn yields a higher reactivity (Britto et al., 2002). Indeed, coexpression of αL263C and βD268K yielded receptors that were modified by MTSEA with a second-order rate constant of 417 ± 42 m−1 s−1, which is similar to the fast component of the double exponential reaction with wild-type β-subunits. In turn, when αL263C Cys faces a negative charge at both sites (βD268 and γK268D) the pKa of the Cys will be increased, thus leading to a decrease in deprotonation rate which in turn yields a decreased reactivity, which is confirmed by the inability to modify αL263C in αL263CβγK268Dδ. With both αL263C facing a negative charge, the reactivity might have been rendered very slow and hard to detect. These mutations might help to explain the basis of the biexponential MTSEA reaction rates with αL263C.
In addition, we determined the rate constants in the presence of ACh (Fig. 5) for these 10 positions plus the one that only reacted in the presence of ACh, to investigate whether structural rearrangements occur at these positions during gating. At seven positions, the second-order rate constant in the presence of ACh was significantly different (Student's t test) compared with the rate in the absence of ACh. At two positions, αE262C and αP265C, the rate decreased in the presence of ACh, whereas at five positions the rate increased in the presence of ACh, αT267C, αS269C, αV271C, αP272C, and αI274C.
The timeline of conformational changes from ligand binding to channel gating in this region has also been analyzed by φ value analysis. These studies indicate that residues flanking the ligand-binding site move first, followed by Cys-loop and loop 2 positions, then the M2M3 loop and most of the attached M2 segment, and later by the more cytoplasmic part of M2 (Grosman et al., 2000a,b; Chakrapani et al., 2003, 2004; Mitra et al., 2005; Purohit et al., 2007). Previously, intersubunit disulfide cross-linking in the GABAA receptor demonstrated that the αM2 top is highly mobile in the closed and open state (Horenstein et al., 2001, 2005).
Homology modeling
To investigate the three homology models that we constructed (based on the Torpedo, ELIC, and GLIC structures), we plotted the 16 to Cys mutated positions color-coded by accessibility toward MTSEA on the different models (Fig. 6C,D). Two distinct patterns were observed. In the models based on both bacterial structures (ELIC and GLIC), positions of similar reactivity are distributed at random locations through the entire investigated 16 aa stretch; especially positions of high reactivity that had second-order reaction rate constants ≥10,000 m−1 s−1 (20′, 23′, 24′, and 27′) are found at random, disparate locations, some are at the end of the M2-helix, and most are buried in the M2M3 linker stretch at the interface of the transmembrane and extracellular domains, far away from the channel (Fig. 6C, right; D, right). Interestingly, when the same color-coding is applied on the model based on the eukaryotic Torpedo structure, a nonrandom, structured pattern can be visualized (Fig. 6C, left; D, left). Residues with fast reacting rates all line up on the face of the M2-helix that faces the ion-conducting channel, residues that react slowly are found on the back of this face, away from the channel, facing other transmembrane segments (M1, M3). In this model the M2M3 loop contains both positions of slow reactivity and nonreactivity, in agreement with this region being buried in the interdomain interface. Only in the model based on the Torpedo structure, can a reactivity pattern be observed that corresponds to structural features of the model. Therefore, our current data strongly favor the Torpedo structure as a template for the mouse nAChR.
To further evaluate our different homology models in the M2M3 region, several other published findings in diverse Cys- loop receptor family members are available. In a similar study of the GABAA receptor α1-subunit covering positions 19′ to 35′ using Cys accessibility, the initial observation was made that the α-helix of M2 extended for two more α-helical turns than predicted because the reaction rate pattern indicated a slow and a fast reacting face as well (Bera et al., 2002). This finding was later confirmed by the Torpedo structure and therefore also favors the Torpedo structure as a template for homology modeling.
In a SCAM study of the GlyR α1-subunit for the 19′ to 29′ segment, residues from 19′ to 24′ were accessible toward MTSET, whereas the more C-terminal positions were inaccessible. The overall spread of reaction rate constants was <10-fold and an α-helical pattern was not found. The MTSES modification rates differed by <100-fold and there was also no indication for an α-helical pattern (Lynch et al., 1995, 1997).
In the 5HT3A receptor, Cys at the M2 positions 26′ (A304) and 27′ (I305) could be cross-linked with 5HT3AK81C in loop 2 (β1-β2 loop) indicating their close proximity (Reeves et al., 2005). These positions are in close proximity in our models based on the Torpedo structure and also based on the ELIC structure, however, the separation distances between these residues increases in the GLIC structure.
