Abstract
Adenosine, prostaglandin E2, or increased intracellular cyclic AMP concentration each elicit potent anti-inflammatory events in human neutrophils by inhibiting functions such as phagocytosis, superoxide production, adhesion and cytokine release. However, the endogenous molecular pathways mediating these actions are poorly understood. In the present study, we examined their impact on the gene expression profile of stimulated neutrophils. Purified blood neutrophils from healthy donors were stimulated with a cocktail of inflammatory agonists in the presence of at least one of the following anti-inflammatory agents: adenosine A2A receptor agonist CGS 21680, prostaglandin E2, cyclic-AMP-elevating compounds forskolin and RO 20-1724. Total RNA was analyzed using gene chips and real-time PCR. Genes encoding transcription factors, enzymes and regulatory proteins, as well as secreted cytokines/chemokines showed differential expression. We identified 15 genes for which the anti-inflammatory agents altered mRNA levels. The agents affected the expression profile in remarkably similar fashion, suggesting a central mechanism limiting cell activation. We have identified a set of genes that may be part of important resolution pathways that interfere with cell activation. Identification of these pathways will improve understanding of the capacity of tissues to terminate inflammatory responses and contribute to the development of therapeutic strategies based on endogenous resolution.
Introduction
Neutrophils constitute the majority of circulating leukocytes and are often the first cells to migrate toward inflammatory lesions, where they exert host defense functions including the phagocytosis of cell debris and invading microorganisms, the generation of oxygen-derived reactive agents and the release of proteolytic enzymes [1]. In response to specific stimuli, neutrophils can synthesize and release an array of factors such as anti-microbial proteins and extracellular matrix proteins as well as several cytokines and chemokines and thereby play a major role in orchestrating early stages of the inflammatory response [2]. Although recurrent infections in patients with defective neutrophil function confirm their importance in host defense, these cells also bear enormous destructive capacity and can elicit significant tissue damage. Unchecked activation of neutrophils is associated with pathological states such as ischemia, sepsis, chronic obstructive pulmonary disease and rheumatoid arthritis [3]–[5]. It is therefore of both fundamental and clinical interest to gain understanding of not only the mechanisms that promote neutrophil functions, but also of those that can restrict such activation and bring about the resolution of inflammation.
Adenosine, through activation of the A2A receptor (A2AR) subtype, ranks among the most potent agents limiting the inflammatory activities of neutrophils. One of the first reports on this matter, published more than two decades ago by Cronstein et al. [6], determined that the autacoid inhibited superoxide production resulting from inflammatory stimuli. Interest in adenosine and its receptors has since fuelled major research efforts, which have contributed to increased appreciation of their pivotal importance in limiting inflammation [7]–[9]. High concentrations of extracellular adenosine can be found in vivo in traumatized tissues and this autacoid may have a role in reducing the accumulation of leukocytes at the site of injury [10]. A paramount role for the A2AR subtype in mediating anti-inflammatory activities has been for all practical purposes established in previous studies [11]–[16]. The cyclic-AMP-elevating Gs-protein-coupled A2AR subtype modulates key pro-inflammatory neutrophil functions such as superoxide generation, de-granulation and adhesion (reviewed in [17]). Endogenous adenosine and A2AR agonists have shown to be potent inhibitors of leukotriene and platelet-activating factor synthesis [13], [18]–[20] and in contrast, to stimulate COX-2 expression in neutrophils [21], [22], thus increasing the capacity of these cells to produce prostaglandin E2. This shift in the profile of lipid mediator production from leukotrienes to prostaglandin E2 may contribute to preventing subsequent neutrophil-elicited inflammatory events. Recently, our laboratory reported that A2AR activation had a striking inhibitory impact on the in vitro and in vivo generation of tumor necrosis factor α and several other neutrophil-derived cytokines and chemokines [23], confirming a preeminent role for adenosine in restricting neutrophil activation. Most of the anti-inflammatory activities of this autacoid through A2AR engagement are thought to involve a rise in intracellular cyclic AMP concentration [22], [24], [25]. Prostaglandin E2, acting through its own set of receptors, is also a potent inhibitor of neutrophil inflammatory functions and can, similarly to adenosine, modulate pivotal neutrophil effector functions such as chemotaxis, aggregation, superoxide production, lysozyme release and leukotriene B4 production by raising intracellular cyclic AMP concentration above basal levels [22], [26]–[33]. Adenosine and prostaglandin E2 thus clearly stand out as two major anti-inflammatory signals, while elevated intracellular cyclic AMP concentration, which can be pharmacologically achieved with a combination of the adenylate cyclase activator forskolin and of the phosphodiesterase IV inhibitor RO-20-1724, often appears to accompany their actions. However, the gene activities that control inflammation resolution pathways remain poorly understood.
In the present study, we used DNA microarray technology and real-time PCR to examine the impact of major anti-inflammatory signals, namely A2AR activation, prostaglandin E2 and elevated intracellular cyclic AMP, on the gene expression profile of human neutrophils stimulated by known inflammatory agonists. We have identified a group of genes for which mRNA levels were significantly altered by anti-inflammatory signals. This may indicate their involvement in pivotal molecular signaling pathways associated with the resolution of inflammation.
Results
Gene expression in stimulated human neutrophils
Microarray data is conform to the MIAME guidelines; unsupervised, raw data was deposited in the GEO database (geo@ncbi.nlm.nih.gov), submission number: GSE14465. Initial analysis of the DNA microarray chips using the Affymetrix software indicated that approximately 15,000 of the 54,675 sequences recognized in the array (i.e. 27.5%) are expressed by resting human neutrophils. Comparison between resting and neutrophils stimulated with a mixture of pro-inflammatory agonists for 30 min, revealed 1,152 differentially expressed sequences, of which 401 corresponded to known proteins [34] and are listed in Supplementary Table S1 in descending order of the expression differential magnitude. Using the Kegg pathway database, we selected genes for further examination, based on their potential implication in immune response processes (http://www.genome.jp/kegg/pathway.html). Selected pathways included: cytokine-cytokine receptor interaction, leukocyte transendothelial migration, Jak-STAT signaling pathways, MAPK signaling pathways, natural killer cell-mediated cytotoxicity, apoptosis, Toll-like receptor signaling pathways, T & B cell receptor signaling pathways, arachidonic acid metabolism, insulin signaling pathways, neuroactive ligand-receptor interactions, focal adhesion, ubiquitin-mediated protein lysis, and chronic myeloid leukemia. By this approach, 68 genes were selected and essentially corresponded to transcription factors, enzymes, regulatory elements, cytokines/chemokines and receptors (Supplementary Table S2).
