Abstract
Mammalian SWI/SNF [also called BAF (Brg/Brahma-associated factors)] ATP-dependent chromatin remodeling complexes are essential for formation of the totipotent and pluripotent cells of the early embryo. In addition, subunits of this complex have been recovered in screens for genes required for nuclear reprogramming in Xenopus and mouse embryonic stem cell (ES) morphology. However, the mechanism underlying the roles of these complexes is unclear. Here, we show that BAF complexes are required for the self-renewal and pluripotency of mouse ES cells but not for the proliferation of fibroblasts or other cells. Proteomic studies reveal that ES cells express distinctive complexes (esBAF) defined by the presence of Brg (Brahma-related gene), BAF155, and BAF60A, and the absence of Brm (Brahma), BAF170, and BAF60C. We show that this specialized subunit composition is required for ES cell maintenance and pluripotency. Our proteomic analysis also reveals that esBAF complexes interact directly with key regulators of pluripotency, suggesting that esBAF complexes are specialized to interact with ES cell-specific regulators, providing a potential explanation for the requirement of BAF complexes in pluripotency.
Keywords: BAF complexes, BAF155, Brg
ES cells are pluripotent cells capable of both limitless self-renewal and differentiation into all embryonic lineages. These abilities are conferred by various mechanisms, including transcription factors (1–3), possibly Polycomb complexes (4, 5), microRNAs (6), and histone modification enzymes (7) that work in coordination to maintain the expression of pluripotency genes while repressing lineage-determinant genes. The involvement of such mechanisms in pluripotency has been investigated extensively in recent years (reviewed in ref. 8), but the role of chromatin remodeling enzymes remains unclear.
The mammalian genome encodes about 30 SWI2/SNF2-like ATPases, which are assembled into SWI/SNF-like complexes with ATP-dependent chromatin remodeling activity. Of these, Brg and Brm are alternative ATPases of a family of 2-MDa multisubunit SWI/SNF or BAF complexes and make up the prototypic mammalian SWI/SNF-like chromatin remodeling complexes (9, 10). BAF complexes have been shown to be essential for many aspects of mammalian development (11–13). A role of BAF complexes in pluripotency is suggested by observations that deletion of Brg, BAF155 (or Srg3), and BAF47 (or hSNF5) all lead to peri-implantation lethality and failure of the totipotent cells that give rise to both the inner cell mass and trophoblast to survive and grow (14–16). The catalytic ATPase subunit, Brg, also was recovered in screens for factors essential for nuclear reprogramming (17) and to ES cell morphology (18). In addition, ES cells lacking BAF250 have defects in ES cell maintenance and differentiation (19, 20). However, the mechanism by which BAF complexes help to establish and maintain pluripotency is not understood.
In vitro, BAF complexes use energy generated from ATP hydrolysis to alter DNA-nucleosome contacts (21) and can also exchange histones or generate torsional stress on nucleosomal templates. Although in Saccharomyces cerevisiae, SWI/SNF complexes primarily activate transcription (22, 23), mammalian BAF complexes can both activate and repress genes directly (24). BAF complexes have been found to work with known repressors such as REST (25) and histone deacetylases (26) to mediate gene repression.
BAF complexes consist of 11 core subunits that are essentially non-exchangeable in vitro, several of which are encoded by gene families (10, 27). The diversity of complexes is derived from combinatorial assembly of alternative family members and is postulated to confer functional specificity to BAF complexes. For example, the subunit composition of BAF complexes switches at mitotic exit in the vertebrate nervous system. Premitotic neuronal progenitor BAF complexes are necessary and sufficient for neuronal progenitor proliferation and self-renewal (11). In contrast, postmitotic neuronal BAF complexes are essential for dendritic outgrowth (28). Functionally distinct complexes have also been identified in cardiac progenitors (29). In addition, Polybromo-BAF (PBAF) complexes are defined by incorporation of BAF180 (Polybromo) and BAF200 and are required for heart development (30, 31).
To understand the mechanism underlying the role of BAF complexes in pluripotency, we performed functional and biochemical characterization of BAF complexes in ES cells. Our studies indicate that a specialized esBAF complex is essential for self-renewal and pluripotency.
