Abstract
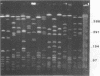
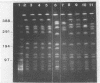
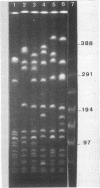
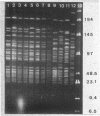
Restriction fragment length polymorphisms in methicillin-susceptible and methicillin-resistant (MRSA) strains of Staphylococcus aureus isolated in the same hospital over a 4-month period were studied by using SmaI and ApaI digestion of genomic DNA and pulsed-field gel electrophoresis. Each of the 20 methicillin-susceptible strains had a unique SmaI pattern, but the 27 MRSA strains showed only seven SmaI patterns. More than half of the SmaI fragments in all of these seven patterns were identical, as were those in the patterns from two unrelated MRSA strains. Digestion with ApaI, which cuts staphylococcus DNA into at least twice as many fragments, confirmed the results obtained with SmaI. Lastly, the plasmid contents of MRSA strains showing identical SmaI and ApaI electrophoretic patterns were not identical. These results are interpreted as supporting the hypothesis that all MRSA strains arose from a single clone and emphasize the need to use several methods in epidemiological investigations of MRSA outbreaks.
Full text
PDF
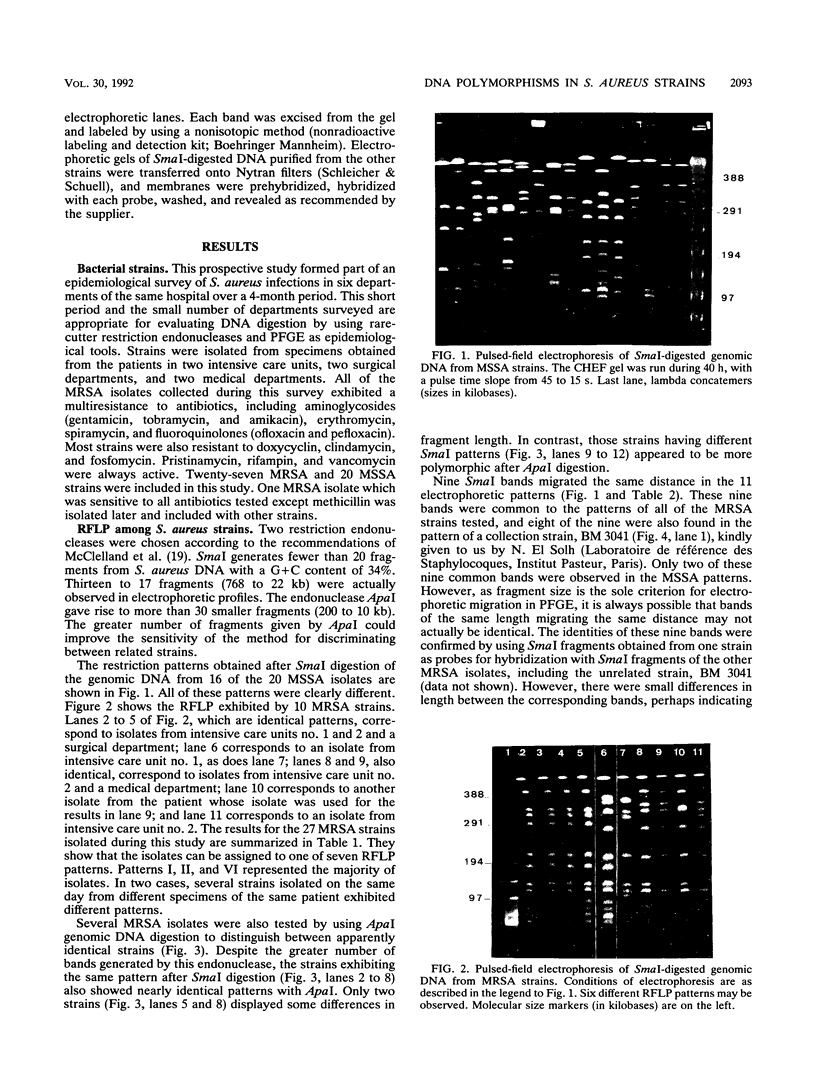
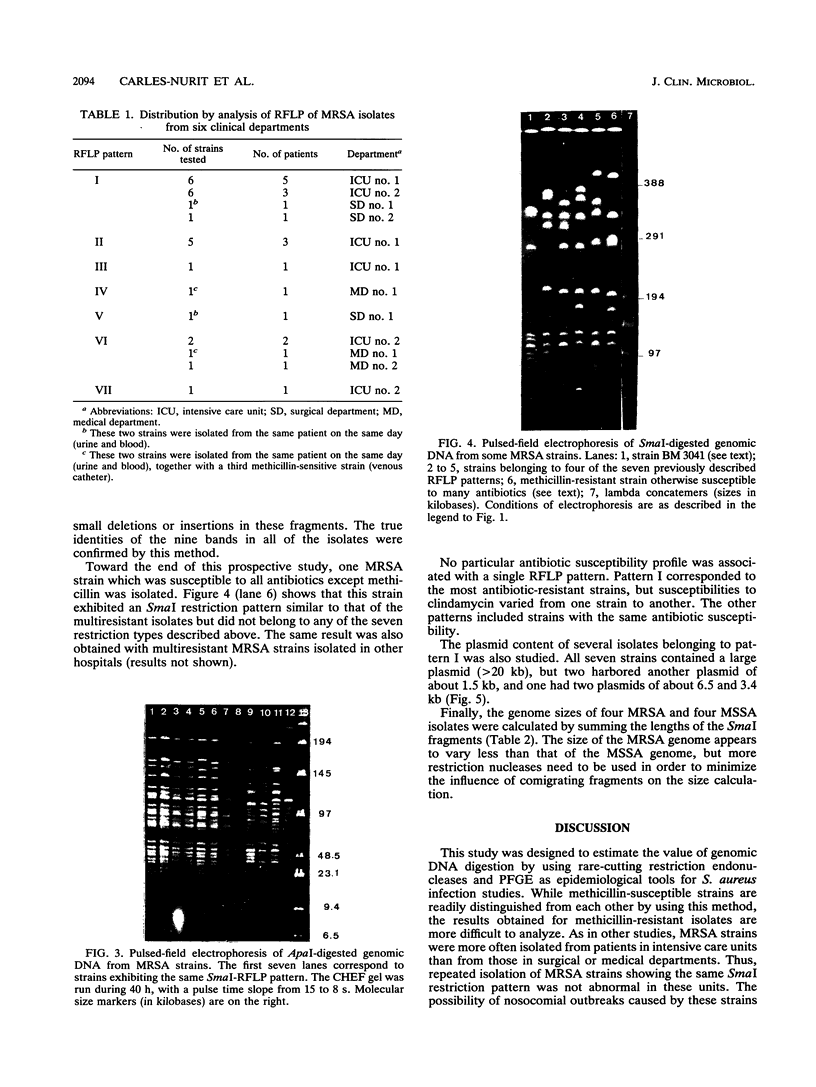


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allardet-Servent A., Bouziges N., Carles-Nurit M. J., Bourg G., Gouby A., Ramuz M. Use of low-frequency-cleavage restriction endonucleases for DNA analysis in epidemiological investigations of nosocomial bacterial infections. J Clin Microbiol. 1989 Sep;27(9):2057–2061. doi: 10.1128/jcm.27.9.2057-2061.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeit R. D., Karakawa W. W., Vann W. F., Robbins J. B. Predominance of two newly described capsular polysaccharide types among clinical isolates of Staphylococcus aureus. Diagn Microbiol Infect Dis. 1984 Apr;2(2):85–91. doi: 10.1016/0732-8893(84)90002-6. [DOI] [PubMed] [Google Scholar]
- Archer G. L., Mayhall C. G. Comparison of epidemiological markers used in the investigation of an outbreak of methicillin-resistant Staphylococcus aureus infections. J Clin Microbiol. 1983 Aug;18(2):395–399. doi: 10.1128/jcm.18.2.395-399.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer A. W., Kirby W. M., Sherris J. C., Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966 Apr;45(4):493–496. [PubMed] [Google Scholar]