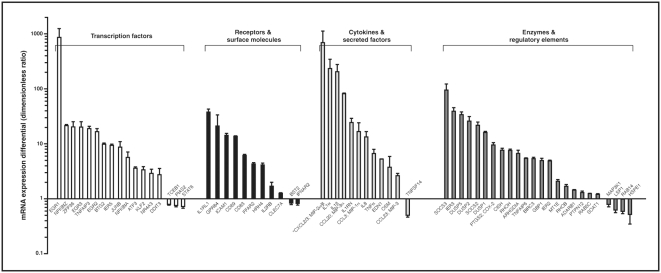
We sought to validate the gene chip results using real-time PCR for analysis of the selected genes. This analysis corroborated a significant differential expression between resting and stimulated cells for 64 of the 68 genes. Integrated real-time PCR results are presented in Figure 1. Overall, these results provided strong corroboration of the gene chip assays, in terms of both the identification of differentially expressed genes and the magnitude of the differential expression. The majority of these genes were up-regulated in stimulated cells, with increases reaching 800-fold in some cases. Among these are members of the early growth response family of transcription factors, the IL-1 receptor-like 1, and cytokines/chemokines IL-1α/β, CXCL8 (IL-8), CCL20/23 and CXCL2/3, as well as suppressor of cytokine signaling (SOCS) 3. Other up-regulated genes encode for a number of acute phase proteins such as IER2, 3 and 5, receptors GPR84 and ICAM1, as well as for enzymes such as dual-specificity phosphatases (DUSP) 1, 2 and 5. In comparison, only a small number of genes were down-regulated following cell stimulation and the observed decreases were of relatively modest magnitude. This constitutes, to our knowledge, a first comprehensive gene expression profiling for inflammatory neutrophils.
Figure 1. Change in levels of mRNA expression by 64 genes in human neutrophils due to stimulation with inflammatory agonists.
Cells were stimulated as described in Materials and Methods for 30 min at 37°C. Values are ratios of mRNA levels (stimulated cells/un-stimulated cells) as determined by real-time PCR, averaged±SEM for six independent experiments performed under identical conditions with a different single donor of cells. *Selected MIP-2 primers do not discriminate between the highly homologous alpha and beta isoforms.
Genes modulated by anti-inflammatory agents
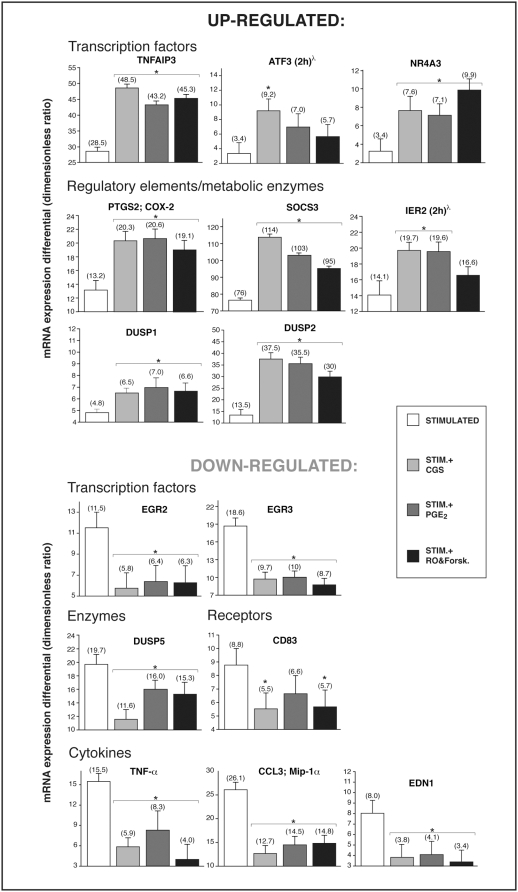
We used the A2AR agonist CGS 21680, PGE2, or the cAMP-elevating compounds RO 20-1724 (phosphodiesterase IV inhibitor) and forskolin (adenylate cyclase activator), which are potent anti-inflammatory agents known to modulate neutrophil activation [17], in order to determine their impact of the gene expression profile of stimulated neutrophils. Analysis by gene chips revealed that, of the 64 genes differentially expressed in stimulated cells, 28 appeared influenced by at least one of these agents (Supplementary Table S3). Several genes behaved as predicted; the inducible cyclooxygenase COX-2 being up-regulated, while TNF-α and MIP-1α down-regulated by A2AR activation, which is in line with earlier findings [21], [23]. These gene chip results were then confronted with real-time PCR experiments performed with new samples from six different donors, which confirmed significant differential expression for 15 of the 28 genes (Figure 2). A2AR engagement, PGE2 or cAMP-elevating agents each increased mRNA expression of immunomodulatory transcription factors NR4A3, ATF3, TNFAIP3 and IER2, of the enzyme COX-2, of dual-specificity phosphatases 1 and 2 and of the regulatory element SOCS3. Conversely, a number of genes were down-regulated by the anti-inflammatory treatments, notably the pro-inflammatory cytokines, TNF-α, macrophage inflammatory peptide-1α (CCL3/MIP-1α), endothelin-1, members of the early-growth response family of transcription factors (EGR2, EGR3) and the DUSP5 enzyme (Supplementary Table S4). Remarkably, the three distinct anti-inflammatory approaches each had a comparable overall impact on the gene expression profile.
Figure 2. Regulation of genes by anti-inflammatory agents in stimulated human neutrophils.
Cells were pretreated with CGS 21680 (1 µM), PGE2 (10 µM) or a mixture of 10 µM RO-20-1724 and 50 µM forskolin, then stimulated as described in Materials and Methods for 30 min at 37°C, or for 2 h where indicated (λ). Top panels show genes that are up-regulated by the anti-inflammatory treatments and bottom ones show genes that are down-regulated.Values are ratios of mRNA levels (treated cells/un-stimulated cells) as determined by real-time PCR, averaged±SEM for six independent experiments performed under identical conditions with a different single donor of cells. *Significantly different from samples stimulated in the absence of any anti-inflammatory agent.
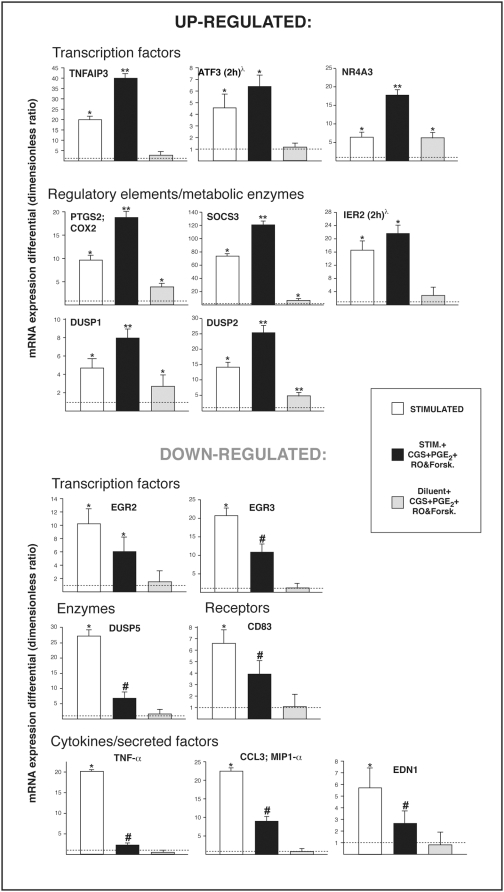
Gene chips and real-time PCR showed similar effects of PGE2 or pharmacological elevation of intracellular cAMP on most of the genes affected by A2AR engagement, suggesting that even when distinct receptors are engaged, signaling pathways eventually merge and cAMP-dependent processes take part in a central anti-inflammatory response. In order to address this point specifically, we next stimulated neutrophils in the simultaneous presence of all three types of anti-inflammatory agent. Messenger RNA levels of the 15 genes identified earlier were determined by real-time PCR. This experiment produced essentially the same result as obtained with each anti-inflammatory strategy alone (Figure 3), further advocating for an important role of these genes in limiting cell activation. Indeed, no additive or synergistic effect was obtained for the majority of the genes. The exceptions were NR4A3 and DUSP5, for which the simultaneous presence of the anti-inflammatory agents proved more potent than any individual agent. Overall, these results support the concept of a relative redundancy between the distinct anti-inflammatory agents and more specifically their participation in a central and largely cAMP-dependent cellular immunomodulatory response.
