Abstract
Background
The liver fluke Opisthorchis viverrini is classified as a class I carcinogen due to the association between cholangiocarcinoma and chronic O. viverrini infection. During its feeding activity within the bile duct, the parasite secretes several cathepsin F cysteine proteases that may induce or contribute to the pathologies associated with hepatobiliary abnormalities.
Methodology/Principal Findings
Here, we describe the cDNA, gene organization, phylogenetic relationships, immunolocalization, and functional characterization of the cathepsin F cysteine protease gene, here termed Ov-cf-1, from O. viverrini. The full length mRNA of 1020 nucleotides (nt) encoded a 326 amino acid zymogen consisting of a predicted signal peptide (18 amino acids, aa), prosegment (95 aa), and mature protease (213 aa). BLAST analysis using the Ov-CF-1 protein as the query revealed that the protease shared identity with cathepsin F-like cysteine proteases of other trematodes, including Clonorchis sinensis (81%), Paragonimus westermani (58%), Schistosoma mansoni and S. japonicum (52%), and with vertebrate cathepsin F (51%). Transcripts encoding the protease were detected in all developmental stages that parasitize the mammalian host. The Ov-cf-1 gene, of ∼3 kb in length, included seven exons interrupted by six introns; the exons ranged from 69 to 267 bp in length, the introns from 43 to 1,060 bp. The six intron/exon boundaries of Ov-cf-1 were conserved with intron/exon boundaries in the human cathepsin F gene, although the gene structure of human cathepsin F is more complex. Unlike Ov-CF-1, human cathepsin F zymogen includes a cystatin domain in the prosegment region. Phylogenetic analysis revealed that the fluke, human, and other cathepsin Fs branched together in a clade discrete from the cathepsin L cysteine proteases. A recombinant Ov-CF-1 zymogen that displayed low-level activity was expressed in the yeast Pichia pastoris. Although the recombinant protease did not autocatalytically process and activate to a mature enzyme, trans-processing by Fasciola hepatica cathepsin L cleaved the prosegment of Ov-CF-1, releasing a mature cathepsin F with activity against the peptide Z-Phe-Arg-NHMec >50 times that of the zymogen. Immunocytochemistry using antibodies raised against the recombinant enzyme showed that Ov-CF-1 is expressed in the gut of the mature hermaphroditic fluke and also in the reproductive structures, including vitelline glands, egg, and testis. Ov-CF-1 was detected in bile duct epithelial cells surrounding the flukes several weeks after infection of hamsters with O. viverrini and, in addition, had accumulated in the secondary (small) bile ducts where flukes cannot reach due to their large size.
Conclusions/Significance
A cathepsin F cysteine protease of the human liver fluke O. viverrini has been characterized at the gene and protein level. Secretion of this protease may contribute to the hepatobiliary abnormalities, including cholangiocarcinogenesis, observed in individuals infected with this parasite.
Author Summary
Opisthorchiasis, oriental liver fluke infection, is a food-borne parasitic disease that afflicts millions of residents in northern Thailand and Laos. Related infections occur in North Asia, including China and Korea. This kind of liver fluke infection is the consequence of eating certain uncooked or undercooked freshwater fish contaminated with the larvae of the parasite Opisthorchis viverrini. Whereas the infection can cause disease in the bile ducts and liver, infection with the oriental fluke can lead to the development of a liver cancer, cholangiocarcinoma (bile duct cancer). Our recent studies have begun to focus on products and metabolites from the parasite that are carcinogenic. Many proteolytic enzymes are known to be secreted by parasites. This report centers on a specific category of protease, termed cathepsin F. We determined here that O. viverrini expresses a cathepsin F in its gut and in other organs. In the liver fluke, cathepsin F likely plays a role in digesting ingested human cells. The gene encoding the parasite enzyme shows evolutionary relatedness to a similar gene in humans. The fluke cathepsin F also is released from the parasite into livers of infected mammals, where it appears to contribute to inflammation surrounding the parasite. In this regard, it may be involved in early events that lead to bile duct cancer.
Introduction
Opisthorchis viverrini is an important human food-borne pathogen endemic in mainland Southeast Asia, predominantly Northeast Thailand [1],[2]. Infection with this liver fluke parasite causes opisthorchiasis, which is associated with a number of hepatobiliary abnormalities, including cholangitis, obstructive jaundice, hepatomegaly, cholecystitis, cholelithiasis and cholangiocarcinoma. O. viverrini infection induces pathological changes including epithelial desquamation, epithelial and adenomatous hyperplasia, goblet cell metaplasia, inflammation, periductal fibrosis and granuloma formation [3]. Experimental and epidemiological findings implicate O. viverrini infection in the etiology of cholangiocarcinoma (CCA), cancer of the bile ducts (reviewed in [1]). O. viverrini is one of only two metazoan pathogens of humans that is considered a Group 1 carcinogen [4],[5].
A number of studies suggest that inflammation of the bile ducts caused by O. viverrini infection and induction of endogenous nitric oxide are important factors for cholangiocarcinogenesis [6],[7]. Other studies have related cell proliferation induced by O. viverrini, its antigens and metabolites, and exposure to exogenous carcinogens such as nitrates, nitrites and even N-nitroso compounds found in fermented or preserved foods such as pla-ra (traditional Thai fermented fish), as factors involved with parasite-associated cholangiocarcinogenesis [8]. The pathogenesis of O. viverrini-mediated hepatobiliary changes, which in turn can lead to CCA, can be attributed to greater or lesser degree to mechanical irritation caused by the liver fluke suckers and to the action of molecules excreted or secreted by the parasite [1].
There may be informative analogies to be drawn between O. viverrini-induced CCA and the development of stomach cancer caused by infection with CagA-positive strains of Helicobacter pylori. In the latter situation, the CagA antigen is delivered by the H. pylori bacterium into gastric epithelial cells, where it undergoes tyrosine phosphorylation. Phosphorylated CagA activates SHP-2 tyrosine phosphatase, causing morphological transformation of the infected cell to the hummingbird phenotype. CagA also destabilizes the E-cadherin/beta-catenin complex to elicit aberrant activation of the beta-catenin signal. These events in signal dysregulation underlie stomach cell metaplasia [9],[10]. Whereas our understanding of cholangiocarcinogenesis is less advanced than with H. pylori-associated gastric adenocarcinoma, CCA—in like fashion to gastric adenocarcinoma—is an epithelial cell adenocarcinoma associated with a gastrointestinal tract pathogen.
In like fashion to CagA-associated stomach cancer, it is conceivable that products secreted or released by O. viverrini into the neighboring bile duct epithelia might promote cholangiocarcinogenesis. The parasite-released mediators might down-regulate apoptosis and/or they may stimulate epithelial cell growth. Accordingly, we have begun to examine the secretome of O. viverrini proteome with a particular interest in proteolytic enzymes since they are prominent components of ES of helminth parasites at large [11]–[13]. Recently we reported the biochemical characterization of cysteine protease activities in extracts of several developmental stages of O. viverrini [14]. We demonstrated that O. viverrini expresses clan CA-like cysteine protease activity with elevated expression in the metacercariae, suggesting that this enzyme activity might participate in larval excystation during mammalian infection. The proteolytic activity was also detected in excretory/secretory (ES) products of sexually mature parasites [14].
In the present report, we have identified a transcript and its genome locus encoding a cathepsin F-like cysteine protease from O. viverrini, which we termed Ov-CF-1. Analysis of the genomic structure of the Ov-cf-1 gene revealed conserved exon/intron boundaries with the gene encoding human cathepsin F, although the human gene exhibits a more complex structure including a zymogen with a cystatin domain found within the pro-segment that is absent from the O. viverrini gene. Phylogenetic analysis revealed that the deduced Ov-CF-1 protease was an orthologue of cathepsin F cysteine proteases, a clade distinct from the cathepsin L proteases described in several related liver and blood flukes. Nevertheless, characterization of a functionally active recombinant form of Ov-CF-1 shows that this enzyme cleaves diagnostic peptides (Z-Leu-Arg-NHMec, Z-Phe-Arg-NMHec) that are also cleaved by cathepsin L. Ov-CF-1 is secreted by adult O. viverrini and immunocytochemical studies localized the protease in the cecum of the adult stage of the parasite. Liberation of Ov-CF-1 protease by adult parasites residing in the bile duct may contribute to the pathologies associated with O. viverrini-induced hepatobiliary abnormalities.
Materials and Methods
Opisthorchis viverrini; Genomic DNA and RNA Extraction; RT-PCR
Metacercariae of O. viverrini were obtained by digestion with pepsin of flesh of naturally infected cyprinoid fish collected from an endemic area of Khon Kaen province, Thailand. About 100 metacercariae of O. viverrini were used to infect hamsters, Mesocricetus auratus, by stomach intubation, as described [15]. Infection of hamsters with liver flukes and maintenance of hamsters was carried out at the animal facility of the Faculty of Medicine, Khon Kaen University, using procedures approved by the Animal Ethics Committee of Khon Kaen University. The hamsters were euthanized six weeks after infection, after which the adult O. viverrini flukes were perfused from the bile ducts with phosphate-buffered saline (PBS), pH 7.2. The worms were washed with sterile saline, after which genomic DNA (gDNA) was extracted using kits from Gentra Systems (Minneapolis, MN) for long range PCR or Bio-Rad (Hercules, CA) for inverted PCR templates. Eggs of O. viverrini were recovered from tissue culture medium where they had been discharged from adult worms [16]. Total RNA was isolated from adults, eggs and metacercariae of O. viverrini using Trizol [16],[17]. Contaminating gDNA was removed by treatment of RNA with DNase I (Promega, Madison, WI). For reverse transcription-PCR (RT-PCR), first-strand cDNA was produced with an oligo (dT) primer from 1.0 µg of total RNA using SUPERSCRIPT III reverse transcriptase (Invitrogen, Carlsbad, CA) at 42°C for 60 min. One µl of the resultant cDNA was employed as template in the presence of primers specific for Ov-cf-1 (5′-TCGGACCAGTATTGGACCAAG -3′ and 5′-TACGCTGGAAAGCACACAACG), with thermal cycling conditions of 30 sec denaturation at 94°C, 30 sec annealing at 55°C and 30 sec extension at 72°C for 30 cycles. Control RT-PCR reactions were performed without reverse transcriptase to ensure that amplified products were derived from cDNA and not contaminating genomic DNA. PCR products were subjected to electrophoresis through 1% agarose and visualized under UV light after staining with ethidium bromide.
