Abstract
Although protein prenylation is widely studied, there are few good methods for isolating prenylated proteins from their non-prenylated relatives. We report that crosslinked agarose (e.g., Sepharose) chromatography media that has been chemically functionalized with β-cyclodextrin (β-CD) is extremely effective in affinity chromatography of prenylated proteins. In this study, a variety of proteins with C-terminal prenylation target (“CAAX box”) sequences were enzymatically prenylated in vitro with natural and non-natural prenyl diphosphate substrates. The prenylated protein products could then be isolated from starting materials by gravity chromatography or fast protein liquid chromatography (FPLC) on a β-CD-Sepharose column. One particular prenylation reaction—farnesylation of a mCherry-CAAX fusion construct—was studied in detail. In this case, purified farnesylated product was unambiguously identified by electrospray mass spectrometry. In addition, when mCherry-CAAX was prenylated with a non-natural, functional isoprenoid substrate, the functional group was maintained by chromatography on β-CD-Sepharose, such that the resulting protein could be selectively bound at its C terminus to complementary functionality on a solid substrate. Finally, β-CD-Sepharose FPLC was used to isolate prenylated mCherry-CAAX from crude HeLa cell lysate, as a model for purifying prenylated proteins from cell extracts. We propose that this method could be generally useful to the community of researchers studying protein prenylation.
Keywords: Cyclodextrin, Sepharose, Prenylation, Affinity chromatography, FPLC
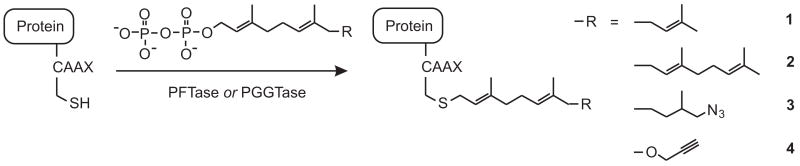
Prenylation is a post-translational protein modification that involves the covalent addition of a farnesyl (C15) or geranylgeranyl (C20) isoprenoid to a C-terminal recognition sequence [1, 2]. Proteins are prenylated by prenyltransferases, and two of these prenyltransferases—protein farnesyltransferase (PFTase) 1 and protein geranylgeranyltransferase type I (PGGTase-I)—recognize a 4-amino acid “CAAX box” sequence (where C is cysteine, A is an aliphatic amino acid, and X is typically serine, methionine or leucine) and catalyze the addition of an isoprenoid to the CAAX cysteine [3]. In eukaryotic cells, a primary function of protein prenylation is to direct proteins to the cell membrane [4, 5]. Prenyl groups are also specifically recognized by other proteins involved in protein trafficking and signal transduction [6]. Because protein prenylation is important for the function of Ras oncoproteins, prenyltransferases have become therapeutic targets for the treatment of some cancers [7]. Prenyltransferases tolerate some non-natural isoprenoid substrates, and this tolerance has recently been exploited to engineer proteins with bioorthogonal functional groups by using prenyltransferases in conjunction with modified isoprenoids bearing these functional groups. Using this approach, proteins bearing a C-terminal CAAX box have been modified with prenyl azides [8–13], alkynes [14–16], fluorophores [17, 18], photoaffinity groups [19], biotin [20], and other functional modifications [21], all under catalysis by prenyltransferases. In principle, this chemically modified fusion tag strategy has the advantage that the CAAX box tag is extremely small, innocuous, and easily inserted at the C-terminus of proteins.
Despite the importance of prenylation to eukaryotic cell biology, and the potential of prenylation as a functional fusion tag strategy, there are few methods for separating prenylated proteins from their non-prenylated relatives. The most common method involves cloud-point extraction of protein from aqueous solution with surfactants such as Triton X-114 [22], whereby alkylated proteins partition into surfactant droplets as the protein-surfactant mixture is warmed above its cloud point. This method usually provides prenylated protein products with only moderate yields and purities. Other methods, including gel electrophoresis [23], partitioning onto phospholiposomes or vesicle membranes [24, 25], and gel permeation chromatography of prenylated product complexed to a isoprenoid-binding protein partner [26], are complex and also suffer from low yields.
Francis and coworkers recently reported that proteins modified with hydrophobic dyes could be separated from unmodified proteins by affinity chromatography, using β-cyclodextrin (β-CD)-modified agarose as the stationary phase [27]. This method relies on the strong interaction between the hydrophobic dye and the interior cavity of the cyclodextrin in aqueous solution. Because β-CD also has a strong affinity for long-chain alkyl groups [28], we postulated that a β-CD-functionalized chromatography support might similarly bind protein-bound prenyl groups. In this Article, we report that proteins modified with either natural or non-natural prenyl groups can be rapidly separated from non-prenylated proteins by affinity chromatography on β-CD-modified Sepharose. Experiments indicate that an association between the column-bound cyclodextrin and protein prenyl groups is primarily responsible for retention of the prenylated protein on the support. Interestingly, we find that free isoprenoid is not retained by the support under the conditions tested, meaning that the method can be used to separate prenylated proteins from excess prenyl substrate. Cell debris, including membrane components, also does not interfere with the capture of the prenylated protein on the stationary phase, so that the method can be used to isolate prenylated proteins from cell extracts. Prenylated proteins that have been captured by the support can be eluted using a salt gradient. Overall, we report that β-CD-modified Sepharose can be used as a support in batch, gravity, or pressure chromatography to provide prenylated proteins with high yield and purity.
Materials and methods
Reagents and equipment
N-Hydroxysuccinimidyl ester (NHS)-activated Sepharose 4 Fast Flow resin was purchased from GE Healthcare (Piscataway, NJ). Recombinant human H-ras was purchased from Calbiochem (San Diego, CA). [3H]-farnesyl diphosphate ([3H]1-PP) was purchased from American Radiolabeled Chemicals (St. Louis, MO). Farnesyl diphosphate (1-PP) [29], geranylgeranyl diphosphate (2-PP) [29], azido-10,11-dihydrofarnesyl diphosphate (3-PP) [9], propargyloxygeranyl diphosphate (4-PP) [14], yeast PFTase [30], human PGGTase-I [31], eGFP-CVIA [9], CDC42 [8], GST-CIIS [32], and tris(benzyltriazolylmethyl)amine (TBTA) [33] were prepared as previously described. Ultrapure water was generated by a Milli-Q water purification system (Millipore Inc., Billerica, MA, R > 12 MΩ·cm−1).
Preparation of β-CD-Sepharose column
β-CD-NH2 was synthesized as previously reported [27], by sequentially adding 1 equiv each of 1,1-carbonyldiimidazole (CDI) and ethylenediamine to β-CD in pyridine. Conversion was confirmed by 1H NMR spectroscopy (300 MHz, Varian Unity) and by electrospray ionization (ESI) MS (reflectron ESI-BioTOF II, Bruker Daltonics, Billerica, MA).
To prepare Sepharose-NHS resin for conjugation with β-CD-NH2, 10 mL of resin suspension (w/18 μmol NHS/mL) was drained and washed twice with 10 mL of cold 1.0 mM HCl and 10 mL of 15 mM HEPES buffer (pH 7.5), alternatively [9]. The drained resin was then directly transferred with stirring into a vial containing 6.0 mL of β-CD-NH2 solution (8 mg/mL in 15 mM HEPES buffer, pH 7.5). After 4 h of stirring at room temperature, 5.0 mL of ethanolamine was added to block unreacted NHS functional groups on the resin. After incubating overnight, a 1.0 mL aliquot of the β-CD-Sepharose suspension was washed with 15 mM HEPES buffer (pH 7.5) and transferred into a Tricorn 10/20 column (GE Healthcare). The resulting β-CD-Sepharose column was connected to the Agilent 1100 HPLC instrument for gradient fast protein liquid chromatography (FPLC). The column was washed with 1.0 M NH4Cl (adjusted to pH 7.0 with 1 M NaOH; solvent B) for 60 min at a flow rate of 0.3 mL/min, and then equilibrated to 80 mM NH4Cl (adjusted to pH 7.0 with 1 M NaOH; solvent A) prior to use.
