Summary
Little is known about the molecular mechanics of the late events of translation initiation in eukaryotes. We present a kinetic dissection of the transition from a pre-initiation complex (PIC) after start codon recognition to the final 80S initiation complex (IC). The resulting framework reveals that eIF5B actually accelerates the rate of ribosomal subunit joining and this acceleration is influenced by the conformation of the GTPase active site of the factor mediated by the bound nucleotide. eIF1A accelerates joining through its C-terminal interaction with eIF5B, and eIF1A release from the initiating ribosome, which occurs only after subunit joining, is accelerated by GTP hydrolysis by eIF5B. Following subunit joining, GTP hydrolysis by eIF5B alters the conformation of the final IC and clears a path to promote rapid release of eIF1A. Our data, coupled with previous work, indicate that eIF1A is present on the ribosome throughout the entire initiation process and plays key roles at every stage.
Keywords: eukaryotic translation initiation, protein synthesis, eIF1A, eIF5B, subunit joining
Introduction
Translation initiation prepares the ribosome to begin reading an mRNA to make the corresponding protein. The main tasks are to load the methionyl initiator tRNA (Met-tRNAi) into the ribosomal peptidyl (P) site, bind the mRNA, locate the initiation codon, and join the small and large ribosomal subunits to form the final IC. Bacteria accomplish these tasks with the help of three initiation factors (IFs) 1. Eukaryotic translation initiation is significantly more complex, requiring at least 23 different polypeptides to accomplish the same fundamental goals 2; 3. In the current model of eukaryotic initiation, eukaryotic initiation factors (eIFs) 1 and 1A bind cooperatively to the 40S ribosomal subunit and induce a structural change in the ribosome converting it to an ‘open’ state, which accelerates the binding of the eIF2•GTP•Met-tRNAi ternary complex (TC) 4–9. At this point, in concert with eIF3 and eIF4F, the resulting 43S complex is thought to bind near the 5′-7-methylguanosine cap of an mRNA and scan along the message in search of the AUG start codon 10. eIF5, a GTPase activating protein (GAP), triggers GTP hydrolysis by eIF2 11–14 and, upon identification of the start codon, a conformational change in the complex decreases the affinity of eIF1 for the initiating ribosome 15; 16. Dissociation of eIF1 triggers release of Pi from eIF2, making GTP hydrolysis irreversible and committing the complex to completion of initiation at that site on the mRNA 17; 18. The complex is also thought to revert to a closed form at this stage that is no longer able to move freely along the message 8; 10; 15; 19.
eIF2•GDP can dissociate from the IC at this stage, with complete dissociation of the factor dependent on ribosomal subunit joining 20. A second GTPase initiation factor, eIF5B, in its GTP-bound form promotes joining of the large (60S) ribosomal subunit to the initiating 40S complex 21. 80S IC formation induces eIF5B to hydrolyze GTP, which reduces its affinity for the ribosome and allows the factor to dissociate 22; 23, resulting in an 80S complex that is capable of entering the elongation phase of translation.
eIF1A and eIF5B are orthologs of bacterial IF1 and IF2, respectively, and provide an evolutionary link among translation initiation processes in all domains of life 24; 25. IF1 and IF2 cooperatively bind to the 30S ribosomal subunit 26–28, with IF1 blocking the A site and directing IF2-mediated fMet-tRNAi loading into the P site 29; 30. Like IF1, eIF1A is thought to bind the A site of the 40S subunit 25; 31; 32. IF2 is a GTPase and, like eIF5B, also plays a role in subunit joining 27; 33; 34.
Until recently, in contrast to their bacterial counterparts, eIF1A and eIF5B were thought to act at temporally distant steps in the initiation pathway. However, recent genetic, biochemical, and structural studies have shown that the factors interact through their respective C-termini 35–37, an interaction unique to eukaryotes due to the lack of one or both interacting domains in other kingdoms 37. We previously explored the role of this interaction in translation initiation and determined that deletion of the interacting domain on either eIF1A or eIF5B reduces the efficiency of subunit joining without inhibiting previous steps in the pathway 38. In addition, we probed the role of GTP hydrolysis by eIF5B in eIF1A release from ICs and showed that blocking GTP hydrolysis led to accumulation of eIF1A in 80S complexes 39, suggesting that eIF1A release is coupled to release of eIF5B. However, because these experiments lacked kinetic resolution, it remained unclear at which point in the initiation pathway eIF1A dissociates during normal translation initiation and what role GTP hydrolysis plays in its release. For example, it is possible that eIF1A is normally released just prior to subunit joining and that blocking GTP hydrolysis leads to accumulation of 80S-associated eIF1A via an off-pathway event such as binding of eIF1A to the stalled complex. It was also not clear from our previous work whether GTP hydrolysis by eIF5B plays an active role in removing eIF1A or merely a passive one, allowing release of eIF5B which in turn clears the way for eIF1A to dissociate from the complex. To answer these questions and further elucidate the timing of the late steps of translation initiation we employed a reconstituted S. cerevisiae translation system containing an unstructured model mRNA to dissect the kinetic parameters of these late steps and the mechanisms employed by eIF1A and eIF5B.
Results and Discussion
Joining of naked 40S and 60S subunits
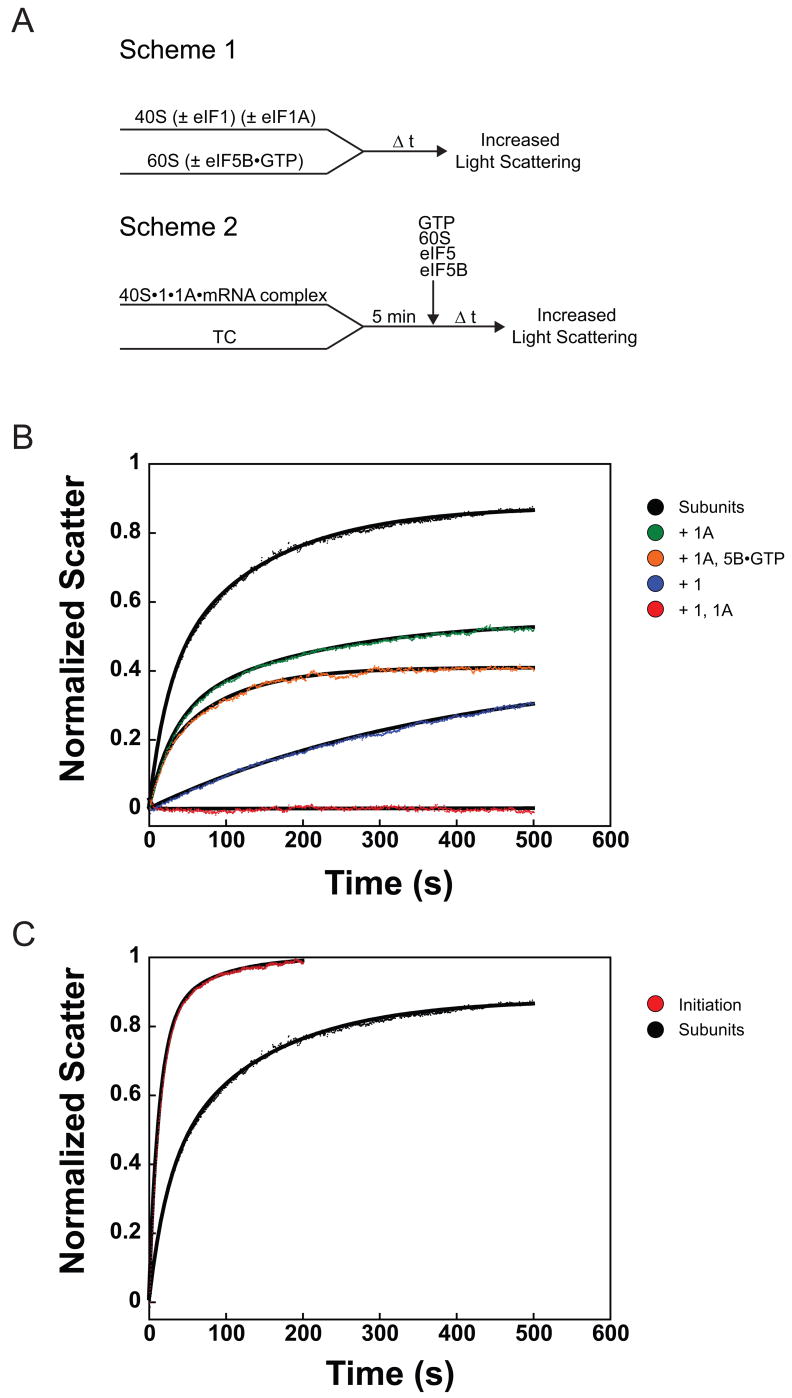
To set a baseline for exploring the mechanism of ribosomal subunit joining during the initiation process, we first determined the rate of subunit joining as it occurs off the initiation pathway, in the absence of the initiation factors and other components. Previous studies of the association of naked eukaryotic 40S and 60S subunits using light scattering techniques yielded second-order rate constants ranging from 2 × 105 to 2 × 107 M−1 s−1 depending on the source of ribosomes (Artemia, wheat germ or rabbit) and presence or absence of eIF3 40; 41. These experiments were conducted at higher Mg2+ concentrations (6 - 9 mM) however, and association of ribosomal subunits is stabilized by high Mg2+ concentrations. We therefore used a stopped-flow apparatus to monitor joining of yeast 40S and 60S ribosomal subunits at physiological Mg2+ concentration (3 mM) by following the increased intensity of light scattered by the newly formed 80S ribosomes over time (Figure 1(a), Scheme 1). When 40S and 60S subunits were rapidly mixed in the absence of all other components, 80S ribosomes were formed with biphasic kinetics with first- and second-phase rate constants of 0.042 ± 0.002 s−1 and 0.007 ± 9×10−4 s−1, respectively (Figure 1(b), black and Table 1). The biphasic nature of subunit joining could be due to 1) two different conformations of 40S subunits that undergo joining with different rates; 2) the existence of subunits as both monomers and dimers 42, with dissociation of dimers responsible for the slow phase; 3) a slow step after initial subunit encounter that pulls the equilibrium forward.
Figure 1.
Ribosomal subunit joining within the initiation pathway and with naked ribosomal subunits monitored by light scattering. (a) Experimental designs. Scheme 1: Assembly of 80S ribosomes outside of the initiation pathway. Scheme 2: Assembly of 80S ICs. TC: eIF2•GTP•Met-tRNAi ternary complex. (b) Association of 40S and 60S ribosomal subunits alone and in the presence of eIF1 and/or eIF1A. Subunit joining with ribosomal subunits only (black): k1 = 0.042 ± 0.002 s−1, α1 = 0.37 ± 0.04, k2 = 0.007 ± 9×10−4 s−1, α2 = 0.51 ± 0.04. Plus eIF1A (green): k1 = 0.031 ± 0.002 s−1, α1 = 0.28 ± 0.02; k2 = 0.007 ± 0.001 s−1, α2 = 0.30 ± 0.04. Plus eIF1 (blue): k = 0.003 ± 6×10−4 s−1, α = 0.39 ± 0.06. In the presence of both eIF1 and eIF1A (red), subunit joining was completely prevented. Plus eIF1A and eIF5B•GTP (orange): k = 0.014 ± 0.001 s−1, α = 0.38 ± 0.01. (c) Initiation pathway-dependent ribosomal subunit joining (red): k1 = 0.076 ± 0.004 s−1, α1 = 0.77 ± 0.06, k2 = 0.019 ± 0.006 s−1, α2 = 0.23 ± 0.06. Joining of subunits alone (black) shown for comparison. Data are means ± average error. All amplitudes are normalized to subunit joining in the complete initiation system (C, red curve). In all experiments the 40S and 60S concentrations were 50 and 100 nM, respectively.
Table 1.
