Abstract
The study of induced pluripotency is complicated by the need for infection with high-titer retroviral vectors, which results in genetically heterogeneous cell populations. We generated genetically homogeneous ‘secondary’ somatic cells that carry the reprogramming factors as defined doxycycline (dox)-inducible transgenes. These cells were produced by infecting fibroblasts with dox-inducible lentiviruses, reprogramming by dox addition, selecting induced pluripotent stem cells and producing chimeric mice. Cells derived from these chimeras reprogram upon dox exposure without the need for viral infection with efficiencies 25- to 50-fold greater than those observed using direct infection and drug selection for pluripotency marker reactivation. We demonstrate that (i) various induction levels of the reprogramming factors can induce pluripotency, (ii) the duration of transgene activity directly correlates with reprogramming efficiency, (iii) cells from many somatic tissues can be reprogrammed and (iv) different cell types require different induction levels. This system facilitates the characterization of reprogramming and provides a tool for genetic or chemical screens to enhance reprogramming.
It has recently been shown that mouse1–4 and human5–8 fibroblasts can be reprogrammed to a pluripotent state through retroviral introduction of four transcription factors such as Oct4, Sox2, Klf4 and c-Myc. Reprogramming can also be achieved in the absence of the tumorigenic factor c-Myc, although with decreased efficiency9,10. Nevertheless, with these approaches only a very small fraction of cells infected with all four factors will eventually reprogram11. The random viral infection results in genetic heterogeneity in the infected cell culture that likely contributes to the low observed frequency of induced pluripotent stem (iPS) cell formation. Therefore, reprogrammed cells must be selected for by the reactivation of endogenous pluripotency genes1–3 or based on morphological criteria11,12. The reprogramming process has been shown to require ~10 to 12 d of sustained transgene expression after viral transduction and to follow a sequential activation of pluripotency markers, with initial activation of alkaline phosphatase and stage-specific embryonic antigen (SSEA1) followed by reactivation of the endogenous Oct4 (Pou5f1) and Nanog genes, after which the cultures are able to sustain the pluripotent state in the absence of transgene activity13,14.
The cellular and genetic heterogeneity of randomly infected fibroblasts complicates the exploration of important molecular events occurring during reprogramming and limits the scalability required for high-throughput analyses. To overcome these problems, we developed a system to generate genetically identical cell populations amenable to reprogramming without any further genetic interference. To this end, primary fibroblasts were infected with dox-inducible lentiviruses encoding the four reprogramming factors Oct4, Sox2, Klf4 and c-Myc. After blastocyst injection, chimeric mice were generated consisting of tissue types clonally derived from reprogrammed fibroblasts. From these mice we derived homogeneous donor cell populations harboring preselected vector integrations permissible for reprogramming, allowing for robust and simple dox-induced reprogramming of primary cell types without the need for direct viral transduction of the reprogramming factors. This technology facilitates the generation of large numbers of genetically identical donor cells and should be advantageous for genetic or chemical screening to improve reprogramming. In addition, the same approach could be used to screen for small molecules that replace each of the four factors by genetic deletion of one particular factor in the pluripotent, reprogrammed fibroblasts15. Furthermore, this tool is not limited to fibroblast cultures but can in principle be similarly applied to all other somatic cell types, providing an attractive way to induce genes in cell types that are difficult to infect with retroviruses, such as lymphocytes or intestinal epithelial cells.
RESULTS
Genetically identical cells to induce reprogramming
To generate cell populations homogeneous with respect to the number and location of proviral integrations, we used a dox-inducible transgene system16,17 and constructed dox-inducible lentiviral vectors encoding the four reprogramming factors. Mouse embryonic fibroblasts (MEFs) containing both a reverse tetracycline transactivator and a phosphoglycerate kinase (Pgk1) promoter–driven puromycin resistance gene targeted to the ROSA26 locus (ROSA26-M2rtTA) in addition to a green fluorescent protein gene (GFP) targeted to the endogenous Nanog locus (NGFP) were infected with the four lentiviruses. Similarly, we infected ROSA26-M2rtTA MEFs harboring the Oct4 cDNA under control of the tetracycline operator targeted to the type I collagen locus16 and a neomycin (neo) resistance gene in the endogenous Nanog locus1,18 (NNeo) with dox-inducible lentiviruses encoding Klf4, Sox2 and c-Myc (Fig. 1a).
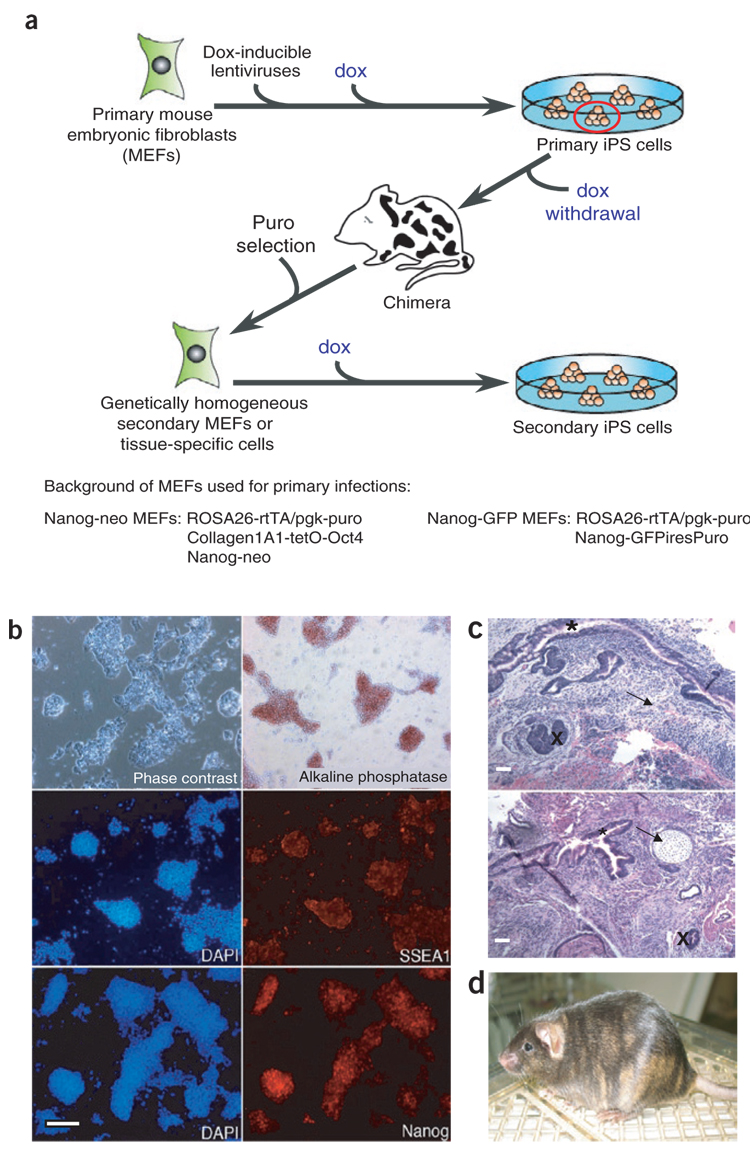
Figure 1. Generation of genetically homogeneous cell cultures for epigenetic reprogramming.