Homology and RACE-PCR Approaches
To search for transcripts encoding Clan CA, Family C1 cysteine proteases of O. viverrini, degenerate primers were designed to target conserved domains of papain-like cysteine proteases. The degenerate forward primer, 5′-TGYGGNTCNTGYTGGGCDTTYTCN targeted the conserved CGSCWAFV residues and the reverse primer, 5′-CCARCTRTTYTTBACDATCCARTA targeted the conserved YWIVKNSW residues (Figure 1), where B = C/G/T; D = A/G/T; N = A/C/G/T; R = A/G; Y = C/T. These primers were employed to amplify target sequences from cDNA of adult O. viverrini. In addition, specific primers Ov-cf-spf1 (5′-TGTGGTTCGTGTTGGGCATTTTCT) and Ov-cf-spr1 (5′-CCAACTGTTTTTGACCGTCCAGTA) targeting the same regions were designed based on the nucleotide sequence of Clonorchis sinensis cathepsin F (DQ909018). PCR was performed as follows; 95°C for 5 min followed by 35 cycles of 94°C for 30 sec, 55°C for 30 sec, 72°C for 30 sec and finally, 72°C for 7 min. Fragments of cDNA sequence encoding an O. viverrini cysteine protease were obtained by using rapid PCR amplification of cDNA ends (RACE); 5′- and 3′-RACE were performed using specific primers Ov-cf-spf1 and Ov-cf-spr1 (above) paired with linker specific primers (Invitrogen). Subsequently, a full length transcript termed Ov-cf-1 was obtained by PCR using primers designed from nucleotide sequences obtained from 5′- and 3′-RACE products.
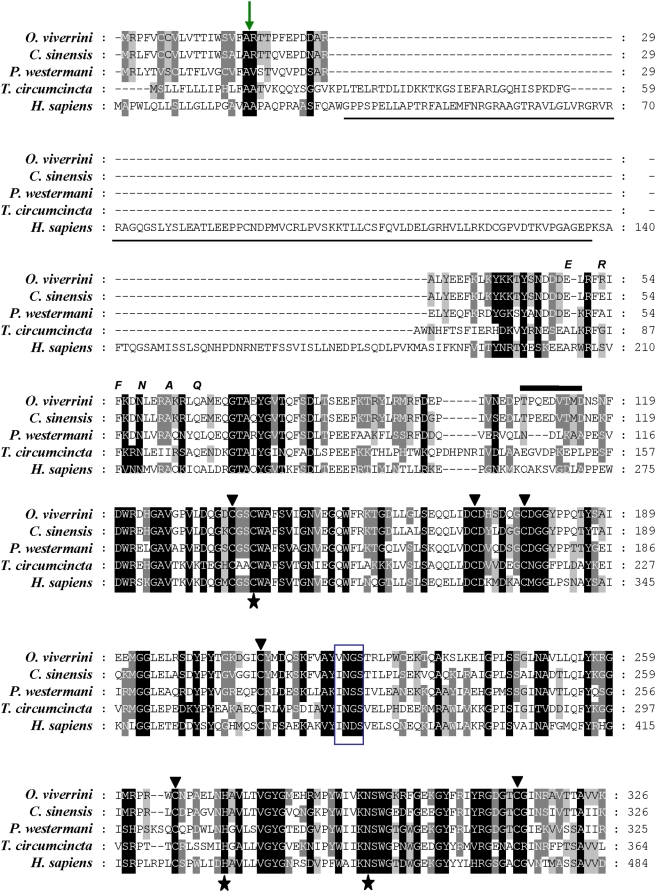
Figure 1. Multiple sequence alignment of deduced amino acids of the cathepsin F from Opisthorchis viverrini Ov-CF-1 (AAV69023).
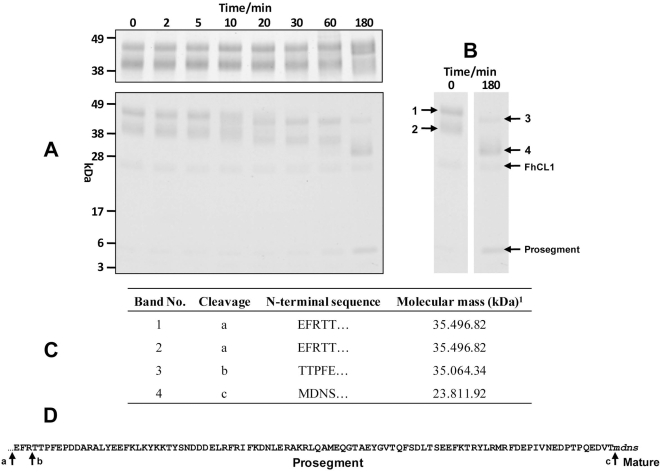
The alignment includes other members of the Clan CA, family C1 peptidase family, including CsCF-6 from Clonorchis sinensis (ABK918111) and orthologues from Paragonimus westermani (AAY81944), Teladorsagia circumcincta (ABA01328) and Homo sapiens (NP_003784). (The T. circumcincta orthologue was included as a representative of a nematode helminth sequence, while human cathepsin F sequence was included to highlight the large prosegment with a cystatin domain, underlined, that exists in some cathepsins F.) Identical residues in more than 50% of sequences are presented in black boxes. Conserved substitutions are identified by grey boxes. The conserved active site cysteine, asparagine and histidine residues are highlighted (stars). The protease-susceptible region at the junction of the prosegment and mature domain is indicated by the thick black line. This region contains the cleavage sites for release of the prosegment from the mature domain and includes the FhCL1 cleavage site for Ov-CF-1, VT↓MDNSNFD, and the cleavage site in human cathepsin F, DL↓APPEWD, determined by X-ray crystallography [50]. The thin (green) arrow indicates the position of signal peptide cleavage site. The arrow heads indicate six cysteine residues likely involved in disulfide bridges. The box indicates a conserved, putative N-linked glycosylation site, NGS in the mature enzyme domain. The position of the ERFNAQ propeptide motif, conserved in cathepsin F, is indicated.
Inverse and Long Range PCR Approaches
Flanking and 5′-UTR sequences upstream of the Ov-cf-1 gene were amplified using inverse PCR (iPCR) [18]. O. viverrini genomic DNA (gDNA) was digested with Hind III, after which the fragments (50 ng/µl) were circularized using T4 DNA ligase (New England Biolabs, Ipswich, MA). One hundred ng of circularized gDNA fragments was employed as template for amplification of the putative 5′-UTR of the gene. Primers IPCRCPFw (5′-CAGGCTCGAATGGGGTAGTTCTGGC) and IPCRCPRe (5′-GCCCGGGCACTATACGAGGAGTTCA) were derived orientated in reverse direction to the mRNA of Ov-cf-1 (AY821800). iPCR reactions were carried out in 100 µl in 50 mM KCl, 10 mM Tris-HCl (pH 9.0 at 25° C), 0.1% Triton X-100, 1.5 mM MgCl2, 0.5 mM dNTP, 0.5 U PfuTurboCx Hotstart DNA polymerase (Promega), with thermal cycling conditions of 94° C for 2 min, followed by 30 cycles of 94° C for 60 sec, 58° C for 60 sec and 72°C for 2 min, and a final extension at 72°C for 10 min. Products were cloned into plasmid TOPO XL PCR (Invitrogen) and nucleotide sequence of inserts determined. In addition, toobtain a genomic fragment spanning the entire gene locus, the Ov-cf-1 gene locus was amplified from gDNA of O. viverrini with the primers specific for the 5′- (CPFw, 5′-GACCTTTCGTGTGTTGCGTGTT) and 3′-termini (CPRe, 5′-GGGCAAAATATCAACATGGAG) of the transcript (GenBank AY821800) using LongRange PCR DNA polymerase (Qiagen, Tubingen, Germany). These amplifications were performed in 50 µl reaction volumes using 100 ng gDNA, 1× LongRange PCR buffer, 1× Q-solution, 0.4 µM of each primer, 2.5 mM MgCl2, 500 µM of each dNTP, 2 units of LongRange PCR DNA polymerase. After an initial denaturation step at 93°C for 3 min, 40 thermal cycles of 93°C for 15 sec, 50°C for 30 sec, 68°C for 15 min were carried out, followed b a final step of 68°C for 7 min. PCR products were purified (Gel DNA extraction kit, Real Genomics, Real Biotech Corporation, Taipei, Taiwan) and ligated to the vector pCR4-TOPO (Invitrogen). The ligation products were used to transform E. coli competent cells (One shot Mach1-T1; Invitrogen) which were cultured in the presence of ampicillin. Plasmid DNA was isolated from clones, and the sequence of long range PCR products determined by gene walking with gene specific primers.
Bioinformatics
Bioinformatics analyses to predict potential open reading frames (ORFs), signal peptides, and/or transmembrane domains, conserved domains, and molecular mass were undertaken using ExPASy (http://expasy.org), including the Compute pI/MW tool, Signal P (www.cbs.dtu.dk/services/SignalP), and TMPred 3.0 (www.ch.embrut.org/software/TMPRED_form.html). Assembly of contiguous sequences and multiple alignments were performed with BioEdit software version 7.0.0, http://www.mbio.ncsu.edu/BioEdit/bioedit.html [19]. Analysis of gene loci using BLAST searches at NCBI was undertaken to determine exon and intron structures and boundaries of the O. viverrini cysteine protease gene, upstream regulatory sequences, and identities of unrelated sequences populating the introns. Positions of splice sites were predicted using NetGene2 program, http://www.cbs.dtu.dk/services/NetGene2/ [20]. For phylogenetic analysis, the amino acid sequence of Ov-cf-1 (GenBank AY821800) was aligned with cathepsin F-like and cathepsin L-like, and related cysteine proteases of a variety of eukaryotes using ClustalW algorithms in the BioEdit software suite [21]. Species names and accession numbers of contributing sequences are provided in the phylogram (below). Cathepsin B from Schistosoma japonicum (AY222871) was used to root the tree. Multiple sequence alignments were calculated with the distance matrix method using Prodist and Neighbor in Phylip software packages to construct neighbor joining trees. Trees were analyzed under 1,000 bootstrap resampling values, and drawn with TreeView [22].
Expression and Purification of Recombinant Ov-CF-1
The sequence encoding the proform of Ov-CF-1 was amplified from cDNA of adult O. viverrini using the primers (Ov-cp-f1) 5′-CGCGCGGAATTCAGAACTACCCATTCGAG and the (Ov-cp-r1) 5′-CGCGCGTCTAGACGTTTGACAAGGCTGTAGT, which include restriction sites for EcoR I and Xba I (underlined). PCR products were cloned into the yeast expression vector pPICZα (Invitrogen) using EcoR I and Xba I restriction sites, and the reading frame confirmed by sequencing. The recombinant plasmid was linearized with Pme I and employed to transform the ×33 strain of Pichia pastoris (Invitrogen) by electroporation. Transformed yeasts were selected on 1 mg/ml zeocin (Invitrogen) Yeast Peptone Dextrose (YPD) plates, screened by PCR using Ov-cf-1 gene specific primers, and positive colonies investigated for protein expression by immunoblotting using a monoclonal antibody specific for the hexa-His tag (Invitrogen). A 1.5 litre P. pastoris culture was fermented using a clone that exhibited strong expression. The culture supernatant at 96 hours after induction was concentrated to 200 ml by ultrafiltration through a 10 kDa cut off membrane (Pall Scientific), and buffer exchanged into 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole (binding buffer). Recombinant enzyme was affinity purified on Ni-NTA resin (Qiagen), buffer exchanged into PBS, and protein concentration determined using the bicinchoninic acid assay (Pierce, Rockford, IL).