Summary of measured rate constants and amplitudesa
| Step | Components or Conditions | Rate constant, k (s−1) | Amplitude, α |
|---|---|---|---|
| Subunit Joining | 40S + 60S | k1 = 0.042 ± 0.002 k2 = 0.007 ± 9×10−4 |
α1 = 0.37 ± 0.04 α2 = 0.51 ± 0.04 |
| 40S, 1A + 60S | k1 = 0.031 ± 0.002 k2 = 0.007 ± 0.001 |
α1 = 0.28 ± 0.02 α2 = 0.30 ± 0.04 |
|
| 40S, 1 + 60S | 0.003 ± 6×10−4 | 0.39 ± 0.06 | |
| 40S, 1, 1A + 60S | n.d.b | n.d.b | |
| 40S, 1A + 5B•GTP, 60S | 0.014 ± 0.001 | 0.38 ± 0.01 | |
| Initiationc | k1 = 0.076 ± 0.004 k2 = 0.019 ± 0.006 |
α1 = 0.77 ± 0.06 α2 = 0.23 ± 0.06 |
|
| Initiation, TC•GDPNP | 0.007 ± 0.001 | 0.21 ± 0.01 | |
| Initiation, 1A-ΔDIDDI | 0.013 ± 0.001 | 0.76 ± 0.01 | |
| Initiation, no eIF5B | 0.005 ± 8×10−4 | 0.29 ± 0.02 | |
| Initiation, eIF5B•GDP | 0.006 ± 6×10−4 | 0.64 ± 0.02 | |
| Initiation, eIF5B•GDPNP | k1 = 0.044 ± 0.001 k2 = 0.013 ± 0.003 |
α1 = 0.70 ± 0.03 α2 = 0.27 ± 0.03 |
|
| Initiation, eIF5B-TA | k1= 0.008 ± 9×10−4 k2= 0.001 ± 3×10−4 |
α1=0.34 ± 0.05 α2=0.52 ± 0.02 |
|
| Initiation, eIF5B-TA, HY | 0.004 ± 3×10−4 | 0.48 ± 0.03 | |
|
| |||
| eIF1A-Fl Dissociation from PICs/ICs | 43S•mRNA (post-Pi release) | 0.013 ± 0.001 | 1 |
| 43S•mRNA (post-Pi release), (+) eIF5B•GTP | 0.012 ± 0.001 | 1 | |
| 43S•mRNA (post-Pi release), (+) eIF5B•GTP, 60S | 0.034 ± 0.001 | 1 | |
| 43S•mRNA (post-Pi release), (+) eIF5B•GTP, 60S (no unlabeled eIF1A chase) | 0.03 ± 0.01 | 0.25 ± 0.06 | |
| 80S IC (eIF5B•GTP) | >0.09 | 1 | |
| 80S IC (eIF5B•GDPNP) | k1 ≥0.14 k2= 0.0085 ± 3×10−5 |
α1 = 0.40 ± 0.03 α2 = 0.6 ± 0.03 |
|
| 80S IC (eIF5B-TA•GTP) | k1 ≥0.16 k2 = 0.011 ± 5×10−4 |
α1 = 0.47 ± 0.03 α2 = 0.53 ± 0.03 |
|
| 80S IC (eIF5B-TA, HY•GTP) | k1 ≥0.18 k2 = 0.012 ± 5×10−4 |
α1 = 0.31 ± 0.05 α2 = 0.69 ± 0.06 |
|
|
| |||
| TAMRA-eIF5B Association with ICs | GTP-bound | 0.009 ± 0.002 | 1 |
| GDPNP-bound | 0.0073 ± 5×10−5 | 1 | |
|
| |||
| TAMRA-eIF5B Dissociation from ICs | GTP-bound | k1 = 0.12 ± 0.02 k2 = 0.0025 ± 5×10−4 |
α1 = 0.47 ± 0.03 α2 = 0.54 ± 0.04 |
| GDPNP-bound | 1.6 × 10−4 ± 2×10−5 | 1 | |
Values are the averages of at least two independent experiments and the errors are the average deviations.
Subunit joining under these conditions is undetectable.
Initiation conditions are 40S and 60S subunits, eIF1, eIF1A, TC (eIF2, Met-tRNAi, GTP), mRNA, eIF5 and eIF5B•GTP unless otherwise specified.
Recent biochemical data indicate that eIF1 and eIF1A bind to the subunit interface surface of the 40S subunit 8; 43 and act synergistically to induce a structural transition from a closed state to an open one 7–10. This open state binds TC much more rapidly than the closed state and has been proposed to be able to scan freely along the mRNA in search of the start codon. To investigate how these states and eIF1 and eIF1A affect the ability of the 40S subunit to join to the 60S subunit, we added saturating amounts of eIF1 and/or eIF1A to 40S subunits before addition of 60S subunits and then measured the kinetics of joining. While the presence of saturating eIF1 alone reduced the rate and end-point of 80S formation (10-fold and 2-fold, respectively; Figure 1(b), blue), eIF1A did not affect the kinetics of subunit joining, reducing only the end-point of the reaction 1.5-fold (Figure 1(b), green). Differences in the amplitudes of subunit joining in the presence of eIF1 and/or eIF1A suggest that these factors shift the position of equilibrium in subunit joining to favor free subunits over 80S ribosomes. The effect of eIF1 alone could be because it stabilizes open complex formation to some extent –although not enough to be detected by cryo-EM - or because it presents a steric block to joining in addition to its effect on the conformation of the 40S subunit. The rate constant for subunit joining decreased less than two-fold (from 0.005 to 0.003 s−1) when the concentration of eIF1 was increased from 200 nM to 1 μM, while the amplitude remained constant, indicating that the effect of eIF1 is maximal in this concentration range. The rate of joining was unchanged between 1 and 5 μM, although the amplitude did decrease suggesting that at very high concentrations of eIF1 non-specific binding to rRNA or binding to weak secondary sites on 40S or 60S subunits may come into play.
In contrast to the presence of eIF1 or eIF1A alone, when both factors were included 80S formation was completely inhibited (Figure 1(b), red). This blocking of subunit joining by eIFs 1 and 1A is synergistic, rather than the additive effects of each factor alone, consistent with the previously noted synergistic effect of the factors on open complex formation 8. These data suggest that the open complex, in addition to being highly receptive to TC binding, is extremely refractory to subunit joining, whereas the closed state (the naked 40S subunit or 40S•1A complex) is active for joining. The fact that inhibition of joining by eIF1 alone plateaus between 200 nM and 1 μM argues against the possibility that the effect is entirely due to eIF1 sterically blocking subunit joining, with the presence of eIF1A enhancing this effect by reducing the rate of eIF1 dissociation from the 40S subunit 7; in this case the effect of eIF1 alone would be expected to increase with concentration until subunit joining was completely blocked.
The kinetics of subunit joining during initiation
We next wanted to measure the rate of subunit joining during translation initiation to determine whether it is accelerated by the initiation machinery relative to the rate with naked 40S and 60S subunits. (Note that all observed rate constants compared herein were measured under the same component concentrations, except where noted.) To this end, we formed 43S•mRNA complexes (in the absence of eIF3 and eIF4F) using an unstructured model mRNA (see Materials and Methods) and initiated subunit joining by the rapid addition of eIF5, eIF5B•GTP, and 60S subunits (Figure 1(a), Scheme 2; It should be noted that N-terminally truncated eIF5B (Δ1-395), which fully rescues growth and associated phenotypes of the eIF5B null mutant yeast strain, 35 was used in all experiments). In the context of the initiation pathway both the rate and amplitude of the first-phase of subunit joining are increased approximately 2-fold relative to 40S and 60S subunits alone (k1 = 0.076 ± 0.004 s−1 and relative amplitude (α) = 0.77 ± 0.06; Figure 1(c), red), making subunit joining in the initiation pathway essentially monophasic. This suggests that the heterogeneity existing in the off-pathway joining of ribosomal subunits is resolved by the components of initiation. Interestingly, the second phase rate constant, whether following on- or off- pathway subunit joining, varies identically with 60S subunit concentration (slope = 4.0 × 104 M−1s−1, data not shown) suggesting the same process is responsible for the second phase of ribosomal subunit joining both on and off the initiation pathway.
To verify that we were indeed observing subunit joining within the initiation pathway, we disrupted initiation at prior steps and determined the effects on the rate of subunit joining. Formation of 43S•mRNA complexes containing GDPNP instead of GTP in the TC, which blocks eIF5-triggered GTP hydrolysis by eIF2, resulted in a 10-fold decrease in the rate of subunit joining (k = 0.007 ± 0.001 s−1 vs. 0.076 ± 0.004 s−1 for the uninterrupted pathway) and a 4-fold decrease in amplitude (0.21 ± 0.01 vs. 0.77 ± 0.06; Figure 2(a), blue). Likewise, replacing wild-type eIF1A with a mutant lacking the last five amino acids (eIF1A-ΔDIDDI), which interact with eIF5B and were previously shown to be required for efficient subunit joining 44, resulted in a 5-fold decrease in the rate and a slight decrease in total amplitude of joining, indicating that eIF1A’s C-terminus accelerates eIF5B-mediated subunit joining. (Figure 2(a), gray). Together, these data demonstrate that the light scattering assay monitors initiation pathway-dependent 80S IC formation.
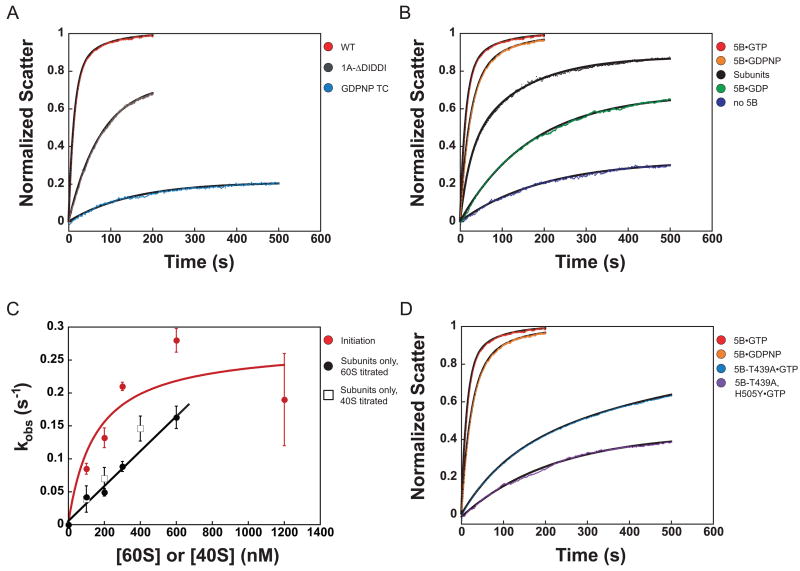
Figure 2.
The initiation pathway accelerates ribosomal subunit joining relative to the intrinsic rate in the absence of factors. (a) Prior steps in the initiation pathway are required for optimal subunit joining. Subunit joining with GDPNP in TC (blue): k = 0.007 ± 0.001 s−1, α = 0.21 ± 0.01. 43S•mRNA complex containing eIF1A-ΔDIDDI (gray): k = 0.013 ± 0.001 s−1, α = 0.76 ± 0.01. Both are fit to single exponentials. Subunit joining in the complete initiation pathway (red) is shown for comparison. (b) The nucleotide state of eIF5B influences subunit joining. Subunit joining with subunits only (black) and in the translation initiation pathway (red), as described above. In the absence of eIF5B (blue), kobs = 0.005 ± 8×10−4 s−1, α = 0.29 ± 0.02. eIF5B•GDP (green): k = 0.006 ± 6×10−4 s−1, α = 0.64 ± 0.02. eIF5B•GDPNP (orange): k1 = 0.044 ± 0.001 s−1, α1 = 0.7 ± 0.03; k2 = 0.013 ± 0.003 s−1, α2 = 0.27 ± 0.03. (c) kobs for subunit joining vs. concentration of 40S or 60S subunits. When only ribosomal subunits are present, kobs increases linearly with 40S (open squares) or 60S (solid circles) concentration in the range tested (slope = 2.5×105 M−1 s−1). When subunit joining occurs in the context of the initiation pathway, kobs varies in a manner best fit by a hyperbola (red; kmax = 0.3 s−1; K1/2 = 150 nM). Data are represented as mean ± average error. (d) GTPase-deficient eIF5B mutants slow subunit joining. eIF5B-T439A•GTP (light blue): k1= 0.008 ± 9×10−4, α1=0.34 ± 0.05, k2= 0.001 ± 3×10−4, α2=0.52 ± 0.02. eIF5B-T439A, H505Y•GTP (purple): kobs = 0.004 ± 3×10−4, α = 0.48 ± 0.03. eIF5B•GTP (red) and eIF5B•GDPNP (orange) are shown for comparison.
eIF5B accelerates subunit joining only in the initiation pathway
In the current model of translation initiation, eIF5B facilitates subunit joining in its GTP-bound state and GTP hydrolysis is required for the factor’s subsequent release from the 80S IC 21–23. Although it is known that eIF5B is required for efficient subunit joining, it remains unclear whether the factor affects the kinetics of the process, the stability of the resulting complex, or both. To determine more specifically the role eIF5B plays in this step, subunit joining was monitored in the presence and absence of eIF5B. As noted above, in the initiation pathway with saturating eIF5B and 100 nM 60S subunits, joining occurs with a first-phase rate constant of 0.076 ± 0.004 s−1 and relative first-phase amplitude 0.77 ± 0.06 (Figure 2(b), red). When eIF5B is omitted from these experiments, subunit joining is reduced over 10-fold in rate (kobs = 0.005 ± 8×10−4 s−1) and over 2-fold in total amplitude (0.29 ± 0.02), indicating that eIF5B accelerates the rate of subunit joining as well as increasing the extent of the process (Figure 2(b), blue).