(a) Scheme for infection of puromycin-resistant, Nanog-GFP or Nanog-neo primary MEFs expressing the reverse tetracycline transactivator (M2rtTA) with dox-inducible lentiviruses encoding the four reprogramming factors followed by induction of reprogramming, primary iPS cell colony selection, dox withdrawal, chimera formation and puromycin selection for iPS cell–derived secondary somatic cells. (b) NNeo secondary MEFs isolated from chimeras undergo complete epigenetic reprogramming. Dox-independent cultures express the pluripotency-associated genes alkaline phosphatase, SSEA1 and Nanog. (c,d) MEF-derived NNeo and NGFP2 secondary iPS cells generate cells of all three germ layers in teratomas (c; arrow, mesoderm; asterisk, endoderm; cross, ectoderm) produced in teratoma formation assays and contribute to chimera formation when injected into blastocysts, as indicated by the presence of iPS cell–derived agouti coat color on a black background (d). Scale bars, 250 µm.
After viral transduction, dox was added to the culture medium to activate the transgenes and initiate the reprogramming process. As expected, Nanog-GFP+ and Nanog-neo–resistant iPS cell colonies appeared, and clonal iPS cell lines were established. All iPS cell lines could be expanded in the absence of dox, exhibited alkaline phosphatase activity and homogeneously expressed the pluripotency markers SSEA1 and Nanog (data not shown). This indicates that these ‘primary’ iPS cell lines had activated their endogenous pluripotency core transcriptional network and no longer relied upon exogenous expression of the four reprogramming factors19. To generate somatic tissues that were composed of genetically homogeneous cells carrying identical proviral insertions known to achieve reprogramming in primary fibroblasts, we injected several of these clonal primary iPS cell lines into blastocysts. The resulting dox-inducible iPS cell chimeras were allowed to gestate until E13.5, at which point MEFs were isolated. Puromycin selection was then used to select against cells derived from the host blastocyst, leaving only iPS cell–derived cells. We will refer to such cells as ‘secondary’ MEFs as they are derived from the primary iPS cells and thus carry a specific set of proviral insertions that is able to reprogram somatic cells (Fig. 1a).
Secondary MEFs were isolated from chimeric iPS cell embryos generated from three distinct, clonal primary iPS cell lines (one Nanog-neo and two Nanog-GFP lines) and were cultured in the presence of dox to determine whether the integrated lentiviral vectors retained competence to mediate epigenetic reprogramming after differentiation in the developing embryo. The addition of dox to these cultures initiated dramatic morphological changes, and ‘secondary’ iPS cell lines were efficiently isolated from these cultures by neo selection or GFP expression and subsequently propagated in the absence of dox. Immunofluorescence demonstrated that secondary iPS cells had reactivated the embryonic stem (ES) cell pluripotency markers alkaline phosphatase, SSEA1 and the endogenous Nanog gene (Fig. 1b and Supplementary Fig. 1 online). The pluripotency of these cell lines was confirmed by their ability to form cells of endodermal, ectodermal and mesodermal lineages in teratoma formation assays and by their ability to contribute to adult chimeric mice upon blastocyst injection (Fig. 1c,d).
Reprogramming kinetics and transgene induction
Although secondary MEFs derived from all three dox-inducible iPS cell lines underwent reprogramming to form secondary iPS cell lines, we noticed differences with respect to their morphological changes and proliferation rates after dox treatment. Initially, MEFs from both Nanog-GFP lines proliferated to form a confluent fibroblastic monolayer after exposure to dox. The cells from Nanog-GFP line 3 (NGFP3) then underwent robust post-confluent proliferation, including growth of cells in suspension, whereas cells from Nanog-GFP line 2 (NGFP2) grew slower, forming discreet, alkaline phosphatase–positive, ES cell–like colonies upon the fibroblastic monolayer (Fig. 2a). The fibroblasts derived from the Nanog-neo line never formed a confluent monolayer upon dox addition, but generated large, three-dimensional colonies. After 12 d of dox administration, iPS cell colonies with ES-cell morphology were readily visible in all three cultures (Fig. 2a, arrows).
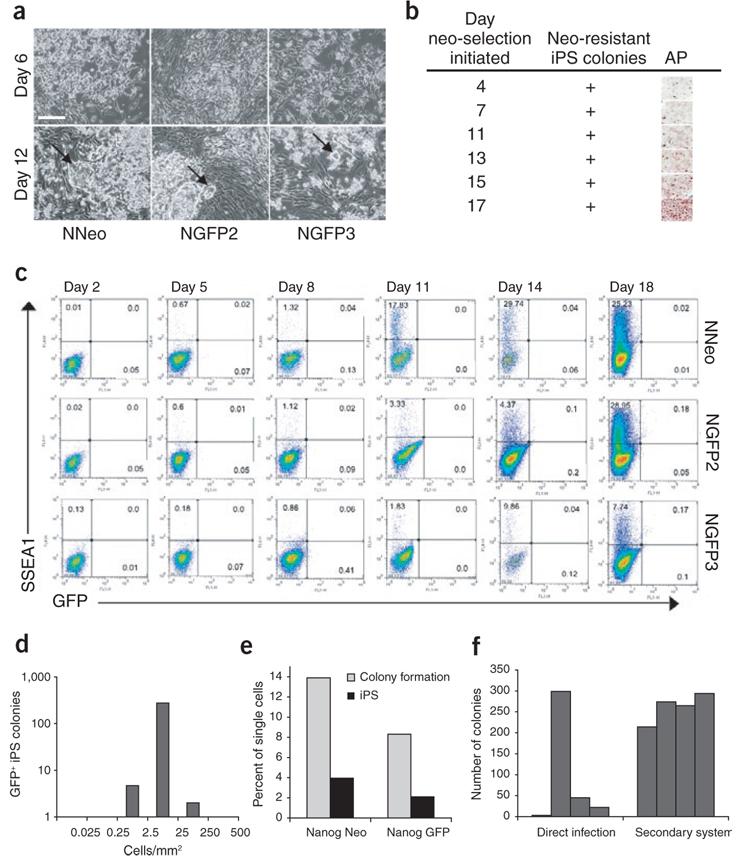
Figure 2. Reprogramming kinetics and efficiencies vary between MEFs from distinct iPS cell lines.
(a) Secondary MEFs from three ‘primary’ iPS cell lines were treated with dox, and reprogramming was monitored visually. The different MEF populations exhibited morphologic differences 6 d after dox administration, but all formed colonies with ES-cell morphology within 12 d (arrows). Scale bar, 250 µm. (b) Neo-resistant and alkaline phosphate–positive colonies were present in NNeo cultures when the drug was added to the media as early as day 4 after dox induction. (c) Flow cytometric analysis for reactivation of SSEA1 and the Nanog-GFP reporter allele (in NGFP2 and NGFP3 lines) over 18 d of dox culture. (d) Secondary NGFP2 MEFs were plated at densities varying from 0.025–500 cells/mm2 followed by dox addition. GFP+ colonies were counted 4 weeks later. (e) Single secondary MEFs were plated in 96-well plates containing a γ-irradiated MEF feeder layer followed by dox induction. The percentage of single cells able to proliferate sufficiently to form a visible colony on the MEF feeder layer (light gray bars) and the percentage of single cells able to form GFP+ or neo-resistant secondary iPS cell colonies (dark gray bars) were scored 4 weeks later. (f) Comparison of the interexperimental variability in iPS cell colony formation efficiency between direct infection and the secondary system. 3 × 105 Oct4-neo MEFs1 derived from a single embryo were infected with the four factors encoded by Moloney-based retroviral vectors on a 10-cm plate, neo selection was initiated on day 6 and resistant colonies were counted on day 20 (left, direct infection). 3 × 104 secondary NGFP2 MEFs derived from one chimeric embryo were plated in a six-well dish, exposed to dox-containing media and GFP+ colonies were counted 3 weeks later (right, secondary system). The bars represent number of colonies in each of the four independent experiments.