Antiserum to Fluke Enzyme, Immunoblots, Immunolocalization
Antiserum against affinity purified Ov-CF-1 was produced by immunization of a male New Zealand White rabbit. Pre-immune blood was collected from the marginal ear vein two days prior to the first injection. For the first immunization, the rabbit was injected subcutaneously with 1.0 mg of recombinant Ov-CF-1 emulsified with an equal volume of Freund's complete adjuvant. The second and third immunizations were carried out with 1.0 mg recombinant protein formulated with Freund's incomplete adjuvant. Immunizations were conducted on days 1, 15 and 29, blood was collected two weeks after the third immunization, and its serum was separated. The specific anti-Ov-CF-1 IgG titer of the rabbit serum was determined by enzyme linked immunosorbent assay, and antiserum was stored at −20°C until needed. Cysteine protease activities were enriched using thiol-sepharose chromatography from ES products and preparations of soluble adult O. viverrini worms (somatic preparation), as detailed [14]. ES and somatic fractions eluted from the thiol-sepharose were sized by SDS-PAGE, electro-transferred to nitrocellulose, and probed with the rabbit antiserum in order to determine the presence of the Ov-CF-1.
O. viverrini adult worms or liver tissue from hamsters infected with O. viverrini (weeks 1–24) were fixed and cut with a microtome into sections of 4 µm [23]. The sections were deparaffinized in xylene, hydrated in a series of ethanol and water, respectively. Endogenous peroxidase was eliminated by incubation of the sectioned tissues in 5% H2O2 in methanol for 30 min. Subsequently, sections were washed in water and PBS, and non-specific staining was blocked by incubation in 5% pre-immunization rabbit serum in PBS for 30 min. Sections were probed with rabbit anti-Ov-CF-1 antiserum or pre-immunization serum from the same rabbit diluted at 1∶100 (v/v) in PBS at 4°C overnight. After rinsing 3×5 min with PBS, sections were incubated with HRP-conjugated goat anti-rabbit IgG for 1 h. Sections were rinsed with PBS, 2×10 min, after which bound antibody was detected using the substrate diaminobenzidine (DAB). Sections were counterstained with Mayer's hematoxylin, dehydrated, cleared in xylene and mounted in Permount. Images of sections were recorded using a digital camera (Nikon DXA 1200 C) fitted to an Olympus model BX40 compound microscope.
Biochemical Analysis, Activation, and Processing of Recombinant Ov-CF-1
To determine whether the recombinant Ov-CF-1 was capable of auto-catalytic activation, 20 µl enzyme (100 µg) was added to 100 µl activation buffer (0.1 M sodium acetate, pH 4.5, 1 mM DTT, 1.25 mM EDTA) and incubated at 37°C. Aliquots of 10 µl were removed at various time points, transferred into tubes containing 1 µl of 1 mM E-64 to halt the enzymatic reaction. The reaction products were analyzed by 4–12% Bis-Tris NuPage gel (Invitrogen) electrophoresis. Trans-processing of recombinant Ov-CF-1 was undertaken by mixing 50 µg purified recombinant enzyme with 5.0 µg of recombinant O. viverrini asparaginyl endopeptidase [17] or an activated recombinant Fasciola hepatica cathepsin L (FhCL1) [24] in 0.1 M sodium acetate, pH 4.5, 1 mM DTT , 1.25 mM EDTA in a total reaction volume of 150 µl. The mixture was incubated for 180 min at 37°C, and samples (10 µl) were removed at intervals for analysis (as above).
N-terminal sequencing was performed on recombinant Ov-CF-1 and samples taken at time 180 min following trans-processing with FhCL1. Following 4–12% Bis-Tris NuPage, proteins were transferred to a polyvinylidene fluoride immobilon-P membrane (Millipore) at 120 mA for 45 min. The membrane was washed with distilled water and stained with 0.025% Coomassie Brilliant Blue R-250 in 40% methanol, 10% acetic acid. Selected protein bands were subjected to 5 cycles of N-terminal (Edman) sequencing at the Biomolecular Research Facility (BRF) at the University of Newcastle (NSW, Australia). To analyze the enzyme specificity of Ov-CF-1 and to monitor the course of activation during the trans-processing experiments, we employed three fluorogenic peptide substrates with different residues at the P2 position, Z-Phe-Arg-NHMec, Z-Leu-Arg-NHMec and Z-Pro-Arg-NHMec (Bachem). Assays were performed in 0.1 M sodium acetate, pH 4.5, 1 mM DTT, 1.25 mM EDTA (200 µl) in 96-well plates. Reaction kinetics were monitored over 150 min at 37°C by measuring the release of the fluorogenic leaving group (NHMec) at 370 nm excitation, 460 nm emission wavelength, using a Bio-Tek KC4 microfluorometer. Reactions with each fluorogenic substrate included (a) no enzyme control, (b) recombinant Ov-CF-1 alone, (c) recombinant FhCL1 alone, and (d) Ov-CF-1 +FhCL1.
Results
A Transcript Encoding a Cathepsin F-like Protease from Adult O. viverrini Liver Fluke
We undertook PCR investigations targeting conserved active site encoding regions of clan CA peptidases employing (a) degenerate primers or (b) primers specific for the C. sinensis cathepsin F gene (DQ909018). The former approach did not amplify products of the expected size whereas DQ909018-specific primers amplified products of ∼500 nt in length. Cloning and sequencing the latter products yielded a single sequence (not shown); using this information for 5′- and 3′-RACE approaches, we cloned and identified a coding sequence (CDS) of 981 bp (AY821800). Sequence analysis showed identity to cysteine proteases of other trematodes, including C. sinensis (85%) and Paragonimus westermani (55%) (Figure 1). The ORF encoded a zymogen with a deduced size of 326 amino acid residues (molecular mass 37,325 Daltons, theoretical pI, 5.45) including a predicted signal peptide of 18 residues in length and a prosegment of 95 residues that encoded a cathepsin F protease. We termed the putative enzyme encoded by this ORF, Ov-CF-1, and the new gene Ov-cf-1. These dimensions were similar to the predicted size of the recently reported cathepsin F, CsCF-6, of the closely related fluke, C. sinensis [13]. The ERFNAQ motif characteristic of cathepsins F and W (similar to the ERFNIN motif of cathepsins L, S and O, etc.) was conserved in the prosegment of Ov-CF-1 (Figure 1). The catalytic triad residues were present as Cys26, His160 and Asn180, numbering from the predicted amino-terminal methionine, Met1, residue of the mature enzyme. Other motifs including six conserved Cys residues likely involved in disulfide bridges were present. The molecular mass of the predicted mature enzyme of 213 amino acids was 23,811.92 Daltons, with a theoretical pI of 6.42. The mature enzyme included a single, predicted glycosylation site, NGS, at residue 109.
BlastP analysis undertaken with the 213 residues of the mature Ov-CF-1 protease as the query matched to peptidase family C1A, superfamily C1. Best matches were cathepsin Fs from C. sinensis and Paragonimus. The next best match was to cathepsin F of zebrafish, Danio rerio, NP_001071036. A TBLASTN search at using the 95 amino acid residues of the prosegment yielded strong matches to the prosegment of cathepsin F-like enzymes from C. sinensis AF093243 [13] and to Paragonimus westermani, DQ016551 [25] and also to schistosome and mammalian cathepsin Fs. (The transcriptome of O. viverrini indicates the presence of other family C1 cysteine proteases including cathepsins B and L [26].)
The Ov-cf-1 Gene Is Organized into Seven Exons
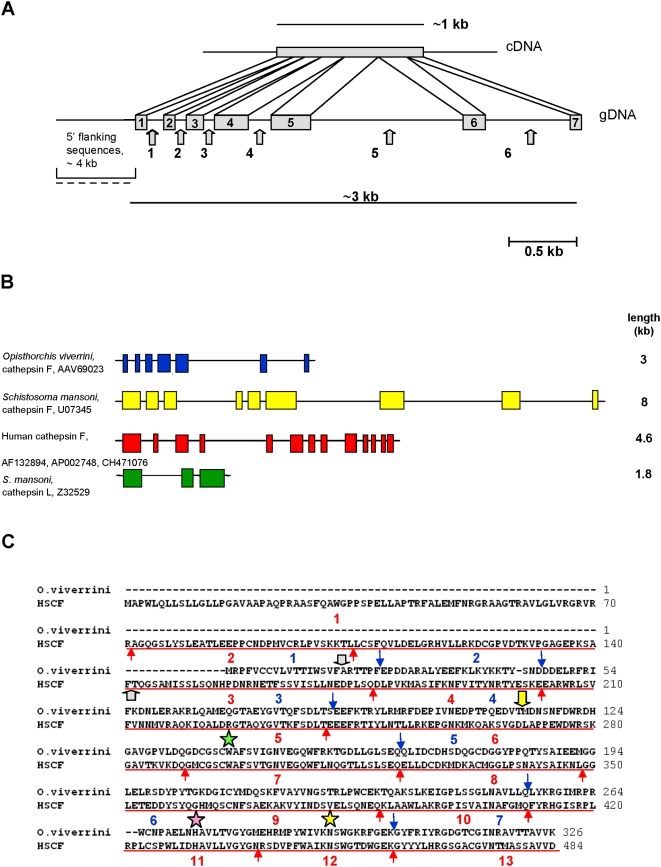
A contiguous sequence of 7,368 bp composed of 5′-UTR and the genomic sequence of O. viverrini cysteine protease was assembled from iPCR and long range PCR fragments (GenBank FJ346536) (Figure S1). The contig extended 4,211 bp upstream of the predicted translation start site (ATG) of the gene. The entire genomic sequence of O. viverrini cysteine protease characterized from start codon referred to mRNA sequence to the end was 3,157 bp. By comparing the genomic and cDNA sequences of Ov-cf-1 and by prediction of the splice sites, a gene structure of seven exons interrupted by six introns was determined for the genomic organization of the Ov-cf-1 gene (Figure 2A). The sizes of exons 1 to 7 were >69, 69, 117, 240, 267, 145 and 113 bp, respectively (Table S1 and Figure 2A–2C). The length of introns 1 to 6 were 132, 43, 46, 192, 1,060 and 664 bp, respectively. The structure of cathepsin F-like genes from several species, cathepsin F from S. mansoni, human cathepsin F, and cathepsin L2 from S. mansoni, was compared with that of the Ov-cf-1 gene (Figure 2B) since these three other genes were likely to be informative in terms of determination of orthology. Ov-cf-1 has 7 exons while S. mansoni CF1 and human cathepsin F have nine and 13 exons, respectively. As illustrated in Figure 2C, the six exon boundaries are conserved exactly or closely when compared with the exon/intron boundaries in human cathepsin F. Human cathepsin F is substantially longer than O. viverrini cathepsin F, 457 aa compared with 326 aa. The catalytic triad Cys, His, Asn residues were located in exons 4, 6 and 6, respectively; in comparison, the catalytic triad C, Cys, His, Asn residues of human cathepsin F are located on exons 6, 11 and 12. Exons 1, 2 and 3 of Ov-cf-1 appear to be orthologous to exons 3, 4 and 5 of human cathepsin F. Further downstream, the human gene is interrupted by more introns than the O. viverrini gene yet the orthology remains apparent. Thus, exons 6–7, 8–10, 11–12, and 13 of human cathepsin F likely are the orthologues of exons 4, 5 and 6, respectively, of Ov-cf-1. As mentioned, the catalytic Cys, His and Asn residues are encoded by exons 7, 11 and 12 in the human cathepsin F gene, and exons 4, 6 and 6 in the Opisthorchis gene, further supporting the orthologous nature of the two genes.
