Abstract
The vitamin D receptor (VDR) regulates a diverse set of genes that control processes including bone mineral homeostasis, immune function and hair follicle cycling. Upon binding to its natural ligand, 1α,25(OH)2D3, the VDR undergoes a conformational change that allows the release of corepressor proteins and the binding of coactivator proteins necessary for gene transcription. We report the first comprehensive evaluation of the interaction of the VDR with a library of coregulator binding motifs in the presence of two ligands, the natural ligand 1α,25(OH)2D3 and a synthetic, non-secosteroidal agonist LG190178. We show that the VDR has relatively high affinity for the second and third LxxLL motifs of SRC1, SRC2 and SRC3 and second LxxLL motif of DRIP205. This pattern is distinct in comparison to other nuclear receptors. The pattern of VDR-coregulator binding affinities was very similar for the two agonists investigated, suggesting that the biologic functions of LG190178 and 1α,25(OH)2D3 are similar. Hairless binds the VDR in the presence of ligand through a LxxLL motif (Hr-1), repressing transcription in the presence and absence of ligand. The VDR binding patterns identified in this study may be used to predict functional differences among different tissues expressing different sets of coregulators, thus facilitating the goal of developing tissue and gene specific vitamin D response modulators.
The vitamin D receptor (VDR), which binds 1α,25-dihydroxyvitamin D3 (1α,25(OH)2D3), contains several functional domains, including a ligand-binding domain (LBD), that mediates ligand-dependent gene regulation (1). A critical step in 1α,25(OH)2D3 action is the induction of a LBD conformational change to form activation function 2 (AF-2)(2), a hydrophobic cleft formed by three helices and a short COOH-terminal amphipathic alpha helix (H12) (3), which serves as a binding surface for coactivators (4). Unliganded nuclear receptor (NR) heterodimers associate with corepressors such as the nuclear receptor corepressor (NCoR) and the silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) (5, 6) and associated histone deacetylases (7, 8). These proteins function as adaptors to convey a repressive signal to the transcriptional apparatus by maintaining a closed chromatin structure with the histone N-terminal ‘tails’ in a charged state tightly associated with DNA (9). Ligand binding promotes the release of corepressors and the binding of coactivators, enhancing the transcription of specific genes (10). Some coactivators, such as the SRC family (11-13), recruit other coregulators with histone acetylase activity and remodel chromatin structure. Other coactivators, such as the DRIP factors (14, 15), interact with the basal transcriptional machinery. In the unliganded state, helix 12 projects away from the globular core of the LBD, while in the liganded state this helix contacts the LBD globular core domain to create an AF-2 surface through which coactivator proteins can dock (16, 17). Upon 1α,25(OH)2D3 binding, VDR localizes at the vitamin D response elements of target genes, recruits coactivators, which recruit histone acetyltransferase to modify histone or bridge the gap between the VDR and the transcription machinery (18). Both coactivators and corepressors can interact with overlapping surfaces on the LBD (19). It has been proposed that ligand-dependent exchange between corepressors and coactivators is caused by a difference in the length of the interacting motifs that can be accommodated by the two conformations of the binding pocket (20). Therefore, a ligand that displaces the AF2 helix from its active position would be expected to facilitate the interaction with corepressor proteins and to repress transcriptional activity (21). Coactivator proteins that bind to AF-2 contain one or more of the consensus sequence LxxLL (L is leucine and x is any amino acid) which forms an amphipathic alpha-helix (Figure 1, A) (22). This helix fits into the hydrophobic cleft of the liganded receptor (17). Receptor-specific binding of coactivators containing the LxxLL motif is governed by amino acid residues flanking the binding site, including in human VDR the conserved residues E420 and K246 (23). Mutation of these residues and adjacent hydrophobic amino acids abolishes both 1α,25(OH)2D3-activated transcription and coactivator interactions with the VDR (24, 25).
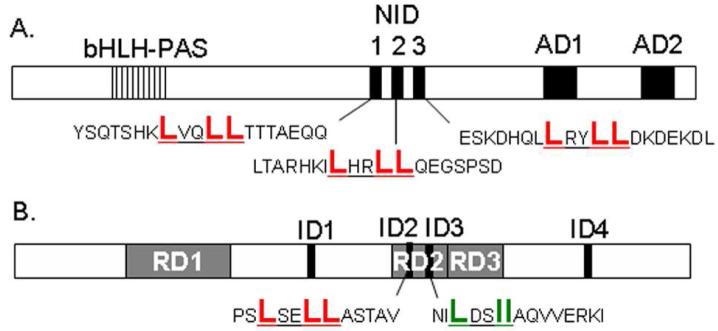
Figure 1. Structural Composition of Coregulator SRC1 and Hr.
