Abstract
Kaposi’s sarcoma-associated herpesvirus (KSHV) ORF57 (MTA, mRNA transcript accumulation) is a multifunctional regulator of the expression of viral lytic genes. KSHV ORF57 is expressed during viral lytic infection and is essential for virus production. Like its homologues in the herpesvirus family, ORF57 promotes the accumulation (stabilization) and export of viral intronless RNA transcripts by a mechanism which remains to be defined. The ORF57-Aly/REF interaction plays only a small role in viral RNA export. Although other members of the family generally inhibit the splicing of cellular RNAs, KSHV ORF57 and EBV EB2, in sharp contrast, stimulate viral RNA splicing for the expression of viral intron-containing genes. The functions of KSHV ORF57 are independent of transcription and of other viral proteins; instead, these functions always rely on cellular components and occur in various protein-RNA complexes. ORF57 may synergize with KSHV ORF50 to transactivate a subset of viral promoters by an unknown mechanism. Thus, some functions of ORF57 have been conserved while others have diverged from its homologues as ORF57 adapted over evolution to KSHV biology and pathogenesis.
Keywords: Kaposi’s sarcoma-associated herpesvirus, Gene expression, ORF57, RNA splicing, Post-transcriptional regulation, Protein-RNA interaction, RNA export, Review
2. INTRODUCTION
Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV-8), is a human gammaherpesvirus initially discovered in AIDS-related Kaposi’s sarcoma (KS) (1). Epidemiological studies later showed that KSHV is indispensable for the development of all forms of KS (2) and other lymphotropic malignancies, including primary effusion lymphoma (PEL), also known as body cavity-based B-cell lymphoma, and multicentric Castleman’s disease (3,4).
KSHV, like other herpesviruses, has a large DNA genome (about 165 kb) with the capacity to encode up to 90 different viral products (5). During infection, KSHV undergoes two distinguishable viral life cycles. The latent infection is characterized by restricted expression of only a few virus genes, which play important roles in segregating and maintaining the viral episomal genome (6,7). Viral lytic infection features a massive, time-regulated cascade of lytic gene expression, leading to viral genome replication and assembly of infectious virions. The viral lytic cycle can be initiated in latently infected cells by various chemicals or by exposing the latently infected cells to stress (8-11).
Like other herpesviruses, KSHV replicates in the nucleus, where a virus-encoded transactivator, ORF50 (Rta, replication and transcription activator), initiates transcription of viral genes from responsive viral promoters. However, viral transcripts have to be processed before they can be used for protein translation. This includes RNA 5′ capping, splicing, polyadenylation, and export. Incorrectly processed transcripts are recognized by cellular RNA surveillance and eliminated. To facilitate this process, the herpesvirus family has evolved specific posttranscriptional regulatory proteins. KSHV ORF57 is one of these regulatory proteins.
ORF57 is essential for the expression of many KSHV genes (12,13) and affects almost every step of viral gene expression. However, the major function of ORF57 is regulating RNA processing at the posttranscriptional level. ORF57 promotes the expression of KSHV intronless genes, independently of transcription, by promoting the accumulation and presumably nuclear export of their transcripts (14-18). In contrast to its homologues, which have an inhibitory effect on RNA splicing, ORF57 surprisingly stimulates intron removal and the expression of several viral intron-containing RNAs (12). Moreover, KSHV ORF57 functions as a transcriptional activator or co-activator of several viral genes (15,19,20), depending on experimental conditions. This review will summarize our current knowledge of KSHV ORF57.
3. ORF57 GENE STRUCTURE AND EXPRESSION
3.1. ORF57 gene structure
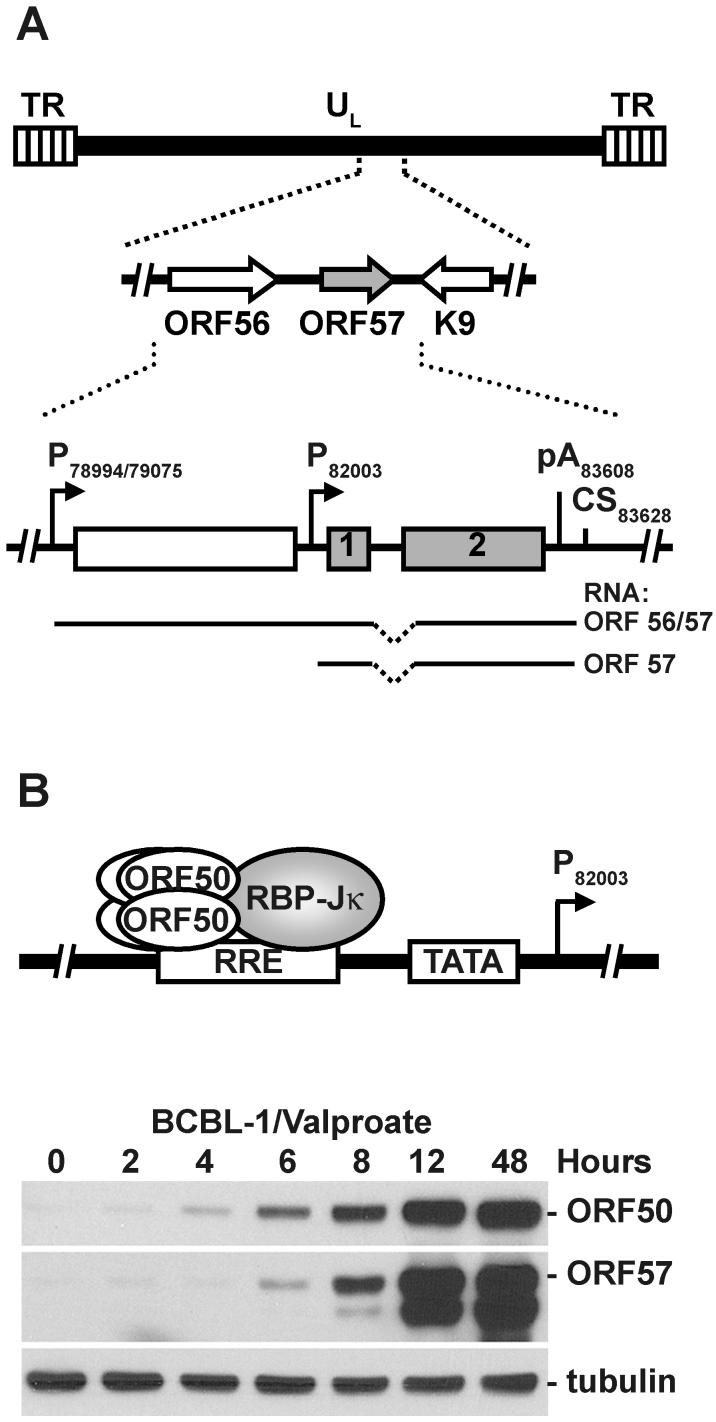
The gene that encodes ORF57 is positioned in a unique long region of the KSHV genome and is flanked by the ORF56 (primase) and K9 (viral interferon regulatory factor [vIRF]) genes (Figure 1A). The ORF56 and ORF57 ORFs lie side by side in the same orientation on the sense strand of the virus genome; each has its own promoter to initiate its transcription, but they share the same polyadenylation signal downstream of the ORF57 ORF. Thus, the ORF57 transcript is monocistronic, whereas the ORF56 transcript is bicistronic, with ORF57 as its 3′ untranslated region. In contrast, the K9 gene is positioned in the opposite direction to ORF57 on the antisense strand of the KSHV genome.
Figure 1.
Gene structure and expression of KSHV ORF57. (A) Schematic diagram of ORF57 gene structure in the context of the KSHV genome. The KSHV genome consists of a unique long region (UL, heavy line) flanked by a terminal repeat (TR, striped boxes) on each end. KSHV ORF57 (grey arrow in the insert) is located between the ORF56 and K9 genes (empty arrows). ORF56 and ORF57 are each transcribed independently from separate promoters (arrows with P, designated by the position of the transcription start site in the virus genome), but share the same polyadenylation signal (pA) and cleavage site (CS) for polyadenylation. The open boxes represent the ORF of each gene. The ORF57 ORF is divided into two exons (numbers in the boxes) by a small intron (dashed line). The intron is removed from both monocistronic ORF57 and bicitronic ORF56/57 pre-mRNAs (heavy lines) by RNA splicing. The numbers are nucleotide positions in the KSHV genome (GenBank accession number U75698). (B) Kinetic production of KSHV ORF50 and ORF57 in BCBL-1 cells during lytic induction. ORF50 (also known as Rta), a major viral transactivator that functions as a multimer, transactivates ORF57 expression by direct binding to specific DNA sequences in the ORF57 promotor (RRE, Rta-response element) in association with cellular recombination signal binding protein-Jkappa (RBP-Jkappa). Western blot analysis (lower panel) shows the kinetics of ORF50 and ORF57 expression in KSHV latently infected B-cells (BCBL-1) after induction of viral lytic replication by valproic acid. ORF50 appears several hours earlier than ORF57 after the lytic induction. Tubulin was used as a loading control.
