Summary
Microsporidia are obligate, intracellular eukaryotic pathogens that infect animal cells, including humans [1]. Previous studies suggested the microsporidia might share a common ancestor with fungi [2–7]. However, the exact nature of this phylogenetic relationship is unclear due to the unusual features of microsporidial genomes, which are compact with a reduced number of highly divergent genes [8]. As a consequence, it is unclear whether microsporidia evolved from a specific fungal lineage, the identity of that lineage, or whether microsporidia are a sister group to all fungi. Here we present evidence to address this controversial question that is independent of sequence-based phylogenetic reconstruction, but rather based on genome structure. In the zygomycete basal fungal lineage, the sex locus is a syntenic gene cluster that governs sexual reproduction in which a high mobility group (HMG) transcription factor gene is flanked by a triose phosphate transporter (TPT) and an RNA helicase gene [9]. Strikingly, microsporidian genomes harbor a sex-related locus with the same genes in the same order (TPT, HMG, RNA helicase). Genome-wide analysis of synteny reveals multiple other loci where microsporidia and zygomycetes are conserved to the exclusion of all other fungal lineages with sequenced genomes. These findings support the hypothesis that microsporidia are true fungi that descended from a zygomycete ancestor, and suggest the microsporidia may have a genetically controlled sexual cycle.
Results and discussion
Microsporidia are unusual eukaryotic obligate intracellular pathogens that infect a wide variety of animals and cell types [1]. The National Institute of Allergy and Infectious Diseases (NIAID) of the NIH includes microsporidia on the list of priority pathogens for biodefense research and the Environmental Protection Agency has also issued a caution concerning these water-borne pathogens. Microsporidia occur globally and infect the human gastrointestinal tract as a common cause of self-limited diarrheal disease in immunocompetent individuals or more chronic disease in immunocompromised hosts. Infections of other tissues, including the eye, the lung and the brain, also commonly occur [10]. Beyond humans, a variety of microsporidian species infect a wide range of animals, especially insects and fish, and in some cases are used for pest control [11, 12]. Infection by microsporidia involves an unusual mechanism in which eversion of the polar tube produces a syringe-like structure that pierces the target cell through which the infectious contents of the spore are rapidly injected into the host [13].
Within microsporidia over 1,200 species in 150 genera have been described [1], however the phylogenetic placement of the group as a whole has been long debated. Based on an apparent absence of mitochondria, they were hypothesized to represent an ancient eukaryotic lineage [14, 15]. However, they are now known to harbor a mitochondrial remnant, the mitosome [16–19]. Phylogenetic studies also now argue against an early origin for microsporidia, suggesting instead that they are either derived from fungi or a sister group to the fungi [4, 6, 7, 20]. However, because the microsporidian genomes that have been studied encode few genes (< 2,000) and the sequences of these genes are highly divergent [15, 21], molecular phylogenetic reconstructions are difficult and notoriously unreliable [6, 8]. Accordingly, to correctly define the relationship between microsporidia and fungi, other kinds of molecular characters may prove more useful than reconstructing phylogenies from divergent gene sequences. In this study we have examined a different kind of data that is not inherently affected by the accelerated rates of substitution common to microsporidian gene sequences. Specifically, we have examined genome-wide synteny and from this data demonstrate that: 1) microsporidia are true fungi, 2) they are specifically related to zygomycetes, and 3) they harbor genetic regulatory components that could function in sex determination and sexual reproduction.
Fungi undergo diverse, genetically programmed sexual development that is governed by the mating type (MAT) or sex locus [22, 23]. MAT loci in some fungi encode homeodomain proteins, and it has been proposed that microsporidian homeodomain genes may be the MAT/sex locus in Encephalitozoon cuniculi [24]. However, these homeodomain gene pairs differ from fungal MAT loci in several ways: they are co-linear or convergent rather than divergently transcribed, of the same functional class (HD2/HD2) or linked but not adjacent, and in no case is there synteny of flanking genes with known fungal MAT loci [25] (Figure 1S). There are reports suggesting some microsporidian species may have extant sexual cycles based solely on morphological observations and inferences [26, 27]. However, there is no molecular or genetic evidence suggesting that microsporidia undergo sexual reproduction, and limited genomic evidence (beyond that noted above for homeodomain gene homologs) for conserved machinery involved in sex in other organisms [28]. Thus, it has been unclear whether microsporidia undergo sexual reproduction and, if so, how it is genetically controlled.