We next wanted to explore whether eIF5B could accelerate off-pathway subunit joining. The 40S•eIF1A complex was mixed with eIF5B and 60S subunits and light scattering was monitored to determine if eIF5B accelerates joining in this context. The addition of eIF5B did not increase the rate or amount of 80S complex formation relative to that in the absence of eIF5B (Figure 1(b), compare orange and green). Similarly, addition of eIF5B did not affect subunit joining in the presence of either eIF1 alone or with eIF1 and eIF1A (data not shown). These data indicate that the stimulatory actions of eIF1A and eIF5B on subunit joining require the other components and intermediate steps in the initiation pathway.
To investigate the dependence of the observed rate constant for subunit joining on the concentration of 60S subunits, we monitored the rate of joining both in the initiation pathway and with naked ribosomal subunits with increasing concentrations of 60S subunits. The observed rate constant of joining of naked subunits varies linearly with concentration of 60S (fast phase rate constant; Figure 2(c), black). For simple binding, kobs = kon * [60S] + koff 45. The slope of 2.5 × 105 M−1 s−1 is the second order rate constant (kon) for the association of the subunits. Titration of 40S subunits yielded nearly identical results (Figure 2(c), open squares). At all concentrations of 60S subunits tested, the amplitude of on-pathway subunit joining remained constant, while the amplitude of joining of the naked subunits increased only 1.5-fold between 100 nm and 600 nM 60S subunits, indicating that the concentration of 60S subunits was above the Kd for subunit-subunit association in all cases (data not shown). In the context of the initiation pathway, however, at all concentrations of 60S subunits tested, the kobs for joining was greater than the kobs for joining of the naked subunits (Figure 2(c)). Also, in contrast to the joining of the naked subunits, the observed rate constant for initiation-dependent subunit joining appears to have a hyperbolic dependence on the concentration of 60S subunits, reaching an extrapolated maximum of approximately 0.3 s−1 at ~1.5 μM 60S subunits (Figure 2(c), red). This suggests that at higher concentrations of 60S subunits, the rate of subunit joining may become limited by a first-order event. Rate constants in this range are sufficient to support the estimated rate constant for initiation in vivo in eukaryotes, which is approximately 0.2 s−1 46.
The conformation of eIF5B influences the rate of ribosomal subunit joining
Although GTP hydrolysis by eIF5B is not required for ribosomal subunit joining, it is unknown how the nucleotide state of the factor affects the rate of the process. Subunit joining was therefore monitored in the presence of eIF5B bound to GTP, the non-hydrolyzable analog GDPNP, or GDP. The addition of eIF5B•GDP increased the amplitude of subunit joining 2-fold, relative to the absence of eIF5B (0.64 ± 0.02 vs. 0.29 ± 0.02, respectively) but did not affect the rate (k = 0.006 ± 6×10−4 s−1 vs. 0.005 ± 8×10−4 s−1 in the absence of eIF5B; Figure 2(b), compare green and blue). A 3-fold increase in the concentration of eIF5B•GDP increased the rate constant for subunit joining by only 50% (data not shown), indicating that the low rate is not due to reduced affinity of the GDP-bound state of the factor for the initiating ribosome. In the presence of GDPNP, eIF5B stimulated subunit joining to nearly the same level observed for GTP, with a first phase rate constant of 0.044 ± 0.001 s−1, less than 2-fold slower than with GTP (Figure 2(b), orange). The difference between GTP- and GDPNP-bound eIF5B could be due to a small amount of contaminating GDP in the GDPNP or to a difference in conformation between eIF5B•GTP and eIF5B•GDPNP.
We previously reported that a change from T to A at position 439 in yeast eIF5B creates a GTPase-deficient mutant with reduced dissociation from the 80S IC, resulting in a severe slow-growth phenotype 23. A second-site suppressor mutation of H to Y at position 505 does not restore GTPase activity, but instead reduces the factor’s affinity for the 80S complex allowing dissociation of the factor following subunit joining, and supporting near wild-type levels of growth 23. When wild-type eIF5B•GTP was replaced with the T439A mutant (bound to GTP), subunit joining was reduced to a level similar to that in the presence of eIF5B•GDP (k1= 0.008 ± 9×10−4; α1=0.34 ± 0.05; k2= 0.001 ± 3×10−4; α2=0.52 ± 0.02;) (Figure 2(d), light blue). Interestingly, substitution of the intragenic suppressor mutant eIF5B-T439A, H505Y•GTP did not restore the rate of subunit joining to wild-type levels, but instead reduced it further (Figure 2(d), purple). A 3-fold increase in the concentration of either eIF5B mutant did not alter the kinetics of subunit joining, indicating that their concentrations in these experiments were saturating and that reduced binding of the mutant factors to the complex was not responsible for the kinetic defects observed (data not shown). The lowered rates of subunit joining supported by these mutants and WT eIF5B•GDP are most likely caused by changes in the structure of the factor from the optimal eIF5B•GTP state, suggesting that the conformation of eIF5B is both critical for its ability to facilitate subunit joining and very sensitive to changes in the GTPase active site.
eIF1A dissociation from the initiating ribosome occurs after subunit joining
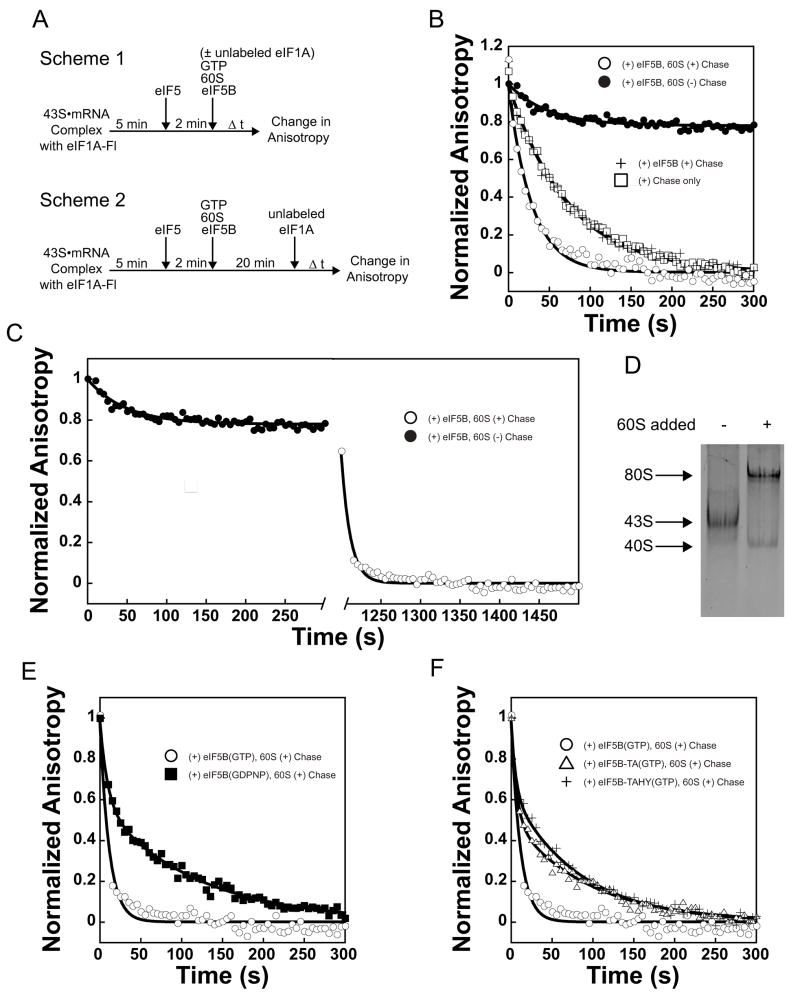
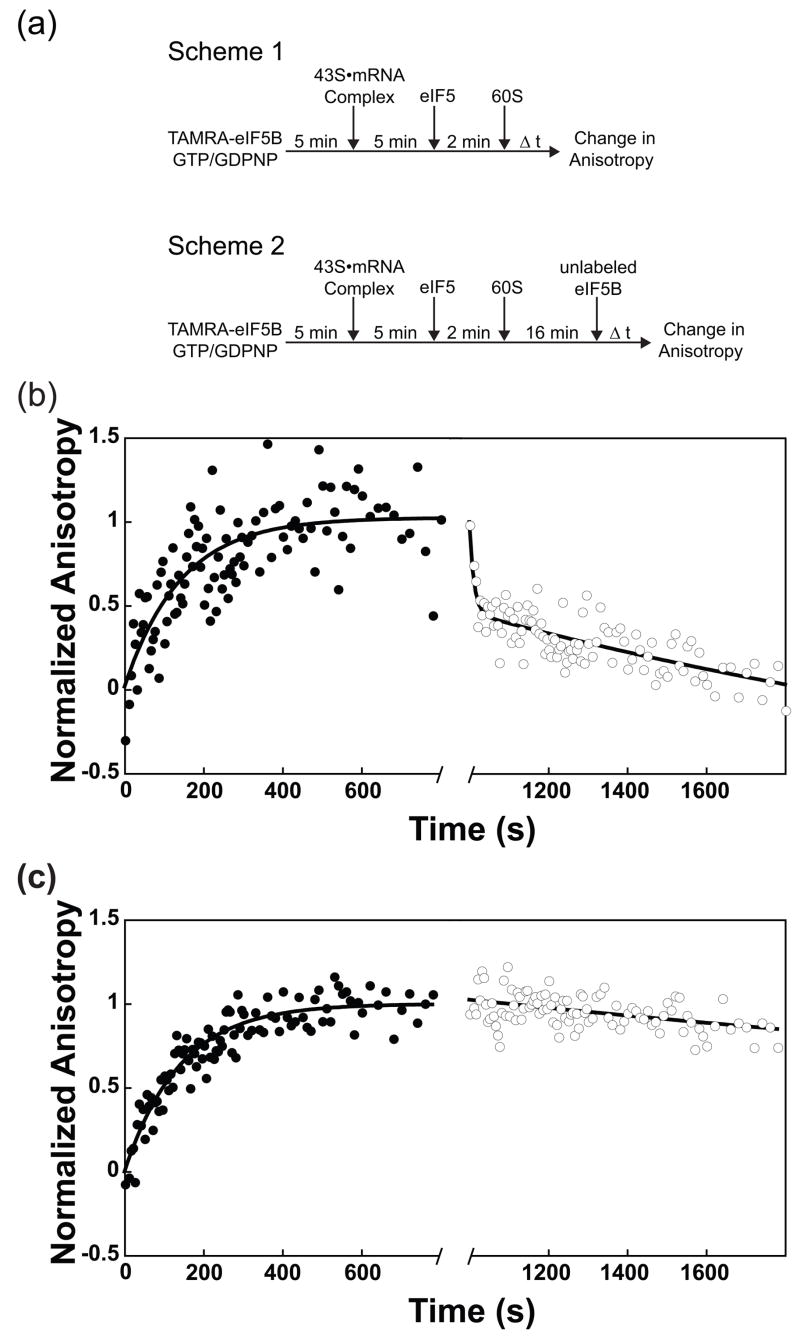
The integrity of eIF1A’s C-terminus, in conjunction with its binding pocket in the C-terminal domain of eIF5B, is important for joining of the 60S ribosomal subunit to the initiating 43S•mRNA complex 38; 39, implying that eIF1A remains bound to the initiating ribosome at least until its encounter with eIF5B. One possible role for this interaction would be that it allows eIF5B to remove eIF1A from the complex prior to subunit joining, possibly in order to clear the subunit-subunit interface. However, it is not known when eIF1A actually leaves the complex during initiation. To determine the timing of eIF1A release from the initiating ribosome, we fluorescently labeled the single cysteine-containing eIF1A mutant E124C via reaction with maleimide-conjugated fluorescein (see Methods and Supplementary Figure S1). Using this labeled factor (eIF1A-Fl), we constructed 43S•mRNA PICs (Figure 3(a), Scheme 1) and then chased these with excess unlabeled eIF1A, either alone or in the presence of additional components. The decrease in fluorescence anisotropy of eIF1A-Fl over time was then followed to monitor irreversible dissociation of the labeled factor. In the absence of additional components, eIF1A-Fl dissociated from the PIC with a rate constant of 0.013 ± 0.001 s−1 (Figure 3(b), squares). Addition of 0.5 μM eIF5B•GTP did not alter the rate of eIF1A-Fl dissociation (Figure 3(b), +), indicating that eIF5B does not facilitate removal of eIF1A prior to subunit joining.
