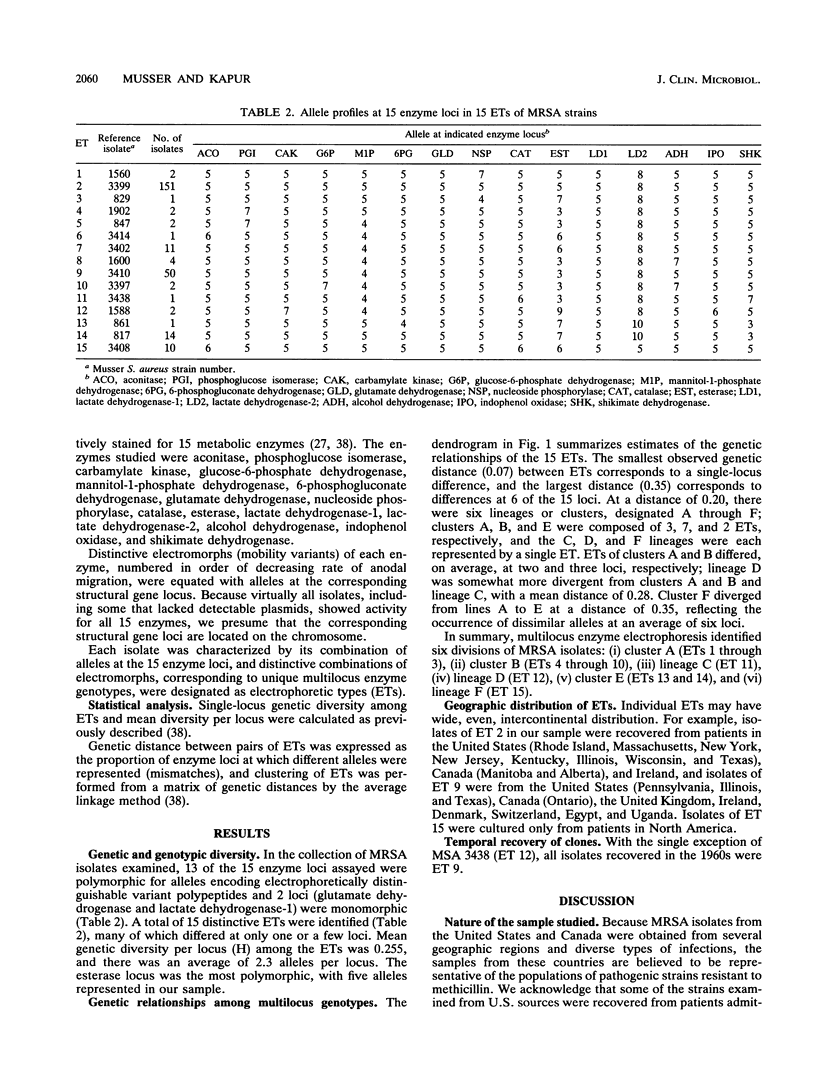
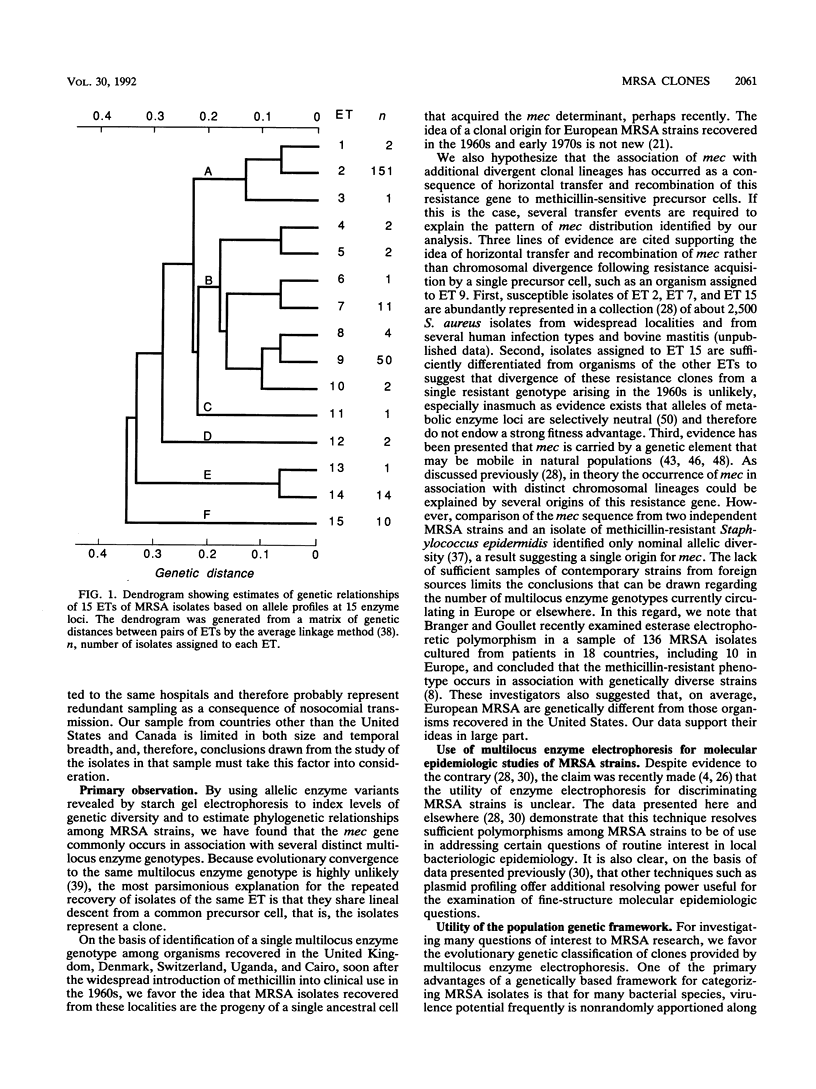
Abstract
Genetic relationships among 254 isolates of Staphylococcus aureus resistant to methicillin recovered between 1961 and 1992 from nine countries on four continents were determined by analyzing electrophoretically demonstrable allelic variation at 15 chromosomal enzyme loci. Fifteen distinctive electrophoretic types, marking clones, were identified. The mec gene is harbored by many divergent phylogenetic lineages representing a large portion of the breadth of chromosomal diversity in the species, a result that is interpreted as evidence that multiple episodes of horizontal transfer and recombination have contributed to the spread of this resistance determinant in natural populations. Isolates recovered in the United Kingdom, Denmark, Switzerland, Egypt, and Uganda in the 1960s are of a single multilocus enzyme genotype and probably are progeny of an ancestral methicillin-resistant clone. There is geographic variation in the frequency of recovery of the common methicillin-resistant clones, an observation that may in part explain reported regional differences in natural history correlates of resistant organisms.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Archer G. L., Mayhall C. G. Comparison of epidemiological markers used in the investigation of an outbreak of methicillin-resistant Staphylococcus aureus infections. J Clin Microbiol. 1983 Aug;18(2):395–399. doi: 10.1128/jcm.18.2.395-399.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Kayser F. H. Characterization of an isogenic set of methicillin-resistant and susceptible mutants of Staphylococcus aureus. Eur J Clin Microbiol. 1986 Dec;5(6):697–701. doi: 10.1007/BF02013308. [DOI] [PubMed] [Google Scholar]
- Blumberg H. M., Rimland D., Kiehlbauch J. A., Terry P. M., Wachsmuth I. K. Epidemiologic typing of Staphylococcus aureus by DNA restriction fragment length polymorphisms of rRNA genes: elucidation of the clonal nature of a group of bacteriophage-nontypeable, ciprofloxacin-resistant, methicillin-susceptible S. aureus isolates. J Clin Microbiol. 1992 Feb;30(2):362–369. doi: 10.1128/jcm.30.2.362-369.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvet A., Fournier J. M., Audurier A., Branger C., Orsoni A., Girard C. Epidemiological markers for epidemic strain and carrier isolates in an outbreak of nosocomial oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1990 Jun;28(6):1338–1341. doi: 10.1128/jcm.28.6.1338-1341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce J. M. Methicillin-resistant Staphylococcus aureus. Detection, epidemiology, and control measures. Infect Dis Clin North Am. 1989 Dec;3(4):901–913. [PubMed] [Google Scholar]
- Branger C., Goullet P. Esterase electrophoretic polymorphism of methicillin-sensitive and methicillin-resistant strains of Staphylococcus aureus. J Med Microbiol. 1987 Nov;24(3):275–281. doi: 10.1099/00222615-24-3-275. [DOI] [PubMed] [Google Scholar]
- Branger C., Goullet P. Genetic heterogeneity in methicillin-resistant strains of Staphylococcus aureus revealed by esterase electrophoretic polymorphism. J Hosp Infect. 1989 Aug;14(2):125–134. doi: 10.1016/0195-6701(89)90115-1. [DOI] [PubMed] [Google Scholar]
