Abstract
Background
Glycerol nucleic acid (GNA) has an acyclic phosphoglycerol backbone repeat-unit, but forms stable duplexes based on Watson-Crick base-pairing. Because of its structural simplicity, GNA is of particular interest with respect to the possibility of evolving functional polymers by in vitro selection. Template-dependent GNA synthesis is essential to any GNA-based selection system.
Principal Findings
In this study, we investigated the ability of various DNA polymerases to use glycerol-nucleoside triphosphates (gNTPs) as substrates for GNA synthesis on DNA templates. Therminator DNA polymerase catalyzes quantitative primer-extension by the incorporation of two glyceronucleotides, with much less efficient extension up to five glyceronucleotides. Steady-state kinetic experiments suggested that GNA synthesis by Therminator was affected by both decreased catalytic rates and weakened substrate binding, especially for pyrimidines. In an attempt to improve pyrimidine incorporation by providing additional stacking interactions, we synthesized two new gNTP analogs with 5-propynyl substituted pyrimidine nucleobases. This led to more efficient incorporation of gC, but not gT.
Conclusions
We suggest that directed evolution of Therminator might lead to mutants with improved substrate binding and catalytic efficiency.
Introduction
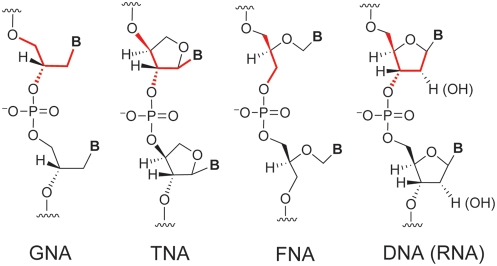
The ribose and 2-deoxyribose components of the backbones of RNA and DNA have long been thought to play a critical role in stabilizing Watson-Crick base pairing and maintaining fidelity in polymerase-mediated information transfer. Having survived billions of years of evolution on Earth, ribose has been considered to be one of the indispensable building blocks of contemporary life [1]. However, recent studies from the Eschenmoser group of nucleic acid analogs with altered sugar-phosphate backbones demonstrate dramatically that D-ribofuranose is not a chemical requirement for the ability of a prebiotically plausible nucleic acid to function as the genetic information carrier [2]. Indeed, a nucleic acid with a simpler backbone structure, such as (3′→2′)-α-L-threose nucleic acid (TNA, Figure 1), might have been an evolutionary progenitor of DNA or RNA [3].
Figure 1. Structures of GNA, TNA, FNA, and DNA (RNA).
The backbone similarity is highlighted in red.
Recently, the Meggers group synthesized S-Glycerol nucleic acid (GNA), a nucleic acid analog with an acyclic, phosphoglycerol repeating unit (Figure 1) [4], [5]. Like TNA, GNA is a nucleic acid with a simplified backbone that can form stable duplexes. As a first step toward understanding the evolutionary potential of GNA, we have recently shown that Bst DNA polymerase can catalyze faithful (but limited) DNA synthesis on a GNA template even though the DNA product cannot form a stable duplex with the GNA template [6]. This surprising observation suggests that, in contrast to common belief [7], stable heteroduplex formation is not required for genetic information transfer between two nucleic acid systems—a short base-paired region stabilized by the enzyme appears to be sufficient.
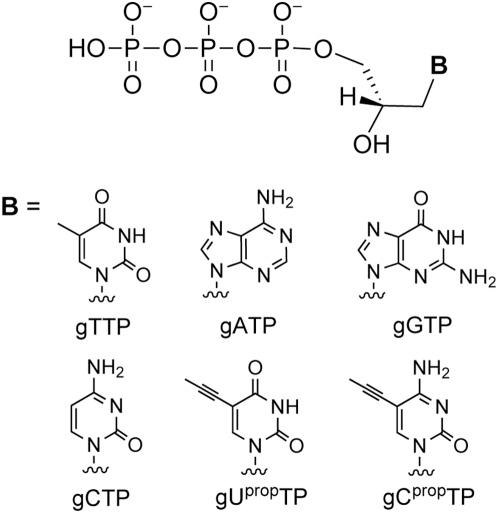
In addition to exploring GNA molecules as a genetic information carrier, we are also interested in probing their ligand binding and catalytic abilities. We envision that an in vitro selection strategy, similar to that used in RNA aptamer or artificial ribozyme selection [8], can be used to isolate functional GNA constructs. This selection strategy requires a polymerase that can synthesize GNA in a template-directed fashion. We have previously synthesized the four canonical glycerol-nucleoside triphosphates (gNTPs with A, G, T, or C, Figure 2) and demonstrated that several DNA polymerases can incorporate a single glyceronucleotide onto a DNA primer/template using gNTPs as the substrates [9]. Incorporation of a second glyceronucleotide was not observed in that study, which we attributed to the poorly constrained nature of the acyclic glycerophosphate backbone of GNA and/or the unstable nature of a GNA/DNA duplex [9].
Figure 2. Structures of glyceronucleoside triphosphates (gNTPs).
In the present study, we show that certain DNA polymerases can incorporate at least two GNA residues onto a growing primer. Kinetic analysis of Therminator-catalyzed single glyceronucleotide incorporation suggested that pyrimidine glyceronucleotides were weakly bound substrates. In an attempt to improve pyrimidine incorporation, we synthesized two 5-propynyl substituted gNTP analogs, to improve the base-stacking interaction between the incoming gNTP and the terminal nucleotide of the primer (Figure 2). This modification led to improved efficiency of incorporation of gCTP, but not of gTTP.
Results
Polymerase Screens
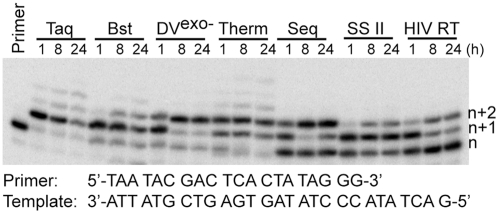
Using a primer extension assay, we screened a number of polymerases (including Taq, Bst, DeepVent (exo-), Therminator, Sequenase, Superscript II, and HIV-1 reverse transcriptase) for their ability to catalyze GNA synthesis on a DNA template. In order to enhance our chances of observing primer-extension by more than just one base as previously observed [9], we used long incubations under a variety of conditions. We found that a number of polymerases were able to incorporate more than one GNA monomer at a 1∶1 enzyme∶primer/template ratio (Figure 3). The Therminator polymerase utilized most of the primer and generated a significant fraction of +2 extended products within 1 h (Figure 3). After 8 h, Deep Vent exo- polymerase had a higher yield of +2 extended primer, but Therminator appeared to synthesize small amounts of +3, +4, and +5 products. We therefore decided to characterize primer-extension and substrate utilization by Therminator in greater detail. Raising the assay temperature from 55°C to 75°C (Therminator's optimum temperature) resulted in decreased efficiency of GNA incorporation. Therefore, all the subsequent kinetic studies were performed at 55°C. Neither the addition of Mn2+ nor substitution of adenosine with diaminopurine-2′-deoxyriboside in the template significantly improved the efficiency of GNA synthesis by this enzyme, using the four canonical gNTPs.