Figure 3.
eIF1A is present in 80S complexes and its dissociation is accelerated by GTP hydrolysis by eIF5B. (a) Experimental designs. Scheme 1: Assembly of 80S ICs with or without chase of excess unlabeled eIF1A included with 60S subunits. Scheme 2: Assembly of 80S ICs followed by chase of excess unlabeled eIF1A. (b) Measuring the rate constants for dissociation of eIF1A-Fl from post-AUG-recognition 43S•mRNA complexes. Anisotropy of eIF1A-Fl in the absence of other factors: rfree = 0.1526 ± 3.5 × 10−3; in 43S•mRNA•eIF5 complexes: r43S = 0.2075 ± 7.4 × 10−3. Addition of eIF1A chase without eIF5B or 60S subunits (□): k = 0.013 ± 0.001 s−1. Addition of eIF1A chase with eIF5B (+): 0.012 ± 0.001 s−1. Addition of eIF1A chase with both eIF5B and 60S subunits (○): k = 0.034 ± 0.001 s−1. When eIF5B and 60S subunits are added in the absence of chase, eIF1A dissociates only slightly (k = 0.03 ± 0.01, = 0.25 ± 0.06, ●). (c) Fluorescence anisotropy change of eIF1A-Fl during formation of 80S ICs (●, as in Figure 4(b)), and with addition of unlabeled eIF1A chase after 20 minutes (○): kobs > 0.09 s−1; dead time of the experimental set-up. Anisotropy of eIF1A-Fl bound to 80S complexes formed with eIF5B•GTP: r80S-5B•GTP = 0.1969 ± 6.8 × 10−3. (d) Fluorescence scan of a native gel showing eIF1A-Fl binds to 43S•mRNA complexes and 80S complexes. 80S complexes were formed with eIF5B•GDPNP. Formation of 80S ICs also results in the appearance of a band corresponding to eIF1A-Fl bound to free 40S subunits. (e) eIF1A-Fl dissociates with biphasic kinetics from complexes formed with eIF5B•GDPNP (k1 ≥0.14; α1 = 0.40 ± 0.03; k2= 0.0085 ± 3 × 10−5; α2 = 0.6 ± 0.03, ■). The first phase most likely represents dissociation from the 40S subunits produced in reactions in which 80S ICs are formed (Fig. 3d). Dissociation of eIF1A-Fl from ICs formed with eIF5B•GTP (○) is shown for comparison. Anisotropy of eIF1A-Fl bound to 80S complexes formed with eIF5B•GDPNP: r80S-5B•GDPNP = 0.2159 ± 1.2 × 10−3. (f) GTPase-deficient eIF5B-T439A•GTP (△) slows eIF1A-Fl dissociation from 80S ICs relative to WT eIF5B (k1 ≥0.16, α1 = 0.47 ± 0.03; k2 = 0.011 ± 5 × 10−4 s−1; α2 = 0.53 ± 0.03; WT eIF5B-GTP, ○). Second-site suppressor mutant eIF5B-T439A, H505Y•GTP does not restore the rate of eIF1A-Fl dissociation relative to eIF5B-T439A (k1 ≥0.18, α1= 0.31 ± 0.05; k2 = 0.012 ± 5 × 10−4 s−1, α2 = 0.69 ± 0.06; +). Anisotropy of eIF1A-Fl bound to 80S complexes formed with eIF5B-TA•GTP: r80S-5B-TA•GTP = 0.2175 ± 1.4 × 10−3; formed with eIF5B-TAHY•GTP: r80S-5B-TAHY•GTP = 0.2210 ± 3.5 × 10−3. Data are represented as mean ± average error.
When both 60S subunits and eIF5B•GTP were included to allow formation of 80S ICs, eIF1A-Fl dissociated 3-fold faster than in the absence of 60S subunits, with a rate constant of 0.034 ± 0.001 s−1 (Figure 3(b), open circles). This rate constant is quite similar to that for subunit joining under these conditions in the presence of eIF1A-Fl (0.047 ± 0.004 s−1; Supplementary Figure S1), suggesting that ribosomal subunit joining triggers rapid eIF1A dissociation. To further test this proposal, we first allowed subunit joining to proceed to completion in the absence of a chase and then added unlabeled eIF1A chase (Figure 3(a), Scheme 2), monitoring eIF1A-Fl anisotropy during both periods. During the formation of 80S ICs only ~20% of the bound eIF1A dissociated (Figure 3(b) and (c), black circles). The rate constant of this limited dissociation was 0.03 s−1, the same as dissociation when 60S subunits were added along with the unlabeled eIF1A chase, suggesting the same process is rate-limiting in both cases. However, addition of excess unlabeled eIF1A chase 20 minutes after initiation of subunit joining, when 80S IC formation was complete, resulted in rapid dissociation of eIF1A-Fl, with a rate constant ≥ 0.09 s−1 (a lower limit due to the dead-time of the experimental setup; Figure 3(c), open circles). These data demonstrate that subunit joining increases the rate of dissociation of eIF1A from the ribosome 10-fold or more (0.013 s−1 prior to subunit joining vs. ≥0.09 s−1 after it), but suggest that the factor can actually bind to an 80S IC with reasonable affinity.
To further demonstrate that eIF1A can bind to an 80S IC after subunit joining, even though its rate of dissociation is significantly increased, native polyacrylamide gel electrophoresis was performed on post-AUG-recognition 43S•mRNA complexes and 80S ICs formed in the presence of eIF1A-Fl. Consistent with the kinetics experiments described above, Figure 3(d) shows that most of the eIF1A-Fl bound to the 43S•mRNA complexes is transferred to 80S complexes when 60S subunits and eIF5B•GDPNP are added.
Dissociation of eIF1A from 80S ICs is accelerated by GTP hydrolysis by eIF5B
GTP hydrolysis by eIF5B has been shown to reduce the factor’s affinity for the newly formed 80S IC, leading to its dissociation 21; 23. However, we previously suggested that GTP hydrolysis can also affect the dissociation of eIF1A from an initiating ribosome 39. To explore this possibility further, we replaced eIF5B•GTP with eIF5B•GDPNP in our 80S ICs (made with eIF1A-Fl) and again added an excess of unlabeled eIF1A chase 20 minutes after initiation (Figure 3(a), Scheme 2) and measured the decrease in fluorescence anisotropy over time to determine the rate of release of eIF1A-Fl. eIF1A-Fl dissociation was biphasic, with a first phase rate constant ≥ 0.14 s−1 (α = 0.4 ± 0.03) and second phase rate constant of 0.0085 ± 3 × 10−5 s−1 (α = 0.6 ± 0.03) when 80S complexes were formed with eIF5B•GDPNP (Figure 3(e), squares). Based on the appearance of a small fraction of free 40S subunits bound to eIF1A-Fl upon 80S complex formation, as visualized by native gel electrophoresis (Figure 3(d), lane 2), we interpret the first phase as the rate of dissociation of eIF1A-Fl from free 40S ribosomal subunits and the second phase as the rate of dissociation of eIF1A-Fl from 80S ICs. This interpretation is consistent with the previously measured rate constant for dissociation of eIF1A from the 40S ribosomal subunit in the presence of eIF1 (0.27 s−1, 15). Considering only the second phase rate constant, dissociation of eIF1A-Fl from 80S complexes is reduced at least 10-fold with eIF5B•GDPNP relative to the rate with eIF5B•GTP (0.0085 vs. ≥ 0.09 s−1, respectively). In addition, we repeated this experiment in the presence of GTP with the GTPase deficient eIF5B mutant T439A and the second-site suppressor mutant, T439A, H505Y, which also cannot hydrolyze GTP. Although these mutants have a decreased rate of subunit joining, subunit joining had proceeded to completion in the 20 minutes prior to the addition of the unlabeled eIF1A chase (Figure 3(a), Scheme 2). Similar to the case of WT eIF5B•GDPNP, eIF5B-T439A and eIF5B-T439A, H505Y reduced the rate of eIF1A-Fl dissociation 8-fold (k2 = 0.011 ± 5 × 10−4 s−1 and k2 = 0.012 ± 2 × 10−4 s−1, respectively; Figure 3(f), triangles and pluses). Taken together, these data indicate that GTP hydrolysis by eIF5B is required for rapid release of eIF1A from 80S ICs.
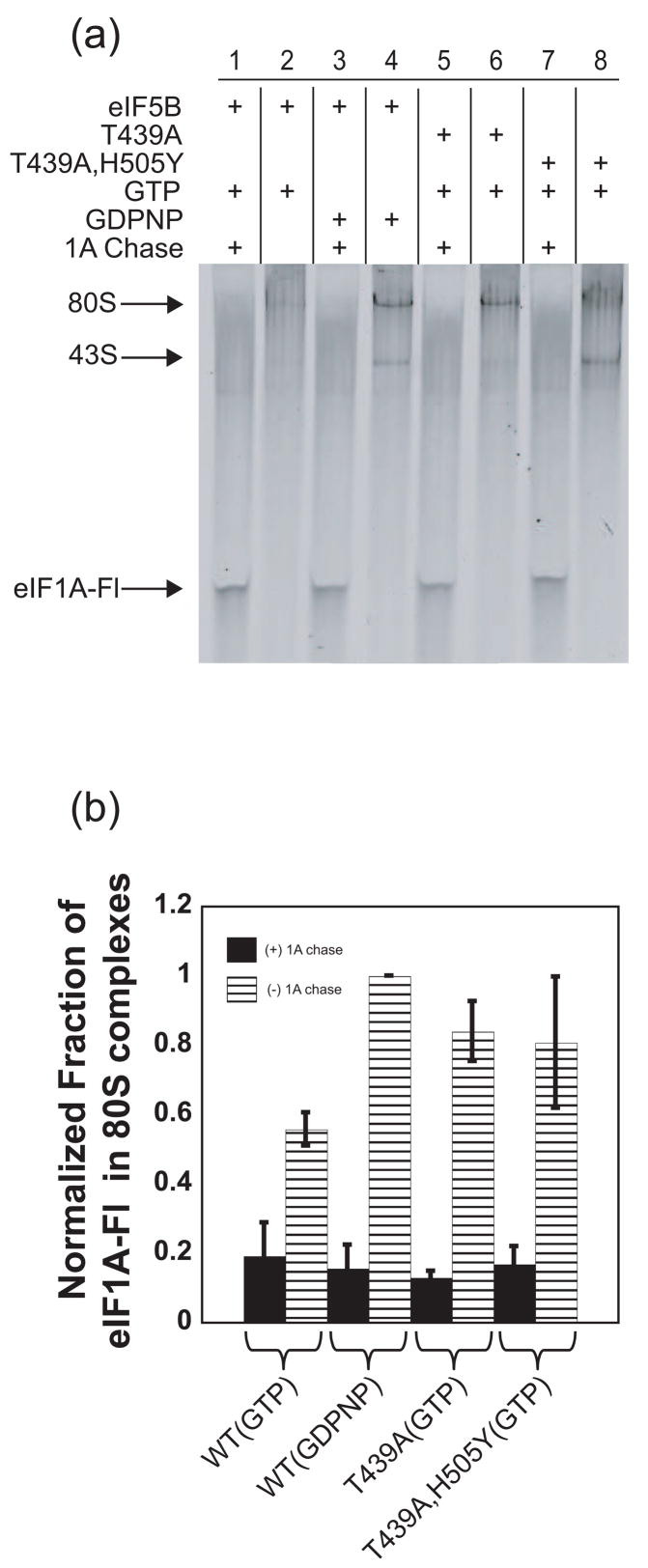
To gain further evidence that eIF5B influences the stability of eIF1A in an 80S IC, 80S ICs (containing eIF1A-Fl) were formed with WT eIF5B•GDPNP or the GTPase deficient eIF5B mutants T439A and T439A, H505Y bound to GTP. These complexes were then subjected to unlabeled eIF1A chase or a mock chase, and analyzed by native polyacrylamide gel electrophoresis as above. In the absence of unlabeled eIF1A chase, 80S ICs formed with WT eIF5B•GDPNP contained nearly 2-fold more eIF1A-Fl than those formed with GTP-bound eIF5B (Figure 4(a), compare lanes 2 and 4; Figure 4(b)). Similarly, as in the anisotropy-based assay above, both eIF5B-T439A and T439A, H505Y behaved like wild-type eIF5B bound to GDPNP, decreasing dissociation of eIF1A-Fl in the absence of eIF1A chase (Figure 4(a), compare lanes 6 and 8 with lane 2; Figure 4(b)). These data support the kinetic experiments above indicating that GTP hydrolysis by eIF5B increases the ability of eIF1A to dissociate from 80S ICs. When complexes were chased for long times (20 min) with unlabeled eIF1A, eIF1A-Fl could dissociate eventually in all cases, as expected (Figure 4(a), odd numbered lanes; Figure 4(b)).