Figure 2. Genome structure of the cathepsin F gene of Opisthorchis viverrini.
(A) Structure of the O. viverrini cathepsin F gene locus, as determined by nucleotide sequence analysis of a ∼3 kb PCR product of genomic DNA of O. viverrini. The top panel shows the size of the cDNA while the bottom panel shows a schematic of positions and relative sizes of the 7 exons and 6 introns (arrowed) that comprise the gene. The 5′-flanking region was generated by inverse PCR. (B) Schematic to compare the structure and exon numbers of the C1 family genes, including the cathepsin F genes of O. viverrini, Schistosoma mansoni, and Homo sapiens, and the cathepsin L gene of S. mansoni. Colored blocks represent exons, thin lines represent introns, and the asterisks identify the exon(s) that encodes the catalytic Cys residue. Database accession numbers are provided for the illustrated sequences. (C) Comparison of the exon/intron structures of O. viverrini cathepsin F and human cathepsin F. The two enzymes were aligned for maximal homology and the amino acid sequences corresponding to an exon for each protein was separated by red (human cathepsin F) or blue (O. viverrini cathepsin F) arrows. Exon numbering is shown below the blocks of amino acid sequences. Positions of the active site triad of Cys, His and Asn residues are indicated with stars. The position of signal sequence cleavage site is indicated with gray arrows, and the position of FhCL1 trans-processing cleavage site between prosegment and mature enzyme indicated with the yellow arrow.
The splice donor and acceptor sites of all six introns in the Ov-cf-1 gene confirmed to the established consensus GT at the 5′-end of the intron and AG at the 3′ end [27] (Table S2). Blastn and tblastx searches were undertaken using the ∼4,200 bp upstream of the start codon, ATG. Several matches were obtained, specifically for query residues 1–250 which gave tblastx matches to a genomic sequence of the nurse shark Ginglymostoma cirratum (AC165195) (tblastx score, 106; E value, 4e-21) and the reverse transcriptase of a Penelope-like retrotransposon from Schistosoma mansoni (BK000685) (score, 100; E value, 2e-19) (Table S1). Similarity searches using tblastx of the intron sequences of ov-cf-1 revealed several matches, including a match of intron five to the gene encoding PHGPx of Clonorchis sinensis and a match for intron six to a trinucleotide repeat containing 6a gene of Mus musculus.
Phylogenetic Analysis of O. viverrini Cysteine Protease
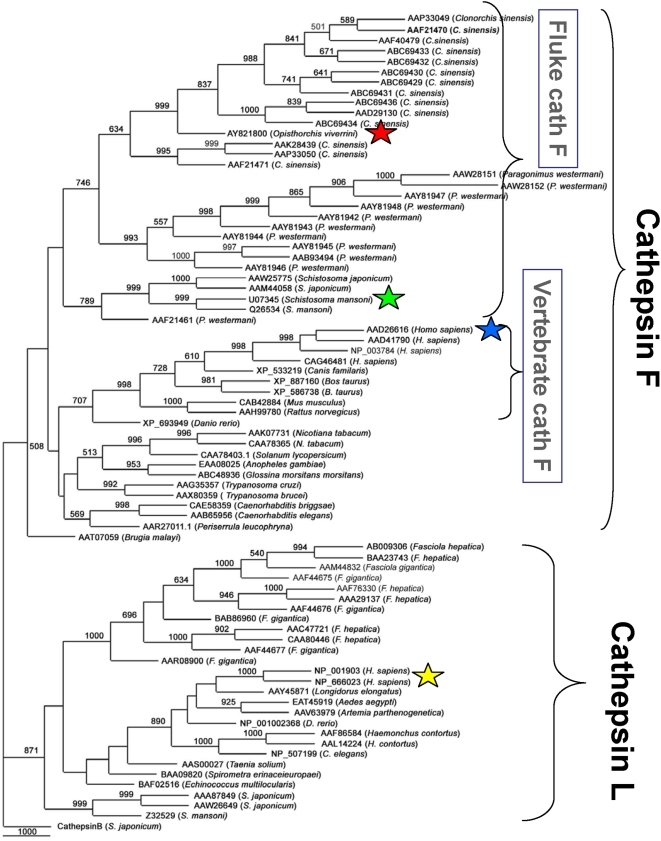
Phylogenetic analysis using >75 related sequences (primarily cathepsin F and cathepsin L like enzymes) confirmed the relationship of Ov-CF-1 with other cathepsin F proteases (Figure 3). Ov-CF-1 clustered with more than ten C. sinensis cysteine protease mRNA sequences. It also clustered with five or more cathepsin F-like sequences from the lung fluke, P. westermani, although the Paragonimus orthologues branched separately from the Opisthorchis and Clonorchis cathepsins F. Further, it clustered with the cathepsins F of the schistosomes S. japonicum and S. mansoni, although the schistosome orthologues branched separately from both the Opisthorchis/Clonorchis and the Paragonimus cathepsins F. Together the branches that included these fluke cathepsins F branched separately from mammalian cathepsins F, including human cathepsin F. The branch that included the mammalian cathepsins F clustered with another branch of cathepsin F-like enzymes from a diverse group of eukaryotes including potato, mosquito, C. elegans and trypanosomes. An unusual cathepsin F-like enzyme from the nematode Brugia malayi (AAT07059) [28] formed a basal branch of the cathepsin F-like sequences. Cathepsin L like sequences from Fasciola species, schistosomes, tapeworms, Haemonchus contortus, Aedes aegypti, human, and other species formed a completely distinct clade to the cathepsins F. The entire tree was rooted with the cathepsin B of S. japonicum.
Figure 3. Neighbor joining tree.
The tree revealed the phylogenetic relationship between the cathepsin F cysteine protease of Opisthorchis viverrini and homologous enzymes from ∼75 other informative species. Species names and GenBank accessions are provided on the branches. Bootstrap values of 1,000 replicates are provided at the nodes of the branches (bootstrap values less than 500 were omitted).
Ov-CF-1 Expressed in Gut of Fluke and Released into Liver of Infected Hamster
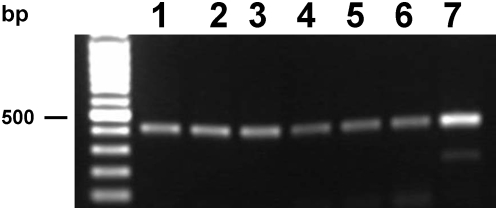
RT-PCR revealed that the Ov-cf-1 mRNA was transcribed in eggs, metacercariae, immature and sexually mature forms of O. viverrini, including one-, two- and three-week-old juveniles and adult worms (Figure 4). Anti-Ov-CF-1 serum was employed to investigate the presence of the cathepsin F in ES products and the organ- and tissue-specific expression of the cysteine protease in sexually mature forms of the O. viverrini liver fluke. Immunoblot analysis of thiol-sepharose enriched cysteine protease activities indicated that Ov-CF-1 was secreted or excreted by adult O. viverrini (Figure S2). A reactive band at ∼30 kDa present in fractions of both ES and somatic fluke extracts. In addition, there were minor bands of reactivity at ∼37 kDa and 23 kDa in the somatic antigen preparation, and 23 kDa in the ES. Given that there is an N-linked glycosylation site in the mature enzyme, the immunoblot findings indicate that the mature enzyme is glycosylated and migrated at ∼30 kDa. We interpret the additionally bands as the unprocessed zymogen (∼37 kDa) and a non-glycosylated form of the mature enzyme (∼23 kDa). Together, the immunoblot findings indicated that the antiserum specifically recognized Ov-CF1.
Figure 4. Transcription of Opisthorchis viverrini cysteine protease mRNA revealed by RT-PCR.
Developmental stages examined: (lane 1), metacercariae (2), juvenile 1 week (3), juvenile 2 weeks (4), juvenile 3 weeks (5), adult worms (6) and O. viverrini cDNA library (7).
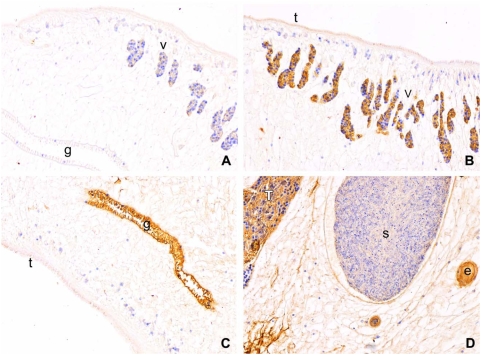
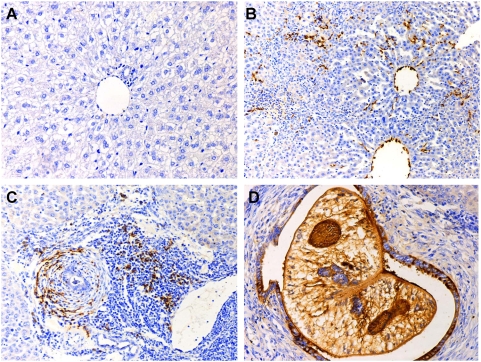
Immunohistochemical localization of Ov-CF-1 revealed strong expression in the gut, vitellaria, egg and testis (Figure 5B–5D). By contrast, control sections of flukes probed with pre-immunization serum showed no or little reactivity (Figure 5A). The intense immunolocalization to the luminal margin of the gut (Figure 5C) indicated strongly that the cathepsin F participates in proteolysis of ingested host tissues. In addition, sections through hamster bile ducts containing adult O. viverrini flukes confirmed the localization of Ov-CF-1 in organs and tissues of the fluke, including the gut and, notably, revealed the presence of Ov-CF-1 in epithelial cells lining the infected bile duct (Figure 6D). Ov-CF-1 also had accumulated in secondary bile ducts too small in internal diameter to include an adult fluke (Figure 6B) and in Kupffer cells and mononuclear cells lining sinuses of the liver (Figure 6C). Control sections from the same infected hamster probed with pre-immunization serum showed no or little reactivity (Figure 6A).