To evaluate the reprogramming kinetics in more detail, we cultured MEFs from the three lines in dox-containing media and used neo resistance to monitor the reactivation of the Nanog-neo reporter gene and flow cytometric analysis to monitor the reactivation of SSEA1 and the Nanog-GFP reporter gene (Fig. 2b,c). All three secondary MEFs exhibited a gradual increase of SSEA1+ cells over the time course, but some differences in timing were observed (Fig. 2c). The NNeo MEFs showed the earliest increase of SSEA1+ cells from 1.3% to 17.8% between days 8 and 11. The NGFP2 MEFs showed a similar increase but at a much later time point (from 4.4% to 29% between days 14 and 18). Likewise, MEFs from the iPS cell line NGFP3 exhibited a slower, gradual activation of SSEA1, reaching about 10% on day 14. The first GFP+ cells were detected as early as day 14 in NGFP2 and on day 18 in NGFP3 MEFs.
To monitor the timing of reactivation of the endogenous Nanog locus in NNeo secondary MEFs, we plated cells and began drug selection at various time points after dox treatment. In contrast to activation of the Nanog-GFP reporter gene ~2 weeks after induction, NNeo MEFs were neo resistant when neo was added to the cultures as early as day 4 (Fig. 2b). This might reflect a faster reactivation of the Nanog locus similar to what we observed for SSEA1 expression in this line (Fig. 2c). Alternatively, neo-resistant colonies may appear earlier because a low level of Nanog gene activation is sufficient to give drug resistance, in contrast to GFP detection, which necessitates higher expression14,20. Although the generation of secondary cells selects for a specific set of proviral integrations, the expression of which is able to induce the formation of iPS cells, the overall kinetics of pluripotency marker activation were similar to that seen in direct infection of MEFs1,13,14. This supports the notion that the reprogramming process requires a series of sequential epigenetic changes11,20.
Next we compared the reprogramming efficiencies of the various secondary MEFs. To determine the optimal plating density, we plated secondary NGFP2 MEFs at densities ranging from 0.025–500 cells/mm2 in dox-containing media and counted GFP+ colonies 4 weeks later. The plating density had a profound effect on iPS cell formation (Fig. 2d). Remarkably, both low and high plating densities completely inhibited GFP+ colony formation. We speculate that paracrine factors might initially be required to facilitate growth, and essential cell proliferation is impeded if cells are contact inhibited before activation of the transgenes.
To stringently determine the reprogramming efficiency in the secondary system, we plated single fibroblasts from the NNeo and NGFP2 lines into 96-well plates containing γ-irradiated MEFs as feeder cells to provide optimal growth support. We observed that only ~14% and ~8% of the seeded cells from the NNeo and NGFP2 MEFs, respectively, had proliferated sufficiently to form distinct colonies after dox administration (light gray bars in Fig. 2e). However, approximately one-quarter of those colonies eventually became neo resistant or GFP+ after 4 weeks in culture, resulting in an overall reprogramming efficiency of ~4% for the NNeo line and ~2% for the NGFP2 line (dark gray bars in Fig. 2e). This is 25–50 times more efficient than what was originally reported for drug resistance–based iPS cell selection1,12 and between 4–8 times more efficient than morphology- based iPS cell selection in cultures of primary infected fibroblasts11.
We next compared the reproducibility of the secondary MEF system with that of direct infections. We infected Oct4-neo MEFs1 with Moloney-based viruses encoding the four reprogramming factors and counted neo-resistant colonies on day 20. Four independent experiments revealed a high degree of interexperimental variability of iPS cell formation using this method (Fig. 2f). In contrast, we noticed a much smaller degree of variability in the secondary system when we counted Nanog-GFP+ colonies from dox-treated NGFP2 MEFs derived from one chimeric embryo in four independent experiments. Although the MEFs for this experiment were derived from the same chimeric embryo, we expect a similar reproducibility between MEFs derived from the same iPS cell line but different chimeras because all these cells would be genetically identical.
To correlate the phenotypic behavior of the three secondary MEF populations with transgene induction, we plated equal numbers of secondary MEFs in the presence or absence of dox for 72 h, at which point the total transcript levels of the four factors were determined by quantitative real-time PCR (qRT-PCR). Surprisingly, both Nanog-GFP lines induced Oct4 at much lower levels than the NNeo line, which expressed Oct4 from the transgene in the collagen 1A1 locus at levels similar to those of ES cells (Fig. 3a). Conversely, Sox2 induction in the Nanog-GFP lines reached levels much closer to those of endogenous Sox2 in ES cells, whereas NNeo expressed Sox2 at substantially lower levels in response to dox. c-Myc (Myc) expression was higher in uninduced MEFs compared with ES cells, and the addition of dox resulted in a dramatic induction of transcript levels in all three secondary MEF lines. In contrast, total Klf4 levels were similar to those in ES cells in all three secondary MEF populations after transgene induction. The observation that total Oct4 levels in dox-treated NNeo secondary MEFs were closest to those of ES cells might explain the faster and more efficient reprogramming kinetics observed in this line (see above). We then determined the expression levels at later stages of reprogramming in NGFP2 MEFs. Sox2, Klf4 and c-Myc were always robustly induced with only little variation, whereas Oct4 expression slowly increased over time (Supplementary Fig. 2a online). This might reflect the selection of cells with higher Oct4 induction over time in culture. Southern blot analysis indicated the genomic integration of one to two c-Myc, one to three Oct4, one to three Sox2, and three to four Klf4 proviruses in the three lines studied (Supplementary Fig. 2b).
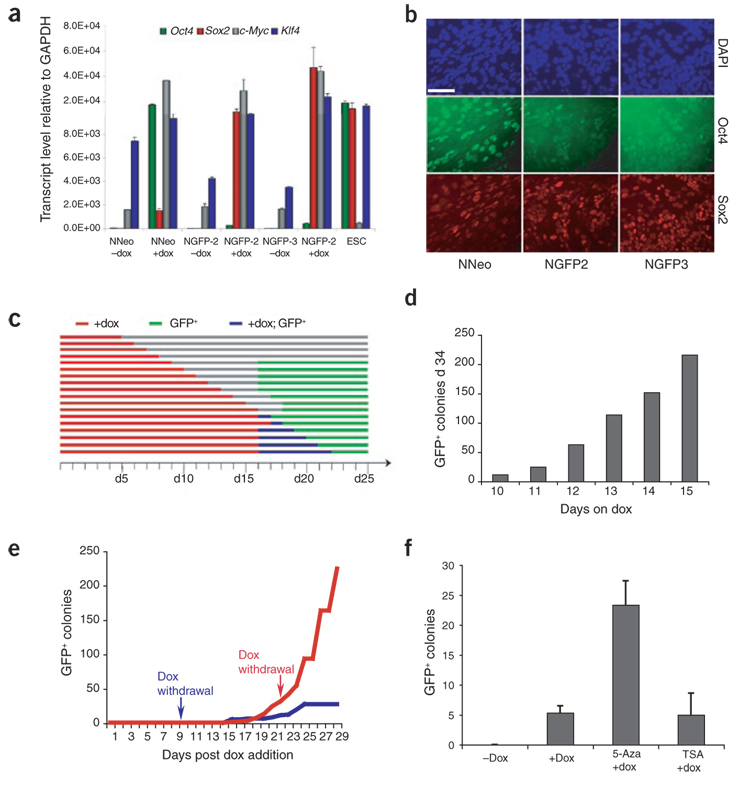
Figure 3. Requirement and expression of four-factor transgenes in secondary MEFs.