Figure 4.
eIF1A-Fl in ICs can be visualized directly in native gels. (a) Fluorescence scan of native gel showing eIF1A-Fl bound to 80S ICs formed in the presence of WT or mutant eIF5B bound to GTP or GDPNP, with or without the addition of excess unlabeled eIF1A chase. Thirty-minute time points are shown and the positions of 43S and 80S complexes are noted. (b) Quantification of the bands in (a). Data in (b) are represented as the mean of three independent experiments ± average error.
These data suggest that promotion of eIF1A release from ICs by eIF5B’s GTPase activity is not a simple, passive event in which dissociation of eIF5B•GDP clears a path for eIF1A to dissociate. The second-site suppressor mutant eIF5B-T439A, H505Y, which rescues the growth defect of the GTPase deficient mutant T439A by allowing release of eIF5B from the 80S IC without restoring GTP hydrolysis 23, does not increase the rate of eIF1A release relative to the parental T439A mutant factor or eIF5B•GDPNP. This result suggests that release of eIF5B alone is not enough to allow rapid dissociation of eIF1A and that GTP hydrolysis by eIF5B actually alters the state of the complex in a way that increases the rate of eIF1A release. The notion that GTP hydrolysis by eIF5B alters the conformation of the 80S IC is consistent with the observation that the stability of Met-tRNAi binding in the complex is decreased by the presence of high concentrations of eIF1A (which forces eIF1A to remain bound) with the T439A, H505Y mutant but not with WT eIF5B (Fig. S3, compare lanes 1 and 2 with 7 and 8; it is not yet clear why the presence of eIF1A in the IC can destabilize Met-tRNAi binding). Together, these data suggest that GTP hydrolysis by eIF5B alters the conformation of the complex with respect to both eIF1A and Met-tRNAi.
The defects in eIF1A release and subunit joining with eIF5B-T439A, H505Y initially seem at odds with the fact that the second-site H505Y mutation partially rescues the growth defect produced by the T439A mutation in the GTPase active site. However, the strain containing the T439A, H505Y double mutant is not without phenotypes. Most strikingly, it fails to upregulate translation of the GCN4 mRNA upon amino acid starvation, a Gcn- phenotype, which is consistent with a defect in subunit joining or efficient release of eIF1A 23; 47. These data do suggest, though, that growth of yeast cells is surprisingly tolerant to defects in the rate of subunit joining, consistent with the fact that removal of the C-terminal DIDDI sequence of eIF1A, which reduces the rate of subunit joining 5-fold in vitro, produces no growth defect at 25 °C in yeast 36, and with the fact that eIF5B is not an essential gene (although the null mutant grows very slowly). These results suggest that subunit joining is not the rate-limiting step in translation initiation in vivo.
Monitoring eIF5B binding to and dissociation from the IC
With a clearer picture of the timing of eIF1A release from an initiating ribosome, we turned our attention to eIF5B recruitment to and release from the IC. We first determined the affinity of eIF5B labeled with the fluorophore TAMRA (see Methods) for different components of the initiation pathway by monitoring the increase in fluorescence anisotropy upon binding of the labeled factor to ribosomal subunits and complexes. In the GDPNP-bound form, TAMRA-eIF5B binds to purified 80S ribosomes with a Kd of 19 ± 2 nM (Table 2). However, when GDPNP is replaced with GDP, the factor binds 35-fold more weakly, with a Kd of 660 ± 80 nM. TAMRA-eIF5B•GDPNP binds to 40S ribosomal subunits, but only weakly, with a Kd ≥1 μM. Binding of the factor to 60S ribosomal subunits was also weak with a Kd ≥1 μM. We were unable to determine the affinity of TAMRA-eIF5B for the 43S•mRNA complex, with either GTP or GDPNP in the TC, because essentially no change in anisotropy was observed upon addition of 100 nM of the complex to GTP- or GDPNP-bound TAMRA-eIF5B, suggesting that eIF5B’s interactions with the initiating ribosome are highly dynamic. We cannot, however, rule out the possibility that full-length eIF5B may have a greater affinity for the initiating ribosome than the N-terminally truncated version used in these studies.
Table 2.
Binding affinities of TAMRA-eIF5B for 40S, 60S and 80S ribosomes
| Subunit or Complex | Kd (nM) | |
|---|---|---|
| TAMRA-eIF5B•GDPNP | 40S | ≥ 1000a |
| 60S | ≥ 1000a | |
| 80Sc | 19 ± 2b | |
|
| ||
| TAMRA-eIF5B•GDP | 80Sc | 660 ± 80b |
Values are lower limits.
Values are the averages of two independent experiments and the errors are the average deviations.
Salt-washed 80S ribosomes, purified as described in methods.
To probe the timing of eIF5B’s recruitment into the IC we monitored the increase in TAMRA-eIF5B anisotropy during the formation of 80S ICs. 43S•mRNA complexes were added to TAMRA-eIF5B in the presence of saturating GTP or GDPNP, followed by the addition of eIF5. (It should be noted that the GTP in the TC in 43S complexes and eIF2•GDP are both inert to exchange with added GTP or GDPNP on the timescales of these experiments 38; 48). 80S complex formation was initiated by addition of 60S ribosomal subunits and fluorescence anisotropy was measured as a function of time (Figure 5(a), Scheme 1). In the presence of either GTP or GDPNP, eIF5B bound to the complexes with similar rate constants (0.009 ± 0.002 s−1 and 0.007 ± 5 × 10−4 s−1, respectively; Figure 5(b) and (c), solid circles), and these rate constants match those for subunit joining under the same conditions (data not shown; the concentration of TAMRA-eIF5B was subsaturating). This is consistent with the data above (Figure 2(b)) showing that eIF5B•GTP and eIF5B•GDPNP increase the rate of subunit joining to similar extents, and also with the current model in which GTP hydrolysis by eIF5B is not required for subunit joining, but is instead essential for factor release from the 80S IC 21; 23.
Figure 5.
eIF5B binding to and release from initiating ribosomal complexes. (a) Scheme 1: Experimental setup to measure the kinetics of TAMRA-labeled eIF5B binding to ribosomal complexes during initiation. Scheme 2: Experimental setup to measure the kinetics of release of TAMRA-eIF5B from the 80S IC following subunit joining. (b) Kinetics of TAMRA-eIF5B•GTP binding to ribosomal complexes during subunit joining. The fluorescence anisotropy of TAMRA-eIF5B•GTP increases during the formation of 80S ICs with a rate constant of 0.009 ± 0.002 s−1 (●). Addition of excess unlabeled eIF5B 16 minutes after initiation of 80S complex formation results in biphasic dissociation of TAMRA-eIF5B from 80S ICs (k1 = 0.12 ± 0.02 s−1, α1 = 0.47 ± 0.03, k2 = 0.0025 ± 5 × 10−4 s−1, α2 = 0.54 ± 0.035; ○). Anisotropy of TAMRA-eIF5B•GTP in the absence of other factors: rfree = 0.2353 ± 1.1 × 10−3; in 80S complexes: rbound = 0.2408 ± 1.8 × 10−3. (c) Kinetics of TAMRA-eIF5B•GDPNP binding to 80S ribosomal complexes during subunit joining. The fluorescence anisotropy of TAMRA-eIF5B•GDPNP increases upon initiation of subunit joining similarly to GTP-bound TAMRA-eIF5B, with a rate constant of 0.0073 ± 5 × 10−5 s−1 (●). However, the rate constant for dissociation of TAMRA-eIF5B•GDPNP from the resulting 80S complexes upon addition of excess unlabeled eIF5B is reduced ~750-fold (k = 1.6 × 10−4 ± 2 × 10−5 s−1; ○). Dissociation of TAMRA-eIF5B•GDPNP was carried out for ~ 4 hours. Plotted data are truncated for clarity, however reported rate constants were determined by fitting the entire data set. Anisotropy of TAMRA-eIF5B•GDPNP in the absence of other factors: rfree = 0.2357 ± 5 × 10−4; in 80S complexes: rbound = 0.2440 ± 1.8 × 10−3. Data are represented as mean ± average error.
We next wanted to address the second part of this model - that eIF5B dissociation from 80S, and hence the transition to translation elongation, is dependent on the ability of the factor to hydrolyze GTP. The inability of eIF5B mutants to hydrolyze GTP (as in the case of eIF5B-T439A discussed above) results in vivo in a significant reduction in growth rate and in vitro (as in the case of eIF5B•GDPNP) in severe inhibition of methionyl-puromycin synthesis 21; 23; 49, a mimic of initial peptide bond formation. To explore eIF5B dissociation from the 80S IC, 80S ICs were formed according to scheme 1 (Figure 5(a)) using TAMRA-eIF5B, and the anisotropy of TAMRA-eIF5B was monitored upon the addition of a chase of excess unlabeled eIF5B (Figure 5(a), Scheme 2) to follow dissociation of the labeled factor. When 80S complexes were formed with GTP-bound TAMRA-eIF5B, dissociation of the factor from the IC occurred in a biphasic manner, with a first-phase rate constant of 0.12 ± 0.02 s−1 and a relative amplitude of 0.47 ± 0.03 and a second phase rate constant of 0.0025 ± 5 × 10−4 s−1 and relative amplitude of 0.53 ± 0.04 (Figure 5(b)). The biphasic nature of this curve can be explained by two populations of eIF5B-TAMRA bound to 80S ICs at the start of dissociation (when unlabeled eIF5B chase is added): 1) TAMRA-eIF5B•GDP which is assumed to dissociate fast; and 2) TAMRA-eIF5B•GTP bound to 80S ICs (from which the original eIF5B•GDP molecules had already departed) which dissociates more slowly. In the latter case, release would be limited by the rate of GTP hydrolysis by the eIF5B•GTP that had bound to this pre-existing 80S complex. In support of this interpretation, the rate constant for GTP hydrolysis by eIF5B bound to pre-formed 80S ribosomes is 0.0017 ± 9 × 10−4 s−1 38, very similar to the rate constant for the slow phase observed here for eIF5B release (0.0025 ± 5 × 10−4 s−1). In contrast, TAMRA-eIF5B•GDPNP dissociated 750-fold more slowly from 80S ICs, with a rate constant of 1.6 × 10−4 ± 2 × 10−5 s−1 (Figure 5(c)), adding further support to the model that GTP hydrolysis by eIF5B serves to destabilize the factor’s binding to the initiating ribosome, allowing it to dissociate rapidly.
Interestingly, the rate of release of eIF1A from an 80S IC formed with eIF5B•GDPNP is faster than dissociation of eIF5B•GDPNP itself, indicating that although GTP hydrolysis by eIF5B accelerates release of eIF1A, eIF1A is not completely trapped in the complex by eIF5B•GDPNP and is able to leave prior to eIF5B dissociation.