Figure 5. Immunolocalization of cathepsin F cysteine protease in adult Opisthorchis viverrini using thin sections of paraffin embedded worms probed with rabbit antiserum.
(A) Representative section spanning the gut, vitellaria, parenchyma and tegument, probed with pre-immunization serum (negative control). Sections of adults, probed with the rabbit anti-cathepsin F serum, the vicinity of the tegument and vitellaria (B), tegument and gut (C), and testis, seminal receptacle, parenchyma and eggs (D). Gut (g), vitelline glands (v), egg (e) and testis (T) all showed strong positive reactions whereas the sperm seminal receptacle (s) was negative. The tegument (t) was faintly positive but the tegumental cells were negative for the cathepsin F cysteine protease. Immunoperoxidase staining, original magnification, ×100.
Figure 6. Immunolocalization of cathepsin F cysteine protease (Ov-CF-1) in Opisthorchis viverrini infected hamster liver.
Thin sections of paraffin embedded liver tissues were probed with rabbit antiserum. (A) Representative section of liver from an uninfected hamster, spanning a portal triad including a secondary bile duct, probed with rabbit anti-Ov-CF-1serum (negative control). Infected hamster liver in the vicinity of the secondary bile ducts too small in internal diameter to include an adult fluke, probed with the rabbit anti-Ov-CF-1 serum (B and C). Immunoperoxidase stain (brown) indicates the presence of Ov-CF-1 in bile ducts epithelial cells (B) and in sinusoidal Kupffer and mononuclear cells (C). Section through bile duct containing an adult O. viverrini, showing strong reactivity to organs and tissues of the fluke (including the gut), and to the epithelial cells lining the infected bile duct (panel D). Immunoperoxidase staining, original magnification, ×100.
Activation, Processing, and Biochemical Analysis of Ov-CF-1
The recombinant Ov-CF-1 purified from yeast culture medium resolved as two major protein bands migrating at 41 kDa and 47 kDa on 4–12% Bis-Tris NuPage gels (Figure 7A). Given the theoretical molecular mass of the recombinant Ov-CF-1 zymogen (including the c-myc and His6 tags) of ∼37.5 kDa, and given that both the 41 kDa and 47 kDa peptides share identical N-termini (EFRTT; Figure 7C), it is likely that the yeast-expressed enzyme exhibits differential addition of N-linked glycans. This is consistent with the smearing observed around both protein bands (Ov-CF-1 contains a single predicted N-linked glycosylation site, NGS, at residue 109; Figure 1). The yeast-expressed Ov-CF-1 zymogen displayed modest activity against the protein substrates hemoglobin and gelatin at acidic pH (4.5–6.0) (not shown). Assays undertaken with the fluorogenic substrate Z-Phe-Arg-NHMec revealed that the enzyme was active over a broad pH range, 4.5–8.0, with optimal activity at pH 5.5 (not shown). However, the apparent activity of the zymogen was very low, which is not surprising since the gel analysis did not reveal evidence of a fully processed and activated enzyme (Figure 7A).
Figure 7. Trans-processing of Ov-CF-1 by Fasciola hepatica cathepsin L1 (FhCL1).
(A) (Top) Purified recombinant Ov-CF-1 was incubated at pH 4.5 for 180 min. Aliquots of the reaction mixtures were removed at time 0, 2, 5, 10, 20, 30, 60 and 180 min and analyzed on 4–12% Bis-Tris NuPage gels (Invitrogen). Recombinant Ov-CF-1 is not capable of auto-activation at pH 4.5. The two bands shown are likely due to differential glycosylation as the recombinant was produced in yeast; (bottom) Purified recombinant Ov-CF-1 (50 µg) was incubated with fully activated mature FhCL1 (5 µg) for up to 180 min in 0.1 M sodium acetate, pH 4.5 (reaction volume 150 µl). Aliquots (15 µl) were removed from the mixtures at time 0, 2, 5, 10, 20, 30, 60 and 180 min and analyzed by 4–12% Bis-Tris NuPage gels. By 180 min marked trans-processing of Ov-CF-1 by FhCL1 had occurred. (B) Profiles of the Ov-CF-1 (time 0 min) and at 180 min of trans-processing with FhCL1 time showing the peptide bands (1–4) that were analyzed by N-terminal sequencing. The position of exogenously added FhCL1 and the released Ov-CF-1 prosegment are also shown. (C) N-terminal sequences obtained for each of the Ov-CF-1 peptides before and after trans-processing by FhCL1. (D) The cleavage sites identified by N-terminal sequences were also mapped onto the primary amino acid sequence of the Ov-CF-1 prosegment. The EF found at the N-terminal was introduced by the EcoR I cloning site used in the pPicZα expression vector.
Both cathepsin L and B cysteine proteases of trematodes such as Schistosoma mansoni and Fasciola hepatica have been shown to auto-catalytically remove the prosegment at low pH to release the fully active mature enzyme (see [29]). In order to investigate whether Ov-CF-1 was likewise capable of auto-catalytic processing, recombinant Ov-CF-1 was incubated for 180 min at pH 4.5, after which the reaction products were analyzed in Coomassie blue-stained gels. Evidence of auto-catalysis of the zymogen was not apparent (Figure 7A, upper panel). When similar reactions were performed in the presence of fluorogenic peptide substrates, no increase in enzyme activity of Ov-CF-1 was detected (see below, also Figure 8A). These data suggested that the recombinant Ov-CF-1 did not auto-catalytically process and activate. Subsequently, we investigated whether Ov-CF-1 could be trans-processed and activated by a functionally active O. viverrini asparaginyl endopeptidase [17] or a fully-activated recombinant cathepsin L cysteine protease from F. hepatica (termed FhCL1). Ov-CF-1 was incubated with asparaginyl endopeptidase or FhCL1 at a ratio of 10∶1, at pH 4.5, 37° C, and the reaction products monitored over 180 min by SDS-PAGE. No trans-processing was obvious with the asparaginyl endopeptidase (not shown); however, in the presence of FhCL1, the 41 kDa and 47 kDa species of Ov-CF-1 were clipped to progressively faster-migrating bands (Figure 7A, lower panel). By 180 min, a prominent band had accumulated at 30 kDa, together with a minor band of 43 kDa and a small peptide migrating at 5 kDa (Figure 7B). N-terminal sequencing confirmed that the 30 kDa band represented a fully mature enzyme generated by cleavage at Val-Thr ↓ Met (Figure 7C and 7D). Peptide sequencing established that the 43 kDa protein was produced by removal of just three residues (EFR) from the N-terminus of the 47 kDa Ov-CF-1 zymogen (Figure 7C and 7D). N-terminal sequencing of the 5 kDa band was inconclusive, but this peptide likely represented a remnant product of the liberated prosegment of Ov-CF-1 (Figure 7B).
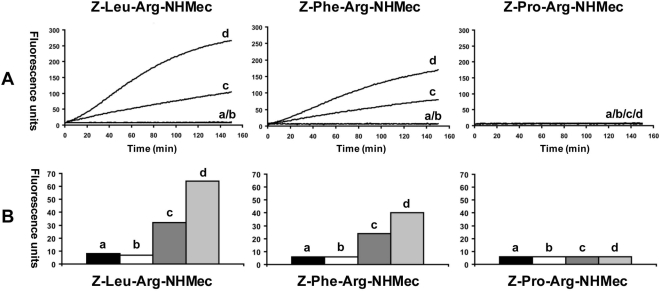
Figure 8. Exogenous activation of Ov-CF-1 by FhCL1 in the presence of peptidyl-NHMec substrates.
(A) Initial rates of hydrolysis of the fluorogenic dipeptide substrates Z-Leu-Arg-NHMec, Z-Phe-Arg-NHMec and Z-Pro-Arg-NHMec measured by monitoring the release of the fluorogenic leaving group (−NHMec) over 150 min at 37°C. (B) Comparison of the hydrolysis of each fluorogenic peptide substrate within the linear range of the reactions (0–30 min) by recombinant Ov-CF-1 and FhCL1; relative fluorescence units are presented. The reactions included negative control (a); recombinant Ov-CF-1 alone (b); recombinant FhCL1 alone (c); Ov-CF-1 +FhCL1 (d).
Figure 8A presents the trans-processing of Ov-CF-1 by FhCL1 in real-time. In these studies, the Ov-CF-1 zymogen displayed very low activity (5–10 relative fluorescent units against both peptide substrates Z-Leu-Arg-NHMec and Z-Phe-Arg-NHMec). This activity did not significantly increase when the zymogen was incubated alone for 150 min at pH 4.5. However, following addition of FhCL1 to Ov-CF-1 the activity of the cathepsin F increased over the course of the experiment (Figure 8), demonstrating that exogenous trans-processing of the Ov-CF-1 prosegment generated a highly active, mature enzyme. By subtracting the activity of the FhCL1 when assayed alone against the substrates from that observed for the mixture of Ov-CF-1 and FhCL1, we estimated that mature Ov-CF-1 was >50 times more active (after 150 min) than its zymogen. From the exogenous activation in the presence of three fluorogenic peptides, Z-Leu-Arg-NHMec, Z-Phe-Arg-NHMec and Z-Pro-Arg-NHMec, we obtained information on the substrate specificity of Ov-CF-1. Ov-CF-1 had a similar specificity for residues in the P2 substrate as FhCL1; both enzymes accommodated the hydrophobic amino acids Phe and Leu but were incapable of cleaving peptides with the bulky residue Pro in the P2 position (Figure 8A and 8B). For FhCL1, this is consistent with the findings of Stack and co-workers [30]. Initial rates of hydrolysis of the substrates during the linear phase (0–30 min) revealed an affinity by Ov-CF-1 for the substrates in the order Z-Leu-Arg-NHMec >Z-Phe-Arg-NHMec (Figure 8B).
Discussion
Cysteine proteases have been characterized in numerous infectious pathogens (e.g., [31]–[33]). In helminth parasites, their functions include excystation, tissue invasion, catabolism of host proteins for nutrition, and immunoevasion [34],[35]. Given the importance of O. viverrini in the etiology of CCA [1], [2], [4]–[6], there is impetus to characterize parasite antigens, including proteases, involved in the host-parasite relationship and pathophysiology of this food-borne fluke. Previously, we have determined the presence of abundant cysteine protease activity using the diagnostic peptide Z-Phe-Arg-NHMec in the ES products of adult O. viverrini and in other developmental stages [14] and shown that the transcriptome encodes numerous proteases [26]. More specifically, these findings demonstrated that O. viverrini expresses clan CA peptidases and suggested that abundant cysteine protease activity was present in metacercariae where it might be involved in cyst excystation during mammalian infection [14].