Figure 3. Primer extension analysis of GNA synthesis mediated by polymerases.
The products of Therminator-catalyzed, single and double glyceronucleotide incorporation were characterized by MALDI-TOF MS. The expected mass difference between a primer extended with one deoxyribonucleotide vs. one glyceronucleotide is 42 Daltons. As shown in Table 1, the observed masses for single glyceronucleotide or deoxyribonucleotide incorporation on Template A are consistent with the calculated values. The identity of the products of two sequential nucleotide incorporations (gTMP+dAMP, gTMP+gAMP, 2 gAMP, or 2 gGMP on Template B) was also confirmed by MALDI-TOF MS (Table 1).
Table 1. MALDI-TOF MS analysis of single and double nucleotide incorporation by Therminator DNA polymerase a.
| XY | NTP substrate | Nucleotide incorporated | Calcd | Obsd | |
| Template A | CA | none | none | 6126.1 | 6126.2 |
| AG | TTP | T | 6430.3 | 6431.5 | |
| TG | dATP | dA | 6439.3 | 6439.5 | |
| CA | dGTP | dG | 6455.3 | 6455.3 | |
| GA | dCTP | dC | 6415.3 | 6415.3 | |
| AG | gTTP | gT | 6388.3 | 6388.3 | |
| AG | gUpropTP | gUprop | 6412.3 | 6411.9 | |
| TG | gATP | gA | 6397.3 | 6396.8 | |
| CA | gGTP | gG | 6413.3 | 6413.4 | |
| GA | gCTP | gC | 6373.3 | 6373.3 | |
| GA | gCpropTP | gCprop | 6411.3 | 6410.8 | |
| Template B | AT | gTTP+dATP | gT+dA | 6701.5 | 6700.0 |
| AT | gTTP+gATP | gT+gA | 6659.5 | 6658.9 | |
| TT | gATP | gA+gA | 6668.5 | 6667.7 | |
| CC | gGTP | gG+gG | 6700.5 | 6699.8 |
Sequences of the primer and the template:
Primer 5′-TAA TAC GAC TCA CTA TAGGG-3′.
Template : 3′-ATT ATG CTG AGT GAT ATC CC XY ACA TCT ATC-5′.
Template B: 3′-ATT ATG CTG AGT GAT ATC CC XY GCA TCT ATC-5′.
Kinetic analysis of glycerol-nucleotide addition to a DNA primer
To obtain insight into the process of GNA synthesis catalyzed by Therminator on DNA templates, we performed steady-state kinetic experiments and measured the k cat and Km values for the four canonical gNTP substrates (Figure 2). The kinetic parameters for addition of a single glyceronucleotide to an all-DNA primer are summarized in Table 2. Because the kinetic experiments were performed at 55°C instead of Therminator's optimum temperature of 75°C, decreased k cat values were observed for regular dNTPs (2–7 s−1 at 55°C, Table 2) compared with those at 75°C (∼15 s−1 calculated from specific activity provided by the manufacturer). Therminator showed similar k cat values for gATP compared to dATP, and for gGTP compared to dGTP, while slower kinetics were observed for gTTP and gCTP (by 5-fold and 3-fold, respectively) compared with TTP and dCTP. All 4 gNTPs showed higher Km values than the corresponding dNTPs, with an ∼20 fold increase for gATP and gGTP and a 200–300 fold increase for gTTP and gCTP. As a result, the decrease of catalytic efficiency (k cat/Km) was much more dramatic for pyrimidine than for purine gNTPs (1000- fold versus only 20-fold). Such discrimination between purine and pyrimidine triphosphate substrates is not observed for threose-nucleoside triphosphates (tNTPs) [10]. One possible explanation for this result is that base-stacking interactions between the incoming nucleoside triphosphate and the primer play a more prominent role in gNTP binding than in dNTP or tNTP binding.
Table 2. Kinetic analysis of single-nucleotide incorporation by Therminator DNA polymerase a.
| XYb | NTP | Km (µM) | k cat (s−1) | k cat/Km (s−1 M−1) |
| TG | dATP | 0.59±0.11 | 3.7±0.5 | 6.2×106 |
| CA | dGTP | 0.47±0.27 | 2.0±0.3 | 4.3×106 |
| AG | TTP | 0.67±0.14 | 4.2±0.4 | 6.4×106 |
| DG | TTP | 0.35±0.10 | 6.8±0.6 | 1.9×107 |
| GA | dCTP | 0.32±0.17 | 3.2±0.8 | 1.0×107 |
| TG | gATP | 14.0±6.7 | 4.7±0.8 | 3.4×105 |
| CA | gGTP | 10.6±4.7 | 2.3±0.1 | 2.1×105 |
| AG | gTTP | 129.4±77.3 | 0.8±0.3 | 5.8×103 |
| DG | gTTP | 54.3±8.3 | 1.6±0.4 | 3.0×104 |
| AG | gUpropTP | 139.5±14.5 | 1.5±0.3 | 1.1×104 |
| DG | gUpropTP | 41.4±4.7 | 1.7±0.2 | 4.1×104 |
| GA | gCTP | 89.5±38.6 | 1.3±0.6 | 1.4×104 |
| GA | gCpropTP | 3.3±0.4 | 1.9±0.2 | 5.8×105 |
Sequences of the primer and the template:
Primer: 5′-TAA TAC GAC TCA CTA TAGGG-3′.
Template: 3′-ATT ATG CTG AGT GAT ATC CC XY ACA TCT ATC-5′.
D denotes 2,6-diaminopurine-2′-deoxyribonucleotide.