(a) Quantitative RT-PCR examining induction of expression of the four reprogramming factors in response to 72 h of dox treatment, relative to GAPDH levels. Error bars represent s.d. (n = 3). (b) Immunofluorescence detecting Oct4 and Sox2 in secondary MEF cultures 72 h after dox induction. Scale bar, 50 µm. (c) NGFP2 secondary MEFs were cultured in the presence of dox for the indicated time (5–22 d, red bars) followed by dox withdrawal. Cultures were monitored daily for the first instance of GFP activation (green bars). Blue bars indicate periods in which GFP+ colonies appeared during dox treatment. (d) NGFP2 MEFs were cultured in the presence of dox for 10–15 d, at which point dox was withdrawn; GFP+ colonies were scored at day 34. (e) NGFP2 MEFs were cultured in the presence of dox for either 9 (blue) or 22 d (red line), and the appearance of GFP+ colonies was scored daily until day 29. Note the appearance of GFP+ colonies as late as 15 d after dox withdrawal (blue line). (f) NGFP3 secondary MEFs were cultured in the presence or absence of dox, dox + 5-Aza, or dox + TSA, and GFP+ colonies were scored 3 weeks later. Error bars represent s.d. (n = 3).
Despite their genetic homogeneity, dox induction resulted in activation of the transgenes that varied at the single-cell level, as determined by immunofluorescence analysis of Oct4 and Sox2 (Fig. 3b). Because not all secondary MEFs induced the transgenes equally in response to dox, we cannot rule out the possibility that a specific stoichiometry of transgene expression is required for reprogramming and occurs in only a subset of the secondary MEFs.
Requirements for transgene expression
To investigate how long the four reprogramming factors must be expressed for stable reprogramming to occur, we plated secondary NGFP2 MEFs at optimal density (see above), exposed them to dox for various periods of time ranging from 5 to 22 d and monitored them daily for GFP fluorescence. The minimum length of dox exposure resulting in GFP+ colonies was 9 d, with the first GFP+ colonies appearing 7 d after dox removal at day 16 (Fig. 3c). Notably, additional exposure to dox did not accelerate the appearance of GFP+ colonies, with GFP appearing between days 16 and 18 regardless of the length of dox administration. Similarly, NNeo secondary MEFs were found to require 11–13 d of dox exposure before stable, neo-resistant secondary iPS cell colonies could be established.
To correlate the duration of transgene expression with overall reprogramming efficiency, we exposed secondary NGFP2 MEFs to dox for 10–15 d and quantified GFP+ colonies on day 34. We found a striking correlation between the length of transgene expression and the number of GFP+ colonies14 (Fig. 3d). We then monitored the appearance of new GFP+ colonies over time in the same dish. Surprisingly, MEFs that were exposed to dox for only 9 d continued to generate GFP+ colonies up to day 25 (15 d after dox withdrawal) (Fig. 3e, blue line). Twenty-two days of dox treatment yielded a much more pronounced increase in GFP+ colony formation over time (Fig. 3e, red line). These findings are consistent with reprogramming being a gradual stochastic process even in this genetically homogeneous system and are in agreement with previous conclusions based upon primary infections11,13,14,20. Furthermore, the reprogramming process continues and can be completed long after the four transgenes have been downregulated in response to dox withdrawal.
We also tested whether the secondary cells could be used to assess the effect of drugs on the efficiency of reprogramming. For this we explored the effects of the DNA demethylating compound 5-Aza-deoxycytidine (5-Aza) and the histone deacetylase inhibitor trichostatin A (TSA). Because of their action on chromatin modifications, both of these small molecules are candidates for improving reprogramming efficiency. Addition of 5-Aza to the medium increased the reprogramming efficiency of MEFs from the NGFP3 line, whereas TSA treatment had no obvious effect on the number of colonies (Fig. 3f).
Reprogramming of other cell types
We sought to determine the range of tissue types amenable to reprogramming by isolating secondary cells from 3–4 month old iPS-cell chimeras generated from the NNeo and NGFP2 lines and examining the reprogramming ability of multiple cell types derived from these chimeras. Some cell types could readily be reprogrammed when isolated from the NGFP2 line, but the same cell types isolated from the NNeo line did not yield iPS cells, suggesting that different cell types require different transgene induction levels, which may result from the different proviral integration sites between the lines studied (Table 1).
Table 1.
Summary of secondary iPS cell generation from multiple tissue and cell types derived from NNeo and NGFP chimeras
| Tissue/cell type | NNeo | NGFP2 |
|---|---|---|
| Neural progenitor | + | N/D |
| Adrenal gland | + | N/D |
| Keratinocyte | + | N/D |
| Muscle | + | N/D |
| Intestinal epithelium | − | + |
| Mesenchymal stem cell | − | + |
| Hematopoietic lineage | − | + |
| MEF | + | + |
| Tail tip fibroblast | − | + |
N/D, not determined.
We initially examined secondary intestinal epithelial cells for their ability to form secondary iPS cells (Fig. 4). Purified intestinal epithelial cells from both secondary NGFP2 and NNeo chimeras responded remarkably quickly to dox treatment and formed spheroids in suspension within 48 h that subsequently adhered to the MEF feeder layer and acquired ES cell–like morphology within 3–4 d (Fig. 4a–c and i–j). Alkaline phosphatase activity, however, was not detected before 10–12 d of culture with dox (Supplementary Fig. 3a online).Using a mechanical fractionation protocol, we found that these colonies formed much more efficiently from fraction 7 (mostly crypt-derived cells) than from earlier fractions (enriched for villus tip–derived cells) (Fig. 4m).
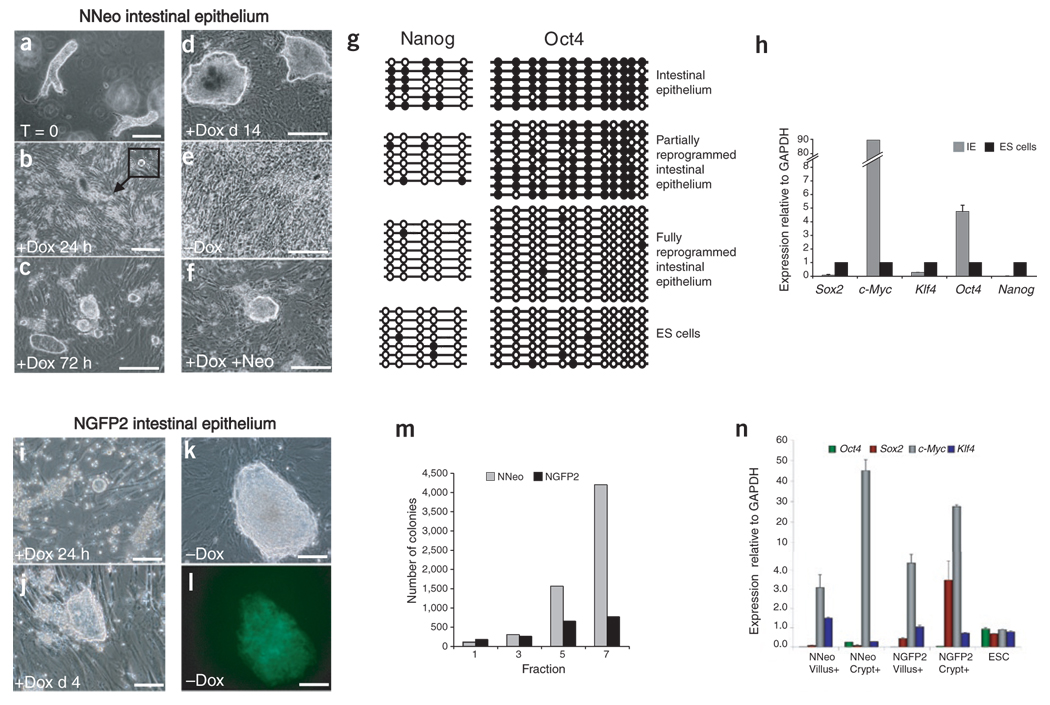
Figure 4. Reprogramming of intestinal epithelial cells.