Mutagenesis, overexpression and purification of mCherry-CVIA
A pRSET-B plasmid containing His6-mCherry was the kind gift of Prof. Roger Tsien (University of California, San Diego). A CVIA tag sequence was inserted at the C-terminus using PCR-based insertion (QuikChange, Stratagene, La Jolla, CA), using the forward-sense insertion primer CATGGACGAGCTGTACAAGtgcgtcatcgcaTAAtggaattcgaagcttgatcc (encoding the end of mCherry, the CVIA tag, the stop codon, and the beginning of the plasmid cloning site), as well as its complement. Successful insertion of the CVIA tag sequence was confirmed by DNA sequencing. The resulting plasmid construct was transformed into BL21(DE3)PlysS supercompetent cells (Novagen, Madison, WI) under heat shock for 30 s at 42 °C. To overexpress the mCherry-CVIA, LB media (250 mL), supplemented with ampicillin (0.10 mg/mL), was inoculated with a single colony of cells containing the mCherry-CVIA plasmid and grown with shaking overnight at 37 °C. The culture (10 mL) was added to 500 mL flasks containing 250 mL of LB, ampicillin (0.10 mg/mL), and chloramphenicol (0.035 mg/mL). After shaking at 37 °C for 2 h, the OD600 of the cells had increased to 0.6, and induction was initiated by the addition of isopropyl-β-D-thiogalactopyranoside (IPTG) to a final concentration of 1.0 mM. The cells were grown with shaking at 37 °C for 5 h after induction. The cells were recovered by centrifugation, and kept frozen at −80 °C after removing supernatant. The next day, wet cells were suspended in lysis buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 10 mM imidazole). After the cells were incubated for 30 min with lysozyme (1.0 mg/mL) and benzonase (1.0 mg/mL), they were lysed by sonication on ice, using six rounds of alternating 30 s pulses and 30 s cooling periods. The lysis extract was centrifuged (10,000 × g) for 30 min at 4 °C, and the supernatant was loaded directly onto a column containing 5 mL of Ni-NTA resin (Qiagen) that had been equilibrated with 50 mL equilibration buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 20 mM imidazole). After loading the supernatant, the column was washed with equilibration buffer, and protein product was eluted with elution buffer (50 mM NaH2PO4, pH 8.0; 300 mM NaCl; 250 mM imidazole).
Prenylation reactions
Enzymatic reactions were conducted in 32 mL volumes, containing 50 mM Tris-HCl adjusted to pH 7.0, 10 mM MgCl2, 10 μM ZnCl2, 5 mM dithiothreitol (DTT), and 2.5 μM protein substrate. This mixture was incubated for 1 h at 25 °C in order to reduce any disulfide-linked protein aggregates before performing the prenylation. Prenyl diphosphate and the appropriate prenyl transferase (either yeast PFTase or PGGTase-I) were then added to final concentrations of 10 nM (with activity 0.089 U/mg) and 10 μM (activity 0.022 U/mg), respectively. The enzymatic reaction incubated at 32 °C for 4 h. In the case of protein prepared only for FPLC analysis, the protein was purified by gel filtration (NAP-5). In the case of samples purified for subsequent MS, the product mixture was concentrated to 500 μL by centrifugal dialysis (Amicon Ultra-15, MWCO 10.0 kDa, Millipore), and was desalted by three rounds of adding 15 mL of 20 mM Tris-HCl (pH 7.5) to the dialysis tube and concentrating to 500 μL, prior to FPLC purification.
FPLC separation of prenylated proteins
FPLC was performed on an Agilent (Santa Clara, CA) 1100 instrument equipped with a diode-array UV/vis detector. The same FPLC method was applied to all protein samples. After sample injection, the mobile phase was maintained at 100% solvent A for 20 min, followed by a linear gradient from 100% A to 100 % B over 50 min, ending with an isocratic elution with 100% solvent B for 20 min. After the run was complete, the column was returned to 100% solvent A.
Prenylation of mCherry-CVIA with [3H]-farnesyl diphosphate
For radiolabeling experiments, 42 μL of 60 mM Tris-HCl, pH 7.0, 12 mM MgCl2, 12 μM ZnCl2, 18 mM dithiothreitol (DTT), and 3.0 μM mCherry-CVIA, was incubated for 1 h at 25 °C. Then, [3H]1-PP (16.7 μM, 1 mCi/mL) and yeast PFTase (activity 0.089 U/mg) were added to final concentrations of 1.0 μM and 10 nM, respectively, to bring the total volume to 50 μL. The reaction tube was incubated at 32 °C for 2 h. Most unreacted [3H]1-PP was removed by gel filtration (NAP-5, GE Healthcare). The entire 50 μL reaction volume was added to the column, and was eluted with 20 mM Tris-HCl, pH 7.5. The first 500 μL to elute from the column was discarded, and then the next 300 μL was collected. This solution was concentrated to 10 μL by centrifugal dialysis (Microcon YM-10, MWCO 10 kDa; Millipore), the dialysis cartridge was washed with 4 × 10 μL, and washes were recombined with the retentate (50 μL total). This sample was injected directly onto the FPLC column as described above. Fractions (1 mL) were collected from the column output, and 500 μL of each fraction was placed in a scintillation vial containing 5 mL of scintillation fluid. The radioactivity of the fractions was then measured using a Beckman LS 3801 scintillation counter.
Analytical mass spectrometry of prenylated proteins
Mass spectrometry of mCherry-CVIA and mCherry-CVIA-1 was performed on a reflectron ESI-micrOTOF-Q (Bruker Daltonics). Each protein was separately subjected to FPLC on β-CD-Sepharose. After collecting eluted protein, the samples were thoroughly washed and concentrated by repeated centrifugal dialysis (Amicon Ultra-15, MWCO 10.0 kDa) with H2O/MeOH (90/10 v/v, 4.0 mL × 7). The washed proteins were further dialyzed (using a Slide-A-Lyzer 2K MWCO cassette, Pierce, Rockford, IL) against H2O/MeOH/AcOH (89/10/1 v/v/v, 2 L × 2) to prepare them for ESI-MS injection. Protein was injected at a concentration of 20–30 μM into a flow of H2O/CH3CN/formic acid (64.9/35.0/0.1 v/v/v) at a flow rate of 0.1 mL/min. Ion series from m/z peaks were deconvoluted using Bruker DataAnalysis software.