KSHV ORF57 is a homologue of herpes simplex virus type 1 (HSV-1) ICP27 (21,22) that is highly conserved among all members of the herpesvirus family. Other members in the family are cytomegalovirus (CMV) UL69 (23), varicella-zoster virus (VZV) ORF4 (24), Epstein-Barr virus (EBV) EB2 (EB2 or SM) (25,26), and herpesvirus saimiri (HVS) ORF57 (27-29). The ORF of KSHV ORF57 was initially predicted to encode a protein of 275 amino acid (aa) residues, with homology to the EBV SM protein (5), in fact, the protein is 51 kDa and is composed of 455 aa residues. The ORF of ORF57 spans two exons that are interrupted by a small intron (14-16). ORF57 initiates its transcription from its promoter with a canonical TATA box (15), and its gene transcription produces an RNA transcript of ∼1.6 kb. The RNA 5′ exon or exon 1 is relatively small, 114 nts, and has four potential initiation codons of which all are in frame. The first ATG, which is in an optimal Kozak context in the 5′ exon, is used for translation of full-length 455-aa ORF57 protein, of which the first 17 aa residues are translated from the 5′ exon. The intron is 109 nt in size and contains consensus splice donor and acceptor sites and utilizes two suboptimal branch points for splicing (30) due to the suboptimal features of the polypyrimidine tract. The 3′ exon or exon 2 is about 1.4 kb long and encodes the remaining 438 aa residues of ORF57. ORF57 polyadenylation is governed by a consensus polyadenylation signal 64 nts downstream of the stop codon, and the transcripts are cleaved at several cleavage sites nearby, but mostly at nt 83628. The observed heterogeneity in the polyadenylation cleavage site is most likely related to the suboptimal content of the downstream GU/U-rich region, which binds cellular cleavage stimulatory factors (CSF) and defines the efficiency of transcript cleavage (30). KSHV ORF57 homologs with a similar gene structure have been found in HVS (27), EBV (25) and murine gammaherpesvirus 68 (MHV-68) (31), but not in other herpesviruses.
3.2. Kinetics of ORF57 expression and localization
ORF57 appears in lytically infected cells very early (Figure 1B) after virus reactivation from latency (19,32,33) and its expression can be blocked by cycloheximide. Therefore, KSHV ORF57 is classified as an early gene. ORF57 expression is highly dependent on KSHV ORF50 (34). KSHV ORF50 or Rta is a viral immediate early gene that controls the expression of several viral early lytic genes by binding to Rta-responsive elements (RRE) in their promoter regions (35-38). Although an RRE in the ORF57 promoter is responsible for ORF50 binding (38,39), ORF50 itself is unable to bind this element and is not sufficient to initiate ORF57 transcription (35). Instead, ORF57 transactivation requires ORF50 in association with the cellular recombination signal binding protein-Jkappa (RBP-Jkappa), a member of the Notch signaling pathway (35).
Expressed ORF57 is found predominantly in the nucleus and has a characteristic punctuate pattern that resembles splicing speckles and colocalizes with the cellular splicing factor SC35 (12,14,17,33). The nuclear localization of ORF57 is driven by three independent nuclear localization signals (NLSs) that form a cluster in the N-terminus (17). Only simultaneous mutation of all three NLSs can prevent localization of ORF57 into the nucleus, although it is not clear why ORF57 needs three NLSs with redundant function. One explanation could be that each has different affinity for different members of the importin family, as was reported for HVS ORF57 (40). This would secure proper localization of ORF57 in the nucleus during viral infection. Multiple NLSs also exist in other homologous proteins, including EBV EB2 (41) and ICP27 (42,43).
KSHV ORF57 shuttles between the nucleus and the cytoplasm (14). The nuclear export of ORF57 homologues is mediated by a leucine-rich nuclear export signal through an interaction with the cellular export factor CRM-1 (44,45). Whether a leucine-rich region in the C-terminus of ORF57 may be responsible for ORF57 export from the nucleus remains to be determined. Overexpression of CRM-1 does not cause an accumulation of KSHV ORF57 in the cytoplasm, as has been observed for other homologues (16), indicating that KSHV ORF57 has a different mechanism of nuclear export than its homologues.
KSHV ORF57 may contain a potential nucleolus-targeting signal (NoLS), KRPR, which overlaps with NLS2. Since only very small fraction of ORF57 is associated with nucleoli in lytically infected PEL cells, the nucleolar localization of ORF57 might be not relevant to virus multiplication or pathogenesis (12). NoLSs have been identified in ORF57 homologues (42,46).
4. ORF57 PROTEIN STRUCTURE AND PUTATIVE MOTIFS
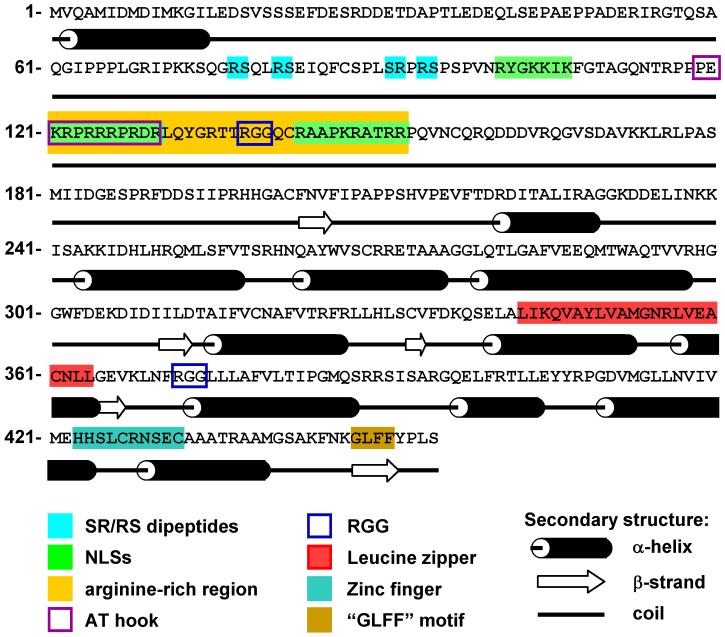
The individual members of the ICP27 family differ in size, and their overall sequence homology is relatively low. KSHV ORF57, EBV EB2, and HVS ORF57 are the three closest members in the family (Figure 2), but share only approximately 30% homology/identity of aa residues. KSHV ORF57 doesn’t have any significant sequence homology to other viral or cellular proteins. Analyses of the ORF57 aa sequence have revealed several putative structural and functional motifs (Figure 3). The N-terminus of KSHV ORF57 contains a long stretch of arginine residues that is similar to arginine or arginine/glycine runs found in other homologues. ORF57 also contains two separate RGG-motifs, which are typical of RNA-binding proteins, and four serine/arginine or arginine/serine dipeptides that are characteristic of SR proteins, the major cellular splicing factors. The three independent NLSs overlap the arginine-rich region in the N-terminus of KSHV ORF57. The regions enriched in arginine residues in other members of the family also bear nuclear and nucleolar localization signals and are involved in RNA-binding (41,47). In KSHV ORF57, each NLS is represented by a short stretch of basic amino acids, mostly arginine and lysine residues (17). In addition to their nuclear localization function, the identified ORF57 NLSs are important in other ORF57 functions, as mutation of individual NLSs significantly decreases the ability of ORF57 to promote the expression of its target gene KSHV ORF59 despite its proper localization in the nucleus. These mutations in KSHV ORF57 prevent the association of ORF57 with ORF59 RNA (17). The N-terminus of KSHV ORF57 might contain an A/T hook motif that overlaps NLS2 (20); this motif is commonly found in chromosomal DNA binding proteins (48).