Synthesis of 5-propynyl substituted glycerol-nucleoside triphosphates
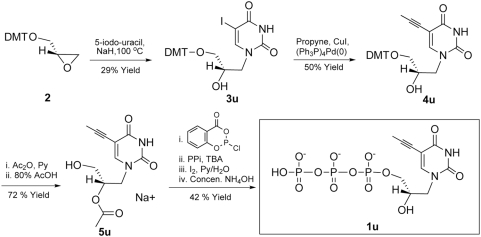
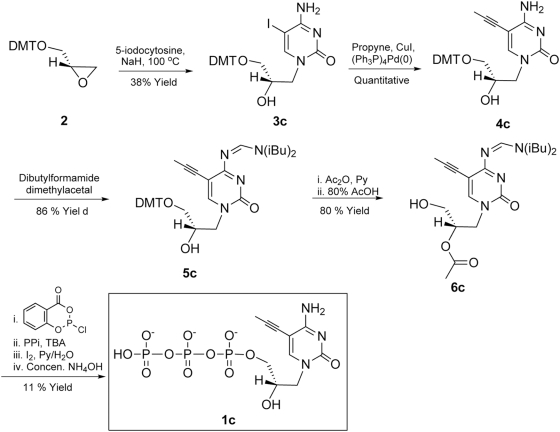
In order to test the hypothesis that poor enzymatic incorporation of pyrimidine gNTPs reflects poor base-stacking, we synthesized the C-5-propynyl substituted pyrimidine nucleoside triphosphates (Figure 2). This co-planar nucleobase modification has been proposed to increase base stacking and hydrophobic interactions between base pairs [11]. In the case of GNA synthesis, we thought that a 5-propynyl group in pyrimidine gNTPs might improve binding to the primer/template complex in the active site of polymerase. The synthetic schemes for the preparation of 5-propynyl substituted gNTPs (gUpropTP and gCpropTP, or 1u and 1c) are shown in Figures 4 and 5. Two key intermediates, the 5-propynyl pyrimidine glycerol-nucleosides (4u and 5c), were prepared from 5-iodo-substituted precursors (3u and 3c) and propyne by Sonogashira coupling (Figure 4 and 5) [12], [13]. A di-butylaminomethylidene group was used to protect the exocyclic amine of 3c instead of an acetyl or benzoyl group in order to avoid a potential cyclization side-reaction involving the amide and the 5-propynyl group [14]. The presence of a propynyl group in 4u and 5c was confirmed by the characteristic chemical shift (4–5 ppm) of the methyl carbon in 13C NMR [15] together with 1H-NMR and ESI-MS analysis. gUpropTP and gCpropTP (1u and 1c) were synthesized from the corresponding nucleosides (5u and 6c) using the one-pot, salicylchlorophosphorin approach developed by Ludwig and Eckstein [16]. The final purified products 1u and 1c were characterized by 1H- and 31P-NMR and by ESI-MS. In addition, 1u and 1c have similar UV absorption profiles to those reported for 5-propynyl-deoxyribonucleosides [15].
Figure 4. Synthesis of 5-propynyluridine glyceronucleoside triphosphate (1u).
Figure 5. Synthesis of 5-propynylcytidine glyceronucleoside triphosphate (1c).
Kinetics of DNA primer-extension using 5-propynyl gNTPs
We measured Km and k cat for Therminator-catalyzed DNA primer-extension, using 5-propynyl gUTP and gCTP as substrates. The results suggest that a stronger base stacking interaction does increase the catalytic efficiency of gNTP incorporation, although the effect is more prominent for gCpropTP than for gUpropTP (Figure 2). Compared to gCTP, gCpropTP had a much lower Km (3.3 µM vs. 89.5 µM) and a slightly faster k cat (1.9 s−1 vs. 1.3 s−1) (Table 2). As a result, the catalytic efficiency (k cat/Km) of Therminator with gCpropTP was 40 fold higher than that with gCTP. In fact, gCpropTP had the lowest Km and highest k cat/Km of all 6 gNTPs. In contrast, the effect of 5-propynyl substitution on gUTP was modest, and gUpropTP showed a similar Km value to gTTP with an ∼2 fold increase in k cat (Table 2). When diaminopurine was used in the template, both gTTP and gUpropTP had a significantly lower Km (54.3 µM and 41.4 µM, respectively) than those measured with the dA-containing template (129.4 µM and 139.5 µM, respectively) with slightly increased k cat (Table 2). These results suggest that both H-bonding and base stacking contribute to binding of gNTPs to the primer/template complex and that the synergy of these two interactions determines the overall catalytic efficiency.
Kinetic analysis of dNMP incorporation on a GNA-terminated primer
We synthesized a primer containing a single GNA monomer (gT) at the 3′-terminus in order to study the effect of the positioning of the nucleophile (2′-OH of GNA vs. 3′-OH of DNA) on the catalytic efficiency of Therminator polymerase. However, kinetic parameters for a 3′-gT-terminated primer were more difficult to obtain than with an all-DNA primer. Under steady-state conditions with a low enzyme∶primer/template ratio, we were not able to observe significant primer-extension even at saturating gNTP concentrations. At a 1∶1 enzyme∶primer/template ratio (1 µM), ∼15% extension of the 3′-gT-terminated primer was observed after 24 h with gATP and gGTP, but not with gTTP and gCTP. This observation suggests that, in the active site of Therminator, the conformation of the terminal GNA nucleotide, especially its 2′-OH group, is not optimal (or is poorly constrained) for nucleophilic attack on the incoming triphosphate. The catalytic efficiency is further reduced by the low affinity of Therminator for gNTP substrates, especially in the case of gTTP and gCTP.
To quantitatively assess the effect of a 3′-terminal gT residue on the catalytic efficiency of continued polymerization, we determined the kinetic parameters for the incorporation of a single deoxyribonucleotide by Therminator, using a 3′-gT primer. Compared with the all-DNA primer (Table 2), the 3′-gT primer caused 2–4 fold decrease in k cat (Table 3). In addition, an increase in Km was also observed, which again was more prominent for pyrimidines (∼80 fold) than for purines (4–8 fold) (Table 3). These results suggest that the presence of gT at the 3′-terminus of the primer may interfere with essential conformational changes of the enzyme during its catalytic cycle, resulting in both slower k cat and increased Km values.
Table 3. Kinetic analysis of single deoxyribonucleotide incorporation by Therminator using a GNA-terminated primer a.
| XY | NTP | Km (µM) | k cat (s−1) | k cat/Km (s−1 M−1) |
| TG | dATP | 4.7±2.0 | 0.7±0.2 | 1.4×105 |
| CA | dGTP | 2.1±1.5 | 1.0±0.1 | 4.7×105 |
| AG | TTP | 50.7±38.5 | 0.9±0.2 | 1.8×104 |
| GA | dCTP | 21.4±7.6 | 1.7±0.6 | 7.8×104 |
Sequences of the primer and the template (the lower case denotes the GNA sequence):
Primer: 5′-TAA TAC GAC TCA CTA TAG GG t.
Template: 3′-ATT ATG CTG AGT GAT ATC CC A XY CAT CTA TC-5′.