A, functional domains of SRC1 including the nuclear interaction domain (NID) and its three nuclear receptor interaction domains (NR boxes, SRC1-1, SRC1-2, SRC1-3). B, functional domains of Hairless including the three transcriptional repression domains (RDs). RD2, comprised of a LxxLL and a ΦxxΦΦ motif, is necessary and sufficient for Hr-VDR interaction.
NR corepressors NCoR and SMRT encode multiple, short receptor interaction domains composed of the sequence ΦxxΦΦ (Φ is leucine or isoleucine and x is any amino acid) (26, 27). This motif is predicted to form an alpha helix, that is one turn longer than that formed by the LxxLL motif. In a manner analogous to the LxxLL-containing motifs, it has been suggested that this helix also binds the AF-2 surface without requiring a docked helix 12 (27). Indeed, deletion of helix 12 enhances corepressor binding (5), suggesting that this helix does not play an active role in nuclear receptor-corepressor recognition.
Another corepressor, Hr, is expressed primarily in brain, epidermis and hair follicles (28) and is known to repress VDR mediated transcription (29, 30). The Hr-VDR interaction is of great interest because null mutations in either protein induces alopecia, in both the mouse or human (31-33). Despite a low sequence identity with other corepressors, Hr functions in a similar manner. For example, it mediates moderate transcriptional repression of liganded TR but strong repression in case of unliganded TR (34). In contrast to TR, Hr strongly represses VDR mediated transcription in the presence of ligand but moderately in the absence of ligand (30). Hr encodes two LxxLL motifs required for Hr/RORα interaction (35) and two ΦxxΦΦ motifs required for Hr/TR interaction (Figure 1, B) (30). In contrast to TR, VDR only interacts physically and functionally with Hr through a region of Hr containing a LseLL motif at position 778-782 (Hr1) and a LdsII motif at position 816-830 (Hr2)(30). As we show in this report only the LseLL motif (Hr1) mediates a ligand dependent binding. Some VDR mediated functions, like its control of hair follicle cycling, are ligand independent, but require Hr suggesting a role for Hr in this VDR regulated function (36, 37).
Ligand selectivity of VDR may affect coregulator recruitment. Published data show that VDR bound to LG190178 is able to recruit coactivator SRC2 like VDR/1α,25(OH)2D3 (38). Unlike 1α,25(OH)2D3, LG190178 does not bind to serum vitamin D binding protein and exhibits no influence on calcium serum levels in mice (39). Both ligands induce HL-60 differentiation into macrophages and inhibit the growth of SK-BR-3 and LNCaP cells. Clearly the interplay between ligand, receptor, and pools of potentially interacting coregulators is important in defining the physiological activity of VDR.
Herein we report the first comprehensive evaluation of the interaction of VDR and coregulator NR boxes using two different biochemical assays: 1) a fluorescence polarization binding assay measuring the equilibrium binding of VDR to a library of fluorescent coregulator peptides, a method that has previously been used to profile other nuclear receptors-coregulator interactions (40-42); and 2) a competition pull-down assay measuring the ability of unlabeled, but otherwise identical, coregulator peptides to inhibit the interaction between VDR and full length SRC2. Additionally, we confirm the binding affinities of one labeled and unlabeled coregulator peptide SRC2-3 using isothermal titration calorimetry (ITC). Because of the difficulties encountered by others in working with a full-length VDR-LBD, we used a mutated form that has no major differences in ligand binding, transactivation, or dimerization with RXRα (43). The binding constants for VDR to a wide range of coregulator NR boxes including LxxLL and LxxII motifs were determined in the presence of 1α,25(OH)2D3 and LG190178.
Experimental Procedures
Reagents
1α,25(OH)2D3 (calcitriol) was purchased from WAKO CHEMICALS USA INC; LG190178 was synthesized using a published procedure (39).
Protein Expression and Purification
The VDR-LBDmt, provided by D. Moras (43), was cloned by PCR (primers 5′-CGCGGATCCAGATCTGACAGTCTGCGGCCCAAG-3′ and 5′-CGCGGATCCAGATCTGACAGTCTGCGGCCCAAG-3′) at the BamHI site of the pMAL-c2X vector (New England Biolabs), in fusion with the maltose binding protein (MBP). The plasmid was expressed in Escherichia coli grown at ambient temperature in 2x LB with 2% glucose. MBP-VDR-LBDmt protein expression was induced with 0.2 mM isopropyl 1-thio-β-D-galactopyranoside for 16 h at 25°C before harvest and cell lysis by freeze-thawing and sonication. Proteins were purified in the absence or presence of ligand 2 μM (1α,25(OH)2D3) or 20 μM (LG190178) using an amylose resin column.
Peptide Library Synthesis
Coregulator peptides were synthesized by the Hartwell Center (St. Jude Children's Research Hospital), purified by RP-HPLC, and analyzed by LC/MS. The peptides containing an N-terminal cysteine were labeled using the thiol-reactive fluorophore, Texas Red maleimide (Molecular Probes): 3 mg of peptide was combined with 1.6 mg of fluorophore in 5 ml of 50% water/DMF. After stirring for 3 h in the dark the reaction mixture was purified by HPLC and analyzed by LC/MS. All fluorescent peptides possessed purities greater than 95%.