Figure 2.
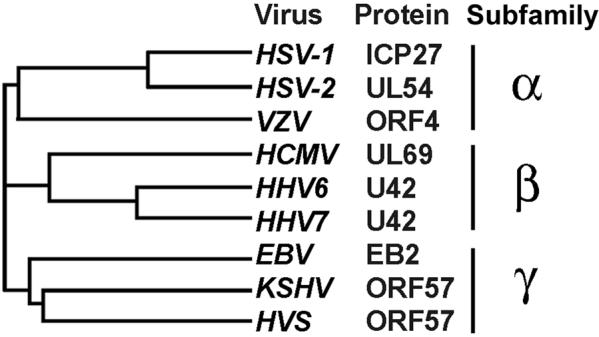
KSHV ORF57 homologues and their phylogenetic relationships. The amino acid sequences of HSV-1 ICP27, HSV-2 UL54, VZV ORF4, CMV UL69, human herpesvirus 6 (HHV6) U42, human herpesvirus 7 (HHV7) U42, EBV EB2, KSHV ORF57, and HVS ORF57 proteins were aligned, and the dendrogram was generated by Clustal W software (www.ebi.ac.uk/clustalw/). The lines represent phylogenetic relationships and evolutionary distance among the homologues.
Figure 3.
Putative secondary structure and functional motifs of KSHV ORF57. The secondary structure displayed under the amino acid sequence of ORF57 was predicted using PSIPRED V2.6 software (http://bioinf.cs.ucl.ac.uk/psipred/) (83). The cylinders represent alpha-helixes, empty arrows indicate beta-sheets, and heavy lines are coiled regions. KSHV ORF57 consists of two structurally distinct regions: the mostly unstructured N-terminal part and the highly structured C-terminal part containing multiple alpha-helixes. Functional motifs (colored) identified or predicted in KSHV ORF57 are serine-arginine and arginine-serine (SR/RS) dipeptides, three nuclear localization signals (NLSs), arginine-rich region, AT hook, arginine-glycine-glycine (RGG) motifs, leucine zipper, zinc finger, and hydrophobic “GLFF” motif.
The C-terminus of KSHV ORF57 is enriched in leucine residues and contains a putative leucine zipper motif, a structure typical of cellular transcription factors. The putative leucine-zipper motif appears to be unique to KSHV ORF57 and is not present in other homologues. Although ORF57 has been shown to homodimerize, the potential leucine zipper in ORF57 is not required for homodimerization and is not critical for ORF57 activity (18). It remains to be shown whether a functional leucine zipper can be formed in vivo for protein-protein interaction. The C-terminal of ORF57 also contains the zinc-finger-like motif H-x-C-x-C, which is conserved in all homologues. Although in ICP27 this motif binds zinc, it is not clear of the relevance of the zinc binding to ICP27 function (49). A hydrophobic “GLFF motif”, characteristic only of ORF57 homologues in gammaherpesviruses, is located at the very end of the C-terminus of ORF57. This motif in other family members is required for transactivation and repression activities (28,50-52), but deletion of the C-terminal half of KSHV ORF57 has only a minimal effect on ORF57 function. In contrast, truncation of KSHV ORF57 from the N-terminus reduces the expression of ORF57-dependent genes, indicating that the domains important for ORF57 are mainly localized at the N-terminal half (17).
KSHV ORF57 protein is phosphorylated in vivo (Majerciak and Zheng, unpublished data). Cellular casein kinase II (CK II or CK2) was proposed to phosphorylate ORF57, because KSHV ORF57 directly interacts with two subunits of CK II (53). However, the phosphorylation site (s) of KSHV ORF57, other kinases involved in the phosphorylation, and the effect of phosphorylation on ORF57 function remain to be determined. ICP27 and EB2 also interact with and are phosphorylated by CK II (54,55) although mutation of several phosphorylation sites in HSV ICP27 did not affect the replication of a mutant virus (56), CK II phosphorylation of EB2 does regulate the production of EBV virus (55).
There are no structural data available for ORF57 or any of its homologs. The secondary structure predicted by several computer programs for ORF57 consists of two distinguishable regions (Figure 3). The N-terminal part represents a surprisingly long unstructured region (approximately aa 1-220) containing only one alpha-helix on the far N-terminus. A large number of cellular proteins without any significant structure in a large (>50 aa) region have recently been predicted and experimentally confirmed. These “intrinsically unstructured proteins” function in many cellular processes, including regulation of transcription and translation, signal transduction, protein phosphorylation, small molecule storage, and assembly of large multiprotein complexes. They also function as chaperones for other proteins or RNA molecules and their defined structure can be obtained only after binding to a ligand (57). In contrast, the C-terminus of KSHV ORF57 is composed of multiple alpha-helixes (10 to 12, depending on the prediction algorithm) and a few beta-sheets. Interestingly, all ORF57 homologues exhibit a similar predicted secondary structure (Majerciak and Zheng, unpublished data). One can speculate that the C-terminal region carries out functions highly conserved in all homologues, whereas the N-terminus deviates within the family to serve the specific needs of each member. One example could be the nuclear localization and nuclear exports signals that are mostly localized in the unstructured region, and whose numbers, positions, and compositions vary greatly among the members in the family.
5. ORF57 IN POSTTRANSCRIPTIONAL REGULATION OF VIRAL GENE EXPRESSION
5.1. ORF57 in expression of viral intronless genes
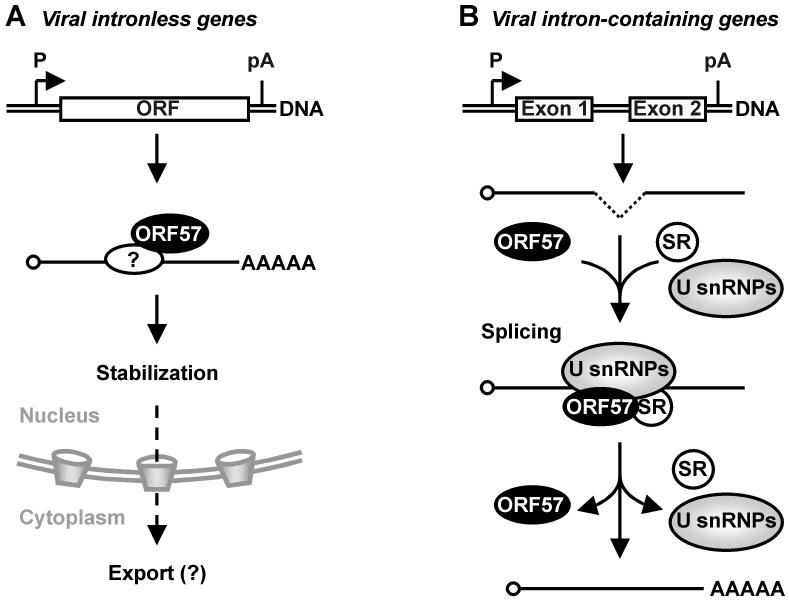
KSHV ORF57 promotes the expression of KSHV intronless genes (Figure 4A). These include several viral early and late genes, including KSHV ORF59 (15,17), PAN (13,15,58), gB, MPC (18) and others. Other KSHV intronless genes, like vGCR or K5, show no dependence on ORF57 expression (15), indicating that ORF57’s effect on the expression of viral intronless genes is selective. However, we do not yet have a complete list of intronless genes affected by ORF57 expression. The effect of ORF57 on the expression of intronless genes is posttranscriptional and independent of the viral promoter activity and polyadenylation (15-17). ORF57 binds the Aly/REF protein, which has a major role in RNA export and was thought to promote the nuclear export of viral RNAs in a CRM1-independent manner (59). Whether ORF57 can function to promote viral RNA export remains largely unknown. However, RNA export has been proposed as a major function of ORF57 by analogy to its homologues from other members in the family.
Figure 4.