Cysteine proteases have been characterized in numerous infectious pathogens (e.g., [31]–[33]). In helminth parasites, their functions include excystation, tissue invasion, catabolism of host proteins for nutrition, and immunoevasion [34],[35]. Given the importance of O. viverrini in the etiology of CCA [1], [2], [4]–[6], there is impetus to characterize parasite antigens, including proteases, involved in the host-parasite relationship and pathophysiology of this food-borne fluke. Previously, we have determined the presence of abundant cysteine protease activity using the diagnostic peptide Z-Phe-Arg-NHMec in the ES products of adult O. viverrini and in other developmental stages [14] and shown that the transcriptome encodes numerous proteases [26]. More specifically, these findings demonstrated that O. viverrini expresses clan CA peptidases and suggested that abundant cysteine protease activity was present in metacercariae where it might be involved in cyst excystation during mammalian infection [14].
Here, we report that O. viverrini expresses a cathepsin F cysteine protease throughout its development and that the enzyme is released from the adult parasites which reside in the bile ducts. The cathepsin F may be responsible for the catalytic activity that we described previously and for which an EST was detected [14],[26]. Earlier studies have shown that both cathepsin L and cathepsin F cysteine proteases are abundant in several flukes, and that these two classes of proteases form distinct but closely related phylogenetic clades [36],[37]. Phylogenetic studies presented here show that the cathepsin F family of O. viverrini is most closely related the numerous cathepsin F-like transcripts expressed by another liver fluke, C. sinensis, which is informative since this parasite is also associated with cancer of the bile duct [38]. Whereas it is not yet clear whether O. viverrini also expresses and secretes cathepsin L, in addition to cathepsin F, transcripts that could encode cathepsin L are not abundant among the presently available O. viverrini ESTs, despite the fact that transcripts corresponding to several other Family C1 proteases, such as cathepsin B, are available [26]. C. sinensis, like S. mansoni [39],[40], S. japonicum [41] and Paragonimus westermani [25],[42],[43], appears to have both cathepsin F and cathepsin L -like enzymes [13]. In conspicuous contrast, Fasciola hepatica and F. gigantica, which also parasitize mammalian bile ducts, have a battery of cathepsin L enzymes yet no recorded cathepsin F [37],[44].
To date, four orthologous cathepsin F-like proteases of fluke parasites of humans have been functionally characterized. These are SmCF from S. mansoni [39],[40], Pw28CCP and related enzymes from P. westermani [25],[42],[43], CsCF-6 from C. sinensis [13] and Ov-CF-1 (this study). Cytochemical studies have shown that the cathepsin L and cathepsin F proteases of schistosomes localize to the gastrodermis of the gut and participate in extracellular digestion of ingested host tissues [40]. Caffrey and colleagues showed that promoter regions of the SmCF gene could drive reporter transgene expression in the gut of adult S. mansoni [45], thus providing strong support for a specific role for this protease in digestion of host blood [40]. CsCF-6 is also predominantly expressed in the gut of the adult C. sinensis where it is likely has a nutritional role [13]. In F. hepatica, cathepsin L1 and L2 are synthesized and secreted from gastrodermal epithelium and are essential for migration of the juvenile fluke through the intestinal wall and liver capsule, and for digestion of ingested blood and other host tissues [37],[44]. RNA interference of cathepsin L transcription blocks penetration of host tissues by the juvenile fluke [46]. Thus the localization of Ov-CF-1 in the gut of adult O. viverrini is consistent with findings in these related trematodes and consistent with a generalized function for both cathepsin F and L proteases in the degradation of host blood and other tissues.
The immunolocalization micrographs revealed that Ov-CF-1 was also expressed in the vitellaria, testis and eggs (Figure 5B and 5D). Transcripts of Ov-CF-1 were also detected in eggs (Figure 4). The role of the hydrolase in the reproductive organs of Opisthorchis has yet to be determined, particularly as reports on the immunolocalization of orthologues of cathepsin F in C. sinensis and P. westermani did not describe localization in reproductive structures [13],[43]. However, it has been reported that SmCL2, a cathepsin L of S. mansoni, is associated with the reproductive organs where it might activate phenol oxidase, an enzyme involved with eggshell formation, or with the seminal fluid [47]. F. hepatica cathepsin L has also been localized to the vitellaria [44]. It is possible that the cathepsin cysteine proteases in the reproductive structures of trematodes are encoded by different transcripts to those present in the gut but the protein products exhibit immunological cross-reactivity.
The immunolocalization investigations also revealed the presence of Ov-CF-1 in the biliary epithelium, and mononuclear cells and Kupffer cells in sinuses of the infected liver, indicating that release of Ov-CF-1 from the flukes into the bile and circulation (blood). It is possible that Kupffer cells and other macrophages phagocytose Ov-CF-1 and present the antigens to T cells, leading to inflammation. In this regard, Ov-CF-1 and other ES components (which are mitogenic in vitro; Sripa, unpublished) may stimulate inflammation and proliferation of biliary cells in the vicinity of the adult O. viverrini parasite. Such phenomena may promote cholangiocarcinogenesis.
Human cathepsin F and L are a papain-like cysteine proteases, with the MEROPS classification Clan CA, Family C1, Subfamily A (C01.018 and C01.032, respectively) [48]. Although the function(s) of cathepsin F has not been fully elucidated, it may play a regulatory role in processing the invariant chain that is associated with MHC class II [49],[50]. Cathepsin L has a wide tissue distribution and is found in lysosomes where its endopeptidase activity is a key catalyst of lysosomal proteolysis, although it also plays specific roles in generation of peptide antigens for the MHC II system and in spermatogenesis. It also has roles in pathological processes including tumor invasion, metastasis and arthritis [49]. Most residues of the prosegment of cathepsin L are conserved in cathepsin F, including the ERFNIN motif (ERFNAQ in cathepsins F and W) [51],[52]. A key structural difference between mammalian cathepsin F and other Family C1 proteases, including cathepsin L, is a cystatin domain within the long prosegment, the physiological role of which has not been established. It has been suggested that the mammalian cathepsin F gene evolved through a gene fusion between an ancestral cystatin and Clan CA cathepsin gene [53] (Figures 1 and 2). Whereas Ov-CF-1 and related fluke enzymes clearly display sequence identity to mature human cathepsin F, none so far reported include the cystatin domain within the prosegment. Therefore, if their genes represent the progeny of the ancestral cathepsin F gene, the hypothesized gene fusion of cathepsin F and cystatin genes must have occurred after the branching event in the tree of life that lead separately to trematodes (a Lophotrochozoan clade) and vertebrates (Deuterostomia).
In human cathepsin F, the two splice sites that interrupt exons 1–3 are conserved between cathepsin F and several cystatin genes [53], providing cogent evidence of a gene fusion during the evolution of human cathepsin F. The genome organization of human cathepsin F includes 13 exons separated by 12 introns; that of Ov-cf-1 is simpler, with seven exons separated by six introns. Because of the absence of a cystatin domain from the fluke cathepsin, exons one and two of the human gene do not have orthologues in the O. viverrini gene. However, downstream of the cystatin sequence, orthology between the human and O. viverrini cathepsin F genes becomes unambiguous; the exon/intron boundaries are strongly conserved between the two genes (Figure 2C). Interestingly, the cathepsin F-like gene Tci-cf-1 from the strongylid nematode Teladorsagia circumcincta, a parasite of the abomasum of sheep, does not encode a cystatin domain in its prosegment, although the prosegment of this nematode cathepsin F is ∼30 amino acids longer than the 95 residue prosegment of Ov-CF-1 and orthologues from other flukes [54]. Cathepsin F is secreted by the parasitic stages of T. circumcincta, including the L4 but not by the free-living larval stages, and is immunogenic. Given that some other nematodes including Caenorhabditis elegans and Brugia malayi do encode a cystatin domain in the prosegment of their cathepsin F orthologues [54], the evolutionary provenance of the cathepsin F/cystatin gene fusion remains obscure.
All cysteine proteases are synthesized as inactive zymogens which include a prosegment [55]. The prosegment lies across the active site, in reverse orientation to normal protein substrates, and prevents premature activation of the zymogen during trafficking and storage. Removal of the prosegment, mediated by protease clipping at the juncture between prosegment and mature enzyme, is essential to allow entry of macromolecular protein substrates into the active site cleft of the hydrolase. Dalton and colleagues proposed that prosegment removal occurs in helminth cathepsins by a two-step route of trans-processing to generate a small pool of activated enzymes which primes a rapid catalytic activation cascade [29]. The initial event might be performed by the same protease or a different protease, such as asparaginyl endopeptidase or cathepsin B or L. Our data showing that recombinant Ov-CF-1 did not undergo autocatalytic activation, despite displaying low-level activity, suggested that native Ov-CF-1 requires trans-processing. Stack and co-workers described how the junction between the prosegment and mature enzyme of human and helminth cysteine proteases consists of a non-conserved, randomly-structured motif that is susceptible to cleavage at several sites, depending on the processing enzyme [24]. Based on the three-dimensional structure of human cathepsin F [50] and multiple sequence alignments with helminth orthologues (Figure 1), we predict that the protease-susceptible region of Ov-CF-1 prosegment is the peptide PTPQEDVTMD. Since this does not include asparagine residues, it is not surprising that Ov-CF-1 was not trans-processed by the O. viverrini asparaginyl endopeptidase, even though this enzyme occurs in the gut of the adult [17]. Several gut-associated cathepsins of schistosomes and Fasciola do possess asparagine residues at the prosegment/mature enzyme juncture and, consequently, are trans-processed by asparaginyl endopeptidase (see [29]). On the other hand, here we have demonstrated trans-processing of Ov-CF-1 in vitro by the cathepsin L protease FhCL1 of F. hepatica. Cleavage of the prosegment occurred at PTPQEDVT↓MD, positioning a Val at P2, which is favored by FhCL1 [16],[56]. This cleavage resulted in a >50-fold increase in Ov-CF-1 activity. The ability of FhCL1 to trans-process Ov-CF-1 suggests that another endogenous Family C1 protease of O. viverrini, e.g. cathepsin B, processes and activates the cathepsin F within the adult gut.