Figure 3. Impact of a combination of anti-inflammatory agents on gene expression in stimulated human neutrophils.
Cells were pretreated with CGS 21680 (1 µM), PGE2 (10 µM), RO-20-1724 (10 µM) and forskolin (50 µM) simultaneously, then stimulated as described in Materials and Methods for 30 min at 37°C, or for 2 h where indicated (λ). Top panels show genes that are up-regulated by the anti-inflammatory treatments and bottom ones show genes that are down-regulated. The dotted line indicates the level of expression observed in un-stimulated cells ( = 1). Values are ratios of mRNA levels (treated cells/un-stimulated cells) as determined by real-time PCR, averaged±SEM for three independent experiments performed under identical conditions with a different single donor of cells. *Significantly higher than in un-stimulated samples. **Significantly higher than in un-stimulated samples and in samples stimulated in the absence of anti-inflammatory agent. “#” Significantly higher than in un-stimulated samples but significantly lower than in samples stimulated in the absence of anti-inflammatory agent.
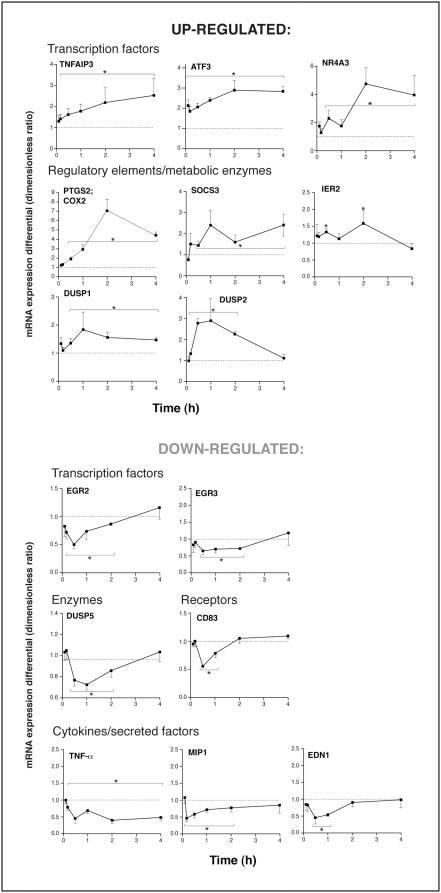
Time-course experiments were undertaken in which cells were stimulated for periods of time ranging from 5 min to 4 h, alone or in presence of the A2AR agonist CGS 21680. Messenger RNA levels for genes of interest were measured by real-time PCR and samples stimulated in the absence or presence of CGS 21680 were compared in a time-matched manner. Depending on the gene, A2AR activation elicited transient (<2 h) or sustained (≥4 h) responses, indicative of gene-specific regulatory processes (Figure 4). However, the impact on gene expression was typically rapid, in most cases becoming apparent in less than 30 minutes.
Figure 4. Kinetics of the effect of A2AR engagement on gene expression.
Cells were pretreated with CGS 21680 (1 µM) then stimulated as described in Materials and Methods for 30 minutes at 37°C. Values are time-matched ratios of mRNA levels (CGS21680-treated cells/stimulated cells) as determined by real-time PCR, averaged±SEM for three independent experiments performed under identical conditions with a different single donor of cells. *Significantly different from cells stimulated in the absence of CGS 21680.
Discussion
The scope of this work encompasses the development of novel therapeutic strategies based on enabling endogenous anti-inflammatory pathways in the treatment of inflammatory conditions such as rheumatoid arthritis, in which unchecked activation of cells can cause significant tissue damage. By profiling gene expression in stimulated neutrophils, we delineated a group of genes that respond to immunomodulatory signals. Gene identification was achieved using a gene chip approach and was corroborated by real-time PCR. In response to three distinct anti-inflammatory approaches, stimulated neutrophils shifted their expression of specific genes. Cells responded to these different anti-inflammatory signals in a strikingly similar fashion, which suggests the engagement of a central endogenous system responsible for uncoupling selected neutrophil inflammatory functions.
The pro-inflammatory, receptor-activating agonists Escherichia coli lipopolysaccharide, granulocyte-macrophage colony-stimulating factor, tumor-necrosis factor α, formyl-methionyl-leucyl-phenylalanine and interleukin 1β were chosen for their importance in inflammatory processes and for their well-documented stimulatory effects on neutrophils [21]–[23], [35]–[37]. In the course of an inflammatory response, it is assumed that recruited neutrophils are more likely to encounter a multitude of extracellular messengers, rather than a single one. Even so, most studies of anti-inflammatory agents have used a single agonist and for that matter, a single anti-inflammatory agent, thereby locking down data interpretation on pathways solicited by that particular agonist only. In this study, we elected to stimulate neutrophils concurrently with a group of agonists chosen for their well-described ability to engage distinct classes of receptors in neutrophils. GM-CSF interacts with a receptor comprising tyrosine kinase activity [38], fMLP signals through seven-transmembrane domain receptors FPR1 and FPRL1 linked to heterotrimeric GTP-binding proteins [39], LPS associates with LPS-binding proteins, then with the Toll-Like receptor 4 and CD14 molecule [40], TNF-α binds to its ceramide-linked receptors TNFRSF1A/TNFR1 and TNFRSF1B/TNFBR [41] and finally, IL-1β engages its own family of receptors (IL-1R1, IL-1R2, IL1-RL1) in the immunoglobulin domain superfamily [42]. This multilateral stimulation, likely closer to what inflammatory cells face, favorably elicits a robust and more comprehensive cellular involvement, a suitable situation for the study of anti-inflammatory processes.
The impact of the anti-inflammatory agents on cell response was clearly multi-pronged, even considering only their effects on gene expression. Genes involved encode transcription factors, enzymes and regulatory factors, receptors and cytokines. It is of interest that some of the genes were actually up-regulated by the anti-inflammatory agents, implying an active cellular reaction rather than a mere response to inhibition. Such up-regulated genes included pivotal transcription factors that have been reported to modulate numerous cell functions. For example, TNFAIP3 displays potent anti-inflammatory properties in a number of different cell types, through the inhibition of NF-kB activation and prevention of TLR-mediated responses [43]–[48]. ATF3 plays a protective role in ischemia-reperfusion injury and in response to stress [49], [50]. NR4A3 prevents NF-kB activation, thereby reducing inflammatory responses such as the generation of pro-inflammatory cytokines. Available evidence also suggests a protective role for this family of transcription factors in atherogenesis [51], [52]. A number of enzymes and regulatory elements were also up-regulated by the anti-inflammatory agents. The inducible cyclooxygenase isoform COX-2 is a pivotal and rate-limiting enzyme in the inflammation-related generation of PGE2, the latter having potent anti-inflammatory activities in leukocytes and other inflammatory cells [21], [22], [35], [53]. Suppressor of cytokine signaling 3 is the main isoform of that family of regulatory factors expressed in neutrophils and functions by inhibiting JAK2 kinase activity [54]–[57]. DUSP 1 and 2 can inactivate mitogen-activated protein kinases by dephosphorylating phosphothreonine and phosphotyrosine residues. Phosphatases of the DUSP family display specificity for different MAP kinases and differ in tissue and sub-cellular distributions, although DUSP 2 is expressed predominantly in hematopoietic tissues [58], [59]. Finally, IER2 attenuates the signaling activity of G proteins by binding to GTP-bound G alpha subunits and by increasing the rate of conversion from GTP to GDP [60]. Clearly, the up-regulation of gene expression by adenosine and PGE2 occurs in a way that can alter cellular programming at several levels.