Posttranscriptional regulation of viral gene expression by KSHV ORF57. (A) ORF57 upregulates the expression of several viral intronless genes. The double line represents the virus genome, P with an arrow indicates a promoter/transcription start site, open box is the coding region or open reading frame (ORF), and pA stands for polyadenylation signal. ORF57 binds to intronless transcripts with a 5′ end cap (open circle) and a 3′ end polyadenylated tail by interacting with unknown cellular factor(s). ORF57 promotes the nuclear accumulation of regulated transcripts presumably by increasing the RNA stability, and in some cases may promote their export from the nucleus. (B) KSHV ORF57 also functions as a viral splicing factor to promote the expression of viral intron-containing transcripts. ORF57 stimulates splicing of primary RNA transcripts (heavy line) containing a suboptimal intron (dashed line) by interacting with components of the cellular splicing machinery, such as SR proteins (serine/arginine-rich proteins) and U snRNPs (uridine-rich small nuclear ribonucleoprotein particles).
Early studies suggested that KSHV ORF57 could mediate the nuclear export of reporter RNAs. In these studies, the chloramphenicol acetyltransferase (CAT) ORF was inserted into the intron region of the pCMV 128 reporter plasmid (59) or downstream of an intron region in a CMV-CAT reporter (16). Because the pre-mRNA transcribed from the pCMV 128 plasmid was inefficiently spliced and a pre-mRNA containing an intron in general would not be exported to the cytoplasm, the ORF57-enhanced CAT activity was interpreted as a result of ORF57-mediated mRNA export (59), because ORF57 also interacts with Aly/REF. However, recent studies in our lab and others indicate that the effect of ORF57 on the expression of viral intronless early genes may be independent of Aly/REF. KSHV ORF59 is an early gene encoding a viral DNA polymerase processivity factor involved in viral DNA replication, and its expression strongly relies on ORF57. The three NLSs of ORF57 are required for ORF59 expression and Aly/REF binding. This enhancement of ORF59 expression by ORF57 is due to the interaction of ORF57 protein with ORF59 RNA, leading to the accumulation of both cytoplasmic and nuclear ORF59 transcripts, independent of Aly/REF. Deletion of the Aly/REF binding domain in ORF57 or knockdown of Aly/REF expression with an siRNA has no effect on the ORF57 enhancement of ORF59 expression (17), indicating that ORF57 possibly stabilizes ORF59 RNA. This observation is consistent with the notion that Aly/REF is dispensable for RNA export in Caenorhabditis elegans (60,61) and Drosophila (62).
Additional evidence of a role for ORF57 in the stabilization of ORF59 comes from studies on polyadenylated nuclear (PAN) RNA in our laboratory and others (13,15,58). KSHV PAN RNA (nut-1, T1.1) is a non-coding RNA with unknown function. PAN accumulates to an unusually high level (∼2.5 × 105 copies) in the nucleus of lytically infected cells. ORF57 enhancement of PAN RNA expression is independent of transcription; it was initially noticed following transient cotransfection (15) and was subsequently confirmed in the context of the KSHV genome (13,58). In either circumstance, the steady-state level of PAN RNA was greatly reduced in the absence of ORF57 in viral lytic induction. Because PAN RNA is exclusively nuclear and cannot be exported to the cytoplasm (63), it is not sensitive to mRNA export regulation. These data strongly imply that KSHV ORF57 promotes the expression of viral intronless early genes by preventing the transcripts from degradation mediated by the cellular RNA surveillance machinery.
5.2. ORF57 is a viral splicing factor that regulates viral RNA splicing
One of the hallmarks of herpesvirus lytic infection is the repression of host gene expression, referred as “virus host shut-off” (VHS) (64,65). ICP27 contributes to VHS by inhibiting RNA splicing (66,67), leading to destabilization of the unspliced cellular RNAs (68,69). A similar inhibitory effect on RNA splicing has been reported for homologous proteins, including EBV EB2 and HVS ORF57 (26,27). However, the suppression of RNA splicing by KSHV ORF57 would be harmful for KSHV gene expression and virus replication, because the KSHV genome encodes more than 30 intron-containing genes that require active splicing during their expression (70).
An investigation of whether KSHV ORF57 has any effect on the expression of intron-containing genes thus resulted in the surprising conclusion that KSHV ORF57 does not suppress the expression of most intron-containing artificial genes (15), a conclusion that contrasts sharply with the concept established from many years of work on KSHV ORF57 homologs in other members of the family. Through systemic analyses of the expression and processing of viral tricistronic ORF50, bicistronic K8, and monocistronic K8.1 pre-mRNAs, which all contain multiple introns, we discovered that KSHV ORF57 functions as a viral splicing factor that promotes viral K8 RNA splicing and the production of a K-bZIP (K8alpha) protein (Figure 4B) (12). This KSHV ORF57 activity for promoting viral K8 RNA splicing is conserved in EBV EB2, but absent in HSV ICP27. By disrupting ORF57 in the virus genome, we also demonstrated a significant reduction of ORF50, K8, and K8.1 expression and accumulation of unspliced ORF50 and K8 pre-mRNAs in lytically induced cells with stable infection of the ORF57-null genome, indicating a defect in the viral RNA splicing of both ORF50 and K8 pre-mRNAs (12). In KSHV-infected cells with viral lytic replication, ORF57 preferentially interacts with intron-containing viral pre-mRNAs, which are in low abundance relative to their corresponding spliced products. This was confirmed by in vitro splicing reactions followed by pulldown assays, in which ORF57 (most likely its N-terminal NLS2 and NLS3 regions) selectively interacted with the intron of the pre-mRNA only in the presence of other cellular proteins. The promotion of RNA splicing by ORF57 is independent of other viral factors and occurs with other viral and nonviral RNAs that undergo inefficient splicing (12).
ORF57 promotes viral RNA splicing by actively associating with multiple splicing components in the spliceosomal complexes in which RNA splicing takes place. KSHV ORF57 associates with the small nuclear RNAs (snRNA) U1, U2, U4, U5, and U6, all of which are essential components of spliceosomes, in isolated ORF57-RNA complexes during KSHV lytic infection or when ORF57 is overexpressed by transient transfection. This association of ORF57 with snRNAs is mediated through an ORF57-Sm protein interaction (12). In addition, KSHV ORF57 interacts with the cellular splicing factors ASF/SF2 and U2AF, two essential factors for bridging 5′ and 3′ splice sites over the intron for splice site recognition by U1 and U2 snRNAs and intron removal. Since ORF57 does not behave like a classical SR protein in in vitro splicing assays, its role in RNA splicing of mediating protein-protein interactions with various cellular splicing factors would be mainly auxiliary. However, the exact function of ORF57 in each step of spliceosome-mediated RNA splicing remains to be investigated.
6. POSSIBLE ROLE OF ORF57 IN VIRAL TRANSCRIPTION
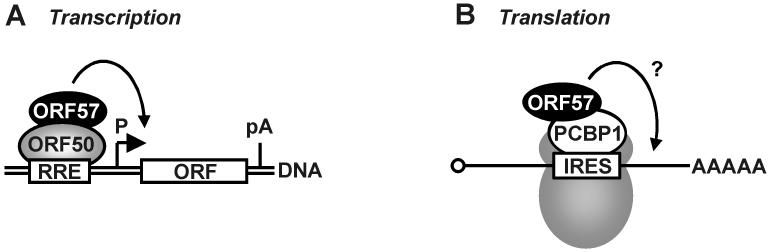
KSHV ORF57 may transactivate specific viral promoters in synergy with KSHV ORF50, a major transactivator in the initiation of viral lytic gene expression (15,19,20). This synergistic effect has been observed in vitro in analyses of viral promoters for ORF50 (19), ORF57 (20), PAN/nut-1, Kaposin (15), ori-Lyt (L) (20), K-bZIP (20), and TK (15,20), but not in promoters that do not respond to ORF50, such as the ISRE and SV40 promoters (19). Among the promoters examined, promoters for PAN/nut-1, ori-Lyt (L), and Kaposin are most responsive (20).
Although the expression of KSHV ORF57 is activated shortly after ORF50 expression in lytic viral induction, the expressed ORF57 may further enhance ORF50-mediated gene expression from other RRE-containing promoters (15). This synergistic effect of ORF57 on viral transcription depends on binding of the ORF50 transactivation domain to the RRE in the promoter (Figure 5A). Interestingly, the co-activation mediated by the ORF50/57 interaction appears to be promoter- or cell line-specific; not all promoters containing an RRE respond to ORF57 co-stimulation at the same level (20). ORF57 interacts with ORF50 (19,20,71) via its N-terminal region (aa 17-215) (19) and the central region (aa 246-484) of ORF50 (20). The ORF50/57 complex is associates with the target promoter in lytically infected cells (19). Although the mechanism by which ORF57 promotes ORF50 activity remains largely unknown, the observation that ORF57 enhanced ORF50 protein levels by 3-fold in a cotransfection assay (15) might imply a posttranscriptional effect of ORF57 on ORF50 expression. In addition, the level of ORF50 expression can be elevated in the presence of ORF57 in the context of the virus genome (12).