Conclusions
The data presented herein provide new insights into the timing and mechanism of the late steps in eukaryotic translation initiation. Our results indicate that eIF1A and eIF5B together accelerate the rate of subunit joining and that this activity depends on the conformation of eIF5B, which is dictated by interactions between its GTPase active site and the bound nucleotide. Hydrolysis of GTP by eIF5B accelerates its release from the ribosome and also induces rapid release of eIF1A from the final 80S IC, which is consistent with recent cryo-EM reconstructions suggesting movement of domain IV of eIF5B upon GTP hydrolysis 50.
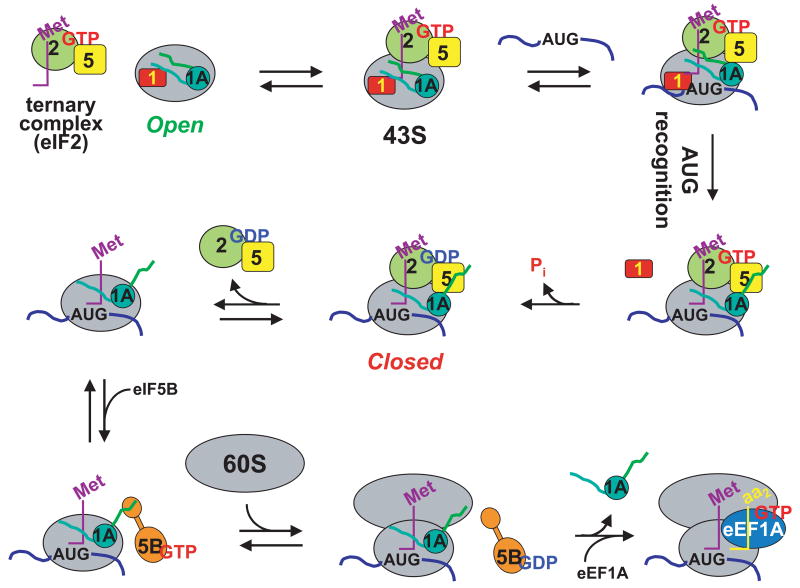
Our data, coupled with previous results, can be incorporated into a model for translation initiation in which eIF1A is present in the ribosomal complex throughout the entire initiation process, playing key roles at every main stage (Figure 6). When eIF1 and eIF1A bind to the 40S subunit at the beginning of initiation they synergistically induce a conformational transition from a closed to an open state 8. The open state was shown to bind TC several orders of magnitude faster than the closed state and is proposed to allow free movement of the complex on the mRNA. Our data demonstrate that this open complex containing both eIF1 and eIF1A is also highly resistant to premature subunit joining. Upon start codon recognition, eIF1 is ejected from the 40S subunit 15; 16, triggering downstream events 13; 14. Release of eIF1 allows rapid release of inorganic phosphate (Pi) from eIF2, driving GTP hydrolysis to completion and converting eIF2 into its inactive, GDP-bound state 17. Release of eIF1 is also expected to cause the complex to revert to the closed state because eIF1A on its own was shown to be insufficient to promote stable open complex formation on the 40S subunit 8. Upon transient dissociation of the (eIF2•GDP)•eIF5 complex, the closed PIC would be in a state competent for eIF5B-mediated subunit joining. Subunit joining would then make dissociation of eIF2•GDP irreversible 20.
Figure 6.
Simplified model of eukaryotic translation initiation: eIF1A is a central organizer of events in the pathway. eIF1A binds to the 40S ribosomal subunit cooperatively with eIF1, inducing a structural change in the subunit resulting in an ‘open’ state that can rapidly bind the eIF2•GTP•Met-tRNAi ternary complex (TC) and is highly resistant to subunit joining. In the open state, the 43S complex binds near the 5′.-end of an mRNA and begins to scan in search of the AUG start codon. Start codon recognition results in formation of a direct or indirect interaction between eIF1A and eIF5. The affinity of eIF1 for the initiating ribosome is reduced upon start codon recognition, leading to the factor’s release from the complex. eIF1 dissociation triggers Pi release from eIF2, making GTP hydrolysis irreversible, and also causes the complex to revert to a closed state. eIF2•GDP has reduced affinity for the initiating ribosome and, presumably together with eIF5, transiently dissociates, leaving the C-terminus of eIF1A free to interact with eIF5B. In its GTP-bound form, eIF5B facilitates subunit joining, and GTP hydrolysis results in dissociation of the factor from the resulting 80S IC. Subunit joining and subsequent GTP hydrolysis by eIF5B trigger eIF1A to vacate the 80S IC, freeing the A site of the 80S ribosome to bind to eukaryotic elongation factor (eEF) 1A•GTP•aminoacyl-tRNA ternary complexes, thus beginning the elongation phase of translation.
Dissociation of eIF1 would also break the thermodynamic interaction between eIF1 and eIF1A on the 40S subunit 7, perhaps allowing eIF1A to interact, either directly or indirectly, with eIF5, an event that occurs when the start codon is recognized and that involves the unstructured C-terminal arm of eIF1A9; 51. It is possible (although only hypothetical at this point) that this interaction moves eIF5 or alters its conformation and that this change in eIF5 (instigated by eIF1 dissociation) is what allows Pi to be released from eIF2. When the (eIF2•GDP)•eIF5 complex leaves the PIC, the interaction between eIF5 and the C-terminal arm of eIF1A will no longer occur, which in turn might free the very end of the C-terminal arm (the C-terminal “hand”; DIDDI) to interact with eIF5B. This interaction, like the conformational state of eIF5B itself, accelerates the rate of subunit joining. Our previous data indicated that this effect was not due simply to enhanced binding of eIF5B to the complex promoted by the interaction 38. Instead, the eIF1A-eIF5B interaction most likely recruits eIF5B and orients the factor within the complex (perhaps also altering its conformation), putting it in the correct position to facilitate joining of the 60S subunit. In this model, the freedom of the C-terminal arm of eIF1A provides a key mechanism for organizing events late in the initiation pathway.
Although eIF3 is not depicted in our model, its five core subunits are essential for growth in S. cerevisiae. eIF3 is thought to assist in both TC and mRNA recruitment to the PIC, and forms interactions with numerous initiation factors as part of the multifactor complex 52. To date, no interaction between eIF3 and eIF5B has been identified, however, and inclusion of eIF3 in in vitro initiation experiments did not alter the rate of initiation-dependent GTP hydrolysis by eIF5B, which is limited by the rate of subunit joining 38. Moreover, inclusion of saturating eIF3 in light scattering experiments did not alter the rate constants for initiation-dependent subunit joining compared with that in the absence of eIF3 (Supplementary Figure S4), indicating that in the yeast reconstituted system, using an unstructured model mRNA, eIF3 does not affect the rate of these steps (although it did reduce the amplitude of the fast phase from 0.77 to 0.44). The reconstituted system used here also lacks the eIF4F complex, a group of initiation factors thought to be responsible for dealing with structures in natural mRNAs. While the current model suggests that eIF4F should influence the early steps of initiation, it will be interesting in the future to see what role, if any, the eIF4F complex plays during ribosomal subunit joining on a natural mRNA message.
Finally, although subunit joining and subsequent GTP hydrolysis by eIF5B increase the rate of eIF1A dissociation from the complex ≥10-fold, eIF1A can still rebind the IC and complete dissociation does not occur under our experimental conditions in the absence of a chase to prevent rebinding of labeled factor. eIF1A is thought to occupy the A site of the ribosome, blocking it and encouraging the ternary complex to bind to the P-site. After initiation, elongation factor eEF1A in its GTP-bound form delivers the aa-tRNA corresponding to the second codon of the mRNA into the A site of the 80S complex. We suggest that eIF1A dissociation is made irreversible in vivo by the binding of the eEF1A ternary complex to the 80S IC and the start of the elongation phase of translation. It is also interesting to note that the concentration of eIF1A in vivo in yeast is lower than the concentrations of the other core factors 53, a situation that makes sense in light of our data as it would minimize competition between eIF1A and eEF1A ternary complexes for the A site of the ribosome after initiation is complete.
Now that we have a clearer picture of the events of the late steps in the initiation pathway and the roles of eIF1A and eIF5B in subunit joining, the stage is set for elucidating the molecular mechanisms underlying their action. The wealth of phenotypically-characterized mutations of both factors in yeast, in conjunction with studies in the reconstituted initiation system using the framework established here, should make possible significant progress on this front.
Materials and Methods
Buffers
1X reaction buffer: 30 mM HEPES-KOH, pH 7.4, 100 mM KOAc, pH 7.4, 3 mM Mg(OAc)2, 2 mM DTT.
Enzyme buffer: 20 mM HEPES-KOH, pH 7.4, 100 mM KOAc, pH 7.4, 2 mM DTT, 10% glycerol.
Reagent preparation
Ribosomal subunits, eIF1, eIF1A, eIF2, eIF3, eIF5, mRNA, [35S]Met-tRNAi and Met-tRNAi were purified as previously described 44. eIF5B was purified essentially as described 44 with slight modifications: following the HiTrap Q HP column, the N-terminal His-tag was not removed by TEV cleavage and instead the protein was further purified on a Superose 12 gel filtration column. The sequence of the unstructured, model mRNA used was 5′-GGAA[UC]7UAUG[CU]10C-3′. This model mRNA obviates the need for mRNA recruitment and remodeling factors (eIF3 and eIF4F), allowing core steps in the pathway to be isolated 49; 54.
Fluorescent Labeling of eIF1A and eIF5B
Rational
Previous studies have made use of intein-mediated protein ligations 55 to fluorescently label eIF1A at its C-terminus 7. Although this modification does not adversely affect early steps in the initiation pathway, we found that it did inhibit ribosomal subunit joining and it was therefore necessary to label the factor at another position. To this end, we changed both of the naturally occurring cysteine residues (positions 51 and 89 in yeast eIF1A) to serine. Importantly, the Cys-less version of eIF1A fully complements the eIF1A-null mutant in vivo, sustaining WT growth rates at temperatures from 25 °C to 30 °C (Supplementary Figure S5). Avoiding areas of high sequence homology and areas in which mutations produce strong phenotypes, we introduced individual cysteines in place of the amino acids K11, P22, Q73, N105 and E124 56. These mutants were then labeled with fluorescein using a thiol-reactive maleimide conjugate. The concentration of fluorophore incorporated agreed very well with the concentration of the protein, suggesting the proteins were labeled to near completion with 1:1 stoichiometry. eIF1A mutant E124C-Fl was chosen for use in later experiments because of its favorable behavior. However, all labeled eIF1A derivatives were capable of binding 40S ribosomes, and showed coupled binding with eIF1 (data not shown; 7). While some eIF1A derivatives had reduced affinities for the 40S subunit (E124C-Fl bound ~20-fold more weakly than WT eIF1A, with a Kd of 90 nM vs. 6 nM for C-terminally labeled eIF1A-TAMRA 7; data not shown), these factors were still capable of efficiently forming both 43S and 80S complexes in the native gel assay. In addition, single-cysteine eIF1A mutants interacted with eIF5 in the 43S complex upon AUG recognition to the same degree as reported previously for the wild-type C-terminally labeled factor 51. Subunit joining in the presence of these mutants also occurred with near wild-type kinetics (Supplementary Figure S1 and data no shown).
As with eIF1A, fluorescent labeling of eIF5B at the C-terminus could disrupt the C-terminal eIF1A-eIF5B interaction, and thus we sought to label eIF5B at a more innocuous position. We chose to label eIF5B using carboxytetramethyl rhodamine (TAMRA) conjugated to a succinimdyl ester, which at neutral pH preferentially targets primary amines and in principle should label the factor primarily at the N-terminus. The N-terminus of eIF5B was an optimal labeling location for several reasons. First, the N-terminal domain of eIF5B can be removed from the protein without significant consequence both in vivo and in vitro 21; 35. Second, the bacterial and archaeal orthologs of eIF5B, IF2 and aIF5B, respectively, lack the N-terminal domain that is found in eukaryotic eIF5B, suggesting that the N-terminus is not necessary for the universally conserved functions of the factor. N-terminally labeled TAMRA-eIF5B allowed efficient formation of 80S complexes and also displayed wild-type kinetics of GTP hydrolysis in the initiation pathway (Figure S2 and data not shown).