Inhibition of key inflammatory factors is also likely pivotal for mediating the potent anti-inflammatory activities of adenosine and PGE2 in neutrophils. Indeed, activities of early-growth-response transcription factors 2 and 3 are believed to have positive involvement in differentiation, mitogenesis and angiogenesis [61], while the phosphatase DUSP5 has been linked positively to immunity through T cell development [62]. In the present study, the only receptor to respond to anti-inflammatory agents (by down-regulation of mRNA) was CD83, which is considered a marker of mature dendritic cells and thought to be involved in the regulation of T- and B-lymphocyte maturation. Although its expression on neutrophils is known, its role is currently unknown [63], [64]. As we reported earlier, TNF-α expression is diminished by adenosine and may well be one of the key targets of this autacoid. Indeed, TNF-α is a pivotal pro-inflammatory cytokine involved in a number of inflammatory and autoimmune diseases, diabetes and cancer [65], [66]. Similarly, MIP-1α is another important cytokine involved in the acute inflammatory state and in the recruitment and activation of neutrophils [67]–[69]. Endothelin 1 is a potent vasoconstrictor that has been linked to graft rejection and to inflammatory events including pain, fever, cell migration and rheumatoid arthritis. It stimulates several mechanisms on neutrophils, including adhesion and migration [70]–[74].
Genes that were found to be affected by A2AR engagement, PGE2 or pharmacological elevation of the intracellular cyclic AMP concentration together provide a first picture of the overall impact these signals have on gene expression. Furthermore, single anti-inflammatory signals affected the expression of the same group of genes, supporting the hypothesis that these genes play a role in the coordination of a cellular response, this role being to limit cell activation. It is therefore possible that the expression profile observed in neutrophils will find similarities in other cell types and tissues, and engage a resolution response. The current picture is still partial; indeed, a number of affected sequences either code for proteins not yet characterized or are altogether not translated. Also, further studies will be necessary to confirm their involvement in resolving inflammation. Nonetheless, genes identified in the present study are likely to provide a better understanding of anti-inflammatory signaling.
In summary, we have identified a series of genes for which expression is altered by major anti-inflammatory signals. All of theses signals affected the gene expression profile in remarkably similar fashion. Characterization of these signaling pathways will improve our understanding of the capacity of tissues to terminate inflammation and may lead to the identification of better therapeutic targets for the treatment of inflammatory diseases associated with unrepressed neutrophil activation.
Materials and Methods
Materials
Anti-inflammatory agents
Compound CGS 21680 (2-[p-(2-carboxyethyl) phenethylamino]-5′-N-ethyl carboxamidoadenosine) was from Research Biochemicals International (Natick, MA, USA). Prostaglandin E2 (PGE2) was purchased from Cayman Chemicals (Ann Arbor, MI, USA). Forskolin and RO 20-1724 were obtained from EMD Chemicals (San Diego, CA, USA).
Inflammatory agonists
Lipopolysaccharide (LPS) from Escherichia coli O111:B4 and formyl-methionyl-leucyl phenylalanine (fMLP) were obtained from Sigma-Aldrich (Oakville, ON, Canada). Recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF), tumor necrosis factor α (TNF-α) and interleukin 1β (IL-1β) were purchased from PeproTech (Rocky Hill, NJ, USA).
Adenosine deaminase was purchased from Roche Applied Science (Laval, QC, Canada).
Neutrophil isolation
Polymorphonuclear leukocytes were isolated as originally described [75] with modifications [22]. Informed consent was obtained in writing and all experiments involving human tissues were approved by the Laval University Ethics Committee. Data collection and analyses were performed anonymously. Briefly, venous blood from healthy volunteers, collected on isocitrate anticoagulant solution was centrifuged (250×g, 10 min) and the resulting platelet-rich plasma was discarded. Leukocytes were obtained following erythrocyte sedimentation in 2% Dextran T-500 (Sigma-Aldrich). Granulocytes were then separated from other leukocytes by centrifugation on a 10 ml cushion of lymphocyte separation medium (Wisent, St-Bruno, QC, Canada). Contaminating erythrocytes were removed by 15 seconds of hypotonic lysis. Purified granulocytes (>95% neutrophils, <5% eosinophils) contained less than 0.1% monocytes, as determined by esterase staining. Viability was greater than 98%, as determined by tryptan blue dye exclusion. The whole cell isolation procedure was carried out at room temperature under sterile conditions.
Cell stimulations
Neutrophils were re-suspended at a concentration of 30×106 cells/ml in Hank's balanced salt solution at 37°C, containing 1% fetal bovine serum, 10 mM HEPES pH 7.4, 1.6 mM Ca2+ and no Mg2+. Adenosine deaminase (0.1 U/ml) was added to cell suspensions 20 min prior to stimulation in order to prevent accumulation of extracellular adenosine in cell suspensions, thus minimizing the modulating effects of adenosine on neutrophil activities [76]. Anti-inflammatory compounds dissolved in dimethylsulfoxide were added to cell suspensions 10 min before stimulation with a mixture of LPS (100 ng/ml), GM-CSF (1.4 nM), TNF-α (100 ng/ml), fMLP (100 nM) and IL-1β (30 nM). Organic solvent concentration was identical in all samples and did not exceed 0.1% (v/v). Stimulations were for 30 min at 37°C, unless indicated otherwise.
RNA isolation
Following stimulation, neutrophil total RNA was isolated using Trizol (Invitrogen, Burlington, ON, Canada) according to the manufacturer's protocol, with modifications [22]. Briefly, a pellet containing 30×106 neutrophils was homogenized in 1 ml Trizol and 200 µl of chloroform were added. After mixing, the sample was centrifuged at 12,000×g for 15 min (4°C) and the upper aqueous phase (450 µl) was transferred to a tube containing an equal volume of isopropanol, mixed thoroughly using a vortex device and centrifuged at 12,000×g for 10 min (4°C). The supernatant was discarded and the precipitated RNA pellet was washed twice using 500 µl of 75% ethanol and centrifuged at 12,000×g for 5 min (4°C). The final pellet was allowed to air-dry for 5–10 min and was then re-suspended in RNAse-free water. RNA was quantitated using a Qubit™ Fluorometer (Invitrogen).