Results
Reduction of Brg Leads to Loss of Self-Renewal in ES Cells.
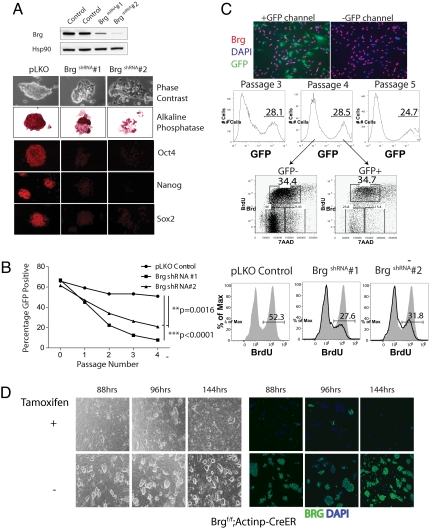
Brg deletion leads to peri-implantation lethality because of the failure of the inner cell mass (ICM) to proliferate and develop (14). Consistent with this finding, we were unable to derive ES cells after Cre-mediated deletion of Brg in ICM cells homozygous for a conditional allele (data not shown), suggesting that Brg is essential for ES cell formation. To circumvent this problem, we depleted Brg in murine E14 ES cells using shRNA-mediated knockdown. Under standard ES culture conditions, BrgshRNA ES cells show a marked reduction in self-renewal. Cell cycle analysis following knockdown by 2 different constructs (Fig. 1A Upper) revealed reduced transition into S phase and decreased cell numbers compared with control (Fig. 1B), in addition to losing colony morphology and alkaline phosphatase staining (Fig. 1A). When co-cultured with wild-type ES cells (Fig. 1B, Left), BrgshRNA ES cells were lost rapidly from the cultures compared with control. This proliferative requirement for Brg is not seen when Brg is removed acutely by genetic deletion in primary mouse embryonic fibroblasts (MEFs, Fig. 1C and ref. 14) or glial cells (11). These findings suggest that Brg has a specific role in ES cell self-renewal.
Fig. 1.
Brg is essential for ES cell self-renewal and proliferation. (A) Immunoblotting of Brg and housekeeping gene Hsp90 protein levels 96 h after transduction of E14 ES cells with shRNA constructs (BrgshRNA#1 and Brg shRNA#2) compared with pLKO control. Colony morphology and alkaline phosphatase, Oct4, Nanog, and Sox staining of BrgshRNA ES cells 10 days following transduction of shRNA contructs. (B Left) Day 3- transduced ES cells (GFP+) were mixed with non-transduced ES cells at a 3:2 ratio. The co-cultures were passaged every 2 d, and the percentage of GFP+ cells (mean value ± SD; p-values by student's t test) was determined by flow cytometry. (Right) BrdU incorporation 96 h following transduction in (A). (C) Brglox/lox primary MEFs were transfected with Cre to mediate deletion of Brg. GFP+ marks transfected and deleted MEF cells (Top). Immunofluorescence for Brg protein. (Middle) Mixed cultures (GFP±) then were passaged serially over 5 d, and BrdU incorporation was performed on passage 4 MEFs (Bottom). (D) Colony morphology of Brglox/lox;Actinp-CreER+ ES cells over a timecourse after addition of either tamoxifen or ethanol vehicle control. (Left) Brightfield and (Right) immunofluorescence for Brg.
To ensure that this reduction in self-renewal was not a nonspecific effect seen with shRNA-mediated knockdown, we derived ES cell lines from Brglox/lox;Actin-CreER transgene+ blastocysts. These ES cell lines resemble wild-type ES cells in marker expression and proliferation kinetics (data not shown) and efficiently delete Brg by 72–88 h after addition of tamoxifen (Fig. 1D). Similar to BrgshRNA ES cells, Brg knockout ES cells lose proliferative capacity immediately upon reduction of Brg protein levels, forming small colonies with flattened morphology indicative of spontaneous differentiation (Fig. 1D).