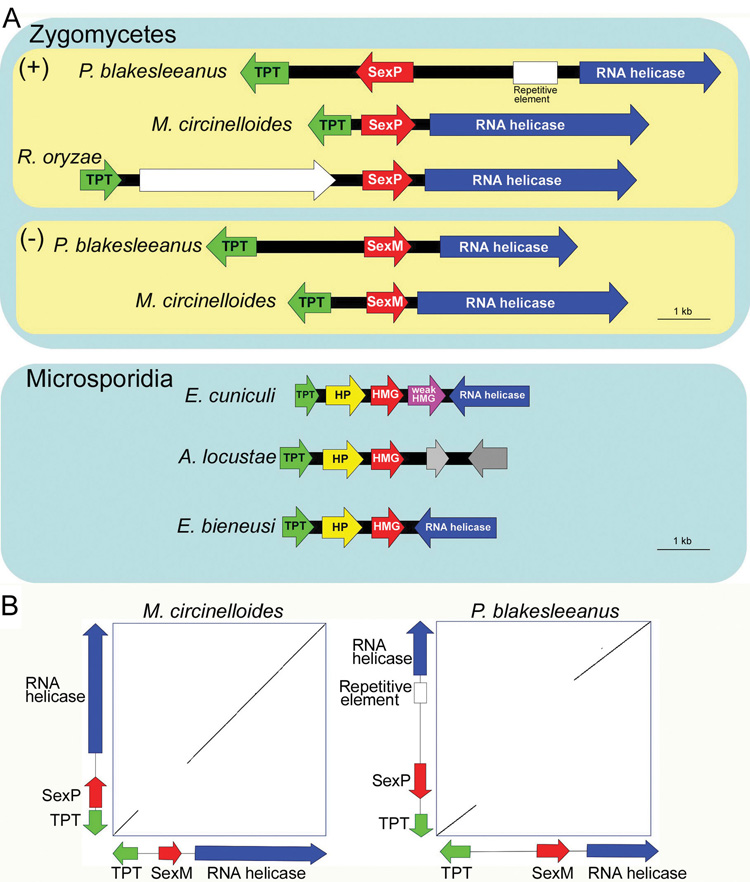
A recent study illustrated that the sex locus of the zygomycete fungi, Phycomyces blakesleeanus and Rhizopus oryzae, represents a different paradigm compared to that of ascomycetes and basidiomycetes. In these species, the sex locus encodes a high mobility group (HMG) domain protein flanked by genes encoding a triose phosphate transporter (TPT) and an RNA helicase [9], forming a microsyntenic genomic locus in zygomycetous fungi (Figure 1A). Plus (+) and minus (−) mating type strains of P. blakesleeanus encode two divergent HMG proteins: SexP and SexM, respectively (Figure 1A). Searching the genome of the zygomycete, Mucor circinelloides (US Department of Energy Joint Genome Institute Mucor genome project) also revealed a sexM homolog in the (−) strain, and this sexM gene is also flanked by TPT and RNA helicase genes (Figure 1A). The corresponding sex locus (+) allele was isolated by PCR amplification and found to contain a sexP gene homolog between TPT and RNA helicase genes (Figure 1A). Two different (+) strains contained sexP and not sexM, whereas four different (−) strains harbored only sexM, consistent with mating type tests (data not shown). In both mating types of M. circinelloides, the sex locus alleles are syntenic with flanking TPT and RNA helicase genes, as in P. blakesleeanus (Figure 1A). In both species, the TPT and RNA helicase genes are highly conserved between the (+) and (−) alleles while the HMG genes (sexP and sexM) and their promoters have diverged (Figure 1B). The promoter regions of the TPT and RNA helicase genes of P. blakesleeanus are conserved in both mating types (Figure 1B), whereas in M. circinelloides, the RNA helicase gene promoter is conserved but the TPT promoter is within the sex locus and diverged (Figure 1B). As a consequence, the TPT genes in the (+) and (−) mating types of M. circinelloides might be differentially regulated during sexual development. The R. oryzae (−) sex locus has not yet been characterized.