- Burnie J. P., Matthews R. C., Lee W., Murdoch D. A comparison of immunoblot and DNA restriction patterns in characterising methicillin-resistant isolates of Staphylococcus aureus. J Med Microbiol. 1989 Aug;29(4):255–261. doi: 10.1099/00222615-29-4-255. [DOI] [PubMed] [Google Scholar]
- Chambers H. F. Methicillin-resistant staphylococci. Clin Microbiol Rev. 1988 Apr;1(2):173–186. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins J. K., Smith J. S., Kelly M. T. Comparison of phage typing, plasmid mapping, and antibiotic resistance patterns as epidemiologic markers in a nosocomial outbreak of methicillin-resistant Staphylococcus aureus infections. Diagn Microbiol Infect Dis. 1984 Jun;2(3):233–245. doi: 10.1016/0732-8893(84)90036-1. [DOI] [PubMed] [Google Scholar]
- Costas M., Cookson B. D., Talsania H. G., Owen R. J. Numerical analysis of electrophoretic protein patterns of methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1989 Nov;27(11):2574–2581. doi: 10.1128/jcm.27.11.2574-2581.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaston M. A., Duff P. S., Naidoo J., Ellis K., Roberts J. I., Richardson J. F., Marples R. R., Cooke E. M. Evaluation of electrophoretic methods for typing methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1988 Jul;26(3):189–197. doi: 10.1099/00222615-26-3-189. [DOI] [PubMed] [Google Scholar]
- Hadorn K., Lenz W., Kayser F. H., Shalit I., Krasemann C. Use of a ribosomal RNA gene probe for the epidemiological study of methicillin and ciprofloxacin resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1990 Sep;9(9):649–653. doi: 10.1007/BF01964265. [DOI] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984 May;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordens J. Z., Hall L. M. Characterisation of methicillin-resistant Staphylococcus aureus isolates by restriction endonuclease digestion of chromosomal DNA. J Med Microbiol. 1988 Oct;27(2):117–123. doi: 10.1099/00222615-27-2-117. [DOI] [PubMed] [Google Scholar]
- Lacey R. W., Grinsted J. Genetic analysis of methicillin-resistant strains of Staphylococcus aureus; evidence for their evolution from a single clone. J Med Microbiol. 1973 Nov;6(4):511–526. doi: 10.1099/00222615-6-4-511. [DOI] [PubMed] [Google Scholar]
- Lee P. K., Kreiswirth B. N., Deringer J. R., Projan S. J., Eisner W., Smith B. L., Carlson E., Novick R. P., Schlievert P. M. Nucleotide sequences and biologic properties of toxic shock syndrome toxin 1 from ovine- and bovine-associated Staphylococcus aureus. J Infect Dis. 1992 Jun;165(6):1056–1063. doi: 10.1093/infdis/165.6.1056. [DOI] [PubMed] [Google Scholar]
- Licitra C. M., Brooks R. G., Terry P. M., Shaw K. J., Hare R. S. Use of plasmid analysis and determination of aminoglycoside-modifying enzymes to characterize isolates from an outbreak of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1989 Nov;27(11):2535–2538. doi: 10.1128/jcm.27.11.2535-2538.