Although decreased proliferation was evident immediately following the reduction of Brg protein, BrgshRNA ES cells lost expression of Oct4, Sox2, and Nanog only after many rounds of cell division, approximately 10 d following transduction of shRNA constructs (Fig. 1A). Similarly, Brglox/lox;Actin-CreER transgene+ ES cells also maintain the expression of these markers for many divisions in the absence of Brg (Fig. S1B) and down-regulate them only after the cells cease to form discernable colonies. Hence, loss of Brg results first in reduced proliferation, self-renewal, and colony morphology, whereas prolonged depletion leads to complete loss of ES cell determinants including Oct4, Sox2, and Nanog expression. To determine if Brg-deficient ES cells retain pluripotency, we induced differentiation of early passage BrgshRNA ES cells (i.e., before the loss of Oct4, Sox2, and Nanog expression) into embryoid bodies. By measuring transcript levels, we observed that they were impaired in ectodermal differentiation and were delayed in mesodermal differentiation (Fig. S1C). Hence, Brg-deficient ES cells were unable to commit to germ lineages properly, reflecting a loss of pluripotency, in addition to being impaired in self-renewal. Consistent with the impairment in both self-renewal and differentiation, we observed increased levels of apoptosis by Annexin V staining in BrgshRNA ES cells (Fig. S1A).
Unique Composition of BAF Components in ES Cells: esBAF.
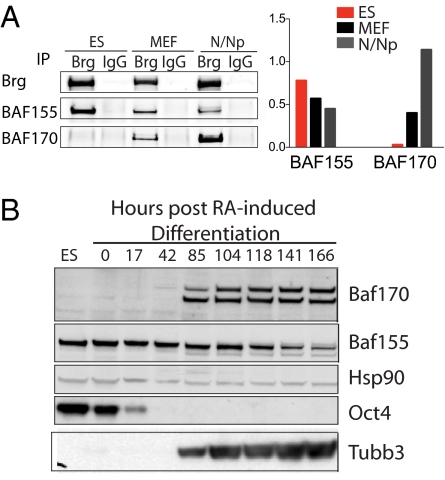
We reasoned that a specialized BAF complex might be involved in mediating the specific role of Brg in ES cell pluripotency. To address this possibility, we affinity purified endogenous BAF complexes from ES cells (> 90% Oct4+ by intracellular flow cytometry, data not shown), MEFs, and P0 mouse brain (neurons/neural progenitors) for comparison using an antibody (clone J1) raised against the exposed C-terminal domain of Brg. This antibody recognizes only Brg and its homologue Brm but not other proteins in nuclear extracts from ES cells, MEFs, and brain (Fig. 2A) (11). The purification was performed under stringent conditions (0.1% SDS and 300 mM NaCl) to minimize any spurious protein associations. Tandem mass spectrometry and sequence database searching were then used to identify proteins in the purified complexes (Fig. 2A) (32). We found 197 proteins from ES cells, 112 proteins from MEFs, and 58 proteins in P0 brain that co-immunoprecipitated with Brg/Brm with probabilities exceeding 95% as determined by ProteinProphet (33) (Table S1). Among these proteins, 28 contributed to > 80% of the total spectra obtained and were found to be common associations in all 3 cell types, including both known and novel components of BAF complexes (Fig. 2B); the remaining proteins were unique to each cell type (Fig. 2B). To determine the predominant composition of known components of the complex in each cell type, we compared spectrum counts for the proteins in each sample. Protein abundance is estimated by the number of spectra acquired for each protein, normalized to account for protein length and normalized to the total spectra in each dataset (see Methods). The predominant BAF complex in each cell type differs significantly in composition (Fig. 2C), particularly when BAF-associated proteins from ES cells (esBAF) are compared with those from neurons and neuronal progenitors. Specifically, the complex purified from ES cells contained Brg, but not Brm, BAF155 but not BAF170, and was enriched for BAF60a and BAF45d. The neuronal-specific subunits, BAF45b, BAF45c, and BAF53b are low to undetectable in esBAF, confirming their specialized role in the central nervous system. The absence of Brm in ES cells is also consistent with the normal early development of Brm knockout mice (34). These studies indicate that BAF complexes in ES cells have a unique subunit composition that is not seen in fibroblasts, brain, and other previously characterized cell lines (35, 36). We refer to this complex as “esBAF” (Fig. 2D).
Fig. 2.