Figure 1. The sex locus is conserved between zygomycetes and microsporidia.
(A) Three zygomycetes and three microsporidia harbor a syntenic sex locus encoding a triose phosphate transporter (TPT), HMG domain protein, and an RNA helicase. In zygomycetes, the orientation of sexP of P. blakesleeanus is opposite to the other HMG protein genes and the P. blakesleeanus (+) allele contains a repetitive element. The TPT gene of R. oryzae is inverted with respect to the P. blakesleeanus and M. circinelloides sex alleles and an additional gene encoding a BTB/Ankyrin/RCC1 repeat protein is present. Microsporidia also have an additional hypothetical protein gene in the extended sex locus, and the RNA helicase and TPT genes are inverted with respect to the zygomycetes. Notably, in A. locustae, the RNA helicase gene lies at a different genomic location.
(B) Dot plot analysis of the sex locus alleles of M. circinelloides and P. blakesleeanus illustrates (+) and (−) specific DNA sequences at the sex locus. X axis is the (+) and Y axis the (−) sex alleles. Sequences shown here are 7,103 bp and 6,511 bp for (+) and (−) strains of M. circinelloides and 9,971 bp and 7,631 bp for (+) and (−) strains of P. blakesleeanus. The unique sequences at the sex locus span 1,541 bp and 1,463 bp for (+) and (−) strains of M. circinelloides and 5,830 bp and 3,494 bp for (+) and (−) strains of P. blakesleeanus. The dot plot was performed with a 17 bp window with no mismatches allowed. HP: hypothetical protein.
Comparison of gene order upstream and downstream of the sexM (M. circinelloides and P. blakesleeanus) or sexP (R. oryzae) genes revealed that only the TPT, HMG, and RNA helicase genes are conserved and syntenic among the three zygomycetes (Figure 2S). More importantly, the synteny of these three genes is not conserved in any other sequenced fungal genome, including representative species of the Ascomycota, Basidiomycota, and Chytridiomycota, where the TPT, HMG, and RNA helicase genes are unlinked and randomly distributed in the genomes (Figure 3S). Thus, the extended sex locus [TPT, HMG (SexP/SexM), RNA helicase] is highly specific to the Zygomycota and possibly to the Mucorales, the sub-phylum to which these species all belong.
Remarkably, given the lack of conservation of the extended sex locus in most fungi, we found that a syntenic gene cluster related to the sex locus is conserved in the genomes of three distantly related microsporidia (Figure 1A). In Encephalitozoon cuniculi and Enterocytozoon bieneusi the gene order conservation extends in both directions (with an additional ORF between TPT and the putative sex locus and inversions of flanking genes), whereas in Antonospora locustae the RNA helicase is no longer linked (Figure 1A). The gene order around the sex related locus of both E. cuniculi and A. locustae was verified by PCR and sequencing. Interestingly, in both R. oryzae and the microsporidians the TPT gene is inverted relative to other zygomycetes, and an additional gene has also been acquired between it and the sex locus (Figure 1A). It is possible that independent inversion events introduced these new genes into the extended sex locus. Structural differences of the sex locus observed in zygomycetes and microsporidia suggest alternative hypotheses for the early steps in the evolution of sex determination and sex chromosomes [9] (Figure 4S).