Immobilization of mCherry-CVIA-azide onto alkyne-functionalized agarose beads
Alkyne-functionalized agarose beads were prepared according to a previously published protocol [9, 14]. To immobilize mCherry-CVIA-3 onto these beads, 30 μL of a 45 μM solution of mCherry-CVIA-3 in 20 mM Tris-HCl, pH 7.5, collected directly from purification on β-CD-Sepharose, was added to 100 μL of beads suspended in 50 mM phosphate buffer (pH 7.3). CuSO4, tris(2-carboxyethyl)phosphine hydrochloride (TCEP), and TBTA were added to this mixture to final concentrations of 1.0 mM, 1.0 mM, and 100 μM, respectively. The reaction was allowed to proceed for 4 h at room temperature. As a negative control, non-prenylated mCherry-CVIA (lacking an azide group) was combined with alkyne-functionalized beads under the same reaction conditions. After the reaction, beads were washed with 1 M NaCl, 50 mM phosphate buffer (pH 7.3, 200 μL × 3). Fluorescence and bright-field images of these agarose beads were obtained with a Nikon Eclipse TE200 inverted microscope. Fluorescence images were obtained with a red filter set (excitation bandpass 540–590 nm, emission bandpass 600–650 nm).
β-CD-Sepharose FPLC purification of prenylated mCherry-CVIA from crude cell lysate
Pelleted HeLa cells (from 0.5 mL suspension culture, National Cell Culture Center) was resuspended in 2.5 mL of freeze-thaw buffer (10 mM Tris, pH 7.8; 0.6 M KCl; 2.8 M glycerol; 6.7 mM pefabloc; 40 μg/mL leupeptin; 40 mg/mL pepstatin; 20 μg/mL aprotinin). The cells were lysed by 5 alternating rounds of flash freezing and thawing in a 30°C water bath followed by brief vortexing. The protein concentration was determined by Bradford assay to be 2.7 mg/mL. Varying amounts of mCherry-CVIA-1 were then doped into aliquots of the crude cell lysate to final concentrations of 8.8%, 2.0%, and 0.4% of total protein concentration. These lysate samples were injected directly onto a β-CD-Sepharose column and analyzed by FPLC as described above. Fractions were collected, desalted and concentrated by centrifugal microdialysis (Microcon YM-10), and analyzed by 15% acrylamide, Tris-glycine SDS-PAGE. The gel was stained with Coomassie Brilliant Blue for imaging.
Results and Discussion
Preparation of β-CD-Sepharose for FPLC
The main objective of this study was to evaluate β-CD-modified Sepharose as an affinity medium for separating prenylated proteins from non-prenylated proteins, and particularly for purifying the products of in vitro prenylation reactions from starting materials. In this study, CAAX-box-containing proteins were prenylated in vitro with either natural or non-natural isoprenoid diphosphates, as shown in Fig. 1. These reactions routinely afford protein product contaminated with unreacted isoprenoid starting material and unmodified protein, and a goal of this study was to use cyclodextrin affinity chromatography to separate these components.
Fig. 1.
Prenylation reactions purified in this study. PFTase and PGGTase-I accept their natural substrates (1-PP and 2-PP, respectively), but also catalyze the addition of isoprenoids containing non-natural functional groups.
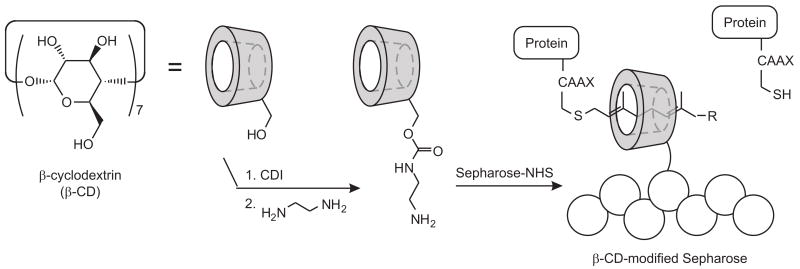
Our method for preparing and using the β-CD-modified Sepharose follows that described by Francis and coworkers [27], and is illustrated in Fig. 2. Amine groups were appended onto β-CD by activating its primary alcohols with carbonyl diimidazole (CDI), and then by condensing the resulting activated carbamate with ethylenediamine. One equivalent of each reagent was used, and because each molecule of β-CD bears seven primary alcohols, this synthesis produced a mixture of singly and multiply modified cyclodextrin as well as unmodified starting material, as verified by ESI-MS (data not shown). Francis reported that this mixture could be directly combined with Sepharose-NHS to produce β-CD-Sepharose that was functionally indistinguishable from Sepharose that had been modified with a pure cyclodextrin amine [27].
Fig. 2.
Preparation and use of β-CD-Sepharose for purification of prenylated proteins. In aqueous buffer, prenylated products are captured by the β-CD-modified support, but non-prenylated proteins bind poorly.
Isolation of prenylated proteins using β-CD-Sepharose as an affinity support
To help evaluate the performance of β-CD-Sepharose in the purification of in vitro prenylation reactions, we altered mCherry, a fluorescent protein recently developed by Tsien and coworkers [34], to serve as a prenylation target. Preliminary fluorescence correlation spectroscopy experiments on mCherry expressed in cells (J.D. Mueller, data not shown) indicate that the protein is monomeric at even high (25 μM) concentrations, and that as a result mCherry would serve as a well-behaved model protein in this study. A DNA sequence corresponding to a CVIA tag was appended to the C-terminus of the mCherry gene via PCR-based insertion. The resulting fusion gene was then subcloned and overexpressed to yield mCherry-CVIA. The emission spectrum and quantum yield of the mCherry-CVIA fusion were indistinguishable from those of mCherry. As an example of how this fusion protein was used, CVIA-tagged mCherry was exposed to PFTase and substrate 1-PP under standard prenylation conditions. Although we expected that this reaction would yield a mixture of farnesylated mCherry-CVIA (mCherry-CVIA-1) and unmodified mCherry-CVIA, these two proteins could not be resolved by SDS-PAGE or by reversed-phase chromatography.