Discussion
Previous studies have shown that certain polymerases are capable of template-directed primer-extension by using as substrates nucleotide analogs with either a shortened (TNA) or an acyclic (flexible nucleic acid, or FNA, Figure 1) sugar moiety [10], [17], [18]. The active sites of some polymerases are clearly flexible enough to accommodate major sugar modifications [19], [20]. However, compared with TNA or FNA, GNA contains both an acyclic and a shortened repeating unit (Figure 1), and it is therefore not surprising that the enzymatic polymerization of gNTPs is quite difficult. In this study, we screened a series of polymerases and identified Therminator as the most efficient GNA polymerase. Not surprisingly, Therminator was also the most efficient polymerase identified in during previous efforts to synthesize TNA or FNA on DNA templates [10], [18].
Our kinetic analysis of primer-extension with gNTP substrates has provided some insight into the particular problems that prevent efficient Therminator-catalyzed GNA synthesis. Analysis of the addition of the first glycerol-nucleotide to a DNA primer revealed that all six gNTPs examined (the four canonical nucleobases plus the two propynyl-pyrimidines) had k cat values that were similar to those of the corresponding natural dNTPs. Decreased catalytic efficiency was mainly due to higher Km values, and thus is most likely due to weaker gNTP binding. The same observation was made previously for TNA synthesis catalyzed by Therminator [10]. These results suggest that altered sugar structure on an incoming NTP does not prevent Therminator from efficiently orienting the triphosphate moiety of the NTP for nucleophilic attack by the 3′-hydroxyl of the DNA primer. The major kinetic difference between gNTPs and threose-nucleoside triphosphates (tNTPs) is in the incorporation of additional nucleotides to the growing primer, where the attacking nucleophile becomes the 2′-OH. In the case of TNA, which has a relatively rigid threose ring, it appears that Therminator can effectively position the 2′-OH for nucleophilic attack. As a result, for both tNTP and dNTP incorporation, the k cat values for a TNA-terminated primer are similar to those obtained with an all-DNA primer [10]. However, with a gT-terminated primer, the efficiency of gNTP incorporation dropped dramatically (Table 3). These results indicate that the main obstacle to continued GNA synthesis is a poorly positioned (or poorly constrained) 2′-OH group at the end of a GNA primer.
Previous studies suggest that a 5-propynyl group on a pyrimidine nucleoside triphosphate is well tolerated by a variety of DNA polymerases [21]–[23]. For example, Kuwahara et al. showed that 5-propynyl dUTP and dCTP were efficiently utilized by several thermophilic polymerases in PCR with high accuracy [24]. However, there are no systematic studies on the effect of the 5-propynyl group on the kinetics of polymerase-mediated (d)NTP incorporation. Wang and Shaw showed that for MMLV reverse transcriptase, 5-propynyl-dUTP had a similar Km (14.1 µM) and a slightly faster k cat (0.023 s−1) compared with TTP (14.4 µM and 0.015 s−1) [23]. Our results show that the 5-propynyl substitution can significantly improve gCTP incorporation by Therminator, but has surprisingly little effect on gTTP incorporation. It appears that the stronger H-bonding of the G:C base-pair acts synergistically with the favorable base stacking interaction induced by the 5-propynyl group. Other effects, such as perturbation of pKa of N4 of cytosine, might also contribute to a better binding of gCpropTP compared to gCTP. As for gTTP and gUpropTP, our results show that the stronger base-pairing interaction with diaminopurine in the template did improve the substrate binding and overall efficiency. However, the 5-propynyl group did not significantly increase the catalytic efficiency of gTMP incorporation in either adenosine- or diaminopurine-containing templates. This observation is consistent with the previous studies by Seela et al., who showed that the duplex stabilizing effect caused by the 5-propynyl group (measured by increase of Tm) was smaller for dT (0.75–1.5°C) than for dC (2–3°C)[25]. Our kinetic analysis suggests that the increased base-stacking interaction caused by the 5-propynyl group in gUpropTP might be offset by a weakening of the Uprop:A base pairing interaction relative to T:A, due to the perturbed basicity of N3 in 5-propynyl-uracil (pK a = 8.7) compared with that in thymine (pK a = 9.8) (F. Seela, Personal Communication).
In addition to DNA templates, we also attempted to identify polymerases that can use GNA templates. Unfortunately, none of the polymerases we studied showed significant activity with gNTP substrates and GNA templates, even when the primer and primer-binding region were composed of DNA. This lack of activity is not surprising given the challenges of binding and pre-organizing a GNA template in the active site, and binding and orienting the gNTP substrate for catalysis. Natural DNA or RNA polymerases have their active sites optimized for binding B-form or A-form duplexes. Recent X-ray structural studies suggest that the conformation of a GNA duplex differs significantly from either B-form or A-form, although the GNA duplex studied also contained a metallo-base-pair [26]. It is likely that significant engineering of the enzyme active site would be required to evolve an efficient GNA-dependent GNA polymerase using the existing Therminator polymerase as a starting point.
Materials and Methods
Materials
Reagents and solvents were purchased from Sigma-Aldrich. DNA oligonucleotides were purchased from IDT and were purified by denaturing polyacrylamide gel electrophoresis. Dibutylformamide dimethylacetal [14], R-3-O-(4,4′-dimethoxytrityl)-glycidol (2) [27] , and gNTPs (A, G, T, C) [9] were prepared according to published procedures. Taq DNA polymerase was purchased from Roche (1 U is defined as the amount of enzyme that incorporates 10 nmol of total dNTPs into acid precipitable DNA within 30 min at 75°C). Therminator, Deep Vent (exo-), and Bst polymerases were purchased from New England Biolabs (1 U is defined as the amount of enzyme that incorporates 10 nmol of total dNTPs into acid insoluble material in 30 min at 75°C for Therminator and Deep Vent (exo-) or at 65°C for Bst). Sequenase was purchased from USB (1 U is defined as the amount of enzyme that catalyzes the incorporation of 1 nmol of nucleotide into acid insoluble form in 30 s at 37°C). Superscript II reverse transcriptase was purchased from Invitrogen (1 U is defined as the amount of enzyme that incorporates 1 nmol of deoxyribonucleotide into acid-precipitable material in 10 min at 37°C using poly(A)oligo(T)1218 as template/primer). HIV-1 reverse transcriptase was purchased from Worthington (1 U is defined as the amount of enzyme that incorporates 1 nmol of tritiated TMP into acid precipitable products using poly(A)/oligo(dT)12–18 as the template/primer in 20 min at 37°C, pH 8.3). Thermostable inorganic pyrophosphatase was purchased from New England Biolabs (1 U is defined as the amount of enzyme that generates 1 µmol of phosphate per minute from inorganic pyrophosphate at 75°C in 50 mM Tricine, 1 mM MgCl2, 0.32 mM PPi, pH 8.5). Flash column chromatography was performed using silica gel from Sigma-Aldrich (Grade 9385, 230–400 mesh) with solvents indicated below. 1H-, 31P-, and 13C-NMR experiments were performed on a Varian 400 MHz spectrometer. Chemical shifts are reported in ppm with reference to TMS (0.00 ppm) for 1H, phosphoric acid (0.00 ppm) for 31P, or CDCl3 (77.16 ppm) for 13C. Coupling constants are reported in Hz. Low-resolution mass spectrometry (LSMS) analysis was performed on a Bruker Daltonics Esquire 6000 mass spectrometer. All reactions were performed under a dry argon atmosphere.