Another study in the GABAA receptor investigated electrostatic interactions between extracellular loop 2 (GABAA α1D57, the residue adjacent to 5HT3AK81) and loop 7 (GABAA α1D149) and the 24′ M2 position (GABAA α1K279) (Kash et al., 2003). In all three homology models D149 is far away from K279, and also the distance between D57 and K279 is unfavorable for electrostatic/cross-linking interactions in either the model based on the Torpedo or the ELIC structure.
Others have found a network of charged interactions between similar positions in these loops (nACh α7) compared with more pairwise interactions (Sala et al., 2005), or no direct evidence for electrostatic interactions (Gly α1) (Absalom et al., 2003). The diverse results indicate that the overall fold and functionality of different Cys-loop receptor members are comparable in this and that there is agreement that loops 2 and 7 together with the M2M3 loop are determinants of coupling ligand binding to channel gating (Lynch et al., 1995, 1997; Rajendra et al., 1995; Campos-Caro et al., 1996; Schofield et al., 2003). However, the precise networks transmitting the gating conformational changes are not entirely identical, and the detailed structure in this region might differ between different family members.
Discussion
All 16 nAChR α-subunit Cys mutants formed functional ion channels when expressed as αβγδ receptors (Figs. 1, 2). The ACh EC50s of the Cys mutants of the prolines at the 23′ (αP265) and 30′ positions (αP272) were not significantly different from wild type. The 23′ proline is absolutely conserved in all and the 30′ proline is highly conserved in cationic Cys-loop receptor subunits. In the related 5HT3A receptor, replacement of the 30′ proline equivalent to ACh αP272 (5HT3AP308, TAIGTPLIGVY) by various natural amino acids yielded receptors that trafficked to the membrane and bound radioactive ligand, but were not functional. Based on unnatural amino acid mutagenesis using proline analogs the authors inferred that cis-trans isomerization at 5HT3AP308 is a molecular switch for channel opening (Lummis et al., 2005). The functionality of the nAChR-αP272C mutant indicates that cis-trans isomerization of a proline at the 30′ position is not a universal requirement for gating in cationic Cys-loop receptors as was recently also shown for the nAChR by mutant cycle analysis (Lee et al., 2008).
We determined the accessibility of the engineered Cys to the positively charged MTSEA. At all 10 positions where MTSEA application in the absence of ACh altered the subsequent ACh-induced current amplitude, we also observed an effect of MTSEA applied in the presence of ACh. One position, αI274C (30′), was only accessible in the presence but not in the absence of ACh. We infer that at these 11 positions MTSEA covalently attached an aminoethanesulfide moiety to the engineered Cys sulfur. Mutants unresponsive to MTSEA application were probed with the negatively charged MTSES. MTSES only induced a significant change in subsequent ACh-induced current amplitudes at one of these positions, αS268C, and sequential application of MTSEA and MTSES demonstrated that MTSEA did not modify this Cys, but MTSES did. This result indicates that the Cys is surface accessible but not reactive with the positively charged MTSEA. αS268 is predicted to face away from the channel and perhaps MTSEA cannot access αS268C, due to electrostatic repulsion at the site or in the access pathway to the Cys.
There are three possible explanations for the positions where MTSEA and MTSES application had no functional effect (αA270C, αL273C, αI274C, αK276C, αY277C): The MTS reagent reacted but has no functional effect, the MTS reagent did not react because of steric constraints and/or electrostatic repulsion along the access pathway to or at the Cys, or local factors prevent deprotonation of the Cys. It is unlikely that electrostatic repulsion prevented reaction by both a positively and a negatively charged reagent. Thus, steric factors are a more likely explanation of lack of reaction. All positions where MTS reagent application had no effect are in the M2M3 loop. In the nAChR structure, this loop is in contact with the β1-β2 and β8-β9 loops from the extracellular domain (Lee and Sine, 2005; Unwin, 2005; Lee et al., 2008). In several Cys-loop receptors, functional coupling between these protein regions has been demonstrated to have a role in transmitting the conformational change from ligand binding in the extracellular domain to channel opening in the transmembrane domain (Kash et al., 2003; Bouzat et al., 2004; Lee and Sine, 2005; Reeves et al., 2005; Unwin, 2005). We infer that this interdomain interface is tightly packed posing steric constraints for MTS reagent accessibility and that the absence of a functional effect is due to absence of reaction rather than silent reaction. We showed previously that a Cys in 5HT3 receptors (5HT3AA304 and 5HT3AI305) at the positions aligned with αS268 and αS269 could be disulfide linked with an engineered Cys in the β1-β2 loop (5HT3AK81), indicating the close interaction of the extracellular end of M2 and the β1-β2 loop (Reeves et al., 2005).