Figure 5.
Other possible roles of KSHV ORF57 in the regulation of viral gene expression. (A) KSHV ORF57 enhances RNA transcription in synergy with a major viral transactivator ORF50 (or Rta). The synergy is dependent on ORF50 binding to a regulatory DNA element, RRE (Rta response element), in the promotor region of affected viral genes. See other details in Figure 4A. (B) Possible involvement of ORF57 in protein translation. KSHV ORF57 was found in association with cellular poly-rC binding protein 1 (PCBP1), which might be involved in IRES (internal ribosome entry site)-initiated translation.
7. POSSIBLE ROLE OF ORF57 IN PROTEIN TRANSLATION
Using a two-hybrid system, Nishimura et al. (33) identified poly (rC)-binding protein 1 (PCBP1, also called hnRNP E1 or alphaCP1) as a protein that interacts and colocalizes with ORF57. This interaction takes place through a PCBP1-binding region of ORF57 (aa 179-205) and an ORF57-binding site of PCBP1 (aa 48-96). PCBP1 is a 356-aa protein and has many functions, possibly including the regulation of alpha-globin mRNA stability (72,73) and the activation and silencing of protein translation (74). Because cotransfection of ORF57 and PCBP1 or transfection of ORF57 alone enhances IRES-mediated translation activity (Figure 5B), and the cotransfection also doubled the level of reporter RNAs, exactly how the proposed ORF57-PCBP1 interaction regulates IRES-mediated protein translation remains to be understood. Interestingly, ICP27 was recently found to associate with polyribosomes and stimulate mRNA translation (75).
8. ORF57 AND RNA INTERACTIONS
KSHV ORF57 is an RNA-binding protein and binds RNAs specifically in the presence of cellular proteins (17). KSHV ORF57 contains an arginine-rich region (aa 122-130) that resembles RNA-binding motifs (ARMs) described in other members of the family, including EBV EB2, CMV UL69, and HVS ORF57 (41,76). Motifs similar to ARMs are present in other viral proteins, such as HIV-1 Tat and Rev or lambda phage N protein (77-79). KSHV ORF57, like its homologues, interacts with RNAs (17,18), but direct binding of ORF57 to an RNA lacks substrate specificity in the absence of cellular extracts (Figure 6) (17), indicating that specific interactions between ORF57 and its RNA targets only occurs in the context of protein-RNA complexes. In KSHV lytic infection, ORF57 is recruited into at least three forms of RNA-protein complexes. For intron-containing viral transcripts, ORF57 assembles into spliceosomal complexes in preferential association with low-abundance viral pre-mRNAs and essential splicing components to facilitate viral RNA splicing. For intronless viral transcripts, ORF57 interacts with RNAs to form a protein-RNA complex that protects the targets from RNA degradation. ORF57 also interacts with RNA export factors and is present in RNA export complexes, but its role in RNA export has not been well characterized. In all three cases, ORF57 interacts with other RNA-binding proteins (see below), some of which are present in all three complexes. Although ORF57 is in close contact with its targeted RNA and can be cross-linked to the bound RNA with ultraviolet light, the RNA sequence for ORF57 interaction has not been identified.
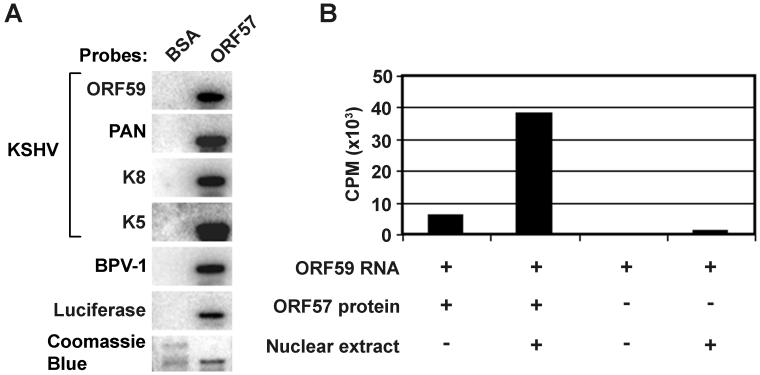
Figure 6.
KSHV ORF57-RNA interaction. (A) Purified recombinant KSHV ORF57 binds nonspecifically to all RNAs analyzed by Northwestern analysis. Baculovirus-expressed FLAG-tagged ORF57 protein was separated in a 12% SDS-PAGE gel, transferred onto a nitrocellulose membrane, renatured, and probed with 32P-labeled individual RNAs as described (76,84). No meaningful difference in RNA binding to ORF57 was observed among those RNAs analyzed, including three ORF57-regulated RNAs (ORF59, PAN, and K8) and three ORF57-nonresponsive RNAs (KSHV K5, bovine papillomavirus type 1 (BPV-1) late, and luciferase RNAs). Purified bovine serum albumin (BSA), which does not bind any RNA, served as a negative protein control. (B) The effect of cellular proteins on the association of KSHV ORF57 with target RNA. The radiolabeled RNA probe was prepared by in vitro transcription from a KSHV ORF59 DNA template. The RNA probe was incubated with baculovirus-expressed ORF57-FLAG protein immobilized on anti-FLAG beads in the presence or absence of HEK-293 cell nuclear extracts. After washes, the amount of bound RNAs was measured with a scintillation counter and expressed as counts per minute (CPM). The amount of ORF59 RNAs associated with ORF57 proteins dramatically increased in the presence of cellular proteins (nuclear extracts) when compared with the RNA-protein binding in the absence of nuclear extracts (Modified with permission from reference (17).
The KSHV ORF57 ARM overlaps with well characterized NLSs. Point mutations in the NLS sequences of ORF57 have a dramatic effect on its RNA binding affinity and on the accumulation of targeted transcripts (17). This is in contrast with the observation that an arginine-rich RNA-binding region of HCMV UL69 is not critical for UL69 function (76).
9. PROTEIN PARTNERS OF ORF57
KSHV ORF57 is a “sticky” protein that interacts with many other proteins to perform the various functions described above. With the exception of the transcriptional synergy with ORF50, ORF57 is more dependent on cellular factors than on viral proteins for its function. This is not surprising, because ORF57 is one of the few viral genes expressed at the early phase of the KSHV lytic cycle. A genome-wide two-hybrid screening of protein-protein interactions between KSHV viral proteins confirmed the binding of ORF57 to KSHV ORF50 protein (71). Some additional interactions of ORF57 with KSHV proteins, including KSHV K-bZIP and others, were also identified (Table 1) (71). Whether these interactions take place in infected cells or have any role in ORF57 function remains to be determined.
Table 1.
Protein partners of KSHV ORF57
| Protein | Function | Reference |
|---|---|---|
| Viral (KSHV) | ||
| ORF57 | (self-interaction) | (53,71) |
| ORF50 | viral transactivator | (19,20,71) |
| K-bZIP (K8) | viral transactivator | (71) |
| ORF23 | tegument protein | (71) |
| ORF61 | ribonucleotide reductase | (71) |
| ORF68 | tegument protein | (71) |
| Cellular | ||
| Aly/REF | RNA export (?) | (17,59) |
| hnRNP K | transcription, RNA processing | (53) |
| CKII alpha/alpha’ | protein kinase | (53) |
| CK II beta | protein kinase | (53) |
| PCBP1 | RNA stability, translation | (33) |
| SR proteins | RNA splicing | (12) |
| Sm proteins | RNA splicing | (12) |
| U snRNPs | RNA splicing | (12) |
| U2AF35 | RNA splicing | (12) |
| ASF/SF2 | RNA splicing | (12) |
The proteins on the list directly interacted or were associated with KSHV ORF57 in a two-hybrid system or protein pull-down and co-immunoprecipitation assays.