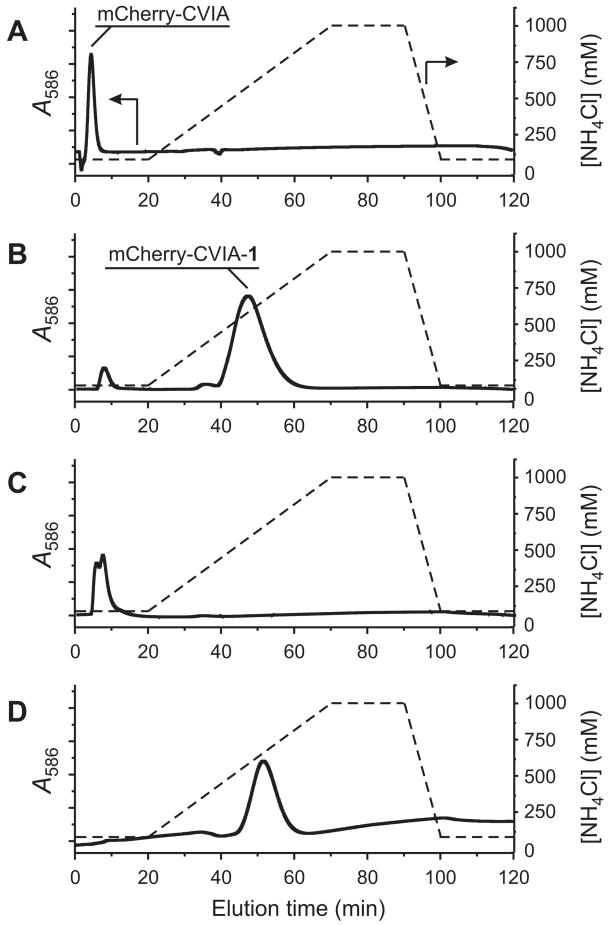
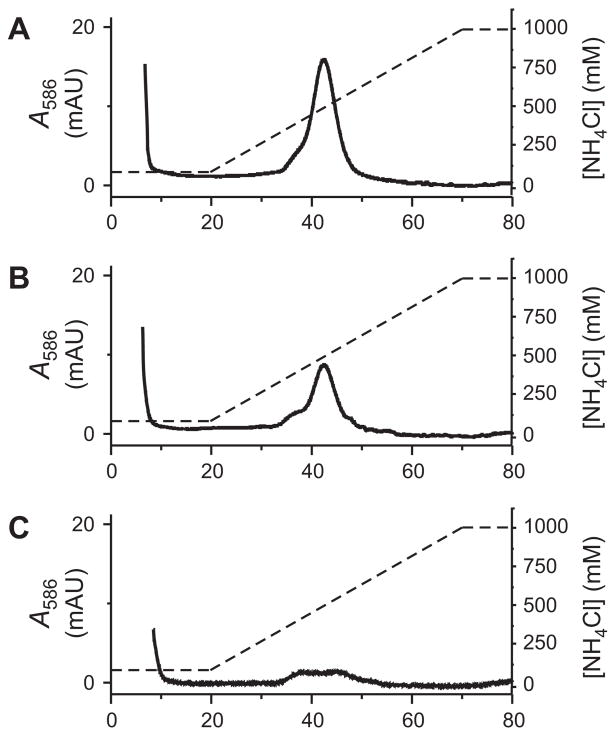
In contrast, when the mixture was injected onto an FPLC column packed with β-CD-Sepharose, we found that prenylated and unmodified mCherry-CVIA were separately eluted from the column using a salt gradient. The FPLC chromatogram of pure, unmodified mCherry-CVIA (monitored by visible absorbance at λmax = 586 nm for mCherry, Fig. 3a) subjected to a NH4Cl gradient showed only one elution peak at low salt concentration (80 mM NH4Cl, ~5 min). In contrast, the chromatogram of the prenylation reaction mixture (Fig. 3b) showed two peaks: one at low salt concentration, identical to that of pure starting material, and a second peak at higher salt concentration (~500 mM NH4Cl, ~50 min), corresponding to prenylated mCherry-CVIA-1. When these two fractions were separately collected from the column, desalted, and re-injected, single peaks appeared at the previously observed retention times, indicating effective separation of the two species (Fig. 3c,d). When fractions were collected from these re-injection runs, > 95% of the originally injected protein was recovered from the column, as quantified by UV/vis spectrophotometry. These features make β-CD-Sepharose a very practical stationary phase for FPLC of prenylated proteins.
Fig. 3.
FPLC analysis, using a β-CD-Sepharose column, of in vitro farnesylation reactions of between mCherry-CVIA and 1-PP, catalyzed by PFTase. (A) Pure mCherry-CVIA, prior to farnesylation. (B) Product mixture (containing both mCherry-CVIA and mCherry-CVIA-1) after prenylation. Based on peak integrations, 95% of mCherry-CVIA was prenylated under these conditions. (C) Fraction collected over t = 6–15 min from run B, reinjected onto the FPLC column. (D) Fraction collected over t = 42–60 min from run B, reinjected onto the FPLC column. Solid lines: Relative optical absorbance of eluent at λ = 586 nm, measured on-line. Dashed lines: Illustration of the NH4Cl salt gradient.
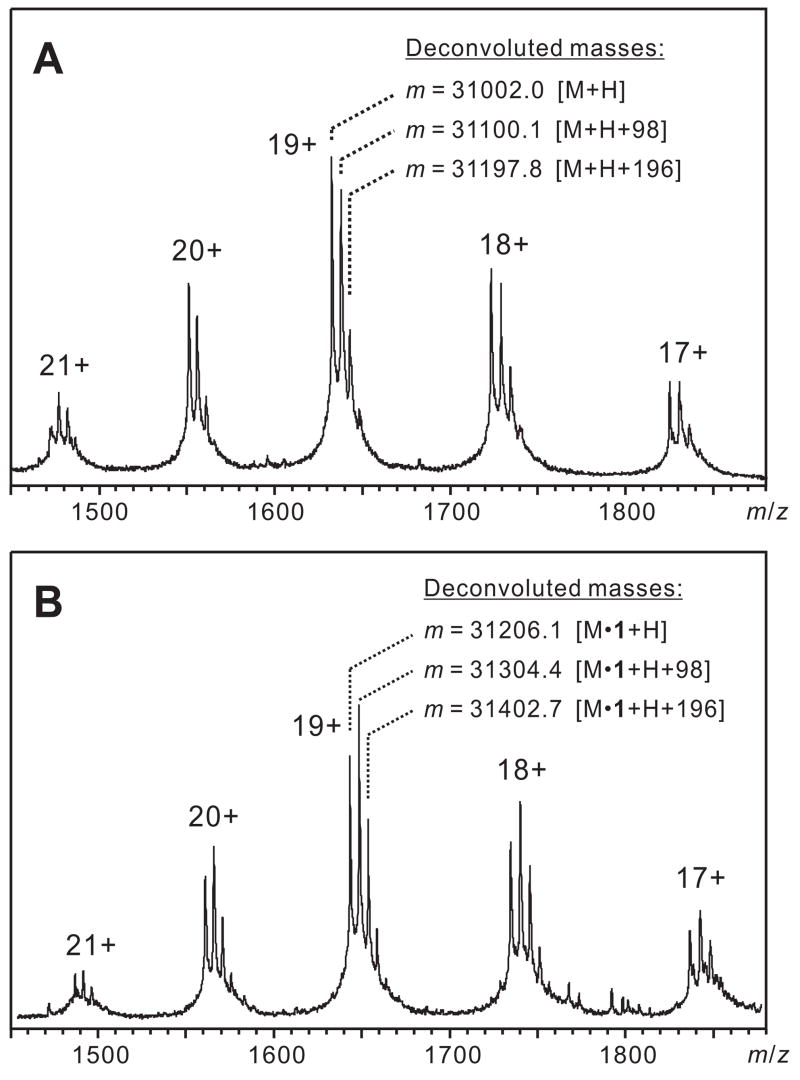
The identities of the two eluted proteins were confirmed by ESI-MS (Figure 4). ESI-MS of unreacted mCherry-CVIA, and of the fraction collected from the first FPLC peak, produced an ion series that was deconvoluted to yield a mass that matched mCherry-CVIA (theoretical [M+H] Mr = 31001.9, deconvoluted m = 31002.0). ESI-MS of the fraction collected from the second FPLC peak gave an ion series that matched mCherry-CVIA-1 (theoretical [M+H] Mr = 31206.2, deconvoluted m = 31206.1).
Fig. 4.
ESI-MS mass spectra of (A) mCherry-CVIA and (B) mCherry-CVIA-1, labeled with deconvoluted masses for each of the three peak series. In addition to an ion series corresponding to parent protein [M+H], the spectra also show ion series [M+H+98] and [M+H+2(98)]. This pattern has been previously observed and ascribed to associated H2PO4- ions [45–47].
We evaluated β-CD-Sepharose as an affinity medium for the purification of prenylated proteins from a variety of in vitro prenylation reactions—including reactions with PFTase and GGTase-I, using natural and non-natural substrates, with a variety of wild-type and engineered CAAX-box targets (Table 1). In all cases, it was possible to separate the prenylated product from unmodified protein using the same gradient FPLC protocol described above, and in all cases, the unmodified and prenylated proteins eluted at nearly identical salt concentrations. The consistency of the method, along with the significantly longer retention time for prenylated proteins, supports our hypothesis that specific interactions between Sepharose-bound cyclodextrin and the hydrophobic prenyl group (as illustrated in Fig. 2) are responsible for the separation in this system.