(a) NNeo secondary intestinal epithelial crypt-villus structures were isolated from chimeras. (b) After 24 h of culture in the presence of dox, spheroids began appearing in suspension (inset). (c) Within 72 h of dox culture, suspended spheroids attached to the γ-irradiated feeder layer and acquired an ES cell–like morphology. (d,e) Colonies continued to grow during 2 weeks of dox treatment (d), but differentiated and became indistinguishable from the feeder layer upon dox withdrawal (e). (f) Dox-dependent intestinal epithelial colonies were neo resistant 2 weeks after dox administration. Scale bars (a–f), 250 µm. (g) Bisulfite sequencing of the endogenous Oct4 and Nanog promoters in freshly isolated NNeo secondary intestinal epithelium, partially reprogrammed dox-dependent cells, fully reprogrammed NNeo iPS cells after infection with Sox2 and Klf4 viruses. (h) Quantitative RT-PCR analyses of expression of the four factors and Nanog revealed that dox-dependent NNeo intestinal epithelial (IE) colonies express Oct4 and c-Myc at high levels compared with ES cells, but Sox2 and Klf4 at very low levels. (i) NGFP2 secondary intestinal epithelial cells also formed spheroids in suspension within 24 h of dox addition and acquired an ES cell–like morphology within 72 h (j). (k,l) NGFP2 intestinal epithelium gave rise to dox-independent secondary iPS cell colonies that express GFP from the endogenous Nanog locus. Scale bars (i,l), 50 µm. (m) EDTA-DTT–based fractionation of intestinal villi from differentiated cells of the tip (fraction 1) to the progenitor cells of the crypt (fraction 7)28 followed by 4 d of dox induction demonstrates that crypt fractions in both NNeo and NGFP2 secondary lines are more efficient at initial colony formation. (n) Quantitative RT-PCR analysis showed that with the exception of Klf4, the transgenes were more efficiently induced in fraction 7 (crypt) than in fraction 1 (villus tip) of the NNeo and NGFP2 intestinal epithelial cells.
Cells derived from NGFP2 chimeras developed into dox-independent iPS cells that expressed endogenous Nanog after ~2 weeks of culture in the presence of dox (Fig. 4k–l and Supplementary Fig. 3b).
In contrast, cells derived from the NNeo chimera became neo resistant after 2 weeks of dox culture, but were unstable and lost their ES cell–like morphology upon dox withdrawal (Fig. 4d–f). Bisulfite sequencing revealed some degree of demethylation of the Nanog promoter but only minimal demethylation of the Oct4 promoter (Fig. 4g), and when injected under the skin of severe combined immunodeficient diabetic (SCID) mice, these cells were unable to generate teratomas in the presence or absence of dox. Quantitative RT-PCR showed that these cells failed to induce Nanog and expressed only very low levels of Sox2 and Klf4 but high levels of Oct4 and c-Myc (Fig. 4h). Additional infection with Sox2 and Klf4 led to the generation of fully reprogrammed, dox-independent iPS cells expressing pluripotency markers and showing complete demethylation of their Oct4 and Nanog promoters (Fig. 4g and Supplementary Fig. 3c).
Comparison of transgene induction levels in NGFP2 and NNeo intestinal epithelial cells 48 h after dox treatment revealed differences in induction levels similar to what was observed in secondary MEFs from these lines (Fig. 4n; compare to Fig. 3a). Intestinal epithelial cells derived from the crypt induced most transgenes more readily than cells from the villus, offering an explanation for their increased colony-formation rate. These findings indicate that the proviral integration sites in the NNeo line, although permissible for reprogramming of MEFs, are not competent to mediate full reprogramming in intestinal epithelial cells, in contrast to those present in NGFP2 cells.
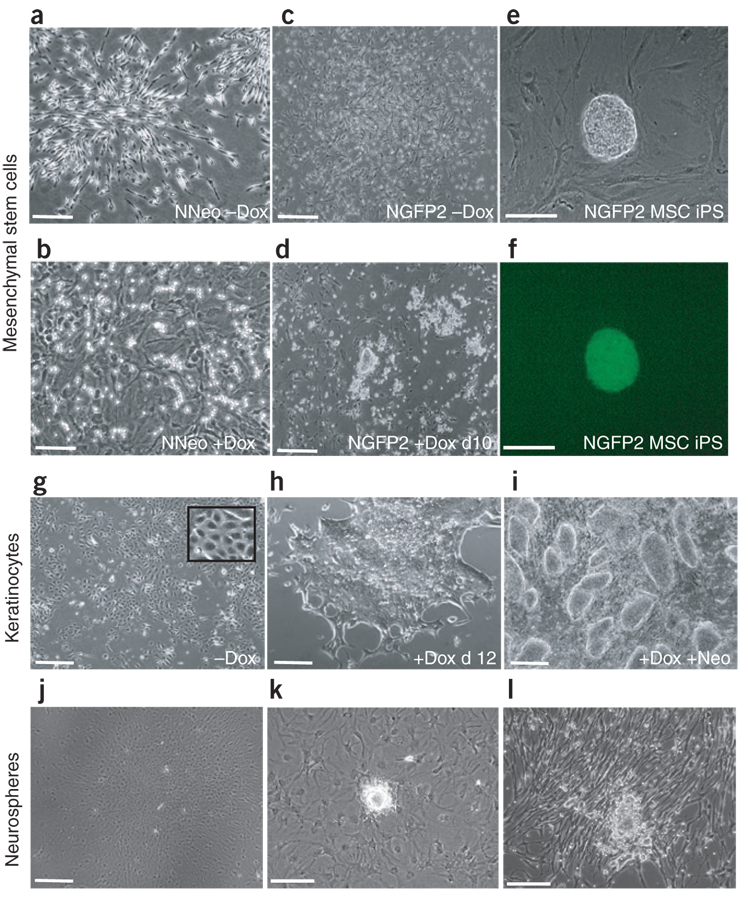
We next compared the reprogramming ability of bone marrow–derived mesenchymal stem cells (MSCs) and tail tip fibroblasts (TTFs) isolated from NNeo and NGFP2 chimeras. These cells represent two mesenchymal populations that are amenable to reprogramming by direct infection1,4,12 (Supplementary Fig. 3d). As with intestinal cells, secondary NGFP2 MSCs and TTFs were capable of generating iPS cells in response to dox, whereas those derived from NNeo chimeras were not (Fig. 5a–f and Supplementary Fig. 3e,f).