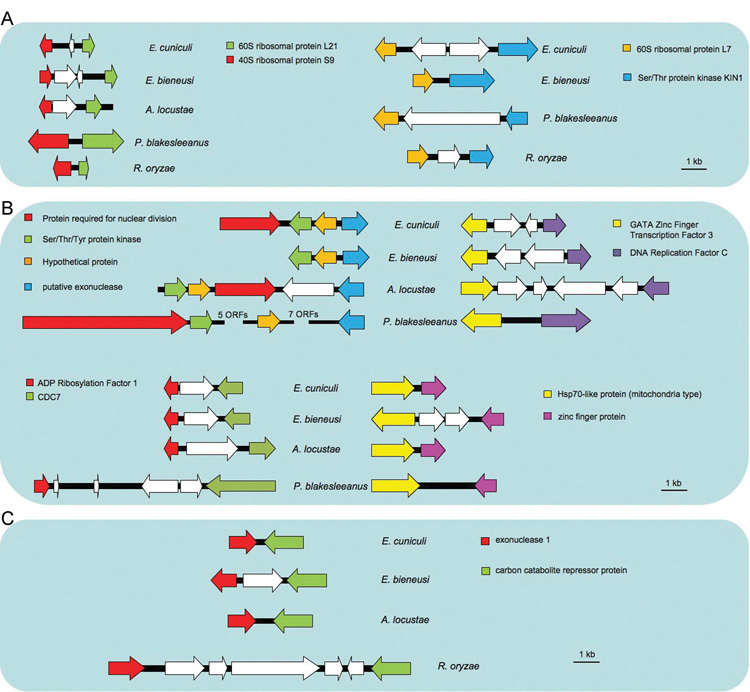
To investigate whether the high degree of genome structure conservation between microsporidia and zygomycetes is restricted to the sex related locus, we also performed whole genome comparisons between fungi and microsporidia. Interestingly, in a relaxed synteny analysis that allows for local changes in gene order or orientation to be accommodated (see experimental procedures and Figure 5S) but still requires a higher level genomic architecture be apparent, microsporidia and the zygomycetes were found to contain a number of conserved pairs or clusters of genes that were not conserved in any other fungal genome (BLAST cut off 1 × e−30) (Figure 2). The number of syntenic gene loci (33 gene pairs or clusters) shared between R. oryzae and E. cuniculi is significant compared to randomized gene orders (17.4 ± 4.4 pairs) (BLAST cut off 1 × e−05) (Figure 5S). On the other hand, by the same analysis, the number of genomic regions exhibiting relaxed synteny that are shared between E. cuniculi and other fungi, including Saccharomyces cerevisiae, Ashbya gossypii, Neurospora crassa, Ustilago maydis, and Cryptococcus neoformans is not significantly different in comparison to the randomized gene orders (data not shown). The only gene pair that was conserved in a broader range of fungi, including the three microsporidia, was RPL21 and RPS9, which is intriguingly conserved across all fungi (Figure 6S) but not in species outside the fungal kingdom, suggesting an ancient functional role for this gene pair within the fungi. The shared, genome-wide preservation of synteny is significant because it is easy to imagine that synteny around the sex locus could be conserved due to functional constraints, and so multiple independent losses of this synteny in other fungal lineages could reflect functional shifts in mating type determination. The striking degree of overall conservation specifically between microsporidia and zygomycetes is not easily explained unless microsporidia and zygomycetes share a recent common ancestor relative to other fungi.
Figure 2. Examples of conserved synteny and relaxed synteny found in zygomycetes and microsporidia.
(A) Conserved synteny among P. blakesleeanus, R. oryzae, and the microsporidia. Syntenic 60S (L21) and 40S (S9) protein genes are conserved throughout fungi (see Supplementary Fig. 6). Another synteny of ribosomal protein and kinase genes is observed in two zygomycete genomes and two microsporidian genomes (E. cuniculi and E. bieneusi).
(B) Conserved synteny between P. blakesleeanus and the three microsporidia.
(C) Conserved synteny between R. oryzae and the three microsporidia. White arrows indicate additional non-syntenic genes.
With the exception of two ribosomal proteins (L21 and S9), none of the syntenic gene clusters shown in panels (A), (B) and (C) were conserved in other ascomycete (A. nidulans), basidiomycete (C. neoformans), or chytridiomycete (B. dendrobatidis) fungi. The genomes of P. blakesleeanus and R. oryzae were compared to the E. cuniculi genome using BLASTp. Homologs in E. bieneusi and A. locustae were identified using tBLASTn.