Table 1.
In vitro prenylation reactions analyzed by β-CD-Sepharose FPLC.
| Protein | Enzyme | Diphosphate Substrate | Retention Time (min) |
|
|---|---|---|---|---|
| Unmodified Protein | Prenylated Protein | |||
| mCherry-CVIA | PFTase | 1-PP | 4.7 | 41.2 |
| mCherry-CVIA | PFTase | 3-PP | 4.3 | 42.9 |
| eGFP-CVIA | PFTase | 3-PP | 8.7 | 42.1 |
| CDC42-CVLL | PGGTase-I | 2-PP | 5.7 | 35.6 |
| CDC42-CVLL | PGGTase-I | 4-PP | 4.8 | 35.0 |
| GST-CIIS | PFTase | 1-PP | 6.7 | 37.2 |
| H-Ras-CVLS | PFTase | 1-PP | 5.9 | 36.6 |
Physical basis of separations with β-CD-Sepharose
We performed a number of control experiments to test the roles of different components of the FPLC method. FPLC columns fabricated with ordinary Sepharose alone, or with Sepharose-NHS that had been reacted with ethanolamine but no β-CD, failed to separate prenylated from non-prenylated proteins. Clearly, resin-bound cyclodextrin is critical to separation. On the other hand, separation did not depend on the specific buffer salt used in the gradient. Ammonium sulfate and chloride, sodium phosphate, and HEPES buffers were all successfully used to elute proteins from the column. Changing the starting salt concentration or the steepness of the salt gradient affected the elution times of both prenylated and non-prenylated proteins, indicating that association of the protein with the solid phase, and not just interaction of the prenyl group with cyclodextrin, affected retention time. This was previously noted by Francis and coworkers for β-CD-Sepharose [27]. One important difference between our method and Francis’ report is that a competitive binder for cyclodextrin was not necessary to displace the protein from the solid phase. Presumably, the specific affinity of cyclodextrin for the prenyl group in this system is just strong enough to strengthen the retention of prenylated proteins on the β-CD-Sepharose, but not so strong that salt-gradient chromatography is insufficient to displace them. This combination of cyclodextrin and matrix affinity has been previously described for cyclodextrin-bonded phases [35].
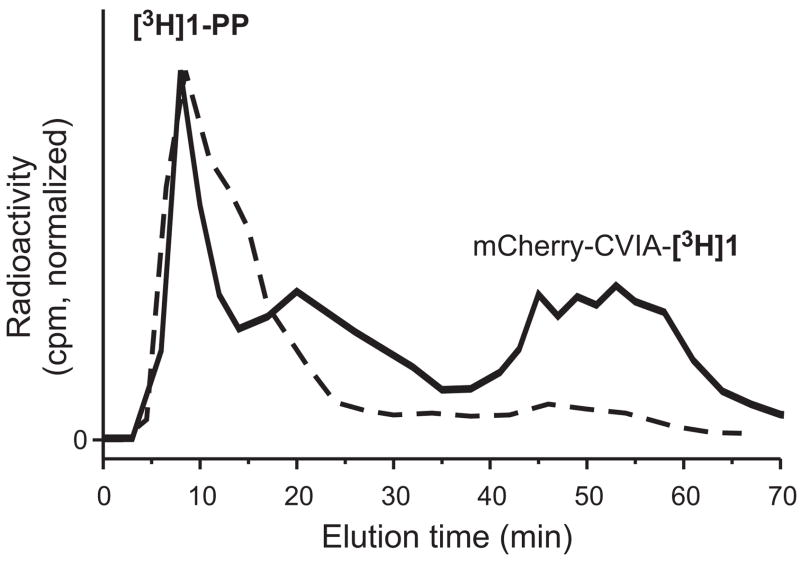
One interesting consequence of this dual affinity for the prenyl group and protein by β-CD-Sepharose is that excess prenyl diphosphate is also separated from prenylated protein by the column. To evaluate this, we used radioactively labeled [3H]1-PP as a substrate in a farnesylation reaction with PFTase and mCherry-CVIA. At the end of the reaction period, the reaction mixture was injected onto the FPLC column and eluted with an NH4Cl gradient. Fractions were collected, and the eluted [3H] label in each fraction was quantitated by scintillation counting. The resulting data are plotted as an FPLC chromatogram in Fig. 5, along with data from a control injection of [3H]1-PP. By itself, [3H]1-PP eluted from the column early, at low salt concentration. The farnesylation reaction mixture showed two radioactive species: one eluting at short retention time, corresponding to unreacted [3H]1-PP; and another at the longer retention time characteristic of prenylated protein, corresponding to mCherry-CVIA-[3H]1. An important conclusion of this experiment is that the β-CD-Sepharose column successfully isolated the prenylated protein product not only from unreacted protein, but also from unreacted prenyl diphosphate.
Fig. 5.
FPLC analysis, using a β-CD-Sepharose column, of (solid line) farnesylation of mCherry-CVIA with PFTase and [3H]1-PP, and (dashed line) [3H]1-PP alone.
Other chromatographic formats
We also tested the use of β-CD-Sepharose as an affinity medium in simpler batch and gravity modes. For example, we packed a cotton-plugged, disposable glass pipette with a slurry of β-CD-Sepharose in 15 mM HEPES, and then loaded onto the column a crude prenylation reaction containing a mixture of prenylated mCherry-CVIA-1 and unreacted mCherry-CVIA. When the column was washed with 100 mM NH4Cl, pH 7.0 buffer, the mCherry-CVIA was eluted from the column (as verified by subsequent FPLC analysis), but the column remained bright red from retained mCherry-CVIA-1. Then, flushing the column with 1.0 M NH4Cl, pH 7.0 resulted in the elution of mCherry-CVIA-1 (again verified by FPLC), leaving behind white, non-fluorescent Sepharose. Spectrophotometry of fractions collected from this column indicated that > 80% of total protein was eluted from the column, and that the extinction and fluorescence spectra of the eluted proteins remained unchanged by the purification.
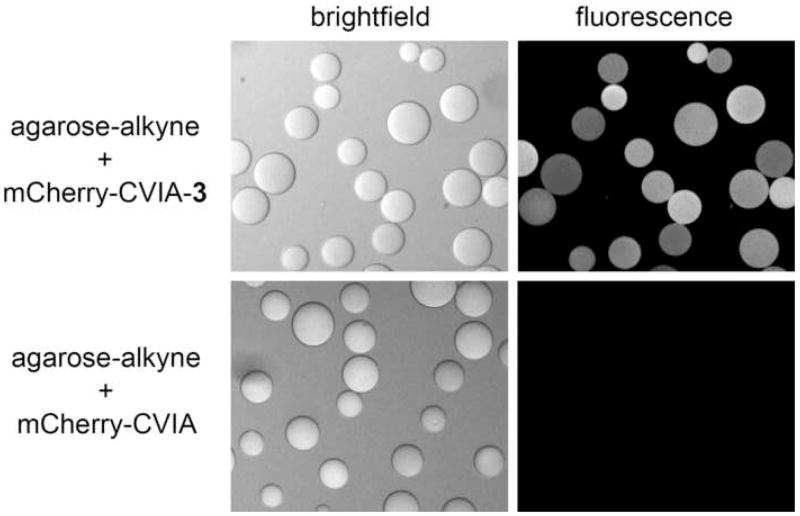
“Clicking” purified mCherry-CVIA-azide to alkyne-functionalized agarose beads
We have previously shown that performing prenylation reactions with non-natural, functional prenyl substrates can be used to add non-biological functionality to the C-terminus of a protein and prepare it for site-specific bioconjugate chemistry [9]. For example, prenylating with substrate 3-PP provided azide-functionalized proteins that could be covalently attached to alkyne-functionalized substrates via Cu(I)-catalyzed, [3+2] Huisgen cycloaddition (“click”) chemistry [36, 37]. In order to demonstrate that C-terminal functionality in these proteins survived purification by β-CD-Sepharose chromatography, we prepared azide-functionalized mCherry-CVIA-3, purified it by FPLC as described above, and then immobilized the protein onto alkyne-functionalized agarose beads via click chemistry. After the immobilization reaction, fluorescence images of the beads (Fig. 6) showed the distinctive red emission from bound mCherry. Negative control experiments, in which unfunctionalized mCherry-CVIA was substituted for mCherry-CVIA-3, showed no fluorescence on the beads under the same reaction conditions. From this data, we argue that β-CD-Sepharose chromatography retains functionality in engineered, prenylated fusion proteins.