Peptide Binding Assay
MBP-VDR-LBDmt protein binding affinities, in presence of 1α,25(OH)2D3 (2 μM) or LG190178 (20 μM) using fluorescently labeled coregulator peptides were measured using a previously described assay (40). The relative doses of 1α,25(OH)2D3 and LG190178 have been described previously (39). Two different protein batches were used for two independent experiments, with each carried out in triplet. Reported values reflect the mean value with associated total error across all experiments.
Competitive Binding Assay
Fluorescently labeled SRC2 was produced by in vitro transcription–translation (TNT kit & FluoroTect™, Promega) from the plasmid pSGT-SRC2. In a 96 well polypropylene plate (Costar 3365) 8 peptides at a time were diluted from 10000-1.0 μM in DMSO. 2 μl of the diluted peptides were added to 91 μl of buffer (25 mM HEPES, 100 mM NaCl, 1 mM DTT, 0.01% NP40, 0.1% BSA [added fresh], 0.5 μM VDRmt on amylose beads, and 1 μM 1α,25 dihydroxyvitamin D3) in a 96 well filter plate (Millipore, Multiscreen HTS BV). After agitation for 2 h (IKA microtiterplate shaker) at room temperature (rt), 7 μl of TNT solution was added to each reaction followed by 2 h of agitation at rt. The filter plate was attached to 96-well plate (Costar 3365) and centrifuged at 50g for 3 minutes followed by the addition of 100 μl of buffer (25 mM HEPES, 100 mM NaCl, 1 mM DTT, 0.01% NP40, 0.1% BSA (added fresh). The filter plate was assembled with another 96-well plate (Costar 3365) and centrifuged at 50g for 3 minutes. For elution of the SRC2-VDR complex 10 μl of a 10 mM maltose solution was added. After a 10 minute incubation period the filter plate was assembled with another 96-well plate (Costar 3365) and centrifuged at 50g for 3 minutes. The elutions were treated with 3 μl of 4x SDS-page loading buffer (invitrogen), sealed and incubated for 30 minutes in an oven at 70°C. After separation using SDS-page the fluorescent bands of SRC2 were visualized using a fluorescence scanner (Typhoon, GE). The bands were integrated using ImageQuant (Molecular Dynamics) and analyzed using Prism (GraphPad). Three independent experiments were carried out for each state. The IC50 values were obtained by fitting data to the following equation (y = min + (max − min)/1 + (x/Kd) Hill slope.
Calorimetric studies
The thermodynamics of the interaction between the SRC2-3 peptides and VDR-LBD were determined using a VP-ITC instrument (Microcal, Northampton, MA). Therefore, a solution of either unlabeled or Texas Red-labeled SRC2-3 peptide (0.42 μM) in the syringe was titrated into a solution of VDR (25 μM) in the cell. 2 μL of the titrant was first injected followed by 25 injections of 10 μL each, with a 300 second delay between injections. The experiments were performed at 25 °C in a buffer containing 20 mM HEPES pH 7.5 and 150 mM NaCl. The binding data were fit to a 1:1 binding model using Origin software (Origin lab, Northampton, MA).
Results
Coregulator Peptide Library
To evaluate VDR coregulator recruitment, the binding between the VDR-LBD and a library of known coregulator peptides consisting of a central LxxLL or ΦxxΦΦ sequence plus up to 8 additional flanking residues at each terminus was measured (Figure 2). To allow attachment of the fluorescent label a non-native cysteine was introduced at the N-terminus of each peptide. Additionally, a cysteine-serine exchange was employed in the case of SRC3-1 and Hr1 to prevent double labeling. Hr1 contains a second cysteine residue at the −3 position of the LxxLL motif which was used for labeling. All peptide probes were synthesized in parallel using Fmoc (9-fluorenylmethyloxycarbonyl) protective group and purified by RP-HPLC. Identity and purity were confirmed using HPLC and MALDI-TOF or LCMS and was greater than 95%.
Figure 2. Amino Acid Sequence of Coregulator Peptides.
Sequence alignment of the coregulator peptides with the N-terminal cysteine label indicated in yellow, the NR box motif LxxLL in red, and the ΦxxΦΦ corepressor motif in green. Note: cysteine-serine exchange was employed in case of SRC3-1 (7) and Hr1 (2) to prevent double labeling.