Figure 5. Reprogramming of other somatic cell types.
(a–b) NNeo mesenchymal stem cells (MSCs) before and after 3 weeks of dox administration. (c–d) NGFP2 MSCs before and after 10 d of dox treatment forming ES cell–like colonies. (e–f) NGFP2 MSCs gave rise to dox-independent iPS cell colonies that express GFP from the endogenous Nanog locus. (g,h) Colonies of dermal keratinocytes from NNeo chimeras with typical epithelial morphology (inset) (g) began to exhibit ES-cell morphology within 12 d of dox treatment (h). (i) These cells fully reprogrammed to form neo-resistant secondary iPS cell colonies. (j) After expansion in serum-free media, plated NNeo-derived neurospheres readily differentiated into astrocytic cells in response to dox- and serum-containing ES cell media. (k,l) When plated neurosphere cells were expanded in adherent conditions with EGF and FGF2 for another 3 weeks and then exposed to dox-containing media, iPS cell–like colonies appeared both in ES cell (k) and serum-free media (l). Scale bars (a–d) and (g–j), 250 µm; (e–f) and (k–l), 50 µm.
Cells isolated from the epidermis of NNeo chimeras were first propagated in the absence of dox in growth conditions optimized for keratinocytes21. Homogeneous epithelial cultures were obtained (Fig. 5g), and dox was added to the media. Clusters of epithelial cells proliferated and changed their morphology over time. After 12 d the medium was changed to dox-containing ES cell medium (Fig. 5h), and 7 d later neo was added. Neo-resistant cells growing in tight colonies resembled ES cells (Fig. 5i) and were passaged onto γ-irradiated feeder cells, at which point the cultures were maintained in the absence of dox and expressed endogenous Nanog (Supplementary Fig. 4a online).
We then tested whether neural progenitor cells could be reprogrammed in this system. Brains from NNeo chimeras were dissected, and a tissue block around the lateral ventricles was dissociated into single cells and plated onto uncoated culture dishes in epidermal growth factor (EGF)- and fibroblast growth factor (FGF)2-containing serum-free media (N3EF) in the presence of puromycin to select for secondary cells. Four weeks later neurospheres had formed that were subsequently plated onto polyornithine/laminin-coated dishes in either ES cell or N3EF media containing dox to activate the lentiviral transgenes. As expected for neural precursors, the cells exposed to the serum-containing ES cell media differentiated into flat astrocytic cells and stopped dividing (Fig. 5j). In contrast, the cells plated in N3EF media continued to proliferate robustly, resembling undifferentiated neuroepithelial cells. Three weeks later these proliferating cells were split and plated in either ES cell or N3EF media containing dox. The cells exposed to serum mostly adopted a flat morphology, whereas in N3EF the cells maintained a bipolar morphology. In contrast to the previous passage however, small ES cell–like colonies appeared in both conditions over the next 2 weeks (Fig. 5k,l). When passaged onto γ-irradiated feeder MEFs, neo-resistant, dox-independent iPS cell lines expressing endogenous Nanog were readily established (Supplementary Fig. 4b).
In addition, we also succeeded in generating secondary iPS cell lines from cells explanted from the kidney, adrenal gland, and muscle of NNeo chimeras. These tissues were dissected, dissociated in trypsin and plated in ES cell media containing dox. After 6–12 d in the presence of dox, colonies with ES-cell morphology appeared that ultimately became neo resistant, dox independent and had activated Nanog (Supplementary Fig. 4c–e).
DISCUSSION
Reprogramming of the somatic epigenome to a pluripotent, embryonic state through ectopic expression of the four transcription factors Klf4, Sox2, c-Myc and Oct4 is a slow and inefficient process. The current method for induction of reprogramming is through retroviral gene delivery, which results in heterogeneous cell populations with proviral integrations that vary in both number and genomic location, offering an explanation for the variability and inefficiency of direct reprogramming.
Here we describe a system for reprogramming genetically homogeneous cell populations. Reprogramming with dox-inducible lentiviral vectors and subsequent chimera formation yields tissues comprised of genetically homogeneous cells that harbor identical proviral integrations and re-express the reprogramming factors upon exposure to dox. This strategy selects for cells that carry the correct number of proviruses inserted at genomic loci that are favorable to drug-induced activation and eliminates the heterogeneity inherent in de novo viral infection of target cells. Surprisingly, the timing of reprogramming in this system was similar to that of directly infected primary fibroblasts. The minimum length of time that dox was required to initiate reprogramming ranged from 9 to 13 d. This timescale is consistent with the 10- to 14-d time frame observed in cells that have been directly infected with vectors13,14. We also observed that when dox was withdrawn from the cultures as early as day 9, GFP+ secondary iPS cell colonies continually appeared for the next several weeks in the absence of dox. These results support the notion that reprogramming is driven by a stochastic sequence of epigenetic modifications requiring a minimum period of transgene expression.
The observed reprogramming efficiency of secondary MEFs was as high as 4%, which is similar to the reprogramming efficiency of mature B cells22, vastly higher than the estimated 0.1% efficiency using de novo infection and drug selection, and about eightfold higher than what has been reported using morphological selection criteria1,11,12. It has been well documented that iPS cells derived from infected MEFs carry on average 15 different proviral copies, suggesting strong selection for the small fraction of the infected cells that carry the ‘correct’ number of proviruses or that express the four factors with the appropriate stoichiometry for successful reprogramming. Thus, the reprogramming frequency of secondary MEFs would be expected to be higher because these cells have been clonally derived from infected cells that carried the ‘correct’ combination of proviruses. If so, why would 4% but not most, or all, dox-treated secondary cells give rise to secondary iPS cells? We consider two non–mutually exclusive explanations. (i) It has been established that genetically identical subclones of directly infected MEFs become reprogrammed at significantly different times or not at all11,20. As discussed previously, this suggests that reprogramming involves a sequence of stochastic events such that cells carrying an identical number of proviral copies will activate the endogenous pluripotency genes at different times. (ii) Our data also show that dox treatment does not activate the proviruses uniformly in all cells but rather that differences in induction levels exist between individual cells. Because of these variegated expression levels, only a fraction of secondary MEFs may achieve high enough expression levels of the factors or the correct relative expression levels between the factors and therefore be capable of generating secondary iPS cells.
Whereas reprogramming is induced by viral transduction of the four factors, the maintenance of the pluripotent state depends on reestablishment of the autoregulatory loop involving the activation of the four endogenous pluripotency factors Oct4, Nanog, Sox2 and Tcf3 (refs. 20,23) and silencing of exogenous factors. Similarly, secondary MEFs were capable of being fully reprogrammed to a pluripotent state that was maintained in the absence of transgene expression.
We also used the secondary system to examine the reprogramming potential of several additional adult somatic cell types. iPS cells could be derived from many other tissues, including brain, epidermis, intestinal epithelium, mesenchymal stem cells, tail tip fibroblasts, kidney, muscle and adrenal gland through dox treatment, indicating that the proviruses were appropriately activated in cell types other than MEFs. This demonstrates that the four reprogramming factors can mediate epigenetic reprogramming in cells with different developmental origins and epigenetic states and highlights the usefulness of the secondary system for the study of reprogramming in a broad range of cell types. Although special care was taken to avoid other contaminating cell types, we cannot unequivocally demonstrate the cells of origin of iPS cells from these various tissue types. Genetic lineage tracing experiments have in fact demonstrated that iPS cells can be derived from liver and pancreas cells after transduction with Oct4, Sox2, c-Myc and Klf4 (refs. 24,25). However, not all cell types are permissive to reprogramming by these four factors. We have shown that reprogramming of mature but not of immature B cells requires the transduction of an additional factor (c/EBP-alpha) or the inhibition of the B cell–specific transcription factor Pax5 (ref. 22). It is possible that additional and as yet unknown factors are required to reprogram certain cell types. One practical advantage of the system described here is that cell types including those that might be refractory to ex vivo culture and retroviral infection, such as intestinal epithelial cells, can be studied.