To explain the genome data in any other way (e.g., if microsporidia are the sister group to all fungi), one would have to postulate that the genomes of certain members of both microsporidia and zygomycetes were somehow special in having independently retained a high proportion of the gene order present in the ancestor of microsporidia and all fungi, which was lost multiple times independently in other fungal lineages. Indeed, it has been demonstrated that gene order evolves slowly in microsporidian genomes [21, 29], however the extended conservation in synteny that we document is not consistent with the idea that the same is true of zygomycetes. If that were true, multiple syntenic regions should be shared by all zygomycete and microsporidian genomes, but aside from the sex and RPL21/RPS9 loci, this is not the case (figure 2). Instead, gene order is typically conserved in all three microsporidia, but only one or two of the zygomycetes. In addition, the overall level of synteny between different zygomycetes does not suggest the same level of conservation found in microsporidia. This is all consistent with slow rearrangements in microsporidian genomes [21, 29], but not consistent with a slow rate of rearrangements in zygomycete genomes. There is accordingly no evidence that the gene order retained by zygomycetes and microsporidia is ancestral to all fungi.
Support for either explanation will be bolstered significantly by the additional sampling of basal fungal genomes. If other species of chytrids or ‘zygomycetes’ not specifically related to Mucorales are subsequently found to share a significant fraction of the synteny described here, then it will suggest the synteny may be derived from the ancestor of all fungi. If, on the other hand, no conservation is detected in these genomes, it further supports the relationship between microsporidia and Mucorales. Indeed, it is also possible that other subgroups of zygomycetes may share an even closer relationship, such as the Entomophthorales, which were proposed to be closely related to microsporidia based on molecular phylogenies [4]. Whatever the outcome, the availability of additional zygomycete, chytridiomycete, and microsporidian genome sequences that are currently in progress will allow repeated tests of their relationships. However, based on the genomes available now, the close evolutionary relationship between microsporidia and zygomycetes is the most parsimonious explanation of the data.
Sexual reproduction in microsporidia is poorly studied at best and completely unknown in most species, so the identification of a putative sex determining locus in microsporidia has functional implications deserving of further study. Analysis of other isolates of the species described here may reveal the existence of divergent HMG proteins, as in the (+) and (−) mating types of zygomycetes. However, it is also possible they are homothallic. The HMG domain proteins of E. cuniculi and A. locustae each contain two HMG domains whereas SexP and SexM of the zygomycetes contain a single HMG domain. The two microsporidian HMG domains are linked by 24 or 27 amino acid spacers respectively (Figure 7S), suggesting the two domains could interact with DNA independently [30]. If these represent functional correlates of SexP and SexM, and each domain in a single protein confers both SexP and SexM activity, the microsporidia might be homothallic and self-fertile. Maximum likelihood analysis showed that both microsporidian HMG domains are more closely related to SexP than to SexM (Figure 7S). In view of this, we propose three possible models for sexual development of microsporidia: 1) homothallism with a tandem HMG domain protein, 2) heterothallism with divergent HMG alleles that confer opposite mating types, as in zygomycetes, 3) loss of sex [31], in which case the sex related locus is an evolutionary vestige. Opportunities for sexual reproduction in microsporidia could occur during co-infection or involve self-fertile, homothallic mating or interactions between nuclei in species known to be dikaryotic. Of note, the hypothesis of extant sexual cycles is further supported by the finding that a variety of meiotic gene homologs are present in all three of the microsporidian genomes analyzed here (Table 1S), some of which were noted for E. cuniculi in a recent report by other investigators [28].