DNA microarrays
Equal quantities of total RNA obtained from neutrophils of five donors were pooled together and purified on QIAGEN RNeasy column (QIAGEN, Mississauga, ON, Canada). Ten ng of total RNA were converted to cDNA using Superscripts reverse transcriptase (Invitrogen) and T7-oligo-d(T)24 primers (Applied Biosystems, Austin, TX, USA). Second-strand synthesis was performed using T4 DNA polymerase and E. coli DNA ligase and then blunt-ended by T4 polynucleotide kinase. cDNA was purified by phenol-chloroform extraction using phase lock gels (Brinkmann, Westbury, NY, USA), then transcribed in vitro for 16 h at 37°C by using the IVT Labelling Kit (Affymetrix, Santa Clara, CA, USA) to produce biotinylated cRNA. Biotin-labelled cRNA was isolated using the RNeasy Mini Kit column (QIAGEN). Purified cRNA was fragmented to 30–200 nucleotide lengths using a fragmentation buffer. The quality of total RNA, cDNA synthesis, cRNA amplification and cRNA fragmentation was monitored and confirmed by capillary electrophoresis (Bioanalyzer 2100, Agilent Technologies, Santa Clara, CA, USA). Fifteen µg of fragmented cRNA were hybridized for 16 h at 45°C with constant rotation on a Human Genome U133 Plus 2.0 GeneChip Array (Affymetrix). After hybridization, gene chips were processed with the Affymetrix GeneChip Fluidic Station 450 (protocol EukGE-WS2v5_450). Briefly, staining was made with streptavidin-conjugated phycoerythrin (SAPE, Invitrogen) followed by amplification with a biotinylated anti-streptavidin antibody (Vector Laboratories, Burlingame, CA, USA) and by a second round of SAPE. Chips were scanned using a GeneChip Scanner 3000 G7 (Affymetrix) enabled for High-Resolution Scanning. Images were extracted with the GeneChip Operating Software (Affymetrix GCOS v1.4). Quality control of microarray chips was performed using the AffyQCReport software [77]. All Microarray data is conform to the MIAME guidelines; unsupervised, raw data was deposited in the GEO database (geo@ncbi.nlm.nih.gov), submission number: GSE14465.
Interpretation of microarray results
Sequences with a level of expression of 200 and over were considered to be positively expressed. A sequence was considered differentially expressed when the ratio of expression level between experimental conditions was ≥2 (≥two-fold increase) or ≤0.5 (two-fold decrease). Gene identification and expression levels were analyzed using the Gene Set Analysis Toolkit, developed and maintained by members of the Department of Biomedical Informatics and the Department of Biostatistics of the Vanderbilt University Medical Center (http://bioinfo.vanderbilt.edu/webgestalt) [34].
Real-Time PCR
First-strand cDNA synthesis was performed using 1 µg of total RNA with Superscript II (Invitrogen) following the manufacturer's instructions, using 500 ng of random hexamers. Real-time PCR was performed as described elsewhere [78]. Briefly, cDNA amplification was carried out in a Rotor-Gene 3000 operated with Rotor-Gene software version 6.0.19 (Corbett Research, Mortlake, NSW Australia) using 35 cycles of 95°C, 58°C and 72°C for 20 seconds each. Each sample consisted of 40 ng of cDNA, 2 µl of 10× buffer (100 mM Tris, 500 mM KCl, 30 mM MgCl2, 1.5% Triton X-100), 0.5 mM dNTP, 500 nM of primers, 0.1 unit of rTaq DNA polymerase (GE Healthcare, Piscataway, NJ, USA) and SYBR Green I dye (Invitrogen; 1∶30,000 dilution) in a reaction volume of 20 µL. Reaction specificity was ascertained by performing the Melt® procedure (58–99°C, 1°C/5 s) at the end of the amplification protocol, according to the manufacturer's instructions. For each gene of interest, specific primers were designed as described previously [78]. Briefly, primers were selected systematically within the coding region, with a theoretical melting point of 58°C, GC content of 50% (±10%) and length of 18–24 bp, for an average product length of 200 bp. Primers thus designed were all tested with gradient PCR prior to their use in real-time PCR and are listed in Supplementary Table S5.
Experimental design
The following experimental treatment conditions were examined: Unstimulated control cells; stimulated cells; cells stimulated in the presence of either CGS 21680, PGE2, or forskolin & RO 20-1724; cells stimulated in the simultaneous presence of CGS 21680, PGE2 and forskolin & RO 20-1724; unstimulated cells in the simultaneous presence of CGS 21680, PGE2 and forskolin & RO 20-1724.
DNA microarray experiments were repeated twice, each using RNA pooled from neutrophils of five donors. Real-time PCR experiments were repeated six times for cell stimulation in the absence of anti-inflammatory agents and for stimulation in the presence of CGS 21680, PGE2, or forskolin plus RO 20-1724 (each repetition with neutrophils from one of six donors) and repeated three times for the time-course study of CGS 21680 (each repetition with neutrophils from one of three donors),
Statistical analysis
Where applicable, statistical analysis was performed using the Student's non-paired t-test (two-tailed) and differences were considered significant (marked by an asterisk) when p<0.05.
Supporting Information
Gene-chip-based identification of differentially regulated genes in stimulated human neutrophils. Genes are listed in decreasing order of the mRNA expression ratio (stimulated/un-stimulated cells). Cells were stimulated as described in Materials and Methods for 30 min at 37 degrees C. RNA from five donors was pooled for analysis. Results are from one experiment.
(0.09 MB XLS)
Inflammatory genes that are differentially- expressed in stimulated human neutrophils (Genechip results). Cells were stimulated as described in Materials and Methods for 30 min at 37 degree C. RNA from five donors was pooled for analysis. Genes from Supplementary Table S1 were selected for their potential involvement in inflammatory processes, and based on differential regulation in stimulated human neutrophils, as revealed by genechip analysis. Genes are listed in decreasing order of the mRNA expression ratio (stimulated/un-stimulated cells). Results are from one experiment.
(0.15 MB XLS)
Differentially-expressed genes in stimulated human neutrophils treated with anti-inflammatory agents (Genechip results) Cells were pretreated with CGS 21680 (1 uM), PGE2 (10 uM) or a mixture of 10 uM RO-20-1724 and 50 uM forskolin, then stimulated for 30 min with the inflammatory cocktail for 30 min at 37 degree C, as described in Materials and Methods. RNA from five donors was pooled for analysis. Genes are listed in decreasing order of the mRNA expression ratio (stimulated/un-stimulated cells). Results are from one experiment.
(0.16 MB XLS)
List of genes, their associated proteins and protein functions, differentially regulated in stimulated human neutrophils and influenced by anti-inflammatory agents, as confirmed by real-time PCR.
(0.03 MB XLS)
List of PCR primers.