Our drug-inducible reprogramming system demonstrates predictable and highly reproducible kinetics and efficiencies (Fig. 2f) that should facilitate the study of early molecular events leading to epigenetic reprogramming. In addition, the genetic homogeneity of secondary cell types increases the feasibility of chemical and genetic screening to enhance reprogramming efficiency. For example, we demonstrate that the DNA demethylating agent 5-Aza-deoxycytidine substantially enhances reprogramming efficiency. Such screens could also be applied to identify compounds that replace the original reprogramming factors. Because the reprogrammed state is not dependent on the exogenous factors, the transgenes could be genetically excised, and secondary cells that lack a particular reprogramming factor could be generated by chimera formation15.
METHODS
Viral preparation and infection
Construction of lentiviral vectors containing Klf4, Sox2, Oct4 and c-Myc under control of the tetracycline operator and a minimal CMV promoter has been described previously14. Replication-incompetent lentiviral particles were packaged in 293T cells with a VSV-G coat and used to infect MEFs containing M2rtTA and the Pgk1-Puro resistance gene at the R26 locus17, as well as either a neo-resistance or GFP allele targeted to the endogenous Nanog locus1,11. Viral supernatants from cultures packaging each of the four viruses were pooled, filtered through a 0.45 µM filter and mixed 1:1 with ES cell medium (DMEM supplemented with 10% FBS (Hyclone), leukemia inhibitory factor, beta-mercaptoethanol (Sigma-Aldrich), penicillin/streptomycin, L-glutamine and nonessential amino acids (all from Invitrogen) before being applied to MEFs.)
Primary iPS cell isolation, teratoma and chimera formation
Approximately 3 weeks after the addition of dox (Sigma-Aldrich; 2 µg/ml), GFP+ or neo-resistant iPS cell colonies were isolated and expanded in the absence of dox. The NanogGFP2 iPS cell line was picked from the same plate as line NanogGFP1 (described in ref. 22 as MEF-iPS#1 line), whereas line NanogGFP3 was derived from an independent experiment. iPS cell lines were injected into C57/B6 × DBA/1 F1 blastocysts. Blastocysts were placed in a drop of DMEM with 15% FBS under mineral oil. A flat-tip microinjection pipette with an internal diameter of 12–15 mm was used for iPS cell injection using a Piezo micromanipulator. About ten iPS cells were injected into the blastocyst cavity, and blastocysts were placed in KSOM (Specialty Media) and incubated at 37 °C until they were transferred to recipient females. Fifteen injected blastocysts were transferred to the uterine horns of pseudopregnant C57/B6 × DBA/1 F1 females at 2.5 d post coitum. For teratoma generation, 2 × 106 cells were injected subcutaneously into the flanks of recipient SCID mice, and tumors were isolated for histological analysis 3–6 weeks later. All animals were treated in accordance with institutional IACUC guidelines.
Secondary somatic cell isolation and culture
For MEF isolation, chimeric embryos were isolated at E13.5, and the head and internal (including reproductive) organs were removed. The remaining tissue was physically dissociated and incubated in trypsin at 37 °C for 20 min, after which cells were resuspended in MEF media containing puromycin (2 µg/ml) and expanded for two passages before freezing. Secondary MEFs used for the described experiments were thawed and experiments plated 1–2 passages after thawing. Kinetic experiments (Fig. 2) were performed by plating 4 × 104 secondary MEFs per well in six-well plates, and plates were stained or analyzed at the indicated times. Cell density experiments were performed in 12-well plates, and GFP+ cell iPS colonies were scored 4 weeks after dox induction. Single-cell efficiency experiments were performed by plating single secondary MEFs onto a layer of wild-type feeder MEFs in 96-well plates before dox induction (using limiting dilutions, which were confirmed by eye in replicate plates lacking feeder MEFs). iPS cell formation was scored 4 weeks later. Representative experiments from two to three biological replicates are shown. For 5-Aza and TSA experiments, 1 × 106 secondary MEFs were plated in six-well plates (~100 cells/mm2) and pretreated with ES cell media containing 5-Aza (1 µM) or TSA (1 µM) for 48 h. After 48 h, secondary MEFs were cultured in ES cell media plus dox lacking 5-Aza or TSA. MEFS were exposed to 5-Aza or TSA for a second 48-h period between days 8–10 after induction, followed by culture with dox only until scoring GFP+ colonies on day 21.
Somatic organs were isolated from 3- to 4-month-old chimeras. Epidermal keratinocytes were isolated and cultured as previously described21,26. Neural progenitor cells were isolated and cultured as previously described27. Total intestinal epithelium was dissociated using a solution of 3 mM EDTA and 0.05 mM DTT in PBS for 30 min at 25 °C. The musculature was discarded, and purified crypts/villi were plated on γ-irradiated feeder MEFs in the presence of dox. For crypt-villus fractionation, the same EDTA-DTT solution was used, but fractions were collected by gentle shaking for 10, 6, 5, 5, 9, 10 and 25 min (corresponding to fractions 1–7, respectively, with 1 representing the villus tip to 7 representing the crypt) after incubation as described in ref. 28. We plated 8 × 106 epithelial cells from each fraction on a MEF feeder layer in ES cell media containing 2 µg/ ml dox. No growth was observed in cultures lacking dox. Whole marrow was isolated from secondary chimeric mice (or from Coll1-TetO-Oct4, ROSA26-M2rtTA mice16 for direct infections) from the femur and tibia after removal of the condyles at the growth plate by flushing with a syringe and 30-gauge needle containing DMEM + 5% FBS (Hyclone, Thermo Fisher Scientific). Mesenchymal stem cells were selected through differential plating on tissue culture plates for 72 h in α-MEM supplemented with 15% FBS. Colony formation of MSCs in culture was carried out by plating 4 × 106 nucleated cells from freshly isolated whole marrow onto 10-cm plates and allowed to expand for 5 d in the presence of puromycin to eliminate host blastocyst–derived cells, after which dox was introduced to induce reprogramming.
Cultures derived from adrenal glands, muscle and kidneys were dissected, mechanically dissociated and digested in trypsin at 37 °C for 20 min before plating on gelatin-coated culture dishes with ES cell media containing dox.
Antibodies
For flow cytometric analysis we used an APC-conjugated anti-mouse SSEA1 (R&D systems) and an alkaline phosphatase substrate kit, Vector Red substrate kit (Vector Laboratories). For immunofluorescence, cells were fixed in 4% paraformaldehyde, and we used mouse monoclonal antibodies against SSEA1 (Developmental Studies Hybridoma Bank), goat anti-Sox2 (R&D Systems), mouse anti-Oct4 (Santa Cruz) and rabbit anti Nanog (Bethyl). Fluorophore-labeled, appropriate secondary antibodies were purchased from Jackson ImmunoResearch.