Fig. 6.
Conjugation reactions of alkyne-modified agarose beads with (top) azide-functionalized mCherry-CVIA-3, and (bottom) non-functionalized mCherry-CVIA. Fluorescence images were acquired using identical illumination and imaging settings.
Isolating prenylated proteins from crude cell extracts using β-CD-Sepharose
Although the data shown above illustrates β-CD-Sepharose chromatography of overexpressed prenylated proteins, the method would also be extremely useful for isolation of endogenously prenylated proteins from cells. A number of recent reports have highlighted chemical methods for proteomic analysis of lipidated proteins,[13, 38–42] and these methods would be complemented by a reliable, chromatographic pretreatment that could enrich lipidated protein products from whole cell extracts. To test β-CD-Sepharose chromatography for this application, we analyzed model samples in which crude HeLa cell lysates were spiked with different amounts of fluorescent, farnesylated mCherry-CVIA-1. These samples were injected directly onto the β-CD-Sepharose FPLC column; no attempt was made to separate membrane and cytosolic fractions, or to otherwise enhance the proportion of farnesylated protein analyzed on the column. Chromatograms from these separations are shown in Fig. 7, in which elution of material from the column is again monitored by absorbance at λ = 586 nm. Most of the injected lysate eluted from the column immediately. We attribute the extremely high extinction signal at early (0–10 min) retention times to scattering from lysate debris and other proteins that traveled through the column with minimal binding to the β-CD-Sepharose support. At higher salt concentrations and longer retention times, we observed a peak consistent with elution of mCherry-CVIA-1 from the β-CD-Sepharose. The intensity of this peak correlated directly with the fraction of mCherry-CVIA-1 combined with the lysate. We were able to distinguish eluted, prenylated mCherry at fractions as low as ~0.5% of total protein using our analytical method; lower fractions of prenylated protein could potentially be fractionated from cell samples by β-CD-Sepharose FPLC, but we were unable to test this. However, given that prenylated proteins can make up as much as 2% of total protein in a cell,[43] this technique would be useful for practical preparation of prenylated proteins from cell lysates even at the high fractions we evaluated in this study.
Fig. 7.
β-CD-Sepharose FPLC of crude Hela cell lysate, spiked with different fractions of mCherry-CVIA-1 (relative to the total amount of protein in the lysate): (A) 8.8%; (B) 2.0%; (C) 0.4%.
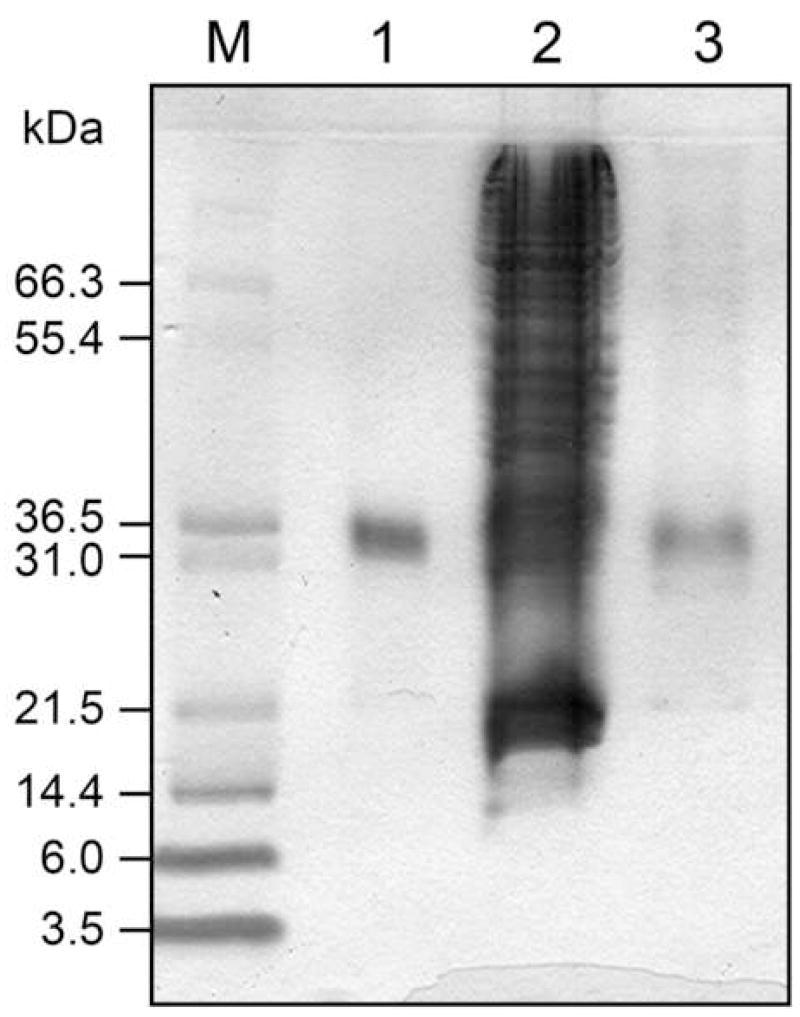
In order to confirm the identity and purity of the eluted protein, we collected the material that eluted from the column from 37–50 min and subjected it to SDS-PAGE analysis (Fig. 8). Lane 3, corresponding to the eluted material, shows that β-CD-Sepharose FPLC successfully enhanced the fraction of prenylated mCherry in the collected protein relative to the remainder of the HeLa cell protein. There are still some other, weak protein bands in lane 3, which may correspond to endogenous lipidated protein from the cell lysate captured by the column. Regardless, this model experiment demonstrates that affinity chromatography on β-CD-Sepharose can be used to isolate prenylated from practical cell samples.
Fig. 8.
SDS PAGE analysis of mCherry-CVIA-1, purified from HeLa cell lysate by β-CD-Sepharose FPLC. Lane 1: pure mCherry-CVIA-1 (1 μg), prior to FPLC; lane 2: 8.8% mCherry-CVIA-1 in HeLa cell lysate (27 μg total); lane 3: fraction collected from FPLC, elution time 37–50 min.