Endogenous BAF complexes in ES cells have a distinctive subunit composition. (A) Endogenous BAF complexes were affinity purified from E14 ES cells and primary MEFs with an anti-Brg/Brm antibody (J1) and were subjected to tandem mass spectrometry. (Left) Immunoblotting of whole nuclear extracts using J1 antibody; (Right) silver stain analysis of purified complexes. (B) Summary of numbers of common and cell-type specific proteins that co-purified with Brg/Brm in ES cells, MEFs, and P0 mouse brain (neurons/neuronal progenitors). Proteins detected in all 3 samples include novel and known components of BAF complexes. A protein is considered specific to a cell type only if it has a probability of 0 in all other cell types. (C) Heatmap of the relative abundance of each BAF component across the 3 cell types, as measured by spectral quantitation. N/Np = neurons/neuronal progenitors; yellow = most abundant of 3 cell types; blue = least abundant. (D) Subunit composition of esBAF from analysis in (C). Numbers indicate BAF subunits (i.e., 155 = BAF155).
The Specific esBAF Composition is Critical to Its Function.
BAF155 and BAF170 are 61.7% identical in protein sequence and previously were thought to be ubiquitously present in stoichiometric amounts of all BAF complexes. Western blotting of whole nuclear extracts and affinity-purified BAF complexes confirmed the enrichment of BAF155 and the absence of BAF170 in ES cells compared with MEFs and P0 brain (Fig. 3A). This finding suggested that BAF155 substitutes for BAF170 in esBAF (Fig. 2D). Indeed, we found that at least 2 BAF155 proteins were present per esBAF complex, shown by their ability to cross-immunoprecipitate (Fig. S2A). Differentiation of ES cells into the neuronal lineage by RA treatment resulted in induction of BAF170 with concurrent reduction of BAF155 (Fig. 3B), indicating a developmental switch in the complex with respect to these subunits. Induction of BAF170 during ES cell differentiation has also been reported previously (20). We hypothesized that this configuration confers critical functional specificity to the esBAF complex. To address this functional significance, we first examined the requirement of BAF155 for ES cells.
Fig. 3.
BAF155 and BAF170 developmental switch. (A) Quantitative immunoblotting of J1-purified complexes from ES cells, MEFs, and P0 brain (N/Np) for BAF155 and BAF170 (Left). Levels of 2 proteins were normalized to levels of Brg for comparison between the 3 cell types (Right). (B) Immunoblotting of whole-cell extracts from ES cells treated with RA under nonadherent growth conditions without leukemia inhibitory factor for the indicated time points.
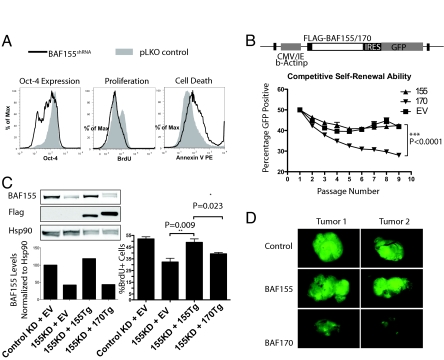
BAF155 knockout embryos die around the time of implantation, and ICMs from knockout blastocysts fails to form in vitro (37). We reasoned that BAF155 depletion also would impair ES cell maintenance. As with the depletion of Brg, shRNA-mediated depletion of BAF155 resulted in decreased proliferation, decreased Oct4 expression, and increased apoptosis of ES cells (Fig. 4A), confirming that BAF155 and Brg work in coordination to maintain ES cells. To address the functional significance of the exclusion of BAF170 from esBAF, we directed the formation of BAF170-containing complexes by transducing ES cells with a lentivirus containing a flag-tagged BAF170 transgene driven by a constitutive actin promoter (Fig. 4B). As controls, ES cells were transduced with BAF155 or empty vector (EV). GFP+ stable transductants then were co-cultured with nontransduced GFP-negative ES cells. BAF170-transduced ES cells showed an accelerated rate of decline in the percentage of GFP+ cells compared with BAF155- or EV-transduced ES cells (Fig. 4B), revealing a competitive disadvantage in self-renewal. To determine if BAF155 and BAF170 are functionally redundant in esBAF complexes, we investigated whether BAF170 could rescue BAF155 deficiency. We reduced the expression of endogenous BAF155 using RNAi directed against the 3′-UTR of the BAF155 gene while stably expressing either BAF155 or BAF170 (Fig. 4C). The BAF155 transgene rescued the proliferative defect caused by BAF155 knockdown, but BAF170 did not restore proliferation (Fig. 4C), suggesting that BAF170-containing complexes could not replace endogenous BAF155-containing complexes. We noticed that expression of BAF170 in ES cells decreased the endogenous levels of BAF155 protein (Fig. S2B), suggesting that BAF155 molecules were excluded from complexes and consequently degraded. We also assayed the ability of BAF170-expressing ES cells to form teratomas in SCID mice compared with control and BAF155-expressing ES cells. After 30 d of in vivo development, tumors arising from control and BAF155-expressing ES cells were wholly GFP+ (i.e., transgene positive). In contrast, tumors arising from BAF170-expressing ES cells were composed largely of GFP-negative cells derived from the very small percentage of nontransduced cells in the transplanted population (Fig. 4D). This result indicates that BAF170-expressing ES cells have a competitive disadvantage in giving rise to differentiated tissue types in the tumor, reflecting a loss of pluripotency.