The second-order rate constants for MTSEA modification were determined in the absence and presence of ACh (Figs. 4, 5). In the absence of ACh the rate constants varied by 4 orders of magnitude: αI264C (16.3 ± 2.1 m−1 s−1) reacted slowest and αS269C (93,800 ± 9100 m−1 s−1) fastest. Based on the rate constants, the reacting Cys can be divided into two groups: Cys reacting with a second-order rate constant faster than 10,000 m−1 s−1 and those reacting slower than 1000 m−1 s−1. All four mutants that reacted with a fast rate were located in the first half of the 16 aa segment that we investigated. When these 8 residues (20′ to 27′) are plotted on an α-helical wheel, the fast reacting positions are on one face of the helix, whereas all slow reacting ones are on the backside (Fig. 6E,F). We infer that these eight residues have an α-helical secondary structure. A similar study in the GABAA receptor previously suggested that the α-helical portion of M2 extended for two more helical turns than indicated by hydropathy plots (Bera et al., 2002). The Torpedo nAChR structure subsequently confirmed that the α-helical portion of M2 extended for two helical turns above the membrane (Unwin, 2005). Two distinct methods, SCAM in nAChR and GABAAR and cryo-electron-microscopy, both agree that the M2 α-helix extends for two α-helical turns past 20′. However, the prokaryotic structures ELIC and GLIC do not show α-helical content past 21′ (Hilf and Dutzler, 2008, 2009; Bocquet et al., 2009). Our current experimental data therefore indicate that these structures are not necessarily good templates for the M2M3 loop region of the eukaryotic channel. We can infer regions that undergo conformational change during gating by comparing the reaction rate constants in the presence and absence of ACh. In the activated states the presence of agonist destabilizes the closed state, thus increasing the probability of the channel to open. High concentrations of agonist induce desensitization. The desensitized states have high agonist affinities but no measurable ion conductance. Because in the presence of ACh nAChR undergo transitions between open and desensitized states we cannot distinguish between reagent reacting in either of these states when MTSEA is applied in the presence of ACh. For simplicity, we assume that reaction of MTSEA in the absence of ACh occurs in the closed state, and that states with agonist bound, open and desensitized, are activated states. Gating refers to channel opening and closing events upon ligand binding. Although amine compounds as simple as tetramethylammonium act as agonists on nAChR (Zhang et al., 1995; Akk and Steinbach, 2003), in our study MTSEA application did not produce macroscopic currents in wild-type or mutant receptors.
Second-order rate constants of MTSEA modification determined in the presence of ACh (activated state) were significantly different from those in the absence of ACh at seven positions indicating conformational changes in the respective regions during gating. At positions 20′ and 23′ toward the extracellular end of M2 the rate decreases in the presence of ACh, whereas at positions 25′ to 29′ the rate constants increase in the presence of ACh. Of the eight M2M3 loop residues following M2, only three are accessible in the closed and four in the activated state. At three positions, the rate of reaction is significantly increased in the activated state, however, slow in both cases. The nonaccessibility of five of eight positions together with the slow rate constants indicate tight packing in closed and activated states.