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuhashi M., Song M. D., Ishino F., Wachi M., Doi M., Inoue M., Ubukata K., Yamashita N., Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986 Sep;167(3):975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monzon-Moreno C., Aubert S., Morvan A., Solh N. E. Usefulness of three probes in typing isolates of methicillin-resistant Staphylococcus aureus (MRSA). J Med Microbiol. 1991 Aug;35(2):80–88. doi: 10.1099/00222615-35-2-80. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Arbeit R. D. Epidemiologic and clinical utility of typing systems for differentiating among strains of methicillin-resistant Staphylococcus aureus. Infect Control Hosp Epidemiol. 1991 Jan;12(1):20–28. doi: 10.1086/646234. [DOI] [PubMed] [Google Scholar]
- Musser J. M., Schlievert P. M., Chow A. W., Ewan P., Kreiswirth B. N., Rosdahl V. T., Naidu A. S., Witte W., Selander R. K. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc Natl Acad Sci U S A. 1990 Jan;87(1):225–229. doi: 10.1073/pnas.87.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opal S. M., Mayer K. H., Stenberg M. J., Blazek J. E., Mikolich D. J., Dickensheets D. L., Lyhte L. W., Trudel R. R., Musser J. M. Frequent acquisition of multiple strains of methicillin-resistant Staphylococcus aureus by healthcare workers in an endemic hospital environment. Infect Control Hosp Epidemiol. 1990 Sep;11(9):479–485. doi: 10.1086/646215. [DOI] [PubMed] [Google Scholar]
- Pfaller M. A., Wakefield D. S., Hollis R., Fredrickson M., Evans E., Massanari R. M. The clinical microbiology laboratory as an aid in infection control. The application of molecular techniques in epidemiologic studies of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis. 1991 May-Jun;14(3):209–217. doi: 10.1016/0732-8893(91)90034-d. [DOI] [PubMed] [Google Scholar]
- Preheim L., Pitcher D., Owen R., Cookson B. Typing of methicillin resistant and susceptible Staphylococcus aureus strains by ribosomal RNA gene restriction patterns using a biotinylated probe. Eur J Clin Microbiol Infect Dis. 1991 May;10(5):428–436. doi: 10.1007/BF01968023. [DOI] [PubMed] [Google Scholar]
- Prevost G., Jaulhac B., Piemont Y. DNA fingerprinting by pulsed-field gel electrophoresis is more effective than ribotyping in distinguishing among methicillin-resistant Staphylococcus aureus isolates. J Clin Microbiol. 1992 Apr;30(4):967–973. doi: 10.1128/jcm.30.4.967-973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost G., Pottecher B., Dahlet M., Bientz M., Mantz J. M., Piémont Y. Pulsed field gel electrophoresis as a new epidemiological tool for monitoring methicillin-resistant Staphylococcus aureus in an intensive care unit. J Hosp Infect. 1991 Apr;17(4):255–269. doi: 10.1016/0195-6701(91)90270-i. [DOI] [PubMed] [Google Scholar]
- Rhinehart E., Shlaes D. M., Keys T. F., Serkey J., Kirkley B., Kim C., Currie-McCumber C. A., Hall G. Nosocomial clonal dissemination of methicillin-resistant Staphylococcus aureus. Elucidation by plasmid analysis. Arch Intern Med. 1987 Mar;147(3):521–524. [PubMed] [Google Scholar]
- Richardson J. F., Chittasobhon N., Marples R. R. Supplementary phages for the investigation of strains of methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1988 Jan;25(1):67–74. doi: 10.1099/00222615-25-1-67. [DOI] [PubMed] [Google Scholar]