Notwithstanding that cathepsins F and L belong to distinct phylogenetic clades [57] (Figure 3), our studies demonstrated that they share overlapping substrate preferences. Both Ov-CF-1 and FhCL1 accommodated substrates with hydrophobic residues in the P2 position; the P2 residue occupies the S2 subsite of the active site which primarily dictates the specificity of members of Clan CA. Additionally, both enzymes preferred Leu over Phe, whereas neither accommodated the bulky residue, proline. The selectively of FhCL1 for hydrophobic residues is likely the result of specific adaptation to the cleavage of mammalian hemoglobin, a substrate in which 42% of the component amino acids are Leu, Phe, Ala and Val [56]. Our limited substrate specificity analysis, however, has not yet revealed significant differences between the cathepsins F and L, which we suspect do exist. Indeed, we hypothesize that divergence of substrate preferences between cathepsins F and L is of central importance to the virulence and host species range of trematode parasites such as O. viverrini, F. hepatica and S. mansoni. Since phylogenetic divergence of the cathepsin F and L clades must have been driven by positive selection, by extension, elucidation of how this favors degradation of their respective target host macromolecules or tissues will inform our understanding of host-parasite adaptation.
Finally, given the extraordinary linkage between a metazoan parasite and a tumor [1],[4],[5],[38], characterization of the nature and action of secreted proteins of O. viverrini such as cathepsin F may provide insights into liver fluke induced cholangiocarcinogenesis, and indeed fundamental insights into carcinogenesis at large. In addition, cathepsin F may have potential as an intervention target including a vaccine candidate, given recent successes with chemotherapy targeting related enzymes in schistosomes [58] and with vaccines targeting cathepsin L of Fasciola hepatica [59]. Indeed, in view of the recent implementation of an acclaimed vaccination of adolescents against papilloma-virus infection to provide protection from cervical cancer [60], there is the tantalizing prospect that vaccination to prevent O. viverrini infection could provide protection against another infection-related cancer, liver fluke-induced cholangiocarcinoma.
Supporting Information
Genomic DNA sequence of the gene locus encoding the cathepsin F of Opisthorchis viverrini. The sequence includes annotation to reveal the positions of the seven exons interrupted by six introns. The sequence has been assigned GenBank accession FJ346536.
(0.04 MB DOC)
Immunoblot analysis of 10% SDS-PAGE separated somatic (lane 1) and excretory secretory (ES) (lane 2) fractions (fractions eluted from thiol-sepharose, as described [14]) probed rabbit anti-O. viverrini cathepsin F serum (diluted 1∶1000). Anti rabbit IgG-conjugated to horse radish peroxidase (Invitrogen) was employed as the secondary antibody at a dilution of 1∶5000. After removal of the second antibody, signals were developed using a chemiluminescence detection system (Amersham), and captured on X-ray film (Kodak). On the left, positions of molecular size standards are indicated in black colored text. The positions of three reactive bands are indicated with the blue arrows, at 37, 30 and 23 kDa in the somatic antigen fraction (lane 1) and 30 and 23 kDa in the ES (lane 2). The mass of the major band, at 30 kDa, indicates it may be a glycosylated form of the mature enzyme of cathepsin F of O. viverrini.
(0.04 MB PDF)
Opisthorchis viverrini cathepsin F-like cysteine protease gene locus, GenBank accession number, size and identities of exons, introns and flanking regions, as determined by tblastx searches of GenBank nr/nt collection of sequences
(0.05 MB DOC)
Exon and intron boundaries, splice donor and splice acceptor sites in the gene encoding Opisthorchis viverrini cathepsin F cysteine protease, Ov-CF-1.
(0.04 MB DOC)
Footnotes
The authors have declared that no competing interests exist.
This work was supported by Thailand-Tropical Diseases Research Programme (T-2, grant number ID 02-2-HEL-05-054), the Sandler Family Foundation, Faculty of Medicine KKU grant number i50206, and award number UO1AI065871 from the National Institute of Allergy and Infectious Diseases (NIAID) (the content is solely the responsibility of the authors and does not necessarily represent the official views of the NIAID or the National Institutes of Health). AL is supported by a senior research fellowship from the Australian National Health and Medical Research Council. MWR is supported by a University of Technology Sydney Chancellor's Postdoctoral Research Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Sripa B, Kaewkes S, Sithithaworn P, Mairiang E, Laha T, et al. Liver fluke induces cholangiocarcinoma. PLoS Med. 2007;4(7):e201. doi: 10.1371/journal.pmed.0040201. doi:10.1371/journal.pmed.0040201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hotez PJ, Brindley PJ, Bethony JM, King CH, Pearce EJ, et al. Helminth infections: The great neglected tropical diseases. J Clin Invest. 2008;118(4):1311–1321. doi: 10.1172/JCI34261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sripa B, Kaewkes S. Gall bladder and extrahepatic bile duct changes in Opisthorchis viverrini-infected hamsters. Acta Trop. 2002;83(1):29–36. doi: 10.1016/s0001-706x(02)00052-9. [DOI] [PubMed] [Google Scholar]
- 4.IARC. Schistosomes, liver flukes and Helicobacter pylori. IARC working group on the evaluation of carcinogenic risks to humans. Lyon, 7–14 June 1994. IARC Monogr Eval Carcinog Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 5.Parkin DM. The global health burden of infection-associated cancers in the year 2002. Int J Cancer. 2006;118(12):3030–3044. doi: 10.1002/ijc.21731. [DOI] [PubMed] [Google Scholar]
- 6.Haswell-Elkins MR, Satarug S, Elkins DB. Opisthorchis viverrini infection in northeast Thailand and its relationship to cholangiocarcinoma. J Gastroenterol Hepatol. 1992;7(5):538–548. doi: 10.1111/j.1440-1746.1992.tb01035.x. [DOI] [PubMed] [Google Scholar]
- 7.Ohshima H, Bartsch H. Chronic infections and inflammatory processes as cancer risk factors: possible role of nitric oxide in carcinogenesis. Mutat Res. 1994;305(2):253–264. doi: 10.1016/0027-5107(94)90245-3. [DOI] [PubMed] [Google Scholar]
- 8.Migasena P, Reaunsuwan W, Changbumrung S. Nitrates and nitrites in local Thai preserved protein foods. J Med Assoc Thai. 1980;63(9):500–505. [PubMed] [Google Scholar]
- 9.Saadat I, Higashi H, Obuse C, Umeda M, Murata-Kamiya N, et al. Helicobacter pylori CagA targets PAR1/MARK kinase to disrupt epithelial cell polarity. Nature. 2007;447(7142):330–333. doi: 10.1038/nature05765. [DOI] [PubMed] [Google Scholar]
- 10.Kurashima Y, Murata-Kamiya N, Kikuchi K, Higashi H, Azuma T, et al. Deregulation of β-catenin signal by Helicobacter pylori CagA requires the CagA-multimerization sequence. Int J Cancer. 2008;122(4):823–831. doi: 10.1002/ijc.23190. [DOI] [PubMed] [Google Scholar]
- 11.Dvorak J, Mashiyama ST, Braschi S, Sajid M, Knudsen GM, et al. Differential use of protease families for invasion by schistosome cercariae. Biochimie. 2008;90(2):345–358. doi: 10.1016/j.biochi.2007.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Robinson MW, Tort JF, Lowther J, Donnelly SM, Wong E, et al. Proteomics and phylogenetic analysis of the cathepsin L protease family of the helminth pathogen Fasciola hepatica: expansion of a repertoire of virulence-associated factors. Mol Cell Proteomics. 2008;7(6):1111–1123. doi: 10.1074/mcp.M700560-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Na BK, Kang JM, Sohn WM. CsCF-6, a novel cathepsin F-like cysteine protease for nutrient uptake of Clonorchis sinensis. Int J Parasitol. 2008;38(5):493–502. doi: 10.1016/j.ijpara.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Kaewpitoon N, Laha T, Kaewkes S, Yongvanit P, Brindley PJ, et al. Characterization of cysteine proteases from the carcinogenic liver fluke, Opisthorchis viverrini. Parasitol Res. 2008;102(4):757–764. doi: 10.1007/s00436-007-0831-1. [DOI] [PubMed] [Google Scholar]
- 15.Pinlaor S, Sripa B, Sithithaworn P, Yongvanit P. Hepatobiliary changes, antibody response, and alteration of liver enzymes in hamsters re-infected with Opisthorchis viverrini. Exp Parasitol. 2004;108(1–2):32–39. doi: 10.1016/j.exppara.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 16.Suttiprapa S, Loukas A, Laha T, Wongkham S, Kaewkes S, et al. Characterization of the antioxidant enzyme, thioredoxin peroxidase, from the carcinogenic human liver fluke, Opisthorchis viverrini. Mol Biochem Parasitol. 2008;160(2):116–122. doi: 10.1016/j.molbiopara.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laha T, Sripa J, Sripa B, Pearson M, Tribolet L, et al. Asparaginyl endopeptidase from the carcinogenic liver fluke, Opisthorchis viverrini, and its potential for serodiagnosis. Int J Infect Dis. 2008;12(6):e49–e59. doi: 10.1016/j.ijid.2008.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Triglia T, Peterson MG, Kemp DJ. A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res. 1988;16(16):8186. doi: 10.1093/nar/16.16.8186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall TA. BioEdit: a user friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:1167–1169. [Google Scholar]
- 20.Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, et al. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24(17):3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 23.Sripa B, Kaewkes S. Localisation of parasite antigens and inflammatory responses in experimental opisthorchiasis. Int J Parasitol. 2000;30(6):735–740. doi: 10.1016/s0020-7519(00)00054-0. [DOI] [PubMed] [Google Scholar]
- 24.Stack CM, Donnelly S, Lowther J, Xu W, Collins PR, et al. The major secreted cathepsin L1 protease of the liver fluke, Fasciola hepatica: a leu-12 to pro-12 replacement in the nonconserved C-terminal region of the prosegment prevents complete enzyme autoactivation and allows definition of the molecular events in prosegment removal. J Biol Chem. 2007;282(22):16532–16543. doi: 10.1074/jbc.M611501200. [DOI] [PubMed] [Google Scholar]
- 25.Na BK, Kim SH, Lee EG, Kim TS, Bae YA, et al. Critical roles for excretory-secretory cysteine proteases during tissue invasion of Paragonimus westermani newly excysted metacercariae. Cell Microbiol. 2006;8(6):1034–1046. doi: 10.1111/j.1462-5822.2006.00685.x. [DOI] [PubMed] [Google Scholar]