Fig. 4.
ES cells require a specific esBAF composition with respect to BAF155 and BAF170. (A) ES cells were transfected with BAF155 shRNA or pLKO control constructs. After 96 h, Oct4 protein levels, cell cycle status, and cell death were determined by flow cytometry. (B) BAF155 (155), flag-BAF170 (170), or control EV-transduced GFP+ cells were mixed with nontransduced E14 ES cells at a 1:1 ratio and were passaged every 2 d. The percentage of GFP+ cells (mean ± SD, p value from Student's t test) was determined by flow cytometry. (C) ES cells first were transduced with BAF155 transgene (Tg), BAF170 Tg, or EV controls as in (B). Stable cell lines were then transfected or transduced with control shRNA construct or shRNA construct targeting the 3′ UTR of endogenous BAF155 (BAF155KD) but not BAF155Tg. (Left) Resulting levels of BAF155 protein were normalized to Hsp90. (Right) Proliferation subsequently was assayed by BrdU incorporation (mean ± SD, indicated p-values from t test). (D) Teratomas from BAF155, BAF170, or EV-transduced GFP+ ES cells formed for 30 d in SCID mice.
Proteins Critical to Establishing and Maintaining Pluripotency Interact Directly with esBAF Complexes.
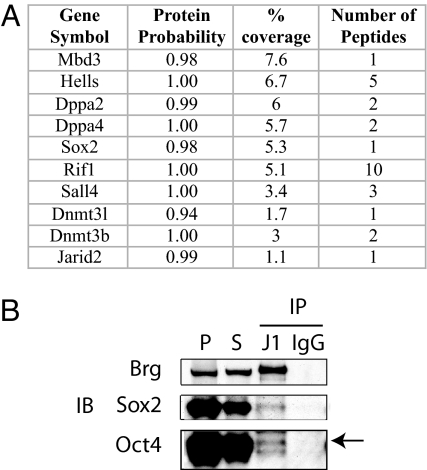
The discovery that ES cells have a specific chromatin remodeling complex suggests that this complex presents unique composite surfaces capable of interacting with regulators specific to ES cells. Indeed, the genes encoding the majority of proteins found to interact only with BAF complexes purified from ES cells (Fig. 2B, n = 127; see Table S1 for a full list) are ES specific and are positively co-regulated with Oct4 during differentiation (Fig. S3), including genes that enhanced the frequency of induced pluripotent stem cell formation such as Sox2, Dppa2, Dppa4, and Sall4 (38) (Fig. 5A). Indeed, we were able to detect physical interaction between Brg and Oct4 by co-immunoprecipitation on chromatin (Fig. 5B). Importantly, we did not find peptides from general transcription factors or mediator subunits despite the high level of expression of these proteins. Rather we found that the esBAF complex interacted specifically with a number of proteins found selectively in ES cells. This specificity indicates that the esBAF complex is tailored for interactions with proteins expressed in the pluripotent state and that esBAF might function in coordination with master regulators such as Oct4 and Sox2 to regulate the ES cell transcriptional circuitry.