- Ryffel C., Tesch W., Birch-Machin I., Reynolds P. E., Barberis-Maino L., Kayser F. H., Berger-Bächi B. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene. 1990 Sep 28;94(1):137–138. doi: 10.1016/0378-1119(90)90481-6. [DOI] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Levin B. R. Genetic diversity and structure in Escherichia coli populations. Science. 1980 Oct 31;210(4469):545–547. doi: 10.1126/science.6999623. [DOI] [PubMed] [Google Scholar]
- Song M. D., Wachi M., Doi M., Ishino F., Matsuhashi M. Evolution of an inducible penicillin-target protein in methicillin-resistant Staphylococcus aureus by gene fusion. FEBS Lett. 1987 Aug 31;221(1):167–171. doi: 10.1016/0014-5793(87)80373-3. [DOI] [PubMed] [Google Scholar]
- Stewart G. C., Rosenblum E. D. Genetic behavior of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol. 1980 Dec;144(3):1200–1202. doi: 10.1128/jb.144.3.1200-1202.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausbaugh L. J., Jacobson C., Sewell D. L., Potter S., Ward T. T. Methicillin-resistant Staphylococcus aureus in extended-care facilities: experiences in a Veterans' Affairs nursing home and a review of the literature. Infect Control Hosp Epidemiol. 1991 Jan;12(1):36–45. doi: 10.1086/646236. [DOI] [PubMed] [Google Scholar]
- Thomson-Carter F. M., Pennington T. H. Characterisation of methicillin-resistant isolates of Staphylococcus aureus by analysis of whole-cell and exported proteins. J Med Microbiol. 1989 Jan;28(1):25–32. doi: 10.1099/00222615-28-1-25. [DOI] [PubMed] [Google Scholar]
- Trees D. L., Iandolo J. J. Identification of a Staphylococcus aureus transposon (Tn4291) that carries the methicillin resistance gene(s). J Bacteriol. 1988 Jan;170(1):149–154. doi: 10.1128/jb.170.1.149-154.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Nonoguchi R., Song M. D., Matsuhashi M., Konno M. Homology of mecA gene in methicillin-resistant Staphylococcus haemolyticus and Staphylococcus simulans to that of Staphylococcus aureus. Antimicrob Agents Chemother. 1990 Jan;34(1):170–172. doi: 10.1128/aac.34.1.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubukata K., Yamashita N., Konno M. Occurrence of a beta-lactam-inducible penicillin-binding protein in methicillin-resistant staphylococci. Antimicrob Agents Chemother. 1985 May;27(5):851–857. doi: 10.1128/aac.27.5.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Ochman H., Selander R. K. Multilocus genetic structure in natural populations of Escherichia coli. Proc Natl Acad Sci U S A. 1983 Mar;80(6):1751–1755. doi: 10.1073/pnas.80.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarelli A. J., Roy I., Harding G. P., Couperus J. J. Diversity and stability of restriction enzyme profiles of plasmid DNA from methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1990 Jan;28(1):97–102. doi: 10.1128/jcm.28.1.97-102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Adhami W., Roberts L., Vickery A., Inglis B., Gibbs A., Stewart P. R. Epidemiological analysis of a methicillin-resistant Staphylococcus aureus outbreak using restriction fragment length polymorphisms of genomic DNA. J Gen Microbiol. 1991 Dec;137(12):2713–2720. doi: 10.1099/00221287-137-12-2713. [DOI] [PubMed] [Google Scholar]