The high degree of synteny shared between microsporidian genomes and zygomycete genomes, including the sex related locus, provide a compelling new line of evidence to resolve the enigmatic origin of these unusual eukaryotic intracellular pathogens. These data demonstrate that microsporidia are true fungi that share a close common ancestor with zygomycetes, and perhaps specifically with the Mucorales. The data also suggest a new avenue to examine the difficult problem of sex in microsporidia. At present, there is no direct, functional link between the sex locus related gene cluster in the microsporidia and mating or meiosis. No sexual cycle has been unambiguously defined for the three microsporidian species considered here and inferences about whether other species even have a sexual cycle are limited. However, the finding that they share a surprisingly well conserved syntenic sex locus that governs mating in zygomycetous fungi leads to the reasonable hypothesis that it may also control sex in microsporidia. Future studies should address whether co-infection with genetically distinct isolates leads to the production of recombinant progeny, analysis of if and when the genes encoded by the sex locus related gene cluster are expressed, and analysis of whether other genes are present that might be inferred to play roles in sexual reproduction (such as genes encoding enzymes involved in production or sensing of the zygomycete trisporic acid pheromone or sex/meiosis related functions) [28]. Our findings that microsporidia evolved from a basal fungal lineage further invites speculation about whether the microsporidian polar tube infection structure is related to the sporangiophores of zygomycetous fungi, with implications for its functional roles in sensing extracellular signals and interactions with the host.
Experimental procedures
To identify the sex locus of M. circinelloides, we performed tBLASTn analysis with the amino acid sequence of P. blakesleeanus SexM against the 8× genome sequence of R7B, a (−) strain of M. circinelloides (US Department of Energy Joint Genome Institute Mucor genome project. http://mucorgen.un.es). The DNA sequence was annotated with FGENESH or Orf finder at NCBI. (−) strains (R7B, ATCC1216b, CBS277.49, and NRRL1443) were used to confirm the (−) sex locus. Genomic DNAs of (+) strains (NRRL3631 and ATCC1216a) were used to amplify, identify, and sequence the (+) sex allele.
Comparison of gene order across genomes was carried out starting with the annotated protein set for E. cuniculi [15], R. oryzae (Broad Institute Database for R. oryzae), and P. blakesleeanus (US Department of Energy Joint Genome Institute P. blakesleeanus genome project). Blast libraries were created using the NCBI blast database formatting tool formatdb [32]. The P. blakesleeanus and R. oryzae genome protein sets were compared against the E. cuniculi genome protein set, and each protein set was compared to their own genomic DNA sequence to ascertain the order of genes within the genome. Genes were classified as homologs between these species in cases where BLASPp E-values were less than 1.0 × e−30. Areas sharing relaxed synteny were defined in which two or more gene homologs (1.0 × e−10) were clustered and in the same order but included cases in which orientation differed or in which one or a few genes had been interposed (see Figure 5S for examples)
Mucor circinelloides sex locus sequences have been deposited in GenBank under accession FJ009106 and FJ009107. Microsporidian putative sex locus sequences have been deposited in GenBank under accession FJ008719 and FJ008720.
Supplementary Material
Acknowledgements