ORF57 binds to many cellular proteins, including Aly/REF, CK II, hnRNP K, PCBP1, ASF/SF2, U2AF, Sm, and other SR members (Table 1) (12,33,53,59). As mentioned above, ORF57 binds Aly/REF directly in vitro and forms an ORF57-Aly/REF complex in cells (17,59). This interaction was thought to recruit TAP/NFX1, another important member of the RNA nuclear export network, for the export of viral intronless RNAs (59). However, our lab and others recently demonstrated that the ORF57-Aly/REF interaction plays a little role in regulating viral gene expression (17,18). Both the N- and C-terminal parts of ORF57 directly interact with hypophosphorylated hnRNP K. A proline-rich motif in the central part of hnRNP K binds RNA to regulate RNA transcription, processing, and translation. By interacting with various proteins, hnRNP K might act as a docking platform to facilitate the interactions between these proteins to regulate gene expression and signal transduction (80). ORF57-hnRNP K complexes from infected cells also contain CK II, which has substantial kinase activity (53) ORF57 binds CK II through a direct interaction between its C-terminal part and both the N- and C-terminal CK II alpha/alpha’ (catalytic) subunits, and an interaction between its central region and the C-terminal CK II beta (regulatory) subunit (53). KSHV ORF57 has multiple phosphorylation sites, including a cluster of potential CK II phosphorylation sites at the ORF57 N-terminus (Majerciak and Zheng, unpublished data), and can be phosphorylated by CK II in vitro (53). Since ORF57 phosphorylation in infected cells is not very sensitive to the CK II-specific inhibitor DRB (53), other protein kinases must be also involved in ORF57 phosphorylation.
Other protein partners of ORF57 and their possible roles have been discussed separately in related sections. Because ORF57 is a component of various complexes in viral lytic infection, it is conceivable that many more proteins will be identified in association with ORF57 in the near future.
10. REMARKS AND PERSPECTIVES
KSHV ORF57 is a structural and functional homologue of other proteins in the herpesvirus family. Like its homologues, ORF57 is essential for viral replication and retains several evolutionarily conserved functions that promote viral gene expression during lytic infection (13,18). However, ORF57 differs in many aspects from other members in the family. One major difference is its enhancement of splicing, which was discovered in our laboratory (12). This function is conserved in EBV EB2, but deviates greatly from the prototype family member, ICP27. The latter exercises a VHS function by suppressing cellular RNA splicing. This VHS function in KSHV lytic infection is performed by another viral protein, KSHV ORF37 (SOX) (65,81); similarly, in EBV it is performed by BGLF5 (82). Other important differences are the interaction of ORF57 with Aly/REF and the way it binds RNA. In contrast to many reports, Aly/REF binding plays little role in KSHV ORF57 function (17) and ORF57 does not bind its target RNAs specifically in the absence of cellular nuclear factors (12,17). KSHV ORF57 and EBV EB2 are particularly close members of the family, but although KSHV ORF57 functions to stimulate viral K8 RNA splicing and KSHV ORF59 expression can be replaced by EBV EB2 (12,58), the two proteins are unable to substitute for each other in rescuing virus production.
Although we have learned a great deal in the past 10 years about KSHV ORF57 and its functions, the molecular mechanisms of KSHV ORF57 and its homologues in the regulation of viral gene expression remain largely unknown. Further studies should address several important questions: First, how many viral genes does KSHV ORF57 regulate? There are eight viral genes known to be under ORF57 regulation, including immediate-early (ORF50), early (ORF56, ORF59, PAN/nut-1, and K8), and late (gB, MCP, and K8.1) genes, but two viral genes, K5 and vGCR, do not respond to ORF57 expression. Thus, construction of a genome-wide ORF57 targeting map would definitely clarify how ORF57 targeting might take place in virus lytic infection. One worthwhile notion is that the expression of KHSV latent genes is independent of ORF57 during viral latent infection, but continues into viral lytic infection. How does ORF57 distinguish different transcripts to selectively regulate the lytic but not latent transcripts? The same question also applies to cellular transcripts during viral lytic infection. If ORF57 has no effect on cellular transcripts, how do these transcripts escape from ORF57 recognition? Second, how many protein partners are involved in ORF57-mediated gene expression and which of them are key players for defining ORF57 function? Although ORF57 promotes the expression of viral genes independently of other viral proteins, its function is facilitated by cellular factors. To date, more than 10 cellular proteins (Table 1) have been identified as ORF57 partners. How do these factors work with ORF57 and direct an ORF57-RNA complex to a specific pathway? Third, what role does ORF57 play in viral RNA export? Is ORF57-mediated viral RNA export a separate function of ORF57 or a consequence of ORF57-mediated RNA stability or RNA splicing? A careful study to dissect the individual events could be challenging because RNA transcription, processing, and export are coupled events for all eukaryotic RNAs.
Although expression of KSHV ORF57 in viral lytic infection is activated by KSHV ORF50, ORF57 expression and function may also be subject to autoregulaton (feedback) at the posttranscriptional level. A functional ORF57 may require multiple modifications after protein translation. For example, KSHV ORF57 is a phosphoprotein, and its full function relies on phosphorylation. Understanding what controls the posttranscriptional and posttranslational regulation of KSHV ORF57 will definitely contribute to our understanding of the KSHV life cycle. Together, the answers to all of the questions raised above will not only elucidate the roles of KSHV ORF57 in viral infection and carcinogenesis, but could also shed light on the regulation of RNA processing and gene expression more generally.
11. ACKNOWLEDGMENTS
This work was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health. We thank Ren Sun for providing the gene structure information of MHV-68 ORF57.
12. REFERENCES
- 1.Chang Y, Cesarman E, Pessin MS, Lee F, Culpepper J, Knowles DM, Moore PS. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 2.Schulz TF. Kaposi’s sarcoma-associated herpesvirus (human herpesvirus-8) J.Gen.Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 3.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N.Engl.J.Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 4.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d’Agay MF, Clauvel JP, Raphael M, Degos L, Sigaux F. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 5.Russo JJ, Bohenzky RA, Chien MC, Chen J, Yan M, Maddalena D, Parry JP, Peruzzi D, Edelman IS, Chang Y, Moore PS. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc.Natl.Acad.Sci.U.S.A. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc.Natl.Acad.Sci.U.S.A. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fakhari FD, Dittmer DP. Charting latency transcripts in Kaposi’s sarcoma-associated herpesvirus by whole-genome real-time quantitative PCR. J.Virol. 2002;76:6213–6223. doi: 10.1128/JVI.76.12.6213-6223.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis DA, Rinderknecht AS, Zoeteweij JP, Aoki Y, Read-Connole EL, Tosato G, Blauvelt A, Yarchoan R. Hypoxia induces lytic replication of Kaposi sarcoma-associated herpesvirus. Blood. 2001;97:3244–3250. doi: 10.1182/blood.v97.10.3244. [DOI] [PubMed] [Google Scholar]
- 9.Klass CM, Krug LT, Pozharskaya VP, Offermann MK. The targeting of primary effusion lymphoma cells for apoptosis by inducing lytic replication of human herpesvirus 8 while blocking virus production. Blood. 2005;105:4028–4034. doi: 10.1182/blood-2004-09-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller G, Rigsby MO, Heston L, Grogan E, Sun R, Metroka C, Levy JA, Gao SJ, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi’s sarcoma-associated herpesvirus in patients with HIV-1 infection. N.Engl.J.Med. 1996;334:1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- 11.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat.Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 12.Majerciak V, Yamanegi K, Allemand E, Kruhlak M, Krainer AR, Zheng ZM. Kaposi sarcoma-associated herpesvirus ORF57 functions as a viral splicing factor and promotes the expression of intron-containing viral lytic genes in spliceosome-mediated RNA splicing. J.Virol. 2008;82:2792–2801. doi: 10.1128/JVI.01856-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. Targeted disruption of Kaposi’s sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J.Virol. 2007;81:1062–1071. doi: 10.1128/JVI.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bello LJ, Davison AJ, Glenn MA, Whitehouse A, Rethmeier N, Schulz TF, Clements JB. The human herpesvirus-8 ORF 57 gene and its properties. J.Gen.Virol. 1999;80:3207–3215. doi: 10.1099/0022-1317-80-12-3207. [DOI] [PubMed] [Google Scholar]