Synthesis of S-1-(5-iodouracil-1-yl)-3-O-(4,4′-dimethoxytrityl)-2,3-propanediol (3u)
5-iodouracil (3.6 g, 15 mmol) and sodium hydride (125 mg in 60% mineral oil, 3.0 mmol) were suspended in 25 mL of anhydrous dimethylformamide and the reaction mixture was stirred at room temperature for 30 min. R-3-O-(4,4′-dimethoxytrityl)-glycidol (2) (15 mmol) in 25 mL of anhydrous dimethylformamide was added to the reaction mixture drop-wise over 5 min. The reaction mixture was then heated to 100°C and stirred for 1.5 h. The solvent was then evaporated in vacuo and the resulting pellet was extracted with 50 mL of CH2Cl2 twice. The organic layer was combined, washed twice with saturated sodium bicarbonate (100 mL×2) and once with brine (100 mL), dried over Na2SO4, and evaporated in vacuo. Compound 3u was further purified by silica gel chromatography (0.5%–1% MeOH/CH2Cl2) to afford 3u as amorphous light yellow solid (2.7 g, 4.4 mmol, 29%). 1H NMR (400 MHz, CDCl3) δ: 7.72 (1H, s), 7.41(1H, s), 7.39(1H, s), 7.26–7.32 (6H, m), 6.84(4H, d, J = 6.8), 4.04–4.07(2H, m), 3.78(6H, s), 3.65 (1H, dd, J = 8.4, 14.8), 3.18(2H, m); 13C NMR (100.5 MHz, CDCl3) δ: 160.6, 158.8, 151.1, 150.5, 144.5, 135.6, 130.1, 128.2, 128.0, 127.2, 113.4, 86.7, 69.2, 67.3, 64.5, 55.4, 52.0; LRMS calcd for [M+Na]+ (C28H27IN2NaO6): 637.0811, obsd, 637.0.
Synthesis of S-1-(5-propynyl-uracil-1-yl)-3-O-(4,4′-dimethoxytrityl)-2,3-propanediol (4u)
Compound 3u (1.87 g, 3.05 mmol) was dissolved in 20 mL of anhydrous dimethylformamide. Triethylamine (0.81 mL, 6.0 mmol), CuI (116 mg, 0.61 mmol), and tetrakis(triphenylphosphine)palladium(0) (352 mg, 0.31 mmol) were added to the reaction mixture at room temperature. Propyne gas was then bubbled through the reaction mixture at room temperature for 4 h. The solvent was then evaporated in vacuo and the resulting pellet was extracted twice with CH2Cl2 (50 mL×2). The organic layer was combined, washed twice with saturated sodium bicarbonate (100 mL×2) and once with brine (100 mL), dried over Na2SO4, and evaporated in vacuo. Compound 4u was further purified by silica gel chromatography (0.5% MeOH/CH2Cl2) to afford the purified product as light yellow foam (0.8 g, 1.5 mmol, 50%). 1H NMR (400 MHz, CDCl3) δ: 8.94 (1H, br), 7.46 (1H, s), 7.42(1H, s), 7.40(1H, s), 7.23–7.31 (6H, m), 6.83(4H, d, J = 7.2), 4.04–4.08(2H, m), 3.79(6H, s), 3.61 (1H, dd, J = 8.4, 14.4), 3.16(2H, m), 3.04 (1H, br), 2.02(3H, s); 13C NMR (100.5 MHz, CDCl3) δ: 162.5, 158.7, 150.5, 148.1, 144.6, 135.6, 130.1, 128.1, 128.0, 127.1, 113.4, 100.1, 90.9, 86.6, 70.1, 69.1, 64.6, 55.4, 52.1, 4.8; LRMS calcd for [M+Na]+ (C31H30N2NaO6): 549.2002, obsd, 549.1.
Synthesis of S-1-(5-propynyl-uracil-1-yl)-2-O-acetyl-2,3-propanediol (5u)
Compound 4u (0.2 g, 0.38 mmol) was mixed with 4 mL of 10% acetic anhydride in anhydrous pyridine. The reaction mixture was stirred at room temperature for 15 min and was evaporated in vacuo. The crude product was dissolved in 50 mL CH2Cl2, washed twice with saturated sodium bicarbonate (50 mL×2) and once with brine (50 mL), dried over Na2SO4, and evaporated in vacuo. The pellet was then treated with 5 mL of 80% acetic acid in H2O at room temperature for 30 min. The mixture was then evaporated and the crude 5u was purified by silica gel chromatography (2.5% MeOH/CH2Cl2) to afford 5u as white foam (72 mg, 0.27 mmol, 72%). 1H NMR (400 MHz, CDCl3) δ: 9.61 (1H, br), 7.42 (1H, s), 5.10 (1H, m), 4.17 (1H, dd, J = 4.4, 14.4), 3.88 (1H, dd, J = 6.4, 14.4), 3.73 (2H, m), 2.11 (3H, s), 2.04 (3H, s). 13C NMR (100.5 MHz, CDCl3) δ: 170.5, 162.6, 150.7, 146.8, 101.1, 91.4, 71.9, 69.8, 60.9, 48.6, 21.1, 4.8. LRMS calcd for [M+Na]+ (C12H14N2NaO5): 289.0800, obsd, 289.0.
Synthesis of S-1-(5-propynyl-uracil-1-yl)-2-O-acetyl-3-O-triphospho-2,3-propanediol (1u)
Compound 5u (53 mg, 200 µmol) was rendered anhydrous by evaporation with anhydrous pyridine (1 mL×3) and was dissolved in 0.8 mL of pyridine/dioxane (1∶3). To this mixture, 220 µL of 1 M 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one in dioxane was added and the reaction mixture was stirred at room temperature for 10 min. A solution (0.6 mL) of 0.5 M tetrabutylammonium pyrophosphate in dimethylformamide was then added and the reaction mixture was stirred for another 10 min. A solution (7.5 mL) of 1% iodine in pyridine/H2O (98∶2) was then added to the above mixture. After 15 min, 5% Na2SO3 aqueous solution was added dropwise until the reaction mixture turned from dark brown to light yellow. The final mixture was evaporated in vacuo and treated with concentrated ammonia solution (10 mL) overnight at room temperature. The mixture was then evaporated in vacuo and the crude product 1u was purified by preparative RP-HPLC using a Varian Microsorb 100-8 C18 column (250×21.4 mm) with a linear gradient from 0 to 20% acetonitrile in 5 mM triethylammonium acetate (pH 7.0) over 20 min with a flow rate of 15.0 mL/min. The fraction containing 1u (retention time 4.5–5.5 min) was combined and lyophilized to yield 84.4 µmol of 1u (42% yield) based on the UV absorption (ε291 nm: 11.3 mM−1 cm−1). 1H NMR (400 MHz, D2O) δ: 7.84 (1H, s), 4.04 (3H, m), 4.02 (1H, m), 3.80 (1H, dd, J = 6.2, 14.0), 2.00 (3H, s). 31P NMR (161.8 MHz, D2O) δ: −6.3 (d, J = 19.4), −9.90 (d, J = 19.6), −21.7 (t, J = 19.6). LRMS calcd for [M-H]− (C10H14N2O13P3): 462.9714, obsd, 463.0.