- Beck W. D., Berger-Bächi B., Kayser F. H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986 Feb;165(2):373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989 Oct;219(1-2):263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- Bouvet A., Fournier J. M., Audurier A., Branger C., Orsoni A., Girard C. Epidemiological markers for epidemic strain and carrier isolates in an outbreak of nosocomial oxacillin-resistant Staphylococcus aureus. J Clin Microbiol. 1990 Jun;28(6):1338–1341. doi: 10.1128/jcm.28.6.1338-1341.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branger C., Goullet P., Boutonnier A., Fournier J. M. Correlation between esterase electrophoretic types and capsular polysaccharide types 5 and 8 among methicillin-susceptible and methicillin-resistant strains of Staphylococcus aureus. J Clin Microbiol. 1990 Jan;28(1):150–151. doi: 10.1128/jcm.28.1.150-151.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cookson B. D., Phillips I. Epidemic methicillin-resistant Staphylococcus aureus. J Antimicrob Chemother. 1988 Apr;21 (Suppl 100):57–65. doi: 10.1093/jac/21.suppl_c.57. [DOI] [PubMed] [Google Scholar]
- Crossley K., Landesman B., Zaske D. An outbreak of infections caused by strains of Staphylococcus aureus resistant to methicillin and aminoglycosides. II. Epidemiologic studies. J Infect Dis. 1979 Mar;139(3):280–287. doi: 10.1093/infdis/139.3.280. [DOI] [PubMed] [Google Scholar]
- Hackbarth C. J., Chambers H. F. Methicillin-resistant staphylococci: genetics and mechanisms of resistance. Antimicrob Agents Chemother. 1989 Jul;33(7):991–994. doi: 10.1128/aac.33.7.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Jordens J. Z., Wang F. Methicillin-resistant Staphylococcus aureus from China characterized by digestion of total DNA with restriction enzymes. Epidemiol Infect. 1989 Aug;103(1):183–192. doi: 10.1017/s095026880003048x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman B. J., Tomasz A. Low-affinity penicillin-binding protein associated with beta-lactam resistance in Staphylococcus aureus. J Bacteriol. 1984 May;158(2):513–516. doi: 10.1128/jb.158.2.513-516.