Conclusions
Affinity chromatography of prenylated proteins on β-CD-Sepharose is a robust method for purifying both natural and engineered proteins from in vitro prenylation reactions. The method successfully isolates prenylated proteins from both non-prenylated starting materials as well as unreacted isoprenoid substrate. Although protein-matrix interactions play a role, separation in the system is driven by interactions between protein prenyl groups and column-bound cyclodextrin. Because in vitro prenylation is performed in a variety of contexts—for example, in screening of prenyltransferase inhibitors [44], in proteomics [13], in protein engineering, and in research on prenylation—the method outlined here could be useful to many researchers working with protein prenyltransferases. In addition, given the selectivity of β-CD for large hydrophobic groups, the method might also be adapted to isolating proteins with other membrane-directing hydrophobic modifications, including myristoyl or palmitoyl groups in addition to the farnesyl and geranylgeranyl groups analyzed here, from a variety of sources. In general, the simplicity of this technique should make it broadly useful for studies of lipid-modified proteins.
Acknowledgments
T.A.T. and M.D.D. acknowledge support from the National Institutes of Health (CA122603 and GM058842). J.D.M. and Y.C. acknowledge support from the National Institutes of Health (GM64589) and the National Science Foundation (PHY-0346782). We thank Roger Tsien (UCSD) for providing the plasmid for mCherry, Stepan Lenevich for determining concentrations of prenyl diphosphates by NMR, and Dana Reed for help with mass spectrometry experiments.
Footnotes
Abbreviations used: PFTase, protein farnesyltransferase; PGGTase-I, protein geranylgeranyltransferase; β-CD, β-cyclodextrin; NHS, N-hydroxysuccinimidyl; eGFP, enhanced green fluorescent protein; GST, glutathione S-transferase; TBTA, tris(benzyltriazolylmethyl)amine; CDI, 1,1-carbonyldiimidazole; FPLC, fast protein liquid chromatography; ESI, electrospray ionization; LB, Luria-Bertani; IPTG, ισοπροπψλ–β-D-thiogalactopyranoside; NTA, nitrilotriacetic acid; DTT, dithiothreitol; MWCO, molecular weight cut-off; TCEP, tris(2-carboxyethyl)phosphine hydrochloride.
Supporting Information Available: DNA sequencing data for mCherry-CVIA construct.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lane KT, Beese LS. Structural biology of protein farnesyltransferase and geranylgeranyltransferase type I. J Lipid Res. 2006;47:681–699. doi: 10.1194/jlr.R600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 2.Zhang FL, Casey PJ. Protein prenylation: Molecular mechanisms and functional consequences. Annu Rev Biochem. 1996;65:241–269. doi: 10.1146/annurev.bi.65.070196.001325. [DOI] [PubMed] [Google Scholar]
- 3.Roskoski R. Protein prenylation: a pivotal posttranslational process. Biochem Biophys Res Commun. 2003;303:1–7. doi: 10.1016/s0006-291x(03)00323-1. [DOI] [PubMed] [Google Scholar]
- 4.Wright LP, Philips MR. CAAX modification and membrane targeting of Ras. J Lipid Res. 2006;47:883–891. doi: 10.1194/jlr.R600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 5.Silvius JR. Mechanisms of Ras protein targeting in mammalian cells. J Membr Biol. 2002;190:83–92. doi: 10.1007/s00232-002-1026-4. [DOI] [PubMed] [Google Scholar]
- 6.Resh MD. Trafficking and signaling by fatty-acylated and prenylated proteins. Nat Chem Biol. 2006;2:584–590. doi: 10.1038/nchembio834. [DOI] [PubMed] [Google Scholar]
- 7.Gelb MH, Brunsveld L, Hrycyna CA, Michaelis S, Tamanoi F, Van Voorhis WC, Waldmann H. Therapeutic intervention based on protein prenylation and associated modifications. Nat Chem Biol. 2006;2:518–528. doi: 10.1038/nchembio818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duckworth BP, Chen Y, Wollack JW, Sham Y, Mueller JD, Taton TA, Distefano MD. A universal method for the preparation of covalent protein-DNA conjugates for use in creating protein nanostructures. Angew Chem, Int Ed. 2007;46:8819–8822. doi: 10.1002/anie.200701942. [DOI] [PubMed] [Google Scholar]
- 9.Duckworth BP, Xu J, Taton TA, Guo A, Distefano MD. Site-specific, covalent attachment of proteins to a solid surface. Bioconjugate Chem. 2006;17:967–974. doi: 10.1021/bc060125e. [DOI] [PubMed] [Google Scholar]
- 10.Xu J, Degraw AJ, Duckworth BP, Lenevich S, Tann CM, Jenson EC, Gruber SJ, Barany G, Distefano MD. Synthesis and reactivity of 6,7-dihydrogeranylazides: Reagents for primary azide incorporation into peptides and subsequent Staudinger ligation. Chem Biol Drug Des. 2006;68:85–96. doi: 10.1111/j.1747-0285.2006.00420.x. [DOI] [PubMed] [Google Scholar]
- 11.Rose MW, Rose ND, Boggs J, Lenevich S, Xu J, Barany G, Distefano MD. Evaluation of geranylazide and farnesylazide diphosphate for incorporation of prenylazides into a CAAX box-containing peptide using protein farnesyltransferase. J Pept Res. 2005;65:529–537. doi: 10.1111/j.1399-3011.2005.00261.x. [DOI] [PubMed] [Google Scholar]
- 12.Rose MW, Xu J, Kale TA, O’Doherty G, Barany G, Distefano MD. Enzymatic incorporation of orthogonally reactive prenylazide groups into peptides using geranylazide diphosphate via protein farnesyltransferase: Implications for selective protein labeling. Biopolymers. 2005;80:164–171. doi: 10.1002/bip.20239. [DOI] [PubMed] [Google Scholar]
- 13.Kho Y, Kim SC, Jiang C, Barma D, Kwon SW, Cheng J, Jaunbergs J, Weinbaum C, Tamanoi F, Falck J, Zhao Y. A tagging-via-substrate technology for detection and proteomics of farnesylated proteins. Proc Natl Acad Sci U S A. 2004;101:12479–12484. doi: 10.1073/pnas.0403413101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duckworth BP, Zhang Z, Hosokawa A, Distefano MD. Selective labeling of proteins by using protein farnesyltransferase. ChemBioChem. 2007;8:98–105. doi: 10.1002/cbic.200600340. [DOI] [PubMed] [Google Scholar]
- 15.Labadie GR, Viswanathan R, Poulter CD. Farnesyl diphosphate analogues with ω-bioorthogonal azide and alkyne functional groups for protein farnesyl transferase-catalyzed ligation reactions. J Org Chem. 2007;72:9291–9297. doi: 10.1021/jo7017747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gauchet C, Labadie GR, Poulter CD. Regio- and chemoselective covalent immobilization of proteins through unnatural amino acids. J Am Chem Soc. 2006;128:9274–9275. doi: 10.1021/ja061131o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dursina B, Reents R, Delon C, Wu Y, Kulharia M, Thutewohl M, Veligodsky A, Kalinin A, Evstifeev V, Ciobanu D, Szedlacsek SE, Waldmann H, Goody RS, Alexandrov K. Identification and specificity profiling of protein prenyltransferase inhibitors using new fluorescent phosphoisoprenoids. J Am Chem Soc. 2006;128:2822–2835. doi: 10.1021/ja052196e. [DOI] [PubMed] [Google Scholar]