Synthesis of S-1-(5-iodocytosine-1-yl)-3-O-(4,4′-dimethoxytrityl)-2,3-propanediol (3c)
5-iodocytosine (2.37 g, 10.0 mmol) and potassium carbonate (276 mg, 2.0 mmol) were suspended in 20 mL of anhydrous dimethylformamide and the mixture was stirred at room temperature for 20 min. R-3-(4,4′-dimethoxytrityl)-glycidol (2) (15 mmol) in 20 mL of anhydrous dimethylformamide was added to the reaction mixture drop-wise over 5 min at room temperature. The reaction mixture was then stirred at 80°C for 16 h. The solvent was evaporated in vacuo and the resulting pellet was extracted with 50 mL of CH2Cl2 twice. The organic layer was combined, washed twice with saturated sodium bicarbonate (100 mL×2) and once with brine (100 mL), dried over Na2SO4, and evaporated in vacuo. Compound 3c was further purified by silica gel chromatography (1%–3% methanol/CH2Cl2) as light yellow solid (2.3 g, 3.8 mmol, 38%). 1H NMR (400 MHz, CDCl3) δ: 7.63 (1H, s), 7.48 (1H, br), 7.42(1H, s), 7.40(1H, s), 7.20–7.32 (6H, m), 6.83(4H, d, J = 6.8), 5.50 (1H, br), 4.14–4.25 (2H, m), 3.79(6H, s), 3.75 (1H, m), 3.23 (1H, dd, J = 10, 4.8), 3.00 (1H, dd, J = 10, 6.4); 13C NMR (100.5 MHz, CDCl3) δ: 164.2, 158.7, 157.1, 152.8, 144.8, 135.9, 135.8, 130.0, 128.1, 128.0, 127.0, 113.4, 86.5, 69.3, 64.6, 55.9, 55.4; LRMS calcd for [M+Na]+ (C28H28IN3NaO5): 636.0971, obsd, 635.9.
Synthesis of S-1-(5-propynylcytosine-1-yl)-3-O-(4,4′-dimethoxytrityl)-2,3-propanediol (4c)
Compound 3c (1.3 g, 2.0 mmol) was dissolved in 15 mL of anhydrous dimethylformamide. Triethylamine (0.54 mL, 4 mmol), CuI (76 mg, 0.4 mmol), and tetrakis(triphenylphosphine)palladium(0) (231 mg, 0.2 mmol) were added to the solution at room temperature. Propyne gas was then bubbled through the reaction mixture for 4 h with stirring at room temperature. The solvent was then evaporated in vacuo and the resulting pellet was extracted with 50 mL of CH2Cl2 twice. The organic layer was combined, washed twice with saturated sodium bicarbonate (100 mL×2) and once with brine (100 mL), dried over Na2SO4, and evaporated in vacuo. Compound 4c was further purified by silica gel chromatography (2% methanol/CH2Cl2) to yield amorphous light yellow solid (1.1 g, 2.1 mmol, quantitative yield). 1H NMR (400 MHz, CDCl3) δ: 7.74 (1H, br), 7.40–7.42 (3H, m), 7.23–7.31 (6H, m), 6.82(4H, d, J = 6.8), 5.75 (1H, br), 4.19–4.24 (2H, m), 4.11 (1H, m), 3.77 (6H, s), 3.74(1H, dd, J = 14, 6.8), 3.24 (1H, dd, J = 9.6, 5.2), 2.97 (1H, dd, J = 9.6, 6.8), 2.02 (3H, s); 13C NMR (100.5 MHz, CDCl3) δ: 165.5, 158.6, 156.9, 149.1, 144.8, 135.9, 135.8, 130.0, 128.1, 128.0, 127.0, 113.3, 92.5, 91.9, 86.5, 70.2, 69.8, 94.3, 55.3, 4.6; LRMS calcd for [M+H]+ (C31H32N3O5): 526.2342, obsd, 526.2.
Synthesis of S-1-(5-propynyl-4-N-dibutylaminomethylidene-cytosine-1-yl)-3-O-(4,4′-dimethoxytrityl)-2,3-propanediol (5c)
Compound 4c (0.34 g, 0.65 mmol) was dissolved in 7.5 mL of methanol. Dibutylformamide dimethylacetal (0.73 mL, 3.2 mmol) was added drop-wise at room temperature. The reaction mixture was stirred at room temperature for 3 h and was then evaporated in vacuo to afford the crude product as yellow oil. Compound 5c was further purified by silica gel chromatography (1–2% methanol/CH2Cl2) as white foam (0.37 g, 0.56 mmol, 86%). 1H NMR (400 MHz, CDCl3) δ: 8.78 (1H, s), 7.54 (1H,s), 7.42 (2H, d, J = 8.4), 7.20–7.32 (7H, m), 6.83 (4H, d, J = 6.8), 4.30 (1H, d, J = 4.8), 4.24 (1H, dd, J = 2.8, 14.0), 4.08 (1H, m), 3.84 (1H, dd, J = 6.8, 14.0), 3.78 (6H, s), 3.61 (2H, m), 3.34 (2H, t, J = 7.2), 3.27 (1H, dd, J = 5.2, 9.6), 2.97 (1H, dd, J = 6.8, 9.6), 2.02 (3H, s), 1.58–1.69 (4H, m), 1.30–1.41 (4H, m), 0.98 (3H, t, J = 7.2), 0.94 (3H, t, J = 7.6); 13C NMR (100.5 MHz, CDCl3) δ: 171.4, 162.8, 158.6, 158.3, 157.5, 150.1, 144.9, 136.0, 135.8, 130.0, 128.03, 127.99, 126.9, 113.3, 100.3, 89.2, 86.5, 72.6, 70.1, 64.4, 55.3, 54.8, 52.5, 46.1, 31.1, 29.0, 20.2, 19.9, 13.8, 4.6; LRMS calcd for [M+H]+ (C40H49N4O5): 665.3703, obsd, 665.3.