The overall accessibility toward MTS reagents we observed in our study is in good agreement with previous experimental evidence suggesting water filled crevices around all transmembrane helices (Goren et al., 2004) that were consistent with loose packing observed in the electron-microscopy-derived Torpedo nAChR model (Unwin, 2005). In contrast, the recent ELIC crystal structure (Hilf and Dutzler, 2008) that supposedly also depicts the closed state shows a tightly packed transmembrane region (Fig. 6I). The computed water accessibility for most transmembrane residues in ELIC is significantly lower than for Torpedo nAChR (Fig. 6H,I). Several studies have investigated the importance of lipids, of an immobilized lipid layer around the receptor (Marsh and Barrantes, 1978) or of the lipid composition (Criado et al., 1982, 1984), on nAChR functionality. A variety of hydrophobic substances can render nAChR desensitized or nonfunctional (Bouzat and Barrantes, 1993, 1996; Blanton et al., 1999; Nievas et al., 2007). Interestingly, recent photoaffinity labeling showed differences in an M2M3 loop residue in reconstituted receptors compared with receptors in native membranes (Hamouda et al., 2008). Recently, it was shown that Torpedo nAChR are only stable and functional when purified with lipid analog detergents but not in nonlipid analog detergents (Asmar-Rovira et al., 2008). For the electron-microscopy studies of the Torpedo nAChR, the receptors were imbedded in their native lipid environment without detergent treatment (Miyazawa et al., 1999). In contrast, for the ELIC crystal structure, the protein was crystallized with a nonlipid-like detergent that might have destabilized the transmembrane domain leading to a non-native, collapsed conformation. Thus, our data suggest that the Torpedo nAChR structure is a better model for the metazoan Cys-loop receptor transmembrane domain in the closed state. Further experiments are necessary to validate different models for individual Cys-loop receptor members.
Footnotes
This work was supported in part by National Institutes of Health Grant K99NS059841 to M.J. We thank Myles Akabas, Moez Bali, Nicole McKinnon, Rishi Parikh, David Reeves, and Paul Riegelhaupt for helpful discussions and comments on this manuscript. We thank Ithiel Frame and Jarrett Linder for technical assistance.
References
- Absalom NL, Lewis TM, Kaplan W, Pierce KD, Schofield PR. Role of charged residues in coupling ligand binding and channel activation in the extracellular domain of the glycine receptor. J Biol Chem. 2003;278:50151–50157. doi: 10.1074/jbc.M305357200. [DOI] [PubMed] [Google Scholar]
- Akabas MH, Karlin A. Identification of acetylcholine receptor channel-lining residues in the M1 segment of the alpha-subunit. Biochemistry. 1995;34:12496–12500. doi: 10.1021/bi00039a002. [DOI] [PubMed] [Google Scholar]
- Akabas MH, Stauffer DA, Xu M, Karlin A. Acetylcholine receptor channel structure probed in cysteine-substitution mutants. Science. 1992;258:307–310. doi: 10.1126/science.1384130. [DOI] [PubMed] [Google Scholar]
- Akabas MH, Kaufmann C, Archdeacon P, Karlin A. Identification of acetylcholine receptor channel-lining residues in the entire M2 segment of the alpha subunit. Neuron. 1994;13:919–927. doi: 10.1016/0896-6273(94)90257-7. [DOI] [PubMed] [Google Scholar]
- Akk G, Steinbach JH. Activation and block of mouse muscle-type nicotinic receptors by tetraethylammonium. J Physiol. 2003;551:155–168. doi: 10.1113/jphysiol.2003.043885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmar-Rovira GA, Asseo-García AM, Quesada O, Hanson MA, Cheng A, Nogueras C, Lasalde-Dominicci JA, Stevens RC. Biophysical and ion channel functional characterization of the Torpedo californica nicotinic acetylcholine receptor in varying detergent-lipid environments. J Membr Biol. 2008;223:13–26. doi: 10.1007/s00232-008-9107-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera AK, Chatav M, Akabas MH. GABA(A) receptor M2–M3 loop secondary structure and changes in accessibility during channel gating. J Biol Chem. 2002;277:43002–43010. doi: 10.1074/jbc.M206321200. [DOI] [PubMed] [Google Scholar]