Methods
Labeling of eIF1A was performed by dialyzing 216 nmol of protein against 2 L of enzyme buffer lacking DTT. Protein was concentrated to 1 mL and combined with 500 μL of 5 mM fluorescein-5-maleimide (Invitrogen) in DMF and incubated at room temperature for two hours with mild mixing. The sample was then filtered and subjected to ≥ 8 rounds of buffer exchange: ~15-fold dilution of sample with enzyme buffer, followed by concentration on a 10K MWCO concentrator (Millipore). Finally, the sample was dialyzed overnight against 2 L enzyme buffer. Protein concentration agreed very well with the concentration of incorporated fluorophore as measured using the extinction coefficient of the Fluorescein (83,000 cm−1M−1) at 515 nm.
To label eIF5B at its N-terminus with TAMRA, 150 nmols of eIF5B was dialyzed overnight against 2 L of 100 mM NaPO4 buffer, pH 7.0, supplemented with 75 mM KOAc and 2 mM DTT and concentrated to 1 mL final volume. To the protein sample, 100 μL of 1 mg/mL 5-(and-6)-carboxytetramethylrhodamine succinimidyl ester in DMSO was added and the sample was incubated at room temperature for one hour with gentle rocking. The protein-dye solution was then filtered, loaded onto an ~120 mL Superose 12 gel filtration column equilibrated with enzyme buffer and purified as for WT eIF5B.
Activity of Purified Components
Multiple preparations of all reagents were tested (with the exception of eIF1A-Fl), and results were consistent from preparation to preparation. Purified 40S ribosomal subunits were consistently found to be ~100% active for eIF1, eIF1A, TC and mRNA binding 15 and 80S IC formation (data not shown). eIF1 and eIF1A were fully active for binding to 40S subunits as determined by competitive binding assays (7 and data not shown). eIF2 was found to be ≥ 50% active as previously reported 48 and the factor was therefore added in excess to adjust for activity. eIF3 was found to be near 100% active for binding to eIF1, eIF5 and 43S complexes (S. Mitchell and J. Lorsch, unpublished data). Preparations of eIF5B were equally active in ribosome-dependent GTP hydrolysis and subunit joining experiments. 60S subunits were found to be nearly 100% active for formation of 80S ICs in native gel assays following incorporation of stoichiometric amounts of [35S]Met-tRNAi (data not shown). In addition, the equivalence of the measured second-order rate constant for joining of naked 40S and 60S subunits when either 40S or 60S subunits were the component varied (Fig. 2C) indicates that neither contains a substantial amount of inactive particles.
Kinetics of Ribosomal Subunit Joining
Kinetic experiments following ribosomal subunit joining were measured on an SX.180MV-R stopped flow fluorometer (Applied Photophysics) in fluorescence mode. The final concentrations of components were as follows: 100 nM each eIF1, eIF2 and Met-tRNA, 50 nM eIF1A and 40S subunits, 1 μM mRNA, 250 nM eIF5, 500 nM eIF5B, 100–1200 nM 60S subunits and 1 mM GTP•Mg2+. For each reaction, 2X 43S•mRNA complex containing 200 nM each eIF1, eIF2 and Met-tRNAi, 100 nM eIF1A and 40S subunits, 2 μM mRNA, and 20 μM GTP•Mg2+ in 1X reaction buffer was formed by adding preformed TC to the remaining components. 2X initiation mix contained 500 nM eIF5, 1 μM eIF5B, 400 – 3600 nM 60S subunits and 2 mM GTP•Mg2+ in 1X reaction buffer. Both samples were centrifuged in Eppenddorf tubes for 30 minutes at 14000 rpm to remove insoluble contaminants. In the stopped flow, the samples were mixed rapidly and the intensity of 435 nm light scattered at a right angle to the incident beam was measured as a function of time with the PMT set to 293V. Maintaining a consistent PMT voltage between experiments enabled comparison of the amplitudes of 80S complex formation under different reaction conditions. Increasing the concentration of eIF5B 3-fold in these experiments did not significantly affect the rate of subunit joining indicating that its concentration was saturating (data not shown). For subunit joining experiments with naked ribosomal subunits, eIF1 and eIF1A were added at 1 μM final concentration when present. The concentrations of 40S and 60S subunits and eIF5B were as above.
Error values
All experiments were performed at least in duplicate. Reported values are means of independent experiments and errors are mean deviations.
Kinetics of eIF1A Dissociation
Final concentrations were the same as for subunit joining experiments. In a quartz cuvette, TC was combined with 40S subunits, eIF1, eIF1A-E124C-Fl, mRNA and eIF5 in 1X reaction buffer. In the case of concurrent eIF1A chase, 80S complex formation was initiated by adding eIF5B, GTP•Mg2+, 60S subunits and eIF1A (3 μM final) in 1X reaction buffer. In the case of delayed eIF1A chase, 80S complex formation was initiated by adding eIF5B, GTP•Mg2+, and 60S subunits in 1X reaction buffer and eIF1A chase was added to a final concentration of 3 μM at 20 min. Addition of excess unlabeled eIF1A chase did not affect the rate or endpoint of 80S complex formation as monitored by light scattering (data not shown), suggesting that 80S complexes remain intact upon addition of 1A chase. Reactions lacking one or more component were supplemented with the appropriate buffer and performed following the same reaction scheme. Fluorescence anisotropy measurements were performed on a Spex Fluorolog-3 (J.Y. Horiba) by monitoring anisotropy as a function of time at λex = 492 nm and λem = 520 nm. Anisotropy values did not require correction for intensity change because fluorescence intensity differed < 4% between bound and unbound states.
Kinetics of TAMRA-eIF5B binding
43S•mRNA complex was formed by adding TC (formed with 4 μM eIF2 and Met-tRNAi, 200 μM GTP•Mg2+) in 1X reaction buffer to eIF1, eIF1A, 40S subunits (2 μM each) and mRNA (20 μM) in 1 X reaction buffer and incubating for 5 min. The 43S complex was then added to a cuvette containing GTP•Mg2+ and TAMRA-eIF5B in 1X reaction buffer and allowed to incubate for 5 min. eIF5 was then added and, after 2 min, the reaction was initiated by addition of 60S subunits. The final reaction contained 250 nM eIF5, 100 nM each TAMRA-eIF5B, eIF1, eIF1A and 40S subunits, 200 nM eIF2 and Met-tRNAi 100 μM GTP•Mg2+, 120 nM 60S subunits and 1 μM mRNA. Unlabeled eIF5B chase was used at a final concentration of 2 μM, a 20-fold excess over TAMRA-eIF5B. Fluorescence anisotropy was measured as a function of time at λex = 553 nm and λem = 579 nm.
Native gel assays
The native gel assay monitoring 80S complex formation was performed as described, at published concentrations, 38; 44 with slight modifications. To the 3X TC mix, 2.4 μM Met-tRNAi was added along with trace (≤ 1 nM) [35S]Met-tRNAi to form ternary complex. Additionally, eIF1A- E124C-Fl was included in the reaction in place of WT eIF1A. 1.5X 43S complex was formed by addition of 3X TC to 3X factors mix and incubated for 5 min at 26°C. Next, eIF5 was added and allowed to incubate for 2 min followed by initiation of 80S IC formation by addition of eIF5B, GTP•Mg2+, and 60S subunits. 80S complexes were formed for 5 min before addition of either excess unlabeled eIF1A (34 μM final concentration) or enzyme buffer, followed by incubation at 26°C for 30 min. Reactions were run on native polyacrylamide gels and the amount of [35S]Met-tRNAi in 80S complexes was quantified as previously described 38. To quantify the amount of E124C-Fl eIF1A present in 80S complexes, the gel was scanned using a Typhoon (General Electric) scanner and the fraction of E124C-Fl eIF1A in 80S complexes for each experiment was normalized to that in the experiment containing WT eIF5B•GDPNP, in which it was maximal.
Supplementary Material
Figure S1. Subunit joining on the initiation pathway occurs at a similar rate with WT eIF1A and eIF1A-Fl. Subunit joining with eIF1A-Fl (blue) in place of WT eIF1A occurs with a first-phase rate constant of 0.047 ± 0.004 s−1 and relative amplitude of 0.58 and a second phase rate constant of 0.014 ± 0.004 s−1 and relative amplitude of 0.37. This is similar to the rate of subunit joining with eIF5B•GDPNP (k1 = 0.044 ± 0.001 s−1, α1 = 0.7 ± 0.03; k2 = 0.013 ± 0.003 s−1, α2 = 0.27 ± .03; orange) and less than two-fold slower than subunit joining with eIF5B•GTP (k1 = 0.076 ± 0.004 s−1, α1 = 0.77 ± 0.06, k2 = 0.019 ± 0.006 s−1, α2 = 0.23 ± 0.06; red). Subunit joining with 40S and 60S subunits alone is shown for comparison (black).
Figure S2. TAMRA-eIF5B facilitates subunit joining to the same level as WT eIF5B. (a) Native gel showing 80S complexes formed with either WT eIF5B or TAMRA-eIF5B bound to either GTP or GDPNP. (b) Quantification of the bands in (a) determined as the fraction of [35S]Met-tRNAi present in 80S complexes normalized to that in the experiment containing WT eIF5B•GDPNP. Data in B is the average of two independent experiments and the errors are the mean deviation.
Figure S3. Stability of binding of Met-tRNAi in 80S complexes with different versions of eIF5B. (a) The same polyacrylamide gel as in Figure 4(a) but detecting [35S]Met-tRNAi. (b) Quantification of the bands in (a).
Figure S4. Subunit joining in the initiation pathway in the presence of eIF3. Addition of eIF3 to 80S IC formation experiments (black) results in subunit joining with a first-phase rate constant similar to that in the absence of the factor (k1 = 0.081 ± 0.012 s−1 vs. 0.076 ± 0.004 s−1, respectively) and a 1.7-fold decrease in amplitude (α1 = 0.44 ± 0.03 vs. 0.077 ± 0.06, respectively). With eIF3: k2 = 0.039 ± 0.002 s−1, α2 = 0.56 ± .02. Subunit joining along the initiation pathway in the absence of eIF3 is shown for comparison (red).
Figure S5. Cysteine-less eIF1A fully complements the eIF1A-null mutant in vivo. A plasmid containing TIF11 was altered to produce a protein product in which eIF1A’s two native cysteines at positions 51 and 89 were replaced with serine. Because TIF11 is an essential gene in yeast, the plasmid containing cysteine-less tif11 was transformed into yeast lacking a functional chromosomal copy of TIF11, instead possessing TIF11 on a plasmid. Via plasmid shuffle, the plasmid containing wild-type TIF11 was lost, and any growth observed was due to cysteine-less tif11. Cysteine-less eIF1A rescued the eIF1A-null phenotype at both 25°C and 30°C.