Synthesis of S-1-(5-propynyl-4-N-dibutylaminomethylidene-cytosine-1-yl)-2-O-acetyl-2,3-propanediol (6c)
The procedure to synthesize 6c is similar to that of 5u. The yield of 6c from 0.2 g of 5c (0.3 mmol) was 0.11 g (0.24 mmol, 80%). 1H NMR (400 MHz, CDCl3) δ: 8.83 (1H, s), 7.50 (1H, s), 5.03 (1H, m), 4.21 (1H, dd, J = 4, 14.4), 4.07 (1H, dd, J = 5.2, 14.4), 3.63 (1H, dd, J = 4, 12.4), 3.53 (1H, dd, J = 6.4, 12.4), 3.47 (2H, m), 3.18 (2H, d, J = 7.6), 2.11 (3H, s), 2.05 (3H,s), 0.95 (6H, d, J = 6.4), 0.93 (6H, d, J = 6.8); 13C NMR (100.5 MHz, CDCl3) δ: 171.5, 170.2, 159.1, 157.0, 148.8, 101.5, 89.8, 72.7, 72.3, 60.8, 59.5, 54.3, 48.5, 27.5, 26.6, 21.1, 20.4, 20.0, 4.6. LRMS calcd for [M+H]+ (C21H33N4O4): 405.2496, obsd, 405.1.
Synthesis of S-1-(5-propynylcytosine-1-yl)-3-O-triphospho-2,3-propanediol (1c)
The procedure to synthesize 1c is similar to that of 1u. The crude product of 1c after concentrated ammonia deprotection was first purified by anion-exchange liquid chromatography using DEAE A25 resin with a gradient of 0–0.5 M triethylammonium bicarbonate. The fraction containing 1c was combined, evaporated in vacuo, and desalted using the preparative RP-HPLC as described for 1u (retention time 8.5 min). The final yield of 1c was 22.3 µmol (11%) based on the UV absorption (ε295 nm: 7.7 mM−1 cm−1). 1H NMR (400 MHz, D2O) δ: 7.85 (1H, s), 4.10 (2H, m), 4.01 (1H, m), 3.77 (1H, dd, J = 10, 14.4), 3.52 (1H, dd, J = 7.6, 14.4), 2.02 (3H, s). 31P NMR (161.8 MHz, D2O) δ: −5.34 (d, J = 21.5), −9.75 (d, J = 19.1), −21.5 (t, J = 21.1). LRMS calcd for [M-H]− (C10H15N3O12P3): 461.9874, obsd, 462.0.
Enzyme Screen
A DNA primer (5′-TAA TAC GAC TCA CTA TAG GG-3′) was 5′-labeled with 32P and annealed to a DNA template (3′-ATT ATG CTG AGT GAT ATC CCA TAT CAG-5′, the underlined denotes the region base-paired with the primer). The primer extension experiments of GNA synthesis on the DNA template were preformed with 50 nM primer/template, four gNTPs (A, G, T, and C, 100 µM each), 1 U of Thermostable pyrophosphatase, and polymerase (Taq, 2.5 U; Therminator, 1 U; Deep Vent (exo-), 1 U; Bst, 4 U, Sequenase, 6.5 U; Superscript II, 100 U; HIV-1 reverse transcriptase, 5 U) in a final volume of 10 µL containing appropriate buffers supplied by the manufactures. The reaction mixtures were incubated at 55°C for thermophilic enzymes or 37°C for mesophilic enzymes. At 1, 8, and 24 h, an aliquot of 2 µL was removed from the reaction mixture and was analyzed by 20% denaturing polyacrylamide gel electrophoresis.
MALDI-TOF Mass Spectrometry
The single-nucleotide extension reaction mixture (50 µL) contained 20 µM pre-annealed primer/template, 250 µM appropriate dNTP or gNTP, 1 U of thermostable pyrophosphatase, and 1 U of Therminator DNA polymerase. The reactions mixtures were incubated at 55°C for 5 minutes for dNTPs or 8 h for gNTPs. The reaction mixtures were then precipitated by adding 200 µL of ethanol, 50 µL of 2 M ammonium acetate, and 0.5 µL of 3 mg/mL glycogen solution. The samples were redissolved in 0.2 M triethlyammonium acetate buffer (pH 7.0) and were absorbed on C18 Zip Tips (Millipore). Samples were eluted with 1.5 µL of a matrix solution containing a 2∶1 mixture of 52.5 mg/mL 3-hydroxypicolinic acid in 50% acetontrile and 0.1 M ammonium citrate in water. Eluents were directly spotted onto a stainless steel MALDI-TOF plate and were analyzed in positive mode on a Voyager MALDI-TOF mass spectrometer (Applied Biosystems) with an average of 200 scans.
Steady-State Kinetics
Kinetic measurements were performed as previously described using 5′-[32P]-labeled primer and the appropriate template for each dNTP or gNTP [10], [28]. The single nucleotide extension reactions were initiated by mixing 10 µL of 2× dNTP or gNTP solution with 10 µL of a solution containing the remaining reaction components. The final reaction mixture of 20 µL contained 1 µM primer/template, 0.05–10 µM dNTP or 5–500 µM gNTP, 0.5 U of thermostable pyrophosphatase, and 8.2 nM Therminator in 1× Thermopol buffer (20 mM Tris-HCl, 10 mM KCl, 10 mM (NH4)2SO4, 20 mM MgSO4, 0.1% Triton X-100, 100 µM DTT, pH 8.8). The reaction mixtures were incubated at 55°C. At each time point, an aliquot of 4 µL was removed from the mixture and was quenched by mixing with 4 µL of solution containing 8 M urea, 100 mM EDTA, and 0.05% xylene cyanol and bromophenol blue. Single nucleotide extension was analyzed by 20% denaturing polyacrylamide gel electrophoresis and quantified using a PhosphorImager (Molecular Dynamics) as described before [10].