We acknowledge access to the Mucor circinelloides and Phycomyces blakesleeanus genome data established by the US Department of Energy Joint Genome Institute and to the Rhizopus oryzae genome data generated by the Broad Institute/Fungal Genome Initiative. We thank Donna Akiyoshi for the access to the E. bieneusi genome database. The Enterocytozoon bieneusi genome sequence survey was supported by National Institutes of Health grants R21 AI52792 and R21 AI064118. We thank Hilary Morrison and acknowledge the Josephine Bay Paul Center for Comparative Molecular Biology and Evolution for the use of data included in the Antonospora locustae Genome Project funded by NSF award number 0135272. We thank Rytas Vilgalys, Andrii Gryganskyi, Tim James, Alex Idnurm, Raphael Valdivia, Banu Metin, Charles Li, and Naomi Fast for discussions or comments on the manuscript. This work was supported by National Institutes of Health grant AI50113 to J.H. and by Canadian Institutes for Health Research grant MOP-84265 to P.J.K. P.J.K. is a Fellow of the Canadian Institute for Advanced Research and a Senior Scholar of the Michael Smith Foundation for Health Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Keeling PJ, Fast NM. Microsporidia: Biology and evolution of highly reduced intracellular parasites. Annu. Rev. Microbiol. 2002;56:93. doi: 10.1146/annurev.micro.56.012302.160854. [DOI] [PubMed] [Google Scholar]
- 2.Edlind TD, Li J, Visvesvara GS, Vodkin MH, McLaughlin GL, Katiyar SK. Phylogenetic analysis of beta-tubulin sequences from amitochondrial protozoa. Mol. Phylogenet. Evol. 1996;5:359–367. doi: 10.1006/mpev.1996.0031. [DOI] [PubMed] [Google Scholar]
- 3.Keeling PJ, Doolittle WF. Alpha-tubulin from early-diverging eukaryotic lineages and the evolution of the tubulin family. Mol. Biol. Evol. 1996;13:1297–1305. doi: 10.1093/oxfordjournals.molbev.a025576. [DOI] [PubMed] [Google Scholar]
- 4.Keeling PJ. Congruent evidence from alpha-tubulin and beta-tubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genet. Biol. 2003;38:298–309. doi: 10.1016/s1087-1845(02)00537-6. [DOI] [PubMed] [Google Scholar]
- 5.James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Celio G, Gueidan C, Fraker E, Miadlikowska J, et al. Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature. 2006;443:818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 6.Hirt RP, Logsdon JM, Healy B, Dorey MW, Doolittle WF, Embley TM. Microsporidia are related to Fungi: Evidence from the largest subunit of RNA polymerase II and other proteins. Proc. Natl. Acad. Sci. U.S.A. 1999;96:580–585. doi: 10.1073/pnas.96.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gill EE, Fast NM. Assessing the microsporidia-fungi relationship: Combined phylogenetic analysis of eight genes. Gene. 2006;375:103–109. doi: 10.1016/j.gene.2006.02.023. [DOI] [PubMed] [Google Scholar]
- 8.Thomarat F, Vivarès CP, Gouy M. Phylogenetic analysis of the complete genome sequence of Encephalitozoon cuniculi supports the fungal origin of microsporidia and reveals a high frequency of fast-evolving genes. J. Mol. Evol. 2004;59:780–791. doi: 10.1007/s00239-004-2673-0. [DOI] [PubMed] [Google Scholar]
- 9.Idnurm A, Walton FJ, Floyd A, Heitman J. Identification of the sex genes in an early diverged fungus. Nature. 2008;451:193–196. doi: 10.1038/nature06453. [DOI] [PubMed] [Google Scholar]
- 10.Didier ES, Weiss LM. Microsporidiosis: Current status. Curr. Opin. Infect. Dis. 2006;19:485–492. doi: 10.1097/01.qco.0000244055.46382.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lom J, Nilsen F. Fish microsporidia: fine structural diversity and phylogeny. Int. J. Parasitol. 2003;33:107–127. doi: 10.1016/s0020-7519(02)00252-7. [DOI] [PubMed] [Google Scholar]
- 12.Becnel JJ, Andreadis TG. Microsporidia in insects. In: Wittner M, editor. The microsporidia and microsporidiosis. Washington, D.C: ASM Press; 1999. pp. 1–6. [Google Scholar]
- 13.Xu Y, Weiss LM. The microsporidian polar tube: a highly specialised invasion organelle. Int. J. Parasitol. 2005;35:941–953. doi: 10.1016/j.ijpara.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vossbrinck CR, Maddox JV, Friedman S, Debrunner-Vossbrinck BA, Woese CR. Ribosomal-RNA sequence suggests microsporidia are extremely ancient eukaryotes. Nature. 1987;326:411–414. doi: 10.1038/326411a0. [DOI] [PubMed] [Google Scholar]