Flow cytometry
Cells were trypsinized, washed once in PBS and resuspended in fluorescence-activated cell sorting (FACS) buffer (PBS + 5% FBS). 106 cells were stained with 10 µl of APC-conjugated anti-SSEA1 antibody in a 100 µl volume for 30 min; cells were then washed twice in PBS. Cells were then washed once with wash buffer and resuspended in FACS buffer for analysis on a FACS-calibur cell sorter.
Bisulfite sequencing and Southern blotting
Bisulfite treatment of DNA was done using the CpGenome DNA Modification Kit (Chemicon) following the manufacturer’s instructions. The resulting modified DNA was amplified by nested PCR using two forward (F) primers and one reverse (R) primer: Oct4 (F1, 5′-GTTGTTTTGTTTTGGTTTTGGATAT-3′); (F2, 5′-ATGGGTTGAAATATTGGGTTTATTTA-3′); (R, 5′-CCACCCTCTAACCTTAACCTC TAAC-3′) and Nanog (F1, 5′-GAGGATGTTTTTTAAGTTTTTTTT-3′; F2, 5′-AATGTTTATGGTGGATTTTGTAGGT; R, 5′-CCCACACTCATATCAATATAATAAC-3′). The first round of PCR was done as follows: 94 °C for 4 min; five cycles of 94 °C for 30 s, 56 °C for 1 min (−1 °C per cycle), 72 °C for 1 min; and 30 cycles of 94 °C for 30 s, 51 °C for 45 s and 72 °C for 1 min, 20 s. The second round of PCR was 94 °C for 4 min; 30 cycles of 94 °C for 30 s, 53.5 °C for 1 min and 72 °C for 1 min 20 s. The resulting amplified products were gel-purified (Zymogen, Zymo Research), subcloned into the TOPO TA vector (Invitrogen), and sequenced. Southern blotting of genomic DNA was carried out by digesting 10 µg of DNA with SpeI (which cuts once in the lentiviral vector backbone) followed by hybridization with random primed full-length cDNA probes for the four factors.
Quantitative RT-PCR
Total RNA was isolated using Trizol reagent (Invitrogen). Five micrograms of total RNA was treated with DNase I to remove potential contamination of genomic DNA using a DNA Free RNA kit (Zymo Research). One microgram of DNase I–treated RNA was reverse transcribed using a First Strand Synthesis kit (Invitrogen) and ultimately resuspended in 100 µl of water. Quantitative PCR analysis was performed in triplicate using 1/50 of the reverse transcription reaction in an ABI Prism 7000 (Applied Biosystems) with Platinum SYBR green qPCR SuperMix-UDG with ROX (Invitrogen). Primers used for amplification were as follows: Oct4 F, 5′-ACATCGCCAATCAGCTTGG-3′ and R, 5′-AGAACCATACTCGAACCACATCC-3′; c-Myc F, 5′-CCACCAGCAGCGACTCTGA-3′ and R, 5′-TGCCTCTTCTCCACAGACACC-3′; Klf4 F, 5′-GCACACCTGCGAACTCACAC-3′ and R, 5′-CCGTCCCAGTCACAGTGGTAA-3′; Sox2 F, 5′-ACAGATGCAACCGATGCACC-3′ and R, 5′-TGGAGTTGTACTGCAGGGCG-3′; Nanog F, 5′-CCTCCAGCAGATGCAAGAACTC-3′ and R, 5′-CTTCAACCACTGGTTTTTCTGCC-3′. To ensure equal loading of cDNA into qRT-PCR reactions, we amplified GAPDH mRNA using the following: F, 5′-TTCACCACCATGGAGAAGGC-3′; and R, 5′-CCCTTTTGGCTCCACCCT-3′. Data were extracted from the linear range of amplification. All graphs of qRT-PCR data shown represent samples of RNA that were DNase treated, reverse transcribed and amplified in parallel to avoid variation inherent in these procedures. Error bars represent s.d. of the mean of triplicate reactions.
Supplementary Material
Note: Supplementary information is available on the Nature Biotechnology website.
ACKNOWLEDGMENTS
We thank J. Dausman for assistance with animal husbandry. M.W. was supported in part by fellowships from the Human Frontiers Science Organization Program and the Ellison Foundation. C.J.L. was supported by a Ruth L. Kirschstein Fellowship from the US National Institutes of Health. J.H. was supported by a fellowship from the Helen Hay Whitney Foundation. R.J. was supported by grants from the US National Institutes of Health.
Footnotes
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests: details accompany the full-text HTML version of the paper at http://www.nature.com/naturebiotechnology/
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions/
References
- 1.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 2.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 3.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Yu J, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 7.Park IH, et al. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 8.Lowry WE, et al. Generation of human induced pluripotent stem cells from dermal fibroblasts. Proc. Natl. Acad. Sci. USA. 2008;105:2883–2888. doi: 10.1073/pnas.0711983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wernig M, Meissner A, Cassady JP, Jaenisch R. c-Myc is dispensable for direct reprogramming of mouse fibroblasts. Cell Stem Cell. 2008;2:10–12. doi: 10.1016/j.stem.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 10.Nakagawa M, et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 11.Meissner A, Wernig M, Jaenisch R. Direct reprogramming of genetically unmodified fibroblasts into pluripotent stem cells. Nat. Biotechnol. 2007;25:1177–1181. doi: 10.1038/nbt1335. [DOI] [PubMed] [Google Scholar]
- 12.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat. Protocols. 2007;2:3081–3089. doi: 10.1038/nprot.2007.418. [DOI] [PubMed] [Google Scholar]
- 13.Stadtfeld M, Maherali NDTB, Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brambrink T, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hanna J, et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science. 2007;318:1920–1923. doi: 10.1126/science.1152092. [DOI] [PubMed] [Google Scholar]
- 16.Hochedlinger K, Yamada Y, Beard C, Jaenisch R. Ectopic expression of Oct-4 blocks progenitor-cell differentiation and causes dysplasia in epithelial tissues. Cell. 2005;121:465–477. doi: 10.1016/j.cell.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 17.Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis. 2006;44:23–28. doi: 10.1002/gene.20180. [DOI] [PubMed] [Google Scholar]
- 18.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–642. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 19.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaenisch R, Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones PH, Watt FM. Separation of human epidermal stem cells from transit amplifying cells on the basis of differences in integrin function and expression. Cell. 1993;73:713–724. doi: 10.1016/0092-8674(93)90251-k. [DOI] [PubMed] [Google Scholar]
- 22.Hanna J, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aoi T, et al. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008 February 14; doi: 10.1126/science.1154884. published online. [DOI] [PubMed] [Google Scholar]
- 25.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of pancreatic β cells into induced pluripotent stem cells. Curr. Biol. 2008;18:890–894. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rheinwald J. Culture of epithelial and mesothelial cells. In: Baserga R, editor. Cell Growth and Division: A Practical Approach. Oxford: Oxford Press; 1989. pp. 81–94. [Google Scholar]
- 27.Vescovi AL, Galli R, Gritti A. Adult neural stem cells. In: Zigova T, Sanberg PR, Sanchez-Ramos JR, editors. Neural Stem Cells: Methods and Protocols. New York: Humana; 2002. pp. 115–123. [Google Scholar]
- 28.Ferraris RP, Villenas SA, Diamond J. Regulation of brush-border enzyme activities and enterocyte migration rates in mouse small intestine. Am. J. Physiol. 1992;262:G1047–G1059. doi: 10.1152/ajpgi.1992.262.6.G1047. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary information is available on the Nature Biotechnology website.