Acknowledgments
The authors thank Dr. D. Treco, Dr. A. Ricardo, and Dr. J. Ichida for helpful discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: JJC was supported by a fellowship from the Harvard Origins of Life Initiative. This work was supported by NSF grants CHE 0434507 to JWS and MCB 0451488 to LWM. JWS is an Investigator of the Howard Hughes Medical Institute and CHT is a Research Associate of the Howard Hughes Medical Institute. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Benner SA. Understanding nucleic acids using synthetic chemistry. Acc Chem Res. 2004;37:784–797. doi: 10.1021/ar040004z. [DOI] [PubMed] [Google Scholar]
- 2.Eschenmoser A. Chemical etiology of nucleic acid structure. Science. 1999;284:2118–2124. doi: 10.1126/science.284.5423.2118. [DOI] [PubMed] [Google Scholar]
- 3.Schoning K, Scholz P, Guntha S, Wu X, Krishnamurthy R, et al. Chemical etiology of nucleic acid structure: the alpha-threofuranosyl-(3′→2′) oligonucleotide system. Science. 2000;290:1347–1351. doi: 10.1126/science.290.5495.1347. [DOI] [PubMed] [Google Scholar]
- 4.Zhang L, Meggers E. An extremely stable and orthogonal DNA base pair with a simplified three-carbon backbone. J Am Chem Soc. 2005;127:74–75. doi: 10.1021/ja043904j. [DOI] [PubMed] [Google Scholar]
- 5.Zhang L, Peritz A, Meggers E. A simple glycol nucleic acid. J Am Chem Soc. 2005;127:4174–4175. doi: 10.1021/ja042564z. [DOI] [PubMed] [Google Scholar]
- 6.Tsai C-H, Chen J, Szostak JW. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc Natl Acad Sci U S A. 2007;104:14598–14603. doi: 10.1073/pnas.0704211104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang YW, Zhang S, McCullum EO, Chaput JC. Experimental evidence that GNA and TNA were not sequential polymers in the prebiotic evolution of RNA. J Mol Evol. 2007;65:289–295. doi: 10.1007/s00239-007-9017-9. [DOI] [PubMed] [Google Scholar]
- 8.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu Rev Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 9.Horhota AT, Szostak JW, McLaughlin LW. Glycerol nucleoside triphosphates: synthesis and polymerase substrate activities. Org Lett. 2006;8:5345–5347. doi: 10.1021/ol062232u. [DOI] [PubMed] [Google Scholar]
- 10.Horhota A, Zou K, Ichida JK, Yu B, McLaughlin LW, et al. Kinetic analysis of an efficient DNA-dependent TNA polymerase. J Am Chem Soc. 2005;127:7427–7434. doi: 10.1021/ja0428255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Froehler BC, Wadwani S, Terhorst TJ, Gerrard SR. Oligodeoxynucleotides containing C-5 propyne analogs of 2′-deoxyuridine and 2′-deoxycytidine. Tetrahedron Lett. 1992;33:5307–5310. [Google Scholar]
- 12.Hobbs FW. Palladium-catalyzed synthesis of alkynylamino nucleosides—a Universal linker for nucleic acids. J Org Chem. 1989;54:3420–3422. [Google Scholar]
- 13.Robins MJ, Barr PJ. Nucleic-Acid Related-Compounds. 39. Efficient conversion of 5-Iodo to 5-Alkynyl and derived 5-substituted uracil bases and nucleosides. J Org Chem. 1983;48:1854–1862. [Google Scholar]
- 14.Froehler BC, Matteucci MD. Dialkylformamidines—Depurination resistant N-6-protecting group for deoxyadenosine. Nucleic Acids Res. 1983;11:8031–8036. doi: 10.1093/nar/11.22.8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seela F, Budow S, Leonard P. Oligonucleotides forming an i-motif: the pH-dependent assembly of individual strands and branched structures containing 2′-deoxy-5-propynylcytidine. Org Biomol Chem. 2007;5:1858–1872. doi: 10.1039/b704583b. [DOI] [PubMed] [Google Scholar]
- 16.Ludwig J, Eckstein F. Rapid and efficient synthesis of nucleoside 5′-O-(1-thiotriphosphates), 5′-triphosphates and 2′, 3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one. J Org Chem. 1989;54:631–635. [Google Scholar]
- 17.Chaput JC, Szostak JW. TNA synthesis by DNA polymerases. J Am Chem Soc. 2003;125:9274–9275. doi: 10.1021/ja035917n. [DOI] [PubMed] [Google Scholar]
- 18.Heuberger BD, Switzer C. A pre-RNA candidate revisited: Both enantiomers of flexible nucleoside triphosphates are DNA polymerase substrates. J Am Chem Soc. 2008;130:412–413. doi: 10.1021/ja0770680. [DOI] [PubMed] [Google Scholar]
- 19.Kempeneers V, Renders M, Froeyen M, Herdewijn P. Investigation of the DNA-dependent cyclohexenyl nucleic acid polymerization and the cyclohexenyl nucleic acid-dependent DNA polymerization. Nucleic Acids Res. 2005;33:3828–3836. doi: 10.1093/nar/gki695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuwahara M, Obika S, Nagashima J, Ohta Y, Suto Y, et al. Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2′, 4′-bridged nucleosides. Nucleic Acids Res. 2008;36:4257–4265. doi: 10.1093/nar/gkn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakthivel K, Barbas CF. Expanding the potential of DNA for binding and catalysis: Highly functionalized dUTP derivatives that are substrates for thermostable DNA polymerases. Angew Chem-Int Ed. 1998;37:2872–2875. doi: 10.1002/(SICI)1521-3773(19981102)37:20<2872::AID-ANIE2872>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 22.Sawai H, Ozaki AN, Satoh F, Ohbayashi T, Masud MM, et al. Expansion of structural and functional diversities of DNA using new 5-substituted deoxyuridine derivatives by PCR with superthermophilic KOD Dash DNA polymerase. Chem Commun. 2001:2604–2605. [Google Scholar]
- 23.Wang JX, Shaw BR. Synthesis of 5-(1-propynyl)-2′-deoxyuridine 5′-(alpha-p-borano)triphosphate and kinetic characterization as a substrate for MMLV reverse transcriptase. Nucleosides Nucleotides Nucleic Acids. 2005;24:947–950. doi: 10.1081/ncn-200059307. [DOI] [PubMed] [Google Scholar]
- 24.Kuwahara M, Nagashima J, Hasegawa M, Tamura T, Kitagata R, et al. Systematic characterization of 2′-deoxynucleoside-5′-triphosphate analogs as substrates for DNA polymerases by polymerase chain reaction and kinetic studies on enzymatic production of modified DNA. Nucleic Acids Res. 2006;34:5383–5394. doi: 10.1093/nar/gkl637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Seela F. Propynyl groups in duplex DNA: stability of base pairs incorporating 7-substituted 8-aza-7-deazapurines or 5-substituted pyrimidines. Nucleic Acids Res. 2002;30:5485–5496. doi: 10.1093/nar/gkf689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schlegel MK, Essen LO, Meggers E. Duplex structure of a minimal nucleic acid. J Am Chem Soc. 2008;130:8158–8159. doi: 10.1021/ja802788g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L, Peritz AE, Carrikk PJ, Meggers E. Synthesis of glycol nucleic acids. Synthesis. 2005;4:645–653. [Google Scholar]
- 28.Boosalis MS, Petruska J, Goodman MF. DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J Biol Chem. 1987;262:14689–14696. [PubMed] [Google Scholar]