- 26.Laha T, Pinlaor P, Mulvenna J, Sripa B, Sripa M, et al. Gene discovery for the carcinogenic human liver fluke, Opisthorchis viverrini. BMC Genomics. 2007;8:189. doi: 10.1186/1471-2164-8-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Senapathy P, Shapiro MB, Harris NL. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 1990;183:252–278. doi: 10.1016/0076-6879(90)83018-5. [DOI] [PubMed] [Google Scholar]
- 28.Guiliano DB, Hong X, McKerrow JH, Blaxter ML, Oksov Y, et al. A gene family of cathepsin L-like proteases of filarial nematodes are associated with larval molting and cuticle and eggshell remodeling. Mol Biochem Parasitol. 2004;136(2):227–242. doi: 10.1016/j.molbiopara.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 29.Dalton JP, Brindley PJ, Donnelly S, Robinson MW. The enigmatic asparaginyl endopeptidase of helminth parasites. Trends Parasitol. 2009;25(2):59–61. doi: 10.1016/j.pt.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 30.Stack CM, Caffrey CR, Donnelly SM, Seshaadri A, Lowther J, et al. Structural and functional relationships in the virulence-associated cathepsin L proteases of the parasitic liver fluke, Fasciola hepatica. J Biol Chem. 2008;283(15):9896–9908. doi: 10.1074/jbc.M708521200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dalton JP, McKerrow JH, Brindley PJ. Trematode cysteine endopeptidases. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. London: Elsevier; 2004. pp. 1176–1182. [Google Scholar]
- 32.Binford SL, Maldonado F, Brothers MA, Weady PT, Zalman LS, et al. Conservation of amino acids in human rhinovirus 3C protease correlates with broad-spectrum antiviral activity of rupintrivir, a novel human rhinovirus 3C protease inhibitor. Antimicrob Agents Chemother. 2005;49(2):619–626. doi: 10.1128/AAC.49.2.619-626.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drew ME, Banerjee R, Uffman EW, Gilbertson S, Rosenthal PJ, et al. Plasmodium food vacuole plasmepsins are activated by falcipains. J Biol Chem. 2008;283(19):12870–12876. doi: 10.1074/jbc.M708949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williamson AL, Lecchi P, Turk BE, Choe Y, Hotez PJ, et al. A multi-enzyme cascade of hemoglobin proteolysis in the intestine of blood-feeding hookworms. J Biol Chem. 2004;279(34):35950–35957. doi: 10.1074/jbc.M405842200. [DOI] [PubMed] [Google Scholar]
- 35.Perrigoue JG, Marshall FA, Artis D. On the hunt for helminths: innate immune cells in the recognition and response to helminth parasites. Cell Microbiol. 2008;10(9):1757–1764. doi: 10.1111/j.1462-5822.2008.01174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tort J, Brindley PJ, Knox D, Wolfe KH, Dalton JP. Proteinases and associated genes of parasitic helminths. Adv Parasitol. 1999;43:161–266. doi: 10.1016/s0065-308x(08)60243-2. [DOI] [PubMed] [Google Scholar]
- 37.Robinson MW, Dalton JP, Donnelly S. Helminth pathogen cathepsin proteases: it's a family affair. Trends Biochem Sci. 2008;33(12):601–608. doi: 10.1016/j.tibs.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 38.Lim MK, Ju YH, Franceschi S, Oh JK, Kong HJ, et al. Clonorchis sinensis infection and increasing risk of cholangiocarcinoma in the Republic of Korea. Am J Trop Med Hyg. 2006;75(1):93–96. [PubMed] [Google Scholar]
- 39.Smith AM, Dalton JP, Clough KA, Kilbane CL, Harrop SA, et al. Adult schistosoma mansoni express cathepsin L proteinase activity. Mol Biochem Parasitol. 1994;67(1):11–19. doi: 10.1016/0166-6851(94)90091-4. [DOI] [PubMed] [Google Scholar]
- 40.Bogitsh BJ, Dalton JP, Brady CP, Brindley PJ. Gut-associated immunolocalization of the Schistosoma mansoni cysteine proteases, SmCL1 and SmCL2. J Parasitol. 2001;87(2):237–241. doi: 10.1645/0022-3395(2001)087[0237:GAIOTS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 41.Day SR, Dalton JP, Clough KA, Leonardo L, Tiu WU, et al. Characterization and cloning of the cathepsin L proteinases of Schistosoma japonicum. Biochem Biophys Res Commun. 1995;217(1):1–9. doi: 10.1006/bbrc.1995.2737. [DOI] [PubMed] [Google Scholar]
- 42.Yun DH, Chung JY, Chung YB, Bahk YY, Kang SY, et al. Structural and immunological characteristics of a 28-kilodalton cruzipain-like cysteine protease of Paragonimus westermani expressed in the definitive host stage. Clin Diagn Lab Immunol. 2000;7(6):932–939. doi: 10.1128/cdli.7.6.932-939.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang SH, Park JO, Lee JH, Jeon BH, Kim WS, et al. Cloning and characterization of a new cysteine proteinase secreted by paragonimus westermani adult worms. Am J Trop Med Hyg. 2004;71(1):87–92. [PubMed] [Google Scholar]
- 44.Dalton JP, Neill SO, Stack C, Collins P, Walshe A, et al. Fasciola hepatica cathepsin L-like proteases: biology, function, and potential in the development of first generation liver fluke vaccines. Int J Parasitol. 2003;33(11):1173–1181. doi: 10.1016/s0020-7519(03)00171-1. [DOI] [PubMed] [Google Scholar]
- 45.Wippersteg V, Sajid M, Walshe D, Khiem D, Salter JP, et al. Biolistic transformation of Schistosoma mansoni with 5′ flanking regions of two peptidase genes promotes tissue-specific expression. Int J Parasitol. 2005;35(6):583–589. doi: 10.1016/j.ijpara.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 46.McGonigle L, Mousley A, Marks NJ, Brennan GP, Dalton JP, et al. The silencing of cysteine proteases in Fasciola hepatica newly excysted juveniles using RNA interference reduces gut penetration. Int J Parasitol. 2008;38(2):149–155. doi: 10.1016/j.ijpara.2007.10.007. [DOI] [PubMed] [Google Scholar]
- 47.Michel A, Ghoneim H, Resto M, Klinkert MQ, Kunz W. Sequence, characterization and localization of a cysteine proteinase cathepsin L in Schistosoma mansoni. Mol Biochem Parasitol. 1995;73(1–2):7–18. doi: 10.1016/0166-6851(95)00092-f. [DOI] [PubMed] [Google Scholar]
- 48.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36(Database issue):D320–D325. doi: 10.1093/nar/gkm954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brömme D. Cathepsin F. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. London: Elsevier; 2004. pp. 1087–1088. [Google Scholar]
- 50.Somoza JR, Palmer JT, Ho JD. The crystal structure of human cathepsin F and its implications for the development of novel immunomodulators. J Mol Biol. 2002;322(3):559–568. doi: 10.1016/s0022-2836(02)00780-5. [DOI] [PubMed] [Google Scholar]
- 51.Nagler DK, Sulea T, Menard R. Full-length cDNA of human cathepsin F predicts the presence of a cystatin domain at the N-terminus of the cysteine protease zymogen. Biochem Biophys Res Commun. 1999;257(2):313–318. doi: 10.1006/bbrc.1999.0461. [DOI] [PubMed] [Google Scholar]
- 52.Wex T, Levy B, Wex H, Bromme D. Human cathepsins F and W: A new subgroup of cathepsins. Biochem Biophys Res Commun. 1999;259(2):401–407. doi: 10.1006/bbrc.1999.0700. [DOI] [PubMed] [Google Scholar]
- 53.Wex T, Wex H, Bromme D. The human cathepsin F gene—a fusion product between an ancestral cathepsin and cystatin gene. Biol Chem. 1999;380(12):1439–1442. doi: 10.1515/BC.1999.185. [DOI] [PubMed] [Google Scholar]
- 54.Redmond DL, Smith SK, Halliday A, Smith WD, Jackson F, et al. An immunogenic cathepsin F secreted by the parasitic stages of Teladorsagia circumcincta. Int J Parasitol. 2006;36(3):277–286. doi: 10.1016/j.ijpara.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 55.Coulombe R, Grochulski P, Sivaraman J, Menard R, Mort JS, et al. Structure of human procathepsin L reveals the molecular basis of inhibition by the prosegment. EMBO J. 1996;15(20):5492–5503. [PMC free article] [PubMed] [Google Scholar]
- 56.Lowther J, Robinson MW, Donnelly SM, Xu W, Stack CM, et al. The importance of pH in regulating the function of Fasciola hepatica cathepsin L1 cysteine protease. PLoS Negl Trop Dis. 2008;3(1):e369. doi: 10.1371/journal.pntd.0000369. doi:10.1371/journal.pntd.0000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dalton JP, Caffrey CR, Sajid M, Stack C, Donnelly S, et al. Proteases in trematode biology. In: Maule AGMN, editor. Parasitic Flatworms: Molecular Biology, Biochemistry, Immunology and Physiology. Oxford, United Kingdom: CAB International; 2006. pp. 348–368. [Google Scholar]
- 58.Abdulla MH, Lim KC, Sajid M, McKerrow JH, Caffrey CR. Schistosomiasis mansoni: Novel chemotherapy using a cysteine protease inhibitor. PLoS Med. 2007;4(1):e14. doi: 10.1371/journal.pmed.0040014. doi:10.1371/journal.pmed.0040014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McManus DP, Dalton JP. Vaccines against the zoonotic trematodes schistosoma japonicum, Fasciola hepatica and Fasciola gigantica. Parasitology. 2006;133(Suppl):S43–S61. doi: 10.1017/S0031182006001806. [DOI] [PubMed] [Google Scholar]
- 60.Schiller JT, Castellsague X, Villa LL, Hildesheim A. An update of prophylactic human papillomavirus L1 virus-like particle vaccine clinical trial results. Vaccine. 2008;26(Suppl 10):K53–K61. doi: 10.1016/j.vaccine.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic DNA sequence of the gene locus encoding the cathepsin F of Opisthorchis viverrini. The sequence includes annotation to reveal the positions of the seven exons interrupted by six introns. The sequence has been assigned GenBank accession FJ346536.
(0.04 MB DOC)
Immunoblot analysis of 10% SDS-PAGE separated somatic (lane 1) and excretory secretory (ES) (lane 2) fractions (fractions eluted from thiol-sepharose, as described [14]) probed rabbit anti-O. viverrini cathepsin F serum (diluted 1∶1000). Anti rabbit IgG-conjugated to horse radish peroxidase (Invitrogen) was employed as the secondary antibody at a dilution of 1∶5000. After removal of the second antibody, signals were developed using a chemiluminescence detection system (Amersham), and captured on X-ray film (Kodak). On the left, positions of molecular size standards are indicated in black colored text. The positions of three reactive bands are indicated with the blue arrows, at 37, 30 and 23 kDa in the somatic antigen fraction (lane 1) and 30 and 23 kDa in the ES (lane 2). The mass of the major band, at 30 kDa, indicates it may be a glycosylated form of the mature enzyme of cathepsin F of O. viverrini.
(0.04 MB PDF)
Opisthorchis viverrini cathepsin F-like cysteine protease gene locus, GenBank accession number, size and identities of exons, introns and flanking regions, as determined by tblastx searches of GenBank nr/nt collection of sequences
(0.05 MB DOC)
Exon and intron boundaries, splice donor and splice acceptor sites in the gene encoding Opisthorchis viverrini cathepsin F cysteine protease, Ov-CF-1.
(0.04 MB DOC)