(0.16 MB XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work is supported by grants from the Canadian Institutes of Health Research (CIHR, to MP; grants no. MOP-64315 & NSM-72200). MP is the recipient of a Senior Investigator Scholarship from the Fonds de la Recherche en Sante du Quebec (FRSQ) and MSO is the recipient of a studentship award from the FRSQ. Funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Edwards SW. Biochemistry and physiology of the neutrophil. Cambridge University Press; 1994. pp. 1–319. [Google Scholar]
- 2.Girard D. Phenotypic and Functional Changes of Neutrophils Activated by Recently Identified Modulators. Research Signpost; 2007. p. 134. [Google Scholar]
- 3.de Boer WI, Yao H, Rahman I. Future therapeutic treatment of COPD: struggle between oxidants and cytokines. Int J Chron Obstruct Pulmon Dis. 2007;2:205–228. [PMC free article] [PubMed] [Google Scholar]
- 4.Lynch JP, 3rd, Flint A, Miller DM, Fantone JC. Noncaseating pulmonary granulomas associated with small cell carcinoma of the lung. Am J Med. 1985;78:691–696. doi: 10.1016/0002-9343(85)90416-4. [DOI] [PubMed] [Google Scholar]
- 5.Sawyer DW, Donowitz GR, Mandell GL. Polymorphonuclear neutrophils: an effective antimicrobial force. Rev Infect Dis. 1989;11(Suppl 7):S1532–1544. doi: 10.1093/clinids/11.supplement_7.s1532. [DOI] [PubMed] [Google Scholar]
- 6.Cronstein BN, Kramer SB, Weissmann G, Hirschhorn R. Adenosine: a physiological modulator of superoxide anion generation by human neutrophils. J Exp Med. 1983;158:1160–1177. doi: 10.1084/jem.158.4.1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cristalli G, Lambertucci C, Marucci G, Volpini R, Dal Ben D. A2A adenosine receptor and its modulators: overview on a druggable GPCR and on structure-activity relationship analysis and binding requirements of agonists and antagonists. Curr Pharm Des. 2008;14:1525–1552. doi: 10.2174/138161208784480081. [DOI] [PubMed] [Google Scholar]
- 8.Hasko G, Pacher P. A2A receptors in inflammation and injury: lessons learned from transgenic animals. J Leukoc Biol. 2008;83:447–455. doi: 10.1189/jlb.0607359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohta A, Sitkovsky M. Role of G-protein-coupled adenosine receptors in downregulation of inflammation and protection from tissue damage. Nature. 2001;414:916–920. doi: 10.1038/414916a. [DOI] [PubMed] [Google Scholar]
- 10.Cronstein BN, Naime D, Ostad E. The antiinflammatory mechanism of methotrexate. Increased adenosine release at inflamed sites diminishes leukocyte accumulation in an in vivo model of inflammation. J Clin Invest. 1993;92:2675–2682. doi: 10.1172/JCI116884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dianzani C, Brunelleschi S, Viano I, Fantozzi R. Adenosine modulation of primed human neutrophils. Eur J Pharmacol. 1994;263:223–226. doi: 10.1016/0014-2999(94)90547-9. [DOI] [PubMed] [Google Scholar]
- 12.Felsch A, Stocker K, Borchard U. Phorbol ester-stimulated adherence of neutrophils to endothelial cells is reduced by adenosine A2 receptor agonists. J Immunol. 1995;155:333–338. [PubMed] [Google Scholar]
- 13.Krump E, Lemay G, Borgeat P. Adenosine A2 receptor-induced inhibition of leukotriene B4 synthesis in whole blood ex vivo. Br J Pharmacol. 1996;117:1639–1644. doi: 10.1111/j.1476-5381.1996.tb15334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao ZQ, Sato H, Williams MW, Fernandez AZ, Vinten-Johansen J. Adenosine A2-receptor activation inhibits neutrophil-mediated injury to coronary endothelium. Am J Physiol. 1996;271:H1456–1464. doi: 10.1152/ajpheart.1996.271.4.H1456. [DOI] [PubMed] [Google Scholar]
- 15.Thibault N, Harbour D, Borgeat P, Naccache PH, Bourgoin SG. Adenosine receptor occupancy suppresses chemoattractant-induced phospholipase D activity by diminishing membrane recruitment of small GTPases. Blood. 2000;95:519–527. [PubMed] [Google Scholar]
- 16.Visser SS, Theron AJ, Ramafi G, Ker JA, Anderson R. Apparent involvement of the A(2A) subtype adenosine receptor in the anti-inflammatory interactions of CGS 21680, cyclopentyladenosine, and IB-MECA with human neutrophils. Biochem Pharmacol. 2000;60:993–999. doi: 10.1016/s0006-2952(00)00414-7. [DOI] [PubMed] [Google Scholar]
- 17.György Haskó BNC, Csaba Szabó. Adenosine Receptors: Therapeutic Aspects for inflammatory and Immune Disease. CRC Taylor & Francis Group LLC; 2007. p. 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flamand N, Boudreault S, Picard S, Austin M, Surette ME, et al. Adenosine, a potent natural suppressor of arachidonic acid release and leukotriene biosynthesis in human neutrophils. Am J Respir Crit Care Med. 2000;161:S88–94. doi: 10.1164/ajrccm.161.supplement_1.ltta-18. [DOI] [PubMed] [Google Scholar]
- 19.Krump E, Borgeat P. Adenosine. An endogenous inhibitor of arachidonic acid release and leukotriene biosynthesis in human neutrophils. Adv Exp Med Biol. 1999;447:107–115. [PubMed] [Google Scholar]
- 20.Krump E, Picard S, Mancini J, Borgeat P. Suppression of leukotriene B4 biosynthesis by endogenous adenosine in ligand-activated human neutrophils. J Exp Med. 1997;186:1401–1406. doi: 10.1084/jem.186.8.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cadieux J-S, Leclerc P, St-Onge M, Dussault A-A, Laflamme C, et al. Potentiation of Neutrophil Cyclooxygenase-2 by Adenosine: An Early Anti-Inflammatory Signal. J Cell Sci. 2005;118:1437–1447. doi: 10.1242/jcs.01737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pouliot M, Fiset ME, Masse M, Naccache PH, Borgeat P. Adenosine up-regulates cyclooxygenase-2 in human granulocytes: impact on the balance of eicosanoid generation. J Immunol. 2002;169:5279–5286. doi: 10.4049/jimmunol.169.9.5279. [DOI] [PubMed] [Google Scholar]
- 23.McColl SR, St-Onge M, Dussault AA, Laflamme C, Bouchard L, et al. Immunomodulatory impact of the A2A adenosine receptor on the profile of chemokines produced by neutrophils. Faseb J. 2006;20:187–189. doi: 10.1096/fj.05-4804fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinsel JF, Sitkovsky MV. Possible targeting of G protein coupled receptors to manipulate inflammation in vivo using synthetic and natural ligands. Ann Rheum Dis. 2003;62(Suppl 2):ii22–24. doi: 10.1136/ard.62.suppl_2.ii22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flamand N, Surette ME, Picard S, Bourgoin S, Borgeat P. Cyclic AMP-mediated inhibition of 5-lipoxygenase translocation and leukotriene biosynthesis in human neutrophils. Mol Pharmacol. 2002;62:250–256. doi: 10.1124/mol.62.2.250. [DOI] [PubMed] [Google Scholar]
- 26.Fantone JC, Kunkel SL, Ward PA. Suppression of human polymorphonuclear function after intravenous infusion of prostaglandin E1. Prostaglandins Med. 1981;7:195–198. doi: 10.1016/0161-4630(81)90062-8. [DOI] [PubMed] [Google Scholar]
- 27.Fantone JC, Marasco WA, Elgas LJ, Ward PA. Anti-inflammatory effects of prostaglandin E1: in vivo modulation of the formyl peptide chemotactic receptor on the rat neutrophil. J Immunol. 1983;130:1495–1497. [PubMed] [Google Scholar]