Fig. 5.
esBAF interacting proteins. (A) Examples of proteins detected to interact with esBAF complexes. (B) Co-immunoprecipitation of Brg (J1 IP and control IgG IP) with Oct4 and Sox2 on chromatin. P = positive control; S = input. The arrow indicates the band for Oct4 protein.
Discussion
We have demonstrated that ES cells contain a family of functionally and structurally specialized chromatin remodeling complexes, esBAF, that are critical for the self-renewal of ES cells and maintenance of the stem cell state. The requirement for Brg in ES cell maintenance and in blastocysts was reported recently by Knott and coworkers (39), confirming earlier results with gene deletion and studies reported here. In addition, we show that Brg acts in the context of a specialized complex, esBAF. In a companion paper (40), we demonstrated that esBAF complexes participate in the regulation of the core pluripotency transcription circuitry by regulating the expression of members of the core circuitry as well as interacting functionally with Oct4 and Sox2. esBAF complexes also appear to colocalize with Smad1 and Stat3, key effectors of leukemia inhibitory factor and bone morphogenetic protein signaling, providing another potential explanation for the critical requirement for esBAF complexes in ES cells.
We initially assumed that the genetic requirement for BAF complex family members in the formation of the pluripotent state reflected a relatively nonspecific requirement for general transcription. Indeed, our studies of the yeast SWI/SNF complex have demonstrated that it is necessary for activation of most promoters and is required continuously for transcription (41). However, several lines of evidence indicate that the role of SWI/SNF-like BAF complexes in pluripotency is specific and is a product of a distinctive family of complexes. First, in our proteomic studies of purified esBAF complexes, we did not detect general transcription factors. Second, our studies indicate that although ES cells are vitally dependent upon Brg, fibroblasts or glia can proliferate and respond to stimuli in the complete absence of Brg. Third, the requirement for esBAF in ES cells requires a specific subunit composition. Therefore, although expression of BAF155 in ES cells is compatible with self-renewal and pluripotency, expression of the BAF170 subunit (found in BAF complexes in non-pluripotent cells) restricts pluripotency and interferes with self-renewal. Fourth, genome-wide occupancy studies show that although binding of esBAF complexes is widespread and occurs in nearly 1/4 of all annotated genes in ES cells, all known genes that contribute to pluripotency and other ES-specific genes are selective targets (40). Last, esBAF complexes interact directly with key regulators of the pluripotent state such as Sox2, perhaps explaining why they are found at the promoters of genes in the pluripotency network that use Sox2/Oct4 complexes selectively. These lines of evidence support a specific function of esBAF complexes in maintaining and possibly establishing the pluripotent state and indicate that this role is programmatic rather than simply a coincidence of the promoters bound by the remodeling complexes.
The specific subunit composition of esBAF complexes is likely to provide a large and specialized surface area. For example, the unique BAF155 homodimer present in the complex should provide a surface for binding not present in other cells. Indeed, we found that esBAF complexes made unique interactions with proteins not described to date, including several components of the DNA-methylation machinery (Hells, DNMT3b, and DNMT3L), suggesting a role in gene silencing or heterochromatin maintenance. esBAF also interacts with several transcription factors that have been implicated in the maintenance of transcriptional networks in ES cells, such as Sall4 and Sox2. esBAF also interacts with several proteins such as Rif1, Dppa2, and Dppa4 whose molecular functions are unclear but that have been implicated in ES cell maintenance (42, 43). From these observations we propose that the surface of esBAF complexes is tailored for interactions with factors found specifically in ES cells and that through these functional interactions esBAF maintains the pluripotent chromatin landscape.
In conclusion, our studies illustrate how a chromatin remodeling complex specifically tailored to interact physically and functionally with a group of ES cell-specific transcription factors provides specificity, stability, and robustness to the genetic circuitry underlying the essential goals of self renewal and pluripotency. This understanding may be helpful in development of new routes to reprogram differentiated cell types for therapeutic and investigational purposes.
Materials and Methods
Culture of ES Cells.
All experiments were performed with E14Tg2a murine ES cells cultured under standard feeder-free conditions. For details, please refer to SI Methods
Antibodies.
All antibodies used in this study are listed in Table S2.
RNAi and cDNA-Mediated Overexpression.