Initial validation of coactivator peptides binding assay
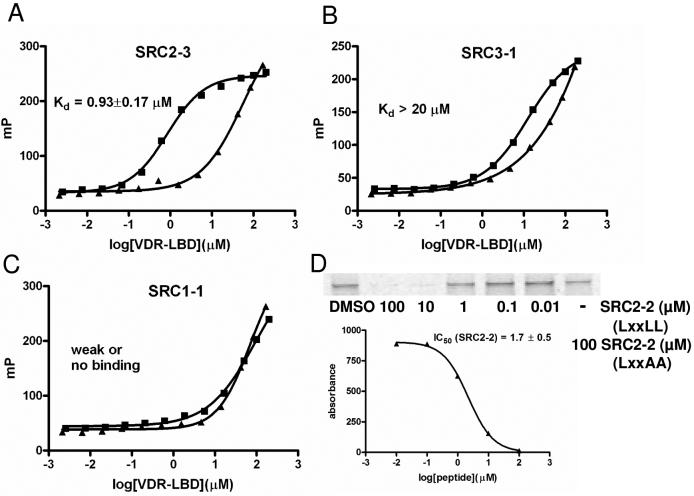
Initial peptide binding studies were carried out with SRC2-2 in the presence of 1α,25(OH)2D3 with both MBP-VDR-LBDwt and MBP-VDR-LBDmt, from the construct developed by D. Moras (43). Control experiments using a MBP protein alone showed no interaction with SRC2-2 (data not shown). SRC2-2 binds to both MBP-VDR-LBDwt and MBP-VDR-LBDmt in a similar saturable dose-dependent manner, with somewhat higher affinity for MBP-VDR-LBDmt observed than for MBP-VDR-LBDwt (supplementary material). We used MBP-VDR-LBDmt for further studies because it was soluble at higher concentrations, which was essential to determine the binding constants (Kd) using fluorescence polarization. To exclude potential perturbing interactions between the label attached to the peptide and MBP-VDR-LBDmt we measured the binding affinity Kd values for labeled and unlabeled coregulator peptide SRC2-3 using isothermal titration calorimetry (ITC) (Table 1 and supplementary material). The Kd values observed for the Texas-Red labeled and unlabeled peptide SRC2-3 were 285 nM and 174 nM, respectively. To demonstrate the ability of unlabeled peptides to inhibit the interaction between VDR-LBD and full length SRC2 exhibiting three LxxLL motifs, we performed pull-down competition assays. In these assays the native peptides (LxxLL) blocked this interaction, whereas altered peptides (LxxAA) did not affect this binding (Figure 3, D). To verify that binding of Hr-1 is not compromised by Cystein/Serine (C/S) exchange, a competitive fluorescence polarization experiment was carried out showing that Hr-1wt and Hr-1C/S equally inhibited the binding between SRC2-3 Texas-Red and MBP-VDR-LBDmt (supplementary material).
Table 1.
Isothermal titration calorimetry using VDR-LBD and coregulator peptide SRC2-3 or SRC2-3 labeled with Texas-Red.
| Interaction | Kd (nM) | n | ΔH (kcal/mole) |
|---|---|---|---|
| VDR-SRC2-3 | 174 ±37 | 1.02±0.02 | −6.7±0.3 |
| VDR-SRC2-3_Texas-Red | 285±18 | 0.93±0.04 | −2.2±0.12 |
Figure 3. VDR-coactivator binding isotherms in the presence and absence of 1α,25(OH)2D3.
■1α,25(OH)2D3; ▲ no ligand. A, High affinity binding between VDR and SRC2-3 in the presence and absence of 1α,25(OH)2D3. B, Low affinity binding between SRC3-1 and VDR in the presence and absence of 1α,25(OH)2D3. C, weak or no binding between SRC1-1 and VDR in the presence and absence of 1α,25(OH)2D3. D, SRC2 full length binding to VDR-LBD is blocked by increasing concentration of SRC2-2 peptide (LxxLL) but not by the mutated SRC2-2 peptide (LxxAA); Quantified SDS-page converted into binding isotherm to determine the IC50 value of SRC2-2.
Coactivator binding assays
The fluorescence polarization assay was executed by maintaining a constant concentration of fluorescently labeled coactivator peptide (10 nM) and a variable concentration of VDR-LBDmt from 0.002—199 μM in the absence and presence of 1α,25(OH)2D3 or LG190178. The data were fit to a sigmoidal dose-response curve and Kd values with 95% confidence intervals are summarized in Figure 4. The competition pull down assay was carried out with labeled SRC2, solid - supported VDR-LBDmt, and unlabeled coregulator peptides. Displaced fluorescent SRC2 was removed by washes and residual SRC2 was quantified after separation by SDS-page (Figure 3, D). The data were fit to a sigmoidal dose-response curve and Kd values with 95% confidence intervals are summarized in Figure 4 and supplementary material.
Figure 4. Coregulator binding patterns and specificity determinants.