- 28.Ham EA, Soderman DD, Zanetti ME, Dougherty HW, McCauley E, et al. Inhibition by prostaglandins of leukotriene B4 release from activated neutrophils. Proc Natl Acad Sci U S A. 1983;80:4349–4353. doi: 10.1073/pnas.80.14.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehmeyer JE, Johnston RB., Jr Effect of anti-inflammatory drugs and agents that elevate intracellular cyclic AMP on the release of toxic oxygen metabolites by phagocytes: studies in a model of tissue-bound IgG. Clin Immunol Immunopathol. 1978;9:482–490. doi: 10.1016/0090-1229(78)90144-7. [DOI] [PubMed] [Google Scholar]
- 30.Marone G, Thomas LL, Lichtenstein LM. The role of agonists that activate adenylate cyclase in the control of cAMP metabolism and enzyme release by human polymorphonuclear leukocytes. J Immunol. 1980;125:2277–2283. [PubMed] [Google Scholar]
- 31.O'Flaherty JT, Kreutzer DL, Ward PA. Effect of prostaglandins E1, E2 and F2alpha on neutrophil aggregation. Prostaglandins. 1979;17:201–210. doi: 10.1016/0090-6980(79)90039-x. [DOI] [PubMed] [Google Scholar]
- 32.Rivkin I, Rosenblatt J, Becker EL. The role of cyclic AMP in the chemotactic responsiveness and spontaneous motility of rabbit peritoneal neutrophils. The inhibition of neutrophil movement and the elevation of cyclic AMP levels by catecholamines, prostaglandins, theophylline and cholera toxin. J Immunol. 1975;115:1126–1134. [PubMed] [Google Scholar]
- 33.Zurier RB, Weissmann G, Hoffstein S, Kammerman S, Tai HH. Mechanisms of lysosomal enzyme release from human leukocytes. II. Effects of cAMP and cGMP, autonomic agonists, and agents which affect microtubule function. J Clin Invest. 1974;53:297–309. doi: 10.1172/JCI107550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pouliot M, Gilbert C, Borgeat P, Poubelle PE, Bourgoin S, et al. Expression and activity of prostaglandin endoperoxide synthase-2 in agonist-activated human neutrophils. Faseb J. 1998;12:1109–1123. doi: 10.1096/fasebj.12.12.1109. [DOI] [PubMed] [Google Scholar]
- 36.McDonald PP, Pouliot M, Borgeat P. Enhancement by GM-CSF of agonist-induced 5-lipoxygenase activation in human neutrophils involves protein synthesis and gene transcription. J Lipid Mediat. 1993;6:59–67. [PubMed] [Google Scholar]
- 37.McColl SR, Beauseigle D, Gilbert C, Naccache PH. Priming of the human neutrophil respiratory burst by granulocyte-macrophage colony-stimulating factor and tumor necrosis factor-alpha involves regulation at a post-cell surface receptor level. Enhancement of the effect of agents which directly activate G proteins. J Immunol. 1990;145:3047–3053. [PubMed] [Google Scholar]
- 38.Hamilton JA. Colony-stimulating factors in inflammation and autoimmunity. Nat Rev Immunol. 2008;8:533–544. doi: 10.1038/nri2356. [DOI] [PubMed] [Google Scholar]
- 39.Le Y, Murphy PM, Wang JM. Formyl-peptide receptors revisited. Trends Immunol. 2002;23:541–548. doi: 10.1016/s1471-4906(02)02316-5. [DOI] [PubMed] [Google Scholar]
- 40.Palsson-McDermott EM, O'Neill LA. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology. 2004;113:153–162. doi: 10.1111/j.1365-2567.2004.01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149–160. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 42.Boraschi D, Tagliabue A. The interleukin-1 receptor family. Vitam Horm. 2006;74:229–254. doi: 10.1016/S0083-6729(06)74009-2. [DOI] [PubMed] [Google Scholar]
- 43.Golovko O, Nazarova N, Tuohimaa P. A20 gene expression is regulated by TNF, Vitamin D and androgen in prostate cancer cells. J Steroid Biochem Mol Biol. 2005;94:197–202. doi: 10.1016/j.jsbmb.2005.01.019. [DOI] [PubMed] [Google Scholar]
- 44.Gon Y, Asai Y, Hashimoto S, Mizumura K, Jibiki I, et al. A20 inhibits toll-like receptor 2- and 4-mediated interleukin-8 synthesis in airway epithelial cells. Am J Respir Cell Mol Biol. 2004;31:330–336. doi: 10.1165/rcmb.2003-0438OC. [DOI] [PubMed] [Google Scholar]
- 45.Kunter U, Daniel S, Arvelo MB, Choi J, Shukri T, et al. Combined expression of A1 and A20 achieves optimal protection of renal proximal tubular epithelial cells. Kidney Int. 2005;68:1520–1532. doi: 10.1111/j.1523-1755.2005.00564.x. [DOI] [PubMed] [Google Scholar]
- 46.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, et al. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem. 2006;281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 47.Patel VI, Daniel S, Longo CR, Shrikhande GV, Scali ST, et al. A20, a modulator of smooth muscle cell proliferation and apoptosis, prevents and induces regression of neointimal hyperplasia. Faseb J. 2006;20:1418–1430. doi: 10.1096/fj.05-4981com. [DOI] [PubMed] [Google Scholar]
- 48.Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 49.Kool J, Hamdi M, Cornelissen-Steijger P, van der Eb AJ, Terleth C, et al. Induction of ATF3 by ionizing radiation is mediated via a signaling pathway that includes ATM, Nibrin1, stress-induced MAPkinases and ATF-2. Oncogene. 2003;22:4235–4242. doi: 10.1038/sj.onc.1206611. [DOI] [PubMed] [Google Scholar]
- 50.Nobori K, Ito H, Tamamori-Adachi M, Adachi S, Ono Y, et al. ATF3 inhibits doxorubicin-induced apoptosis in cardiac myocytes: a novel cardioprotective role of ATF3. J Mol Cell Cardiol. 2002;34:1387–1397. doi: 10.1006/jmcc.2002.2091. [DOI] [PubMed] [Google Scholar]
- 51.Bonta PI, van Tiel CM, Vos M, Pols TW, van Thienen JV, et al. Nuclear receptors Nur77, Nurr1, and NOR-1 expressed in atherosclerotic lesion macrophages reduce lipid loading and inflammatory responses. Arterioscler Thromb Vasc Biol. 2006;26:2288–2294. doi: 10.1161/01.ATV.0000238346.84458.5d. [DOI] [PubMed] [Google Scholar]
- 52.Hisaoka M, Okamoto S, Yokoyama K, Hashimoto H. Coexpression of NOR1 and SIX3 proteins in extraskeletal myxoid chondrosarcomas without detectable NR4A3 fusion genes. Cancer Genet Cytogenet. 2004;152:101–107. doi: 10.1016/j.cancergencyto.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Williams CS, DuBois RN. Prostaglandin endoperoxide synthase: why two isoforms? Am J Physiol. 1996;270:G393–400. doi: 10.1152/ajpgi.1996.270.3.G393. [DOI] [PubMed] [Google Scholar]
- 54.Miao T, Wu D, Zhang Y, Bo X, Xiao F, et al. SOCS3 suppresses AP-1 transcriptional activity in neuroblastoma cells through inhibition of c-Jun N-terminal kinase. Mol Cell Neurosci. 2008;37:367–375. doi: 10.1016/j.mcn.2007.10.010. [DOI] [PubMed] [Google Scholar]
- 55.Pothlichet J, Chignard M, Si-Tahar M. Cutting edge: innate immune response triggered by influenza A virus is negatively regulated by SOCS1 and SOCS3 through a RIG-I/IFNAR1-dependent pathway. J Immunol. 2008;180:2034–2038. doi: 10.4049/jimmunol.180.4.2034. [DOI] [PubMed] [Google Scholar]
- 56.Wu X, Herndon DN, Wolf SE. Growth hormone down-regulation of Interleukin-1beta and Interleukin-6 induced acute phase protein gene expression is associated with increased gene expression of suppressor of cytokine signal-3. Shock. 2003;19:314–320. doi: 10.1097/00024382-200304000-00004. [DOI] [PubMed] [Google Scholar]