RNAi knockdown of Brg and BAF155 proteins was mediated by pLKO lentiviral vectors (OpenBiosystems) encoding short-hairpin constructs driven by human U6 promoter. These constructs were modified by insertion of an IRES-GFP sequence downstream of the puromycin resistance gene. Overexpression of BAF155 and BAF170 was mediated by pRRL.sin19-based lentiviral vectors (kind gift of Irving Weissman) encoding full-length cDNA sequences (Openbiosystem clones 6400646 and 5705438, respectively) under the control of Chicken beta-Actin promoter with CMV immediate/early enhancer.
Affinity Purification and Mass Spectrometry.
Affinity purification and mass spectrometry of endogenous BAF complexes were performed as previously described (11). For details, please refer to SI Methods.
Spectrum Counting and Quantitative Analysis of Mass Spectrometry Data.
The proteins identified in different cell types were combined, and ambiguities resulting from protein isoforms and multiple accession numbers were resolved using in-house software. For each protein, a spectrum count index was calculated as a measure of protein abundance (32). Peptides shared among multiple proteins were apportioned using weighting factors calculated based on the ratio of spectral counts computed for those proteins using only distinct (non-shared) peptides (44). The spectrum counts for each protein were normalized further for protein length and for the total number of identified peptides in each experiment (45). The relative abundance of a protein in a cell type was determined as the percentage of spectrum from that protein belonging to the cell type.
Additional methods can be found in SI Text.
Supplementary Material
Acknowledgments.
We are grateful to Jiang Wu, Andrew Yoo, and Elena Gallo for critical comments on the manuscript. We thank Bong Kim and Laura Hohrmann for their technical assistance. This work was supported by the Howard Hughes Medical Institute (G.R.C.) and the Intramural Research Program of the National Heart, Lung and Blood Institute, National Institutes of Health (Keji Zhao). L.H. is funded by the Agency of Science, Technology and Research of Singapore. B.T.S. was supported by National Institute of Child Health and Human Development Grant T32 HD007249.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812889106/DCSupplemental.
References
- 1.Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang J, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10(3):353–360. doi: 10.1038/ncb1698. [DOI] [PubMed] [Google Scholar]
- 4.Boyer L, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 5.Chamberlain SJ, Yee D, Magnuson T. Polycomb repressive complex 2 is dispensable for maintenance of embryonic stem cell pluripotency. Stem Cells. 2008;26:1496–1505. doi: 10.1634/stemcells.2008-0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134(3):521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 8.Surani M, Hayashi K, Hajkova P. Genetic and epigenetic regulators of pluripotency. Cell. 2007;128(4):747–762. doi: 10.1016/j.cell.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 9.Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366(6451):170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- 10.Wang W, et al. Diversity and specialization of mammalian SWI/SNF complexes. Genes Dev. 1996;10(17):2117–2130. doi: 10.1101/gad.10.17.2117. [DOI] [PubMed] [Google Scholar]
- 11.Lessard J, et al. An essential switch in subunit composition of a chromatin remodeling complex during neural development. Neuron. 2007;55(2):201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chi T. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19(2):169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- 13.de la Serna IL, Carlson KA, Imbalzano AN. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat Genet. 2001;27(2):187–190. doi: 10.1038/84826. [DOI] [PubMed] [Google Scholar]
- 14.Bultman S, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6(6):1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 15.Kim JK, et al. Srg3, a mouse homolog of yeast SWI3, is essential for early embryogenesis and involved in brain development. Mol Cell Biol. 2001;21(22):7787–7795. doi: 10.1128/MCB.21.22.7787-7795.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klochendler-Yeivin A, et al. The murine SNF5/INI1 chromatin remodeling factor is essential for embryonic development and tumor suppression. EMBO Reports. 2000;1(6):500–506. doi: 10.1093/embo-reports/kvd129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansis C, Barreto G, Maltry N, Niehrs C. Nuclear reprogramming of human somatic cells by Xenopus egg extract requires BRG1. Curr Biol. 2004;14(16):1475–1480. doi: 10.1016/j.cub.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 18.Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60–p400 as a regulator of embryonic stem cell identity. Cell. 2008;134(1):162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao X, et al. ES cell pluripotency and germ-layer formation require the SWI/SNF chromatin remodeling component BAF250a. Proc Natl Acad Sci USA. 2008;105(18):6656–6661. doi: 10.1073/pnas.0801802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Z, et al. BAF250B-associated SWI/SNF chromatin-remodeling complex is required to maintain undifferentiated mouse embryonic stem cells. Stem Cells. 2008;26(5):1155–1165. doi: 10.1634/stemcells.2007-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol. 2007;19(3):250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- 22.Breeden L, Nasmyth K. Cell cycle control of the yeast HO gene: cis- and trans-acting regulators. Cell. 1987;48(3):389–397. doi: 10.1016/0092-8674(87)90190-5. [DOI] [PubMed] [Google Scholar]
- 23.Martens JA, Wu PY, Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 2005;19(22):2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chi TH, et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418(6894):195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 25.Battaglioli E, et al. REST repression of neuronal genes requires components of the hSWI. SNF complex. J Biol Chem. 2002;277(43):41038–41045. doi: 10.1074/jbc.M205691200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang HS, et al. Exit from G1 and S phase of the cell cycle is regulated by repressor complexes containing HDAC-Rb-hSWI/SNF and Rb-hSWI/SNF. Cell. 2000;101(1):79–89. doi: 10.1016/S0092-8674(00)80625-X. [DOI] [PubMed] [Google Scholar]
- 27.Olave I. Identification of a polymorphic, neuron-specific chromatin remodeling complex. Genes Dev. 2002;16(19):2509–2517. doi: 10.1101/gad.992102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu J, et al. Regulation of dendritic development by neuron-specific chromatin remodeling complexes. Neuron. 2007;56(1):94–108. doi: 10.1016/j.neuron.2007.08.021. [DOI] [PubMed] [Google Scholar]
- 29.Lickert H, et al. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432(7013):107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- 30.Yan Z, et al. PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 2005;19(14):1662–1667. doi: 10.1101/gad.1323805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, et al. Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 2004;18(24):3106–3116. doi: 10.1101/gad.1238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nesvizhskii A, Vitek O, Aebersold R. Analysis and validation of proteomic data generated by tandem mass spectrometry. Nature Methods. 2007;4(10):787–797. doi: 10.1038/nmeth1088. [DOI] [PubMed] [Google Scholar]
- 33.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Analytical Chemistry. 2003;75(17):4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 34.Reyes JC, et al. Altered control of cellular proliferation in the absence of mammalian Brahma (SNF2alpha) EMBO J. 1998;17(23):6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang W, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15(19):5370–5382. [PMC free article] [PubMed] [Google Scholar]
- 36.Lemon B, Inouye C, King DS, Tjian R. Selectivity of chromatin-remodelling cofactors for ligand-activated transcription. Nature. 2001;414(6866):924–928. doi: 10.1038/414924a. [DOI] [PubMed] [Google Scholar]
- 37.Han D, et al. SRG3, a core component of mouse SWI/SNF complex, is essential for extra-embryonic vascular development. Dev Biol. 2008;315(1):136–146. doi: 10.1016/j.ydbio.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 38.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 39.Kidder BL, Palmer S, Knott JG. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2008 Dec 4; doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 40.Ho L, et al. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc Natl Acad Sci USA. 2009 doi: 10.1073/PNAS.0812888106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biggar SR, Crabtree GR. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J. 1999;18(8):2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loh Y, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38(4):431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 43.Masaki H, Nishida T, Kitajima S, Asahina K, Teraoka H. Developmental pluripotency-associated 4 (DPPA4) localized in active chromatin inhibits mouse embryonic stem cell differentiation into a primitive ectoderm lineage. J Biol Chem. 2007;282(45):33034–33042. doi: 10.1074/jbc.M703245200. [DOI] [PubMed] [Google Scholar]
- 44.Choi H, Fermin D, Nesvizhskii AI. Significance analysis of spectral count data in label-free shotgun proteomics. Molecular & Cellular Proteomics. 2008;7:2373–2385. doi: 10.1074/mcp.M800203-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zybailov B, et al. Statistical analysis of membrane proteome expression changes in Saccharomyces cerevisiae. Journal of Proteome Research. 2006;5(9):2339–2347. doi: 10.1021/pr060161n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.