The equilibrium binding constants (Kd values) for the binding of VDR to coregulator peptides (SRC1, SRC2, SRC3, DRIP205, P300, RIP140-5, NCoR, SMRT and Hr) in the presence and absence of 1α,25(OH)2D3 or LG190178 were determined by fluorescent polarization. IC50 values of peptides determined by a competition pull down assay employing VDR-LBD and full length SRC2 in the presence of 1α,25(OH)2D3. The Kd and IC50 values are color coded: dark green (<3 μM), green (3.1-20.0 μM), light green (>20 μM), grey (>50 μM, weak or no binding).
Coregulator peptides bind VDR-LBDmt in the absence or presence of ligand in three different modes
Binding isotherms for coregulator peptides exhibited three behaviors: saturable binding with a clear high dosage plateau, clear interaction without saturation, and no interaction. The higher affinity group of peptides bound in a ligand-dependent, dose-dependent, and saturable manner where a plateau was reached within the protein concentration range studied (Figure 3, A ■). The Kd values for this class in the presence of ligand were lower than 20 μM as determined by fitting the data to a sigmoidal dose-response curve. Nine coactivator peptides exhibited this mode: SRC1-2, SRC1-3, SRC2-2, SRC2-3, SRC3-2, SRC3-3, DRIP205-2, RIP140-5, and Hr1 (Figure 4, green and dark green color code). The binding affinities of theses peptides in the absence of ligand were dose dependent but exhibited no plateaus at higher protein concentrations (Figure 3, A ▲). Because of the missing isotherm saturation we labeled these interactions as weak or no binding (Figure 4, grey color code and supplementary material). The IC50 values determined by an independent competition pull down assay were similar to Kd values in the presence of ligand measured by FP.
The lower affinity group included peptides exhibiting dose-dependent binding in the presence of ligand but did not display a plateau at the highest protein concentrations used. In comparison with the binding isotherm in the absence of ligand we observed a clear shift indicating ligand-dependent binding (Figure 3, B). For this group of peptides we suggest a Kd value higher than 20 μM, and the group includes SRC3-1, DRIP-1, and P300. (Figure 4, light green color code). The binding constants in the absence of ligand exhibited high values indicative of very weak binding (Figure 4, grey color code and supplementary material). Inhibition of binding between SRC2 and VDR was observed at concentrations of these peptides greater than 50 μM.
The third group of peptides showed identical dose-dependent binding in the presence and absence of ligand and did not display saturation at higher concentrations of peptide. This group included SRC1-1 and SRC2-1 bearing an LxxLL motif and corepressor peptides NCoR, SMRT and Hr2 with LxxIIxxxL/V motifs (Figure 3, C). The mode of binding of these peptides was indicated as weak binding or no binding (Figure 4, grey color code and supplementary material).
Discussion
In this study, we examined the binding of VDR-LBD to a range of target motifs within potential VDR coregulators. Because of the difficulties encountered by others in working with wild-type VDR-LBD, we used a mutated form, which lacks a domain with no apparent function and little homology to other NRs (43-46). Native VDR-LBD and VDR-LBDmt have been shown to function similarly with respect to ligand binding, transactivation, or dimerization with RXRα LBD (43). We confirmed these results by showing that SRC2-2 bound to VDR-LBDmt with high affinity in a ligand dependent manner. We observed that VDR-LBDmt exhibited higher affinity for SRC2-2 than did native VDR-LBD, possibly due to the higher solubility of the former construct under our assay conditions.
Our results verify that VDR binds LxxLL motifs from numerous coactivators in a selective fashion. The Kd values measured for the absolute binding of coregulator motifs to the VDR are similar to those observed for binding to other nuclear receptors (40, 47). In contrast to the estrogen receptor and thyroid receptor, which have the highest affinity for the second NR box of each SRC, VDR binds to the second and third NR box with higher affinities than to the first NR box. Considering all NR boxes of each SRC, we observed the same affinity between VDR/SRC1 and VDR/SRC2. In contrast, SRC3 exhibited a stronger interaction between SRC3-1 and VDR in comparison with SRC1-1 and SRC2-1, whereas SRC3-2 has a weaker interaction with VDR in comparison with SRC1-2 and SRC2-2. This may suggest that SRC3 dominates binding to VDR relative to SRC1 or SRC2. SRC3 has been previously described as a more “general” coactivator, but our results may reveal selectivity for VDR for this protein (48).