- 15.Kirshner JR, Lukac DM, Chang J, Ganem D. Kaposi’s sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J.Virol. 2000;74:3586–3597. doi: 10.1128/jvi.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta AK, Ruvolo V, Patterson C, Swaminathan S. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J.Virol. 2000;74:1038–1044. doi: 10.1128/jvi.74.2.1038-1044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Majerciak V, Yamanegi K, Nie SH, Zheng ZM. Structural and functional analyses of Kaposi sarcoma-associated herpesvirus ORF57 nuclear localization signals in living cells. J.Biol.Chem. 2006;281:28365–28378. doi: 10.1074/jbc.M603095200. [DOI] [PubMed] [Google Scholar]
- 18.Nekorchuk M, Han Z, Hsieh TT, Swaminathan S. Kaposi’s Sarcoma-Associated Herpesvirus ORF57 Protein Enhances mRNA Accumulation Independently of Effects on Nuclear RNA Export. J.Virol. 2007;81:9990–9998. doi: 10.1128/JVI.00896-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malik P, Blackbourn DJ, Cheng MF, Hayward GS, Clements JB. Functional co-operation between the Kaposi’s sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J.Gen.Virol. 2004;85:2155–2166. doi: 10.1099/vir.0.79784-0. [DOI] [PubMed] [Google Scholar]
- 20.Palmeri D, Spadavecchia S, Carroll KD, Lukac DM. Promoter- and cell-specific transcriptional transactivation by the Kaposi’s sarcoma-associated herpesvirus ORF57/Mta protein. J.Virol. 2007;81:13299–13314. doi: 10.1128/JVI.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandri-Goldin RM, Hibbard MK, Hardwicke MA. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J.Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sandri-Goldin RM, Hibbard MK. The herpes simplex virus type 1 regulatory protein ICP27 coimmunoprecipitates with anti-Sm antiserum, and the C terminus appears to be required for this interaction. J.Virol. 1996;70:108–118. doi: 10.1128/jvi.70.1.108-118.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler M, Rice SA, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J.Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Defechereux P, Melen L, Baudoux L, Merville-Louis M-P, Rentier B, Piette J. Characterization of the regulatory functions of varicella-zoster virus open reading frame 4 gene product. J. Virol. 1993;67:4379–4385. doi: 10.1128/jvi.67.7.4379-4385.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cook ID, Shanahan F, Farrell PJ. Epstein-Barr virus SM protein. Virology. 1994;205:217–227. doi: 10.1006/viro.1994.1637. [DOI] [PubMed] [Google Scholar]
- 26.Ruvolo V, Wang E, Boyle S, Swaminathan S. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc.Natl.Acad.Sci.U.S.A. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Whitehouse A, Cooper M, Meredith DM. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J.Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper M, Goodwin DJ, Hall KT, Stevenson AJ, Meredith DM, Markham AF, Whitehouse A. The gene product encoded by ORF 57 of herpesvirus saimiri regulates the redistribution of the splicing factor SC-35. J.Gen.Virol. 1999;80:1311–1316. doi: 10.1099/0022-1317-80-5-1311. [DOI] [PubMed] [Google Scholar]
- 29.Goodwin DJ, Hall KT, Stevenson AJ, Markham AF, Whitehouse A. The open reading frame 57 gene product of herpesvirus saimiri shuttles between the nucleus and cytoplasm and is involved in viral RNA nuclear export. J.Virol. 1999;73:10519–10524. doi: 10.1128/jvi.73.12.10519-10524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majerciak V, Yamanegi K, Zheng ZM. Gene structure and expression of Kaposi’s sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J.Virol. 2006;80:11968–11981. doi: 10.1128/JVI.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackett M, Stewart JP, de VP, Chee M, Efstathiou S, Nash AA, Arrand JR. Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J.Gen.Virol. 1997;78:1425–1433. doi: 10.1099/0022-1317-78-6-1425. [DOI] [PubMed] [Google Scholar]
- 32.Lukac DM, Kirshner JR, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi’s sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J.Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura K, Ueda K, Guwanan E, Sakakibara S, Do E, Osaki E, Yada K, Okuno T, Yamanishi K. A posttranscriptional regulator of Kaposi’s sarcoma-associated herpesvirus interacts with RNA-binding protein PCBP1 and controls gene expression through the IRES. Virology. 2004;325:364–378. doi: 10.1016/j.virol.2004.04.041. [DOI] [PubMed] [Google Scholar]
- 34.Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi’s sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 35.Chang PJ, Shedd D, Miller G. Two subclasses of Kaposi’s sarcoma-associated herpesvirus lytic cycle promoters distinguished by open reading frame 50 mutant proteins that are deficient in binding to DNA. J.Virol. 2005;79:8750–8763. doi: 10.1128/JVI.79.14.8750-8763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deng H, Song MJ, Chu JT, Sun R. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi’s sarcoma-associated herpesvirus) J.Virol. 2002;76:8252–8264. doi: 10.1128/JVI.76.16.8252-8264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song MJ, Brown HJ, Wu TT, Sun R. Transcription activation of polyadenylated nuclear rna by rta in human herpesvirus 8/Kaposi’s sarcoma-associated herpesvirus. J.Virol. 2001;75:3129–3140. doi: 10.1128/JVI.75.7.3129-3140.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukac DM, Garibyan L, Kirshner JR, Palmeri D, Ganem D. DNA binding by Kaposi’s sarcoma-associated herpesvirus lytic switch protein is necessary for transcriptional activation of two viral delayed early promoters. J.Virol. 2001;75:6786–6799. doi: 10.1128/JVI.75.15.6786-6799.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Song MJ, Deng H, Sun R. Comparative study of regulation of RTA-responsive genes in Kaposi’s sarcoma-associated herpesvirus/human herpesvirus 8. J.Virol. 2003;77:9451–9462. doi: 10.1128/JVI.77.17.9451-9462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodwin DJ, Whitehouse A. A gamma-2 herpesvirus nucleocytoplasmic shuttle protein interacts with importin alpha 1 and alpha 5. J.Biol.Chem. 2001;276:19905–19912. doi: 10.1074/jbc.M009513200. [DOI] [PubMed] [Google Scholar]
- 41.Hiriart E, Bardouillet L, Manet E, Gruffat H, Penin F, Montserret R, Farjot G, Sergeant A. A region of the Epstein-Barr virus (EBV) mRNA export factor EB2 containing an arginine-rich motif mediates direct binding to RNA. J.Biol.Chem. 2003;278:37790–37798. doi: 10.1074/jbc.M305925200. [DOI] [PubMed] [Google Scholar]
- 42.Mears WE, Lam V, Rice SA. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J.Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hibbard MK, Sandri-Goldin RM. Arginine-rich regions succeeding the nuclear localization region of the herpes simplex virus type 1 regulatory protein ICP27 are required for efficient nuclear localization and late gene expression. J.Virol. 1995;69:4656–4667. doi: 10.1128/jvi.69.8.4656-4667.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boyle SM, Ruvolo V, Gupta AK, Swaminathan S. Association with the cellular export receptor CRM 1 mediates function and intracellular localization of Epstein-Barr virus SM protein, a regulator of gene expression. J.Virol. 1999;73:6872–6881. doi: 10.1128/jvi.73.8.6872-6881.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sandri-Goldin RM. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boyne JR, Whitehouse A. Nucleolar trafficking is essential for nuclear export of intronless herpesvirus mRNA. Proc.Natl.Acad.Sci.U.S.A. 2006;103:15190–15195. doi: 10.1073/pnas.0604890103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruvolo V, Gupta AK, Swaminathan S. Epstein-Barr virus SM protein interacts with mRNA in vivo and mediates a gene-specific increase in cytoplasmic mRNA. J.Virol. 2001;75:6033–6041. doi: 10.1128/JVI.75.13.6033-6041.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aravind L, Landsman D. AT-hook motifs identified in a wide variety of DNA-binding proteins. Nucleic Acids Res. 1998;26:4413–4421. doi: 10.1093/nar/26.19.4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vaughan PJ, Thibault KJ, Hardwicke MA, Sandri-Goldin RM. The herpes simplex virus immediate early protein ICP27 encodes a potential metal binding domain and binds zinc in vitro. Virology. 1992;189:377–384. doi: 10.1016/0042-6822(92)90720-a. [DOI] [PubMed] [Google Scholar]