- Blanton MP, Cohen JB. Identifying the lipid-protein interface of the Torpedo nicotinic acetylcholine receptor: secondary structure implications. Biochemistry. 1994;33:2859–2872. doi: 10.1021/bi00176a016. [DOI] [PubMed] [Google Scholar]
- Blanton MP, Xie Y, Dangott LJ, Cohen JB. The steroid promegestone is a noncompetitive antagonist of the Torpedo nicotinic acetylcholine receptor that interacts with the lipid-protein interface. Mol Pharmacol. 1999;55:269–278. doi: 10.1124/mol.55.2.269. [DOI] [PubMed] [Google Scholar]
- Bocquet N, Nury H, Baaden M, Le Poupon C, Changeux JP, Delarue M, Corringer PJ. X-ray structure of a pentameric ligand-gated ion channel in an apparently open conformation. Nature. 2009;457:111–114. doi: 10.1038/nature07462. [DOI] [PubMed] [Google Scholar]
- Bouzat C, Barrantes FJ. Modulation of muscle nicotinic acetylcholine receptors by the glucocorticoid hydrocortisone. Possible allosteric mechanism of channel blockade. J Biol Chem. 1996;271:25835–25841. doi: 10.1074/jbc.271.42.25835. [DOI] [PubMed] [Google Scholar]
- Bouzat C, Gumilar F, Spitzmaul G, Wang HL, Rayes D, Hansen SB, Taylor P, Sine SM. Coupling of agonist binding to channel gating in an ACh-binding protein linked to an ion channel. Nature. 2004;430:896–900. doi: 10.1038/nature02753. [DOI] [PubMed] [Google Scholar]
- Bouzat CB, Barrantes FJ. Effects of long-chain fatty acids on the channel activity of the nicotinic acetylcholine receptor. Receptors Channels. 1993;1:251–258. [PubMed] [Google Scholar]
- Brejc K, van Dijk WJ, Klaassen RV, Schuurmans M, van Der Oost J, Smit AB, Sixma TK. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. doi: 10.1038/35077011. [DOI] [PubMed] [Google Scholar]
- Britto PJ, Knipling L, Wolff J. The local electrostatic environment determines cysteine reactivity of tubulin. J Biol Chem. 2002;277:29018–29027. doi: 10.1074/jbc.M204263200. [DOI] [PubMed] [Google Scholar]
- Campos-Caro A, Sala S, Ballesta JJ, Vicente-Agulló F, Criado M, Sala F. A single residue in the M2–M3 loop is a major determinant of coupling between binding and gating in neuronal nicotinic receptors. Proc Natl Acad Sci U S A. 1996;93:6118–6123. doi: 10.1073/pnas.93.12.6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celie PH, van Rossum-Fikkert SE, van Dijk WJ, Brejc K, Smit AB, Sixma TK. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. doi: 10.1016/s0896-6273(04)00115-1. [DOI] [PubMed] [Google Scholar]
- Chakrapani S, Bailey TD, Auerbach A. The role of loop 5 in acetylcholine receptor channel gating. J Gen Physiol. 2003;122:521–539. doi: 10.1085/jgp.200308885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrapani S, Bailey TD, Auerbach A. Gating dynamics of the acetylcholine receptor extracellular domain. J Gen Physiol. 2004;123:341–356. doi: 10.1085/jgp.200309004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Criado M, Eibl H, Barrantes FJ. Effects of lipids on acetylcholine receptor. Essential need of cholesterol for maintenance of agonist-induced state transitions in lipid vesicles. Biochemistry. 1982;21:3622–3629. doi: 10.1021/bi00258a015. [DOI] [PubMed] [Google Scholar]
- Criado M, Eibl H, Barrantes FJ. Functional properties of the acetylcholine receptor incorporated in model lipid membranes. Differential effects of chain length and head group of phospholipids on receptor affinity states and receptor-mediated ion translocation. J Biol Chem. 1984;259:9188–9198. [PubMed] [Google Scholar]
- Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR alpha1 bound to alpha-bungarotoxin at 1.94 A resolution. Nat Neurosci. 2007;10:953–962. doi: 10.1038/nn1942. [DOI] [PubMed] [Google Scholar]
- Dutertre S, Lewis RJ. Toxin insights into nicotinic acetylcholine receptors. Biochem Pharmacol. 2006;72:661–670. doi: 10.1016/j.bcp.2006.03.027. [DOI] [PubMed] [Google Scholar]
- Goren EN, Reeves DC, Akabas MH. Loose protein packing around the extracellular half of the GABAA receptor {beta}1 subunit M2 channel-lining segment. J Biol Chem. 2004;279:11198–11205. doi: 10.1074/jbc.M314050200. [DOI] [PubMed] [Google Scholar]