It has been reported that amino acid residues N-terminal to the LxxLL motif have a major impact on the binding affinities of coregulator peptides (22, 49, 50). In particular, coregulator peptides bearing hydrophobic amino acids at the −1 position of the LxxLL motif exhibited high affinities for ER, PPAR, TR and VDR (22, 40, 51, 52). In the case of the peptides investigated in this study we observed the same correlation. In contrast, we showed that coregulator peptides missing a hydrophobic amino acid residue at the −1 position like SRC1-1, SRC2-1, SRC3-1, P300, NCoR, and SMRT exhibited a low binding affinity towards VDR with the exception of Hr1. The crystal structure of SRC1-2 bound to VDR-LBD illustrates the importance of the isoleucine residue in contact with the hydrophobic surface of AF-2 (Figure 5, A) (53). In overlay, the SRC1-1 structure exhibits a protonated lysine residue destabilizing the interaction between coregulator and VDR. Zella et al. (52) reported that VDR agonist derived LxxLL peptides, originated from a phage display screening, have the proposed sequence L × E/H × H/F P L/M/I. Because of the sequence similarity with DRIP peptides we observed a binding constant of 1.6 μM (Figure 4) for DRIP-2 following the proposed paradigm of a HPMLxxLL sequence. In contrast DRIP-1, bearing a NPILxxLL sequence has a weak interaction with VDR. DRIP205 is known to interact directly with ligand-activated VDR/RXR heterodimers mostly through the second motif, although both motifs are equally required for VDR mediated transcription in cells (54).
Figure 5. VDR-Coactivator Interaction.
A. Crystal structure of VDR-SRC1-2(57) and docked SRC1-1; -1 N-terminal residue of LxxLL is K (SRC1-1) and I (SRC1-2); B. Crystal structure of VDR-DRIP2 (53) and docked Hr1; −1 N-terminal residue of LxxLL is M (DRIP2) and S (Hr1).
Hr1 is part of the coregulator Hairless which represses VDR mediated transcription in the presence and absence of 1α,25(OH)2D3 (30). Hairless interacts with VDR predominantly through the 750-864 domain, exhibiting a coactivator LxxLL motif (Hr1) and a corepressor LxxIIxxxV motif (Hr2) (Figure 1, B). Our investigation of these motifs separately showed that Hr2 exhibits a weak affinity towards VDR in the presence and absence of ligand in contrast to Hr1 which exhibited a 1α,25(OH)2D3-dependent VDR-LBD binding of 5.9 μM (Figure 4). Because Hr1 is bearing a serine residue at the −1 N-terminal position of LxxLL (Figure 5, B) this is the first reported coregulator peptide binding to VDR and bearing a polar residue at this particular position. Peptides with a similar sequence to Hr1 were identified by Zella at al. (52) in the presence of the VDR antagonist ZK15922, but binding of these peptides could not be confirmed by a two-hybrid assay (52). The binding affinities for Hr1 measured in the presence of agonist 1α,25(OH)2D3 and LG190178 were similar for VDR. We can conclude that Hr1 is supporting the interaction between VDR and Hr in the presence of ligand. This was a surprise. Our previous findings in keratinocytes with full length molecules showed that VDR and Hairless bind to each other in the absence of ligand and that this binding is partially displaced by addition of 1α,25(OH)2D3 (29). This suggests that results at least for Hairless obtained with peptides recapitulates only partially the binding properties of full length proteins.
Like Hr2, corepressor SMRT and NCoR peptides exhibited weak interactions with VDR in the presence and absence of ligand, although stronger interactions have been reported for full length proteins (55, 56). The length of a polypeptide has a strong influence on its three dimensional structure which determines binding affinity and solubility. Lower affinities of small peptide mimics in comparison with full-length proteins are not uncommon. The nature of the FP assay does not allow reliable Kd measurements of lower affinity probes due to light scattering caused by protein aggregation at high concentrations. In contrast, high affinity probes exhibiting a saturated signal at higher protein concentrations do not suffer from this interference. Moreover, a recent study showed that NCoR and SMRT dependant repression of 1α,25(OH)2D3 mediated transcription by VDR/RXR heterodimer is achieved through their recruitment by RXR but not by VDR (56), suggesting that further studies involving VDR/RXR heterodimers are needed to determine the affinity of these co-repressors for these nuclear receptors.
In the presence of synthetic agonist LG190178, VDR adopts an agonistic conformation, recruiting all investigated coregulators with the same affinities as VDR/1α,25(OH)2D3. This result is supported by the fact that in the case of VDR, crystal structures with different vitamin D3 analogs in the presence of coregulator peptide exhibit an unaltered AF-2 domain (23, 53, 57, 58). We can conclude that the signal transduction mediated by VDR is similar regardless of which of the two ligands is bound.
The IC50 values determined by a competition pull-down assay confirmed the Kd values determined by the FP measurements. We reported a similar behavior for TR (59) suggesting that although full length SRC2 exhibits multiple NR boxes no assisted binding among them occurs in this biochemical assay.
Presumably there are additional factors that influence NR recruitment of coregulators such as post-translational modifications, structural determinants arising from specific DNA response elements, cooperativity, cellular environment, and additional interaction surfaces on the NR and coregulator proteins, needing more complex models to fully dissect NR-coregulator interactions. These binding patterns draw a first picture of potential functional differences for different ligands in tissues with different sets of coregulators. The use of more ligands should allow us to predict these functional differences, thus facilitating the goal of developing tissue and gene specific vitamin D response modulators.