- 50.Hardwicke MA, Vaughan PJ, Sekulovich RE, O’Conner R, Sandri-Goldin RM. The regions important for the activator and repressor functions of herpes simplex virus type 1 alpha protein ICP27 map to the C-terminal half of the molecule. J.Virol. 1989;63:4590–4602. doi: 10.1128/jvi.63.11.4590-4602.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice SA, Knipe DM. Gene-specific transactivation by herpes simplex virus type 1 alpha protein ICP27. J.Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruvolo V, Sun L, Howard K, Sung S, Delecluse HJ, Hammerschmidt W, Swaminathan S. Functional analysis of Epstein-Barr virus SM protein: identification of amino acids essential for structure, transactivation, splicing inhibition, and virion production. J.Virol. 2004;78:340–352. doi: 10.1128/JVI.78.1.340-352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malik P, Clements JB. Protein kinase CK2 phosphorylation regulates the interaction of Kaposi’s sarcoma-associated herpesvirus regulatory protein ORF57 with its multifunctional partner hnRNP K. Nucleic Acids Res. 2004;32:5553–5569. doi: 10.1093/nar/gkh876. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 54.Wadd S, Bryant H, Filhol O, Scott JE, Hsieh TY, Everett RD, Clements JB. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J.Biol.Chem. 1999;274:28991–28998. doi: 10.1074/jbc.274.41.28991. [DOI] [PubMed] [Google Scholar]
- 55.Medina-Palazon C, Gruffat H, Mure F, Filhol O, Vingtdeux-Didier V, Drobecq H, Cochet C, Sergeant N, Sergeant A, Manet E. Protein kinase CK2 phosphorylation of EB2 regulates its function in the production of Epstein-Barr virus infectious viral particles. J.Virol. 2007;81:11850–11860. doi: 10.1128/JVI.01421-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhi Y, Sandri-Goldin RM. Analysis of the phosphorylation sites of herpes simplex virus type 1 regulatory protein ICP27. J.Virol. 1999;73:3246–3257. doi: 10.1128/jvi.73.4.3246-3257.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat.Rev.Mol.Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 58.Han Z, Swaminathan S. Kaposi’s sarcoma-associated herpesvirus lytic gene ORF57 is essential for infectious virion production. J.Virol. 2006;80:5251–5260. doi: 10.1128/JVI.02570-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malik P, Blackbourn DJ, Clements JB. The evolutionarily conserved Kaposi’s sarcoma-associated herpesvirus ORF57 protein interacts with REF protein and acts as an RNA export factor. J.Biol.Chem. 2004;279:33001–33011. doi: 10.1074/jbc.M313008200. [DOI] [PubMed] [Google Scholar]
- 60.Longman D, Johnstone IL, Caceres JF. The Ref/Aly proteins are dispensable for mRNA export and development in Caenorhabditis elegans. RNA. 2003;9:881–891. doi: 10.1261/rna.5420503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.MacMorris M, Brocker C, Blumenthal T. UAP56 levels affect viability and mRNA export in Caenorhabditis elegans. RNA. 2003;9:847–857. doi: 10.1261/rna.5480803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gatfield D, Izaurralde E. REF1/Aly and the additional exon junction complex proteins are dispensable for nuclear mRNA export. J.Cell Biol. 2002;159:579–588. doi: 10.1083/jcb.200207128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sun R, Lin SF, Gradoville L, Miller G. Polyadenylylated nuclear RNA encoded by Kaposi sarcoma-associated herpesvirus. Proc.Natl.Acad.Sci.U.S.A. 1996;93:11883–11888. doi: 10.1073/pnas.93.21.11883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fenwick ML, Clark J. Early and delayed shut-off of host protein synthesis in cells infected with herpes simplex virus. J.Gen.Virol. 1982;61:121–125. doi: 10.1099/0022-1317-61-1-121. [DOI] [PubMed] [Google Scholar]
- 65.Glaunsinger B, Chavez L, Ganem D. The exonuclease and host shutoff functions of the SOX protein of Kaposi’s sarcoma-associated herpesvirus are genetically separable. J.Virol. 2005;79:7396–7401. doi: 10.1128/JVI.79.12.7396-7401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bryant HE, Wadd SE, Lamond AI, Silverstein SJ, Clements JB. Herpes simplex virus IE63 (ICP27) protein interacts with spliceosome-associated protein 145 and inhibits splicing prior to the first catalytic step. J.Virol. 2001;75:4376–4385. doi: 10.1128/JVI.75.9.4376-4385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sciabica KS, Dai QJ, Sandri-Goldin RM. ICP27 interacts with SRPK1 to mediate HSV splicing inhibition by altering SR protein phosphorylation. EMBO J. 2003;22:1608–1619. doi: 10.1093/emboj/cdg166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hardwicke MA, Sandri-Goldin RM. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J.Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Phelan A, Dunlop J, Clements JB. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J.Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zheng ZM. Split genes and their expression in Kaposi’s sarcoma-associated herpesvirus. Rev.Med.Virol. 2003;13:173–184. doi: 10.1002/rmv.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Uetz P, Dong YA, Zeretzke C, Atzler C, Baiker A, Berger B, Rajagopala SV, Roupelieva M, Rose D, Fossum E, Haas J. Herpesviral protein networks and their interaction with the human proteome. Science. 2006;311:239–242. doi: 10.1126/science.1116804. [DOI] [PubMed] [Google Scholar]
- 72.Kiledjian M, Wang X, Liebhaber SA. Identification of two KH domain proteins in the alpha-globin mRNP stability complex. EMBO J. 1995;14:4357–4364. doi: 10.1002/j.1460-2075.1995.tb00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ostareck-Lederer A, Ostareck DH, Hentze MW. Cytoplasmic regulatory functions of the KH-domain proteins hnRNPs K and E1/E2. Trends Biochem.Sci. 1998;23:409–411. doi: 10.1016/s0968-0004(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 74.Makeyev AV, Liebhaber SA. The poly (C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002;8:265–278. doi: 10.1017/s1355838202024627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Larralde O, Smith RW, Wilkie GS, Malik P, Gray NK, Clements JB. Direct stimulation of translation by the multifunctional herpesvirus ICP27 protein. J.Virol. 2006;80:1588–1591. doi: 10.1128/JVI.80.3.1588-1591.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Toth Z, Lischka P, Stamminger T. RNA-binding of the human cytomegalovirus transactivator protein UL69, mediated by arginine-rich motifs, is not required for nuclear export of unspliced RNA. Nucleic Acids Res. 2006;34:1237–1249. doi: 10.1093/nar/gkl007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Malim MH, Tiley LS, McCarn DF, Rusche JR, Hauber J, Cullen BR. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell. 1990;60:675–683. doi: 10.1016/0092-8674(90)90670-a. [DOI] [PubMed] [Google Scholar]
- 78.Calnan BJ, Biancalana S, Hudson D, Frankel AD. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991;5:201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- 79.Lazinski D, Grzadzielska E, Das A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell. 1989;59:207–218. doi: 10.1016/0092-8674(89)90882-9. [DOI] [PubMed] [Google Scholar]
- 80.Bomsztyk K, Denisenko O, Ostrowski J. hnRNP K: one protein multiple processes. Bioessays. 2004;26:629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 81.Glaunsinger B, Ganem D. Lytic KSHV infection inhibits host gene expression by accelerating global mRNA turnover. Mol.Cell. 2004;13:713–723. doi: 10.1016/s1097-2765(04)00091-7. [DOI] [PubMed] [Google Scholar]
- 82.Rowe M, Glaunsinger B, van LD, Zuo J, Sweetman D, Ganem D, Middeldorp J, Wiertz EJ, Ressing ME. Host shutoff during productive Epstein-Barr virus infection is mediated by BGLF5 and may contribute to immune evasion. Proc.Natl.Acad.Sci.U.S.A. 2007;104:3366–3371. doi: 10.1073/pnas.0611128104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bryson K, McGuffin LJ, Marsden RL, Ward JJ, Sodhi JS, Jones DT. Protein structure prediction servers at University College London. Nucleic Acids Res. 2005;33:W36–W38. doi: 10.1093/nar/gki410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zimmer C, von Gabain A, Henics T. Analysis of sequence-specific binding of RNA to Hsp70 and its various homologs indicates the involvement of N- and C-terminal interactions. RNA. 2001;7:1628–1637. [PMC free article] [PubMed] [Google Scholar]