- 18.Dursina BE, Reents R, Niculae A, Veligodsky A, Breitling R, Pyatkov K, Waldmann H, Goody RS, Alexandrov K. A genetically encodable microtag for chemo-enzymatic derivatization and purification of recombinant proteins. Protein Express Purif. 2005;39:71–81. doi: 10.1016/j.pep.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 19.Turek-Etienne TC, Strickland CL, Distefano MD. Biochemical and structural studies with prenyl diphosphate analogues provide insights into isoprenoid recognition by protein farnesyl yransferase. Biochemistry. 2003;42:3716–3724. doi: 10.1021/bi0266838. [DOI] [PubMed] [Google Scholar]
- 20.Nguyen UTT, Cramer J, Gomis J, Reents R, Gutierrez-Rodriguez M, Goody RS, Alexandrov K, Waldmann H. Exploiting the substrate tolerance of farnesyltransferase for site-selective protein derivatization. ChemBioChem. 2007;8:408–423. doi: 10.1002/cbic.200600440. [DOI] [PubMed] [Google Scholar]
- 21.Krzysiak AJ, Rawat DS, Scott SA, Pais JE, Handley M, Harrison ML, Fierke CA, Gibbs RA. Combinatorial modulation of protein prenylation. ACS Chem Biol. 2007;2:385–389. doi: 10.1021/cb700062b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bordier C. Phase-separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 23.Casey PJ, Solski PA, Der CJ, Buss JE. P21ras is modified by a farnesyl isoprenoid. Proc Natl Acad Sci U S A. 1989;86:8323–8327. doi: 10.1073/pnas.86.21.8323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkielstein CV, Overduin M, Capelluto DGS. Cell migration and signaling specificity is determined by the phosphatidylserine recognition motif of Rac1. J Biol Chem. 2006;281:27317–27326. doi: 10.1074/jbc.M605560200. [DOI] [PubMed] [Google Scholar]
- 25.Gorzalczany Y, Sigal N, Itan M, Lotan O, Pick E. Targeting of Rac1 to the phagocyte membrane is sufficient for the induction of NADPH oxidase assembly. J Biol Chem. 2000;275:40073–40081. doi: 10.1074/jbc.M006013200. [DOI] [PubMed] [Google Scholar]
- 26.Rak A, Niculae A, Kalinin A, Thoma NH, Sidorovitch V, Goody RS, Alexandrov K. In vitro assembly, purification, and crystallization of the Rab geranylgeranyl transferase:substrate complex. Protein Express Purif. 2002;25:23–30. doi: 10.1006/prep.2001.1605. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen T, Joshi NS, Francis MB. An affinity-based method for the purification of fluorescently-labeled biomolecules. Bioconjugate Chem. 2006;17:869–872. doi: 10.1021/bc060130i. [DOI] [PubMed] [Google Scholar]
- 28.Connors KA. Population characteristics of cyclodextrin complex stabilities in aqueous solution. J Pharm Sci. 1995;84:843–848. doi: 10.1002/jps.2600840712. [DOI] [PubMed] [Google Scholar]
- 29.Davisson VJ, Woodside AB, Neal TR, Stremler KE, Muehlbacher M, Poulter CD. Phosphorylation of isoprenoid alcohols. J Org Chem. 1986;51:4768–4779. [Google Scholar]
- 30.Mayer MP, Prestwich GD, Dolence JM, Bond PD, Hong-yu W, Poulter CD. Protein farnesyltransferase: Production in Escherichia coli and immunoaffinity purification of the heterodimer from Saccharomyces cerevisiae. Gene. 1993;132:41–47. doi: 10.1016/0378-1119(93)90512-2. [DOI] [PubMed] [Google Scholar]
- 31.DeGraw AJ, Zhao Z, Strickland CL, Taban AH, Hsieh J, Jefferies M, Xie W, Shintani DK, McMahan CM, Cornish K, Distefano MD. A photoactive isoprenoid diphosphate analogue containing a stable phosphonate linkage: Synthesis and biochemical studies with prenyltransferases. J Org Chem. 2007;72:4587–4595. doi: 10.1021/jo0623033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gomez R, Goodman LE, Tripathy SK, O’Rourke E, Manne V, Tamanoi F. Purified yeast protein farnesyltransferase is structurally and functionally similar to its mammalian counterpart. Biochem J. 1993;289:25–31. doi: 10.1042/bj2890025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chan TR, Hilgraf R, Sharpless KB, Fokin VV. Polytriazoles as copper(I)-stabilizing ligands in catalysis. Org Lett. 2004;6:2853–2855. doi: 10.1021/ol0493094. [DOI] [PubMed] [Google Scholar]
- 34.Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 35.Wilder DR, Tindall GW, Cunningham LJ, Little JL. High-performance liquid chromatographic analysis of sulfonated aromatics using a β-cyclodextrin-bonded phase. J Chromatogr. 1993;635:221–226. [Google Scholar]
- 36.Wang Q, Chan TR, Hilgraf R, Fokin VV, Sharpless KB, Finn MG. Bioconjugation by copper(I)-catalyzed azide-alkyne [3 + 2] cycloaddition. J Am Chem Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 37.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem, Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 38.Kostiuk MA, Corvi MM, Keller BO, Plummer G, Prescher JA, Hangauer MJ, Bertozzi CR, Rajaiah G, Falck JR, Berthiaume LG. Identification of palmitoylated mitochondrial proteins using a bio-orthogonal azido-palmitate analogue. FASEB J. 2008;22:721–732. doi: 10.1096/fj.07-9199com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin DDO, Vilas GL, Prescher JA, Rajaiah G, Falck JR, Bertozzi CR, Berthiaume LG. Rapid detection, discovery, and identification of post-translationally myristoylated proteins during apoptosis using a bio-orthogonal azidomyristate analog. FASEB J. 2008;22:797–806. doi: 10.1096/fj.07-9198com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wirtz J, Kolter T. Novel tools for the proteomic identification of acylated proteins. ChemBioChem. 2007;8:1631–1635. doi: 10.1002/cbic.200700334. [DOI] [PubMed] [Google Scholar]
- 41.Troutman JM, Roberts MJ, Andres DA, Spielmann HP. Tools to analyze protein farnesylation in cells. Bioconjugate Chem. 2005;16:1209–1217. doi: 10.1021/bc050068+. [DOI] [PubMed] [Google Scholar]
- 42.Zhao Y, Zhang W, Kho Y, Zhao Y. Proteomic analysis of integral plasma membrane proteins. Anal Chem. 2004;76:1817–1823. doi: 10.1021/ac0354037. [DOI] [PubMed] [Google Scholar]
- 43.Epstein WW, Lever D, Leining LM, Bruenger E, Rilling HC. Quantitation of prenylcysteines by a selective cleavage reaction. Proc Natl Acad Sci U S A. 1991;88:9668–9670. doi: 10.1073/pnas.88.21.9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pais JE, Bowers KE, Stoddard AK, Fierke CA. A continuous fluorescent assay for protein prenyltransferases measuring diphosphate release. Anal Biochem. 2005;345:302–311. doi: 10.1016/j.ab.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 45.Chowdhury SK, Katta V, Beavis RC, Chait BT. Origin and removal of adducts (molecular mass = 98 u) attached to peptide and protein ions in electrospray ionization mass spectra. J Am Soc Mass Spectrom. 1990;1:382–388. doi: 10.1016/1044-0305(90)85018-H. [DOI] [PubMed] [Google Scholar]
- 46.Stephenson JL, Jr, McLuckey SA. Ion/ion proton transfer reactions for protein mixture analysis. Anal Chem. 1996;68:4026–4032. doi: 10.1021/ac9605657. [DOI] [PubMed] [Google Scholar]
- 47.Fountoulakis M, Vilbois F, Oesterhelt G, Vetter W. Phosphoric acid entrapment leads to apparent protein heterogeneity. Bio/Technology. 1995;13:383–388. doi: 10.1038/nbt0495-383. [DOI] [PubMed] [Google Scholar]