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichiyama S., Ohta M., Shimokata K., Kato N., Takeuchi J. Genomic DNA fingerprinting by pulsed-field gel electrophoresis as an epidemiological marker for study of nosocomial infections caused by methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1991 Dec;29(12):2690–2695. doi: 10.1128/jcm.29.12.2690-2695.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhl S. A., Pattee P. A., Baldwin J. N. Chromosomal map location of the methicillin resistance determinant in Staphylococcus aureus. J Bacteriol. 1978 Aug;135(2):460–465. doi: 10.1128/jb.135.2.460-465.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey R. W., Kruczenyk S. C. Epidemiology of antibiotic resistance in Staphylococcus aureus. J Antimicrob Chemother. 1986 Oct;18 (Suppl 100):207–214. doi: 10.1093/jac/18.supplement_c.207. [DOI] [PubMed] [Google Scholar]
- Lindberg M., Sjöström J. E., Johansson T. Transformation of chromosomal and plasmid characters in Staphylococcus aureus. J Bacteriol. 1972 Feb;109(2):844–847. doi: 10.1128/jb.109.2.844-847.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClelland M., Jones R., Patel Y., Nelson M. Restriction endonucleases for pulsed field mapping of bacterial genomes. Nucleic Acids Res. 1987 Aug 11;15(15):5985–6005. doi: 10.1093/nar/15.15.5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. J Bacteriol. 1989 Feb;171(2):874–879. doi: 10.1128/jb.171.2.874-879.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhinehart E., Shlaes D. M., Keys T. F., Serkey J., Kirkley B., Kim C., Currie-McCumber C. A., Hall G. Nosocomial clonal dissemination of methicillin-resistant Staphylococcus aureus. Elucidation by plasmid analysis. Arch Intern Med. 1987 Mar;147(3):521–524. [PubMed] [Google Scholar]
- Weil M. D., McClelland M. Enzymatic cleavage of a bacterial genome at a 10-base-pair recognition site. Proc Natl Acad Sci U S A. 1989 Jan;86(1):51–55. doi: 10.1073/pnas.86.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccarelli A. J., Roy I., Harding G. P., Couperus J. J. Diversity and stability of restriction enzyme profiles of plasmid DNA from methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 1990 Jan;28(1):97–102. doi: 10.1128/jcm.28.1.97-102.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]