- 15.Katinka MD, Duprat S, Cornillot E, Metenier G, Thomarat F, Prensier G, Barbe V, Peyretaillade E, Brottier P, Wincker P, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414:450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 16.Tsaousis AD, Kunji ERS, Goldberg AV, Lucocq JM, Hirt RP, Embley TM. A novel route for ATP acquisition by the remnant mitochondria of Encephalitozoon cuniculi. Nature. 2008;453:553–556. doi: 10.1038/nature06903. [DOI] [PubMed] [Google Scholar]
- 17.Williams BAP, Hirt RP, Lucocq JM, Embley TM. A mitochondrial remnant in the microsporidian Trachipleistophora hominis. Nature. 2002;418:865–869. doi: 10.1038/nature00949. [DOI] [PubMed] [Google Scholar]
- 18.Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, Delbac F, Vivares CP, Hirt RP, Lill R, Embley TM. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–628. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- 19.Williams BAP, Cali ANN, Takvorian PM, Keeling PJ. Distinct localization patterns of two putative mitochondrial proteins in the microsporidian Encephalitozoon cuniculi. J. Eukaryot. Microbiol. 2008;55:131–133. doi: 10.1111/j.1550-7408.2008.00315.x. [DOI] [PubMed] [Google Scholar]
- 20.Keeling PJ, Luker MA, Palmer JD. Evidence from beta-tubulin phylogeny that microsporidia evolved from within the fungi. Mol. Biol. Evol. 2000;17:23–31. doi: 10.1093/oxfordjournals.molbev.a026235. [DOI] [PubMed] [Google Scholar]
- 21.Slamovits CH, Fast NM, Law JS, Keeling PJ. Genome compaction and stability in microsporidian intracellular parasites. Curr. Biol. 2004;14:891–896. doi: 10.1016/j.cub.2004.04.041. [DOI] [PubMed] [Google Scholar]
- 22.Dyer PS. Evolutionary biology: Genomic clues to original sex in fungi. Curr. Biol. 2008;18:R207–R209. doi: 10.1016/j.cub.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Fraser JA, Heitman J. Fungal mating-type loci. Curr. Biol. 2003;13:R792–R795. doi: 10.1016/j.cub.2003.09.046. [DOI] [PubMed] [Google Scholar]
- 24.Bürglin TR. The homeobox genes of Encephalitozoon cuniculi (Microsporidia) reveal a putative mating-type locus. Dev. Genes Evol. 2003;213:50–52. doi: 10.1007/s00427-002-0287-3. [DOI] [PubMed] [Google Scholar]
- 25.James TY. Analysis of mating-type locus organization and synteny in mushroom fungi: Beyond model species. In: Heitman J, Kronstad JW, Taylor JW, Casselton LA, editors. Sex in Fungi. Washington, DC: ASM Press; 2007. pp. 317–331. [Google Scholar]
- 26.Hazard EI, Brookbank JW. Karyogamy and meiosis in an Amblyospora Sp (Microspora) in the mosquito Culex salinarius. J. Invertebr. Pathol. 1984;44:3–11. [Google Scholar]
- 27.Sweeney AW, Graham MF, Hazard EI. Life cycle of Amblyospora dyxenoides sp. nov. in the mosquito Culex annulirostris and the copepod Mesocyclops albians. J. Invertebr. Pathol. 1988;51:46–57. doi: 10.1016/0022-2011(88)90087-0. [DOI] [PubMed] [Google Scholar]
- 28.Malik S-B, Pightling AW, Stefaniak LM, Schurko AM, Logsdon JM., Jr An expanded inventory of conserved meiotic genes provides evidence for sex in Trichomonas vaginalis. PLoS ONE. 2008;3:e2879. doi: 10.1371/journal.pone.0002879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corradi N, Akiyoshi DE, Morrison HG, Feng X, Weiss LM, Tzipori S, Keeling PJ. Patterns of genome evolution among the microsporidian parasites Encephalitozoon cuniculi, Antonospora locustae and Enterocytozoon bieneusi. PLoS ONE. 2007;2:e1277. doi: 10.1371/journal.pone.0001277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stott K, Tang GSF, Lee K-B, Thomas JO. Structure of a complex of tandem HMG boxes and DNA. J. Mol. Biol. 2006;360:90–104. doi: 10.1016/j.jmb.2006.04.059. [DOI] [PubMed] [Google Scholar]
- 31.Ironside J. Multiple losses of sex within a single genus of microsporidia. BMC Evol. Biol. 2007;7:48. doi: 10.1186/1471-2148-7-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. U.S.A. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.