Supplementary Material
Acknowledgement
We thank Dr. Patrick Rodrigues and Bob Cassell for peptide synthesis at the Hartwell Center of Bioinformatics and Biotechnology, St. Jude Children's Research Hospital, Memphis, TN.
This work was supported by NIH grants AR050023 (D. D. B), AR39448 (D. D. B), DK58080 (R. K. G.), a Cancer Center Support Grant 2P30CA021765 (L. A. A., S. O., R. W. K. and R. K. G.), the American Lebanese and Syrian Associated Charities (ALSAC) and St Jude Children's Research Hospital (SJCRH).
Abbreviations
- 1,25(OH)2D3
1,25 dihydroxyvitamin D3 Calcitriol
- DRIP
vitamin D receptor-interacting protein
- Hr
Hairless
- MBP
Maltose binding protein
- NCoR
Nuclear receptor corepressor
- NR box
Nuclear receptor box
- SMRT
silencing mediator for retinoic acid and thyroid hormone receptors
- SRC
Steroid receptor coactivator
- TR
thyroid hormone receptor
- VDR
Vitamin D receptor
Footnotes
Supporting Information Available
FP isotherms for each Texas Red labeled peptide in the presence and absence 1α,25(OH)2D3 and ITC isotherms of unlabeled and Texas Red labeled SRC2-3 peptide. This material is available free of charge via the Internet at http://pubs.acs.org”.
References
- 1.Carlberg C, Polly P. Gene regulation by vitamin D3. Crit Rev Eukaryot Gene Expr. 1998;8:19–42. doi: 10.1615/critreveukargeneexpr.v8.i1.20. [DOI] [PubMed] [Google Scholar]
- 2.Moras D, Gronemeyer H. The nuclear receptor ligand-binding domain: structure and function. Curr Opin Cell Biol. 1998;10:384–391. doi: 10.1016/s0955-0674(98)80015-x. [DOI] [PubMed] [Google Scholar]
- 3.Danielian P, White R, Lees J, Parker M. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. Embo J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feng W, Ribeiro R, Wagner R, Nguyen H, Apriletti J, Fletterick R, Baxter J, Kushner P, West B. Hormone-dependent coactivator binding to a hydrophobic cleft on nuclear receptors. Science. 1998;280:1747–1749. doi: 10.1126/science.280.5370.1747. [DOI] [PubMed] [Google Scholar]
- 5.Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- 6.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M, Glass C. Polarity-specific activities of retinoic acid receptors determined by a co-repressor. Nature. 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 7.Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, Seto E, Eisenman RN, Rose DW, Glass CK, Rosenfeld MG. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- 8.Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- 9.Belandia B, Parker MG. Nuclear receptors: a rendezvous for chromatin remodeling factors. Cell. 2003;114:277–280. doi: 10.1016/s0092-8674(03)00599-3. [DOI] [PubMed] [Google Scholar]
- 10.Robyr D, Wolffe A, Wahli W. Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol Endocrinol. 2000;14:329–347. doi: 10.1210/mend.14.3.0411. [DOI] [PubMed] [Google Scholar]
- 11.Zhu Y, Qi C, Calandra C, Rao MS, Reddy JK. Cloning and identification of mouse steroid receptor coactivator-1 (mSRC-1), as a coactivator of peroxisome proliferator-activated receptor gamma. Gene Expr. 1996;6:185–195. [PMC free article] [PubMed] [Google Scholar]
- 12.Hong H, Kohli K, Trivedi A, Johnson D, Stallcup M. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci U S A. 1996;93:4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voegel JJ, Heine MJ, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. Embo J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- 14.Yuan C, Ito M, Fondell J, Fu Z, Roeder R. The TRAP220 component of a thyroid hormone receptor- associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci U S A. 1998;95:7939–7944. doi: 10.1073/pnas.95.14.7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rachez C, Lemon B, Suldan Z, Bromleigh V, Gamble M, Naar A, Erdjument-Bromage H, Tempst P, Freedman L. Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature. 1999;398:824–828. doi: 10.1038/19783. [DOI] [PubMed] [Google Scholar]
- 16.Renaud J, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature. 1995;378:681–689. doi: 10.1038/378681a0. [DOI] [PubMed] [Google Scholar]
- 17.Shiau A, Barstad D, Loria P, Cheng L, Kushner P, Agard D, Greene G. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 18.Kim S, Shevde NK, Pike JW. 1,25-Dihydroxyvitamin D3 stimulates cyclic vitamin D receptor/retinoid X receptor DNA-binding, co-activator recruitment, and histone acetylation in intact osteoblasts. J Bone Miner Res. 2005;20:305–317. doi: 10.1359/JBMR.041112. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Lambert MH, Xu HE. Activation of nuclear receptors: a perspective from structural genomics. Structure. 2003;11:741–746. doi: 10.1016/s0969-2126(03)00133-3. [DOI] [PubMed] [Google Scholar]