- Grosman C, Zhou M, Auerbach A. Mapping the conformational wave of acetylcholine receptor channel gating. Nature. 2000a;403:773–776. doi: 10.1038/35001586. [DOI] [PubMed] [Google Scholar]
- Grosman C, Salamone FN, Sine SM, Auerbach A. The extracellular linker of muscle acetylcholine receptor channels is a gating control element. J Gen Physiol. 2000b;116:327–340. doi: 10.1085/jgp.116.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guzmán GR, Ortiz-Acevedo A, Ricardo A, Rojas LV, Lasalde-Dominicci JA. The polarity of lipid-exposed residues contributes to the functional differences between Torpedo and muscle-type nicotinic receptors. J Membr Biol. 2006;214:131–138. doi: 10.1007/s00232-006-0051-0. [DOI] [PubMed] [Google Scholar]
- Hamouda AK, Chiara DC, Blanton MP, Cohen JB. Probing the structure of the affinity-purified and lipid-reconstituted Torpedo nicotinic acetylcholine receptor. Biochemistry. 2008 doi: 10.1021/bi801476j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. X-ray structure of a prokaryotic pentameric ligand-gated ion channel. Nature. 2008;452:375–379. doi: 10.1038/nature06717. [DOI] [PubMed] [Google Scholar]
- Hilf RJ, Dutzler R. Structure of a potentially open state of a proton-activated pentameric ligand-gated ion channel. Nature. 2009;457:115–118. doi: 10.1038/nature07461. [DOI] [PubMed] [Google Scholar]
- Horenstein J, Wagner DA, Czajkowski C, Akabas MH. Protein mobility and GABA-induced conformational changes in GABAA receptor pore-lining M2 segment. Nat Neurosci. 2001;4:477–485. doi: 10.1038/87425. [DOI] [PubMed] [Google Scholar]
- Horenstein J, Riegelhaupt P, Akabas MH. Differential protein mobility of the gamma-aminobutyric acid, type A, receptor alpha and beta subunit channel-lining segments. J Biol Chem. 2005;280:1573–1581. doi: 10.1074/jbc.M410881200. [DOI] [PubMed] [Google Scholar]
- Imoto K, Busch C, Sakmann B, Mishina M, Konno T, Nakai J, Bujo H, Mori Y, Fukuda K, Numa S. Rings of negatively charged amino acids determine the acetylcholine receptor channel conductance. Nature. 1988;335:645–648. doi: 10.1038/335645a0. [DOI] [PubMed] [Google Scholar]
- Jansen M, Akabas MH. State-dependent cross-linking of the M2 and M3 segments: functional basis for the alignment of GABAA and acetylcholine receptor M3 segments. J Neurosci. 2006;26:4492–4499. doi: 10.1523/JNEUROSCI.0224-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Bali M, Akabas MH. Modular design of Cys-loop ligand-gated ion channels: functional 5-HT3 and GABA rho1 receptors lacking the large cytoplasmic M3M4 loop. J Gen Physiol. 2008;131:137–146. doi: 10.1085/jgp.200709896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlin A, Akabas MH. Substituted-cysteine accessibility method. Methods Enzymol. 1998;293:123–145. doi: 10.1016/s0076-6879(98)93011-7. [DOI] [PubMed] [Google Scholar]
- Kash TL, Jenkins A, Kelley JC, Trudell JR, Harrison NL. Coupling of agonist binding to channel gating in the GABA(A) receptor. Nature. 2003;421:272–275. doi: 10.1038/nature01280. [DOI] [PubMed] [Google Scholar]
- Lee WY, Sine SM. Principal pathway coupling agonist binding to channel gating in nicotinic receptors. Nature. 2005;438:243–247. doi: 10.1038/nature04156. [DOI] [PubMed] [Google Scholar]
- Lee WY, Free CR, Sine SM. Nicotinic receptor interloop proline anchors beta1-beta2 and cys loops in coupling agonist binding to channel gating. J Gen Physiol. 2008;132:265–278. doi: 10.1085/jgp.200810014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lummis SC, Beene DL, Lee LW, Lester HA, Broadhurst RW, Dougherty DA. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- Lynch JW, Rajendra S, Barry PH, Schofield PR. Mutations affecting the glycine receptor agonist transduction mechanism convert the competitive antagonist, picrotoxin, into an allosteric potentiator. J Biol Chem. 1995;270:13799–13806. doi: 10.1074/jbc.270.23.13799. [DOI] [PubMed] [Google Scholar]