- 57.Zhang S, Guo D, Jiang L, Zhang Q, Qiu X, et al. SOCS3 inhibiting migration of A549 cells correlates with PYK2 signaling in vitro. BMC Cancer. 2008;8:150. doi: 10.1186/1471-2407-8-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Calvisi DF, Pinna F, Meloni F, Ladu S, Pellegrino R, et al. Dual-specificity phosphatase 1 ubiquitination in extracellular signal-regulated kinase-mediated control of growth in human hepatocellular carcinoma. Cancer Res. 2008;68:4192–4200. doi: 10.1158/0008-5472.CAN-07-6157. [DOI] [PubMed] [Google Scholar]
- 59.Caunt CJ, Rivers CA, Conway-Campbell BL, Norman MR, McArdle CA. Epidermal growth factor receptor and protein kinase C signaling to ERK2: spatiotemporal regulation of ERK2 by dual specificity phosphatases. J Biol Chem. 2008;283:6241–6252. doi: 10.1074/jbc.M706624200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Denecke B, Meyerdierks A, Bottger EC. RGS1 is expressed in monocytes and acts as a GTPase-activating protein for G-protein-coupled chemoattractant receptors. J Biol Chem. 1999;274:26860–26868. doi: 10.1074/jbc.274.38.26860. [DOI] [PubMed] [Google Scholar]
- 61.Liu D, Evans I, Britton G, Zachary I. The zinc-finger transcription factor, early growth response 3, mediates VEGF-induced angiogenesis. Oncogene. 2008;27:2989–2998. doi: 10.1038/sj.onc.1210959. [DOI] [PubMed] [Google Scholar]
- 62.Kovanen PE, Bernard J, Al-Shami A, Liu C, Bollenbacher-Reilley J, et al. T-cell development and function are modulated by dual specificity phosphatase DUSP5. J Biol Chem. 2008;283:17362–17369. doi: 10.1074/jbc.M709887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Breloer M, Fleischer B. CD83 regulates lymphocyte maturation, activation and homeostasis. Trends Immunol. 2008;29:186–194. doi: 10.1016/j.it.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 64.Yamashiro S, Wang JM, Yang D, Gong WH, Kamohara H, et al. Expression of CCR6 and CD83 by cytokine-activated human neutrophils. Blood. 2000;96:3958–3963. [PubMed] [Google Scholar]
- 65.Dunlay SM, Weston SA, Redfield MM, Killian JM, Roger VL. Tumor necrosis factor-alpha and mortality in heart failure: a community study. Circulation. 2008;118:625–631. doi: 10.1161/CIRCULATIONAHA.107.759191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Welsh P, Lowe GD, Chalmers J, Campbell DJ, Rumley A, et al. Associations of proinflammatory cytokines with the risk of recurrent stroke. Stroke. 2008;39:2226–2230. doi: 10.1161/STROKEAHA.107.504498. [DOI] [PubMed] [Google Scholar]
- 67.Gaga M, Ong YE, Benyahia F, Aizen M, Barkans J, et al. Skin reactivity and local cell recruitment in human atopic and nonatopic subjects by CCL2/MCP-1 and CCL3/MIP-1alpha. Allergy. 2008;63:703–711. doi: 10.1111/j.1398-9995.2007.01578.x. [DOI] [PubMed] [Google Scholar]
- 68.Pharoah DS, Varsani H, Tatham RW, Newton KR, de Jager W, et al. Expression of the inflammatory chemokines CCL5, CCL3 and CXCL10 in juvenile idiopathic arthritis, and demonstration of CCL5 production by an atypical subset of CD8+ T cells. Arthritis Res Ther. 2006;8:R50. doi: 10.1186/ar1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.van Gassen KL, de Wit M, Koerkamp MJ, Rensen MG, van Rijen PC, et al. Possible role of the innate immunity in temporal lobe epilepsy. Epilepsia. 2008;49:1055–1065. doi: 10.1111/j.1528-1167.2007.01470.x. [DOI] [PubMed] [Google Scholar]
- 70.Conte Fde P, Barja-Fidalgo C, Verri WA, Jr, Cunha FQ, Rae GA, et al. Endothelins modulate inflammatory reaction in zymosan-induced arthritis: participation of LTB4, TNF-alpha, and CXCL-1. J Leukoc Biol. 2008;84:652–660. doi: 10.1189/jlb.1207827. [DOI] [PubMed] [Google Scholar]
- 71.Gawlik R, Jawor B. [Endothelin-1 in nasal lavage fluid of allergic rhinitis patients–new mediator of allergic rhinitis]. Otolaryngol Pol. 2007;61:567–571. doi: 10.1016/S0030-6657(07)70486-9. [DOI] [PubMed] [Google Scholar]
- 72.Kassuya CA, Rogerio AP, Calixto JB. The role of ET(A) and ET(B) receptor antagonists in acute and allergic inflammation in mice. Peptides. 2008;29:1329–1337. doi: 10.1016/j.peptides.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 73.Kugelmeier P, Nett PC, Zullig R, Lehmann R, Weber M, et al. Expression and hypoxic regulation of the endothelin system in endocrine cells of human and rat pancreatic islets. Jop. 2008;9:133–149. [PubMed] [Google Scholar]
- 74.Lopez Farre A, Riesco A, Espinosa G, Digiuni E, Cernadas MR, et al. Effect of endothelin-1 on neutrophil adhesion to endothelial cells and perfused heart. Circulation. 1993;88:1166–1171. doi: 10.1161/01.cir.88.3.1166. [DOI] [PubMed] [Google Scholar]
- 75.Boyum A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand J Clin Lab Invest. 1968;(Suppl 97):77–89. [PubMed] [Google Scholar]
- 76.Pouliot M. Adenosine and neutrophil functions. In: Hasko GCB, Szabo C, editors. Adenosine Receptors: Therapeutic Aspects for Inflammatory and Immune Diseases. CRC Taylor & Francis; 2006. pp. 89–100. [Google Scholar]
- 77.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 78.Dussault AA, Pouliot M. Rapid and simple comparison of messenger RNA levels using real-time PCR. Biol Proced Online. 2006;8:1–10. doi: 10.1251/bpo114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gene-chip-based identification of differentially regulated genes in stimulated human neutrophils. Genes are listed in decreasing order of the mRNA expression ratio (stimulated/un-stimulated cells). Cells were stimulated as described in Materials and Methods for 30 min at 37 degrees C. RNA from five donors was pooled for analysis. Results are from one experiment.
(0.09 MB XLS)
Inflammatory genes that are differentially- expressed in stimulated human neutrophils (Genechip results). Cells were stimulated as described in Materials and Methods for 30 min at 37 degree C. RNA from five donors was pooled for analysis. Genes from Supplementary Table S1 were selected for their potential involvement in inflammatory processes, and based on differential regulation in stimulated human neutrophils, as revealed by genechip analysis. Genes are listed in decreasing order of the mRNA expression ratio (stimulated/un-stimulated cells). Results are from one experiment.
(0.15 MB XLS)
Differentially-expressed genes in stimulated human neutrophils treated with anti-inflammatory agents (Genechip results) Cells were pretreated with CGS 21680 (1 uM), PGE2 (10 uM) or a mixture of 10 uM RO-20-1724 and 50 uM forskolin, then stimulated for 30 min with the inflammatory cocktail for 30 min at 37 degree C, as described in Materials and Methods. RNA from five donors was pooled for analysis. Genes are listed in decreasing order of the mRNA expression ratio (stimulated/un-stimulated cells). Results are from one experiment.
(0.16 MB XLS)
List of genes, their associated proteins and protein functions, differentially regulated in stimulated human neutrophils and influenced by anti-inflammatory agents, as confirmed by real-time PCR.
(0.03 MB XLS)
List of PCR primers.
(0.16 MB XLS)