Acknowledgments
We thank members of our labs and Alan Hinnebusch for comments on the manuscript. Work on eIF1A was funded by NIH grant GM62128 to JRL and on eIF5B by American Cancer Society grant RSG-03-156-01 to JRL. The work was also supported by the Intramural Research Program of the NIH, NICHD (to TED).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–23. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu Rev Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 3.Pestova TV, Lorsch JR, Hellen CUT. The Mechanism of Eukaryotic Translation Initiation in Eukaryotes. In: Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2007. pp. 87–128. [Google Scholar]
- 4.Thomas A, Spann W, Van Steeg H, Voorma HO, Benne R. Mode of action of protein synthesis initiation factor eIF-1 from rabbit reticulocytes. FEBS Lett. 1980;116:67–71. doi: 10.1016/0014-5793(80)80530-8. [DOI] [PubMed] [Google Scholar]
- 5.Thomas A, Goumans H, Voorma HO, Benne R. The mechanism of action of eukaryotic initiation factor 4C in protein synthesis. Eur J Biochem. 1980;107:39–45. doi: 10.1111/j.1432-1033.1980.tb04621.x. [DOI] [PubMed] [Google Scholar]
- 6.Chaudhuri J, Chowdhury D, Maitra U. Distinct functions of eukaryotic translation initiation factors eIF1A and eIF3 in the formation of the 40S ribosomal preinitiation complex. J Biol Chem. 1999;274:17975–17980. doi: 10.1074/jbc.274.25.17975. [DOI] [PubMed] [Google Scholar]
- 7.Maag D, Lorsch JR. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J Mol Biol. 2003;330:917–24. doi: 10.1016/s0022-2836(03)00665-x. [DOI] [PubMed] [Google Scholar]
- 8.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
- 9.Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. Embo J. 2007;26:1602–14. doi: 10.1038/sj.emboj.7601613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pestova TV, Borukhov SI, Hellen CUT. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 11.Das HK, Das A, Ghosh-Dastidar P, Ralston RO, Yaghami B, Roy R, Gupta NK. Protein synthesis in rabbit reticulocyts. Purification and characterization of a double-stranded RNA-dependent protein synthesis inhibitor from reticulocyte lysates. J biol Chem. 1981;256:6491–6495. [PubMed] [Google Scholar]
- 12.Paulin FE, Campbell LE, O’Brien K, Loughlin J, Proud CG. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr Biol. 2001;11:55–9. doi: 10.1016/s0960-9822(00)00025-7. [DOI] [PubMed] [Google Scholar]
- 13.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–93. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majumdar R, Maitra U. Regulation of GTP hydrolysis prior to ribosomal AUG selection during eukaryotic translation initiation. Embo J. 2005;24:3737–46. doi: 10.1038/sj.emboj.7600844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell. 2005;17:265–75. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 16.Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev. 2007;21:1217–30. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell. 2005;20:251–62. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 18.Yatime L, Mechulam Y, Blanquet S, Schmitt E. Structure of an archaeal heterotrimeric initiation factor 2 reveals a nucleotide state between the GTP and the GDP states. Proc Natl Acad Sci U S A. 2007;104:18445–50. doi: 10.1073/pnas.0706784104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–22. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–36. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–5. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- 22.Lee JH, Pestova TV, Shin BS, Cao C, Choi SK, Dever TE. Initiation factor eIF5B catalyzes second GTP-dependent step in eukaryotic translation initiation. Proc Natl Acad Sci U S A. 2002;99:16689–94. doi: 10.1073/pnas.262569399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin BS, Maag D, Roll-Mecak A, Arefin MS, Burley SK, Lorsch JR, Dever TE. Uncoupling of initiation factor eIF5B/IF2 GTPase and translational activities by mutations that lower ribosome affinity. Cell. 2002;111:1015–1025. doi: 10.1016/s0092-8674(02)01171-6. [DOI] [PubMed] [Google Scholar]
- 24.Kyrpides NC, Woese CR. Universally conserved translation initiation factors. Proc Natl Acad Sci U S A. 1998;95:224–8. doi: 10.1073/pnas.95.1.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roll-Mecak A, Shin BS, Dever TE, Burley SK. Engaging the ribosome: universal IFs of translation. Trends Biochem Sci. 2001;26:705–9. doi: 10.1016/s0968-0004(01)02024-2. [DOI] [PubMed] [Google Scholar]
- 26.Fakunding JL, Hershey JW. The interaction of radioactive initiation factor IF-2 with ribosomes during initiation of protein synthesis. J Biol Chem. 1973;248:4206–12. [PubMed] [Google Scholar]
- 27.Gualerzi CO, Pon CL. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–9. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- 28.Moreno JM, Drskjotersen L, Kristensen JE, Mortensen KK, Sperling-Petersen HU. Characterization of the domains of E. coli initiation factor IF2 responsible for recognition of the ribosome. FEBS Lett. 1999;455:130–4. doi: 10.1016/s0014-5793(99)00858-3. [DOI] [PubMed] [Google Scholar]
- 29.Carter AP, Clemons WM, Jr, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan VV. Crystal Structure of an Initiation Factor Bound to the 30S Ribosomal Subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 30.Moazed D, Samaha RR, Gualerzi C, Noller HF. Specific protection of 16 S rRNA by translational initiation factors. J Mol Biol. 1995;248:207–10. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- 31.Battiste JL, Pestova TV, Hellen CU, Wagner G. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol Cell. 2000;5:109–19. doi: 10.1016/s1097-2765(00)80407-4. [DOI] [PubMed] [Google Scholar]
- 32.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci U S A. 2001;98:7029–36. doi: 10.1073/pnas.111145798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grunberg-Manago M, Dessen P, Pantaloni D, Godefroy-Colburn T, Wolfe AD, Dondon J. Light-scattering studies showing the effect of initiation factors on the reversible dissociation of Escherichia coli ribosomes. J Mol Biol. 1975;94:461–78. doi: 10.1016/0022-2836(75)90215-6. [DOI] [PubMed] [Google Scholar]
- 34.Antoun A, Pavlov MY, Andersson K, Tenson T, Ehrenberg M. The roles of initiation factor 2 and guanosine triphosphate in initiation of protein synthesis. EMBO J. 2003;22:5593–601. doi: 10.1093/emboj/cdg525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi SK, Olsen DS, Roll-Mecak A, Martung A, Remo KL, Burley SK, Hinnebusch AG, Dever TE. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol Cell Biol. 2000;20:7183–91. doi: 10.1128/mcb.20.19.7183-7191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003;22:193–204. doi: 10.1093/emboj/cdg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: A new interaction between old partners. Proc Natl Acad Sci U S A. 2003;100:1535–1540. doi: 10.1073/pnas.0437845100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Acker MG, Shin BS, Dever TE, Lorsch JR. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem. 2006;281:8469–75. doi: 10.1074/jbc.M600210200. [DOI] [PubMed] [Google Scholar]
- 39.Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol Cell Biol. 2007;27:2384–97. doi: 10.1128/MCB.02254-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goss DJ, Rounds D, Harrigan T, Woodley CL, Wahba AJ. Effects of eucaryotic initiation factor 3 on eucaryotic ribosomal subunit equilibrium and kinetics. Biochemistry. 1988;27:1489–1494. doi: 10.1021/bi00405a014. [DOI] [PubMed] [Google Scholar]
- 41.Goss DJ, Rounds DJ. A kinetic light-scattering study of the binding of wheat germ protein synthesis initiation factor 3 to 40S ribosomal subunits and 80S ribosomes. Biochemistry. 1988;27:3610–3. doi: 10.1021/bi00410a012. [DOI] [PubMed] [Google Scholar]
- 42.Kainuma M, Hershey JW. Depletion and deletion analyses of eucaryotic translation initiation factor 1A in Saccharomyces cerevisiae. Biochimie. 2001;83:505–14. doi: 10.1016/s0300-9084(01)01279-2. [DOI] [PubMed] [Google Scholar]
- 43.Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV. Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes & Development. 2003;17:2786–2797. doi: 10.1101/gad.1141803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Acker MG, Kolitz SE, Mitchell SF, Nanda JS, Lorsch JR. Reconstitution of yeast translation initiation. Methods Enzymol. 2007;430:111–45. doi: 10.1016/S0076-6879(07)30006-2. [DOI] [PubMed] [Google Scholar]
- 45.Johnson KA. The Enzymes. In: Sigman DS, editor. Transient-state kinetic analysis of enzyme reaction pathways. Vol. 3. Academic Press; 1992. pp. 1–61. [Google Scholar]
- 46.Palmiter RD. Quantitation of parameters that determine the rate of ovalbumin synthesis. Cell. 1975;4:189–197. doi: 10.1016/0092-8674(75)90167-1. [DOI] [PubMed] [Google Scholar]
- 47.Hinnebusch AG. Translational control of GCN4: gene-specific regulation by phosphorylation of eIF2. In: Hershey JWB, Matthews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor Laboratory Press; Cold Spring Harbor: 1996. pp. 199–244. [Google Scholar]
- 48.Kapp LD, Lorsch JR. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol. 2004;335:923–36. doi: 10.1016/j.jmb.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 49.Lorsch JR, Herschlag D. Kinetic dissection of fundamental processes of eukaryotic translation initiation in vitro. EMBO J. 1999;18:6705–6717. doi: 10.1093/emboj/18.23.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Simonetti A, Marzi S, Myasnikov AG, Fabbretti A, Yusupov M, Gualerzi CO, Klaholz BP. Structure of the 30S translation initiation complex. Nature. 2008 doi: 10.1038/nature07192. [DOI] [PubMed] [Google Scholar]
- 51.Maag D, Algire MA, Lorsch JR. Communication between Eukaryotic Translation Initiation Factors 5 and 1A within the Ribosomal Pre-initiation Complex Plays a Role in Start Site Selection. J Mol Biol. 2006;356:724–37. doi: 10.1016/j.jmb.2005.11.083. [DOI] [PubMed] [Google Scholar]
- 52.Hinnebusch AG. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–62. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 53.Von Der Haar T, McCarthy JE. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol Microbiol. 2002;46:531–44. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 54.Algire MA, Maag D, Savio P, Acker MG, Tarun SZ, Sachs AB, Asano K, Nielsen KH, Olsen DS, Phan L, Hinnebusch AG, Lorsch JR. Development and characterization of a reconstituted yeast translation initiation system. RNA. 2002;8:382–397. doi: 10.1017/s1355838202029527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muir TW, Dolan S, Cole PA. Expressed protein ligation: A general method for protein engineering. Proc Natl Acad Sci USA. 1998;95:6705–6710. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fekete CA, Applefield DJ, Blakely SA, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. The eIF1A C-terminal domain promotes initiation complex assembly, scanning and AUG selection in vivo. Embo J. 2005;24:3588–601. doi: 10.1038/sj.emboj.7600821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Subunit joining on the initiation pathway occurs at a similar rate with WT eIF1A and eIF1A-Fl. Subunit joining with eIF1A-Fl (blue) in place of WT eIF1A occurs with a first-phase rate constant of 0.047 ± 0.004 s−1 and relative amplitude of 0.58 and a second phase rate constant of 0.014 ± 0.004 s−1 and relative amplitude of 0.37. This is similar to the rate of subunit joining with eIF5B•GDPNP (k1 = 0.044 ± 0.001 s−1, α1 = 0.7 ± 0.03; k2 = 0.013 ± 0.003 s−1, α2 = 0.27 ± .03; orange) and less than two-fold slower than subunit joining with eIF5B•GTP (k1 = 0.076 ± 0.004 s−1, α1 = 0.77 ± 0.06, k2 = 0.019 ± 0.006 s−1, α2 = 0.23 ± 0.06; red). Subunit joining with 40S and 60S subunits alone is shown for comparison (black).
Figure S2. TAMRA-eIF5B facilitates subunit joining to the same level as WT eIF5B. (a) Native gel showing 80S complexes formed with either WT eIF5B or TAMRA-eIF5B bound to either GTP or GDPNP. (b) Quantification of the bands in (a) determined as the fraction of [35S]Met-tRNAi present in 80S complexes normalized to that in the experiment containing WT eIF5B•GDPNP. Data in B is the average of two independent experiments and the errors are the mean deviation.
Figure S3. Stability of binding of Met-tRNAi in 80S complexes with different versions of eIF5B. (a) The same polyacrylamide gel as in Figure 4(a) but detecting [35S]Met-tRNAi. (b) Quantification of the bands in (a).
Figure S4. Subunit joining in the initiation pathway in the presence of eIF3. Addition of eIF3 to 80S IC formation experiments (black) results in subunit joining with a first-phase rate constant similar to that in the absence of the factor (k1 = 0.081 ± 0.012 s−1 vs. 0.076 ± 0.004 s−1, respectively) and a 1.7-fold decrease in amplitude (α1 = 0.44 ± 0.03 vs. 0.077 ± 0.06, respectively). With eIF3: k2 = 0.039 ± 0.002 s−1, α2 = 0.56 ± .02. Subunit joining along the initiation pathway in the absence of eIF3 is shown for comparison (red).
Figure S5. Cysteine-less eIF1A fully complements the eIF1A-null mutant in vivo. A plasmid containing TIF11 was altered to produce a protein product in which eIF1A’s two native cysteines at positions 51 and 89 were replaced with serine. Because TIF11 is an essential gene in yeast, the plasmid containing cysteine-less tif11 was transformed into yeast lacking a functional chromosomal copy of TIF11, instead possessing TIF11 on a plasmid. Via plasmid shuffle, the plasmid containing wild-type TIF11 was lost, and any growth observed was due to cysteine-less tif11. Cysteine-less eIF1A rescued the eIF1A-null phenotype at both 25°C and 30°C.