- Lynch JW, Rajendra S, Pierce KD, Handford CA, Barry PH, Schofield PR. Identification of intracellular and extracellular domains mediating signal transduction in the inhibitory glycine receptor chloride channel. EMBO J. 1997;16:110–120. doi: 10.1093/emboj/16.1.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsh D, Barrantes FJ. Immobilized lipid in acetylcholine receptor-rich membranes from Torpedo marmorata. Proc Natl Acad Sci U S A. 1978;75:4329–4333. doi: 10.1073/pnas.75.9.4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. Genetic manipulation of ion channels: a new approach to structure and mechanism. Neuron. 1989;2:1195–1205. doi: 10.1016/0896-6273(89)90304-8. [DOI] [PubMed] [Google Scholar]
- Mitra A, Cymes GD, Auerbach A. Dynamics of the acetylcholine receptor pore at the gating transition state. Proc Natl Acad Sci U S A. 2005;102:15069–15074. doi: 10.1073/pnas.0505090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazawa A, Fujiyoshi Y, Stowell M, Unwin N. Nicotinic acetylcholine receptor at 4.6 A resolution: transverse tunnels in the channel wall. J Mol Biol. 1999;288:765–786. doi: 10.1006/jmbi.1999.2721. [DOI] [PubMed] [Google Scholar]
- Mukhtasimova N, Free C, Sine SM. Initial coupling of binding to gating mediated by conserved residues in the muscle nicotinic receptor. J Gen Physiol. 2005;126:23–39. doi: 10.1085/jgp.200509283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nievas GA, Barrantes FJ, Antollini SS. Conformation-sensitive steroid and fatty acid sites in the transmembrane domain of the nicotinic acetylcholine receptor. Biochemistry. 2007;46:3503–3512. doi: 10.1021/bi061388z. [DOI] [PubMed] [Google Scholar]
- Pascual JM, Karlin A. State-dependent accessibility and electrostatic potential in the channel of the acetylcholine receptor. Inferences from rates of reaction of thiosulfonates with substituted cysteines in the M2 segment of the alpha subunit. J Gen Physiol. 1998;111:717–739. doi: 10.1085/jgp.111.6.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit P, Mitra A, Auerbach A. A stepwise mechanism for acetylcholine receptor channel gating. Nature. 2007;446:930–933. doi: 10.1038/nature05721. [DOI] [PubMed] [Google Scholar]
- Rajendra S, Lynch JW, Pierce KD, French CR, Barry PH, Schofield PR. Mutation of an arginine residue in the human glycine receptor transforms beta-alanine and taurine from agonists into competitive antagonists. Neuron. 1995;14:169–175. doi: 10.1016/0896-6273(95)90251-1. [DOI] [PubMed] [Google Scholar]
- Reeves DC, Jansen M, Bali M, Lemster T, Akabas MH. A role for the β1β2 loop in the gating of 5-HT3 receptors. J Neurosci. 2005;25:9358–9366. doi: 10.1523/JNEUROSCI.1045-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DD, Lewis SD, Ballou DP, Olson ST, Shafer JA. Reactivity of small thiolate anions and cysteine-25 in papain towards methylmethanethiosulfonate. Biochemistry. 1986;25:5595–5601. doi: 10.1021/bi00367a038. [DOI] [PubMed] [Google Scholar]
- Sala F, Mulet J, Sala S, Gerber S, Criado M. Charged amino acids of the N-terminal domain are involved in coupling binding and gating in alpha7 nicotinic receptors. J Biol Chem. 2005;280:6642–6647. doi: 10.1074/jbc.M411048200. [DOI] [PubMed] [Google Scholar]
- Schofield CM, Jenkins A, Harrison NL. A highly conserved aspartic acid residue in the signature disulfide loop of the alpha 1 subunit is a determinant of gating in the glycine receptor. J Biol Chem. 2003;278:34079–34083. doi: 10.1074/jbc.M302416200. [DOI] [PubMed] [Google Scholar]
- Unwin N. Refined structure of the nicotinic acetylcholine receptor at 4A resolution. J Mol Biol. 2005;346:967–989. doi: 10.1016/j.jmb.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Williams DB, Akabas MH. gamma-Aminobutyric acid increases the water accessibility of M3 membrane-spanning segment residues in gamma-aminobutyric acid type A receptors. Biophys J. 1999;77:2563–2574. doi: 10.1016/s0006-3495(99)77091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen J, Auerbach A. Activation of recombinant mouse acetylcholine receptors by acetylcholine, carbamylcholine and tetramethylammonium. J Physiol. 1995;486:189–206. doi: 10.1113/jphysiol.1995.sp020802. [DOI] [PMC free article] [PubMed] [Google Scholar]