- 20.Perissi V, Staszewski L, McInerney E, Kurokawa R, Krones A, Rose D, Lambert M, Milburn M, Glass C, Rosenfeld M. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 1999;13:3198–3208. doi: 10.1101/gad.13.24.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu HE, Stanley TB, Montana VG, Lambert MH, Shearer BG, Cobb JE, McKee DD, Galardi CM, Plunket KD, Nolte RT, Parks DJ, Moore JT, Kliewer SA, Willson TM, Stimmel JB. Structural basis for antagonist-mediated recruitment of nuclear co-repressors by PPARalpha. Nature. 2002;415:813–817. doi: 10.1038/415813a. [DOI] [PubMed] [Google Scholar]
- 22.Heery D, Kalkhoven E, Hoare S, Parker M. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature. 1997;387:733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 23.Vanhooke JL, Benning MM, Bauer CB, Pike JW, DeLuca HF. Molecular structure of the rat vitamin D receptor ligand binding domain complexed with 2-carbon-substituted vitamin D3 hormone analogues and a LXXLL-containing coactivator peptide. Biochemistry. 2004;43:4101–4110. doi: 10.1021/bi036056y. [DOI] [PubMed] [Google Scholar]
- 24.Jurutka PW, Hsieh JC, Remus LS, Whitfield GK, Thompson PD, Haussler CA, Blanco JC, Ozato K, Haussler MR. Mutations in the 1,25-dihydroxyvitamin D3 receptor identifying C-terminal amino acids required for transcriptional activation that are functionally dissociated from hormone binding, heterodimeric DNA binding, and interaction with basal transcription factor IIB, in vitro. J Biol Chem. 1997;272:14592–14599. doi: 10.1074/jbc.272.23.14592. [DOI] [PubMed] [Google Scholar]
- 25.Jimenez-Lara AM, Aranda A. Lysine 246 of the vitamin D receptor is crucial for ligand-dependent interaction with coactivators and transcriptional activity. J Biol Chem. 1999;274:13503–13510. doi: 10.1074/jbc.274.19.13503. [DOI] [PubMed] [Google Scholar]
- 26.Hu X, Lazar M. The CoRNR motif controls the recruitment of corepressors by nuclear hormone receptors. Nature. 1999;402:93–96. doi: 10.1038/47069. [DOI] [PubMed] [Google Scholar]
- 27.Nagy L, Kao H, Love J, Li C, Banayo E, Gooch J, Krishna V, Chatterjee K, Evans R, Schwabe J. Mechanism of corepressor binding and release from nuclear hormone receptors. Genes Dev. 1999;13:3209–3216. doi: 10.1101/gad.13.24.3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cachon-Gonzalez MB, Fenner S, Coffin JM, Moran C, Best S, Stoye JP. Structure and expression of the hairless gene of mice. Proc Natl Acad Sci U S A. 1994;91:7717–7721. doi: 10.1073/pnas.91.16.7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie Z, Chang S, Oda Y, Bikle DD. Hairless suppresses vitamin D receptor transactivation in human keratinocytes. Endocrinology. 2006;147:314–323. doi: 10.1210/en.2005-1111. [DOI] [PubMed] [Google Scholar]
- 30.Hsieh JC, Sisk JM, Jurutka PW, Haussler CA, Slater SA, Haussler MR, Thompson CC. Physical and functional interaction between the vitamin D receptor and hairless corepressor, two proteins required for hair cycling. J Biol Chem. 2003;278:38665–38674. doi: 10.1074/jbc.M304886200. [DOI] [PubMed] [Google Scholar]
- 31.Cichon S, Anker M, Vogt IR, Rohleder H, Putzstuck M, Hillmer A, Farooq SA, Al-Dhafri KS, Ahmad M, Haque S, Rietschel M, Propping P, Kruse R, Nothen MM. Cloning, genomic organization, alternative transcripts and mutational analysis of the gene responsible for autosomal recessive universal congenital alopecia. Hum Mol Genet. 1998;7:1671–1679. doi: 10.1093/hmg/7.11.1671. [DOI] [PubMed] [Google Scholar]
- 32.Li YC, Pirro AE, Amling M, Delling G, Baron R, Bronson R, Demay MB. Targeted ablation of the vitamin D receptor: an animal model of vitamin D-dependent rickets type II with alopecia. Proceedings of the National Academy of Sciences USA. 1997;94:9831–9835. doi: 10.1073/pnas.94.18.9831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshizawa T, Handa Y, Uematsu Y, Takeda S, Sekine K, Yoshihara Y, Kawakami T, Arioka K, Sato H, Uchiyama Y, Masushige S, Fukamizu A, Matsumoto T, Kato S. Mice lacking the vitamin D receptor exhibit impaired bone formation, uterine hypoplasia and growth retardation after weaning. Nat Genet. 1997;16:391–396. doi: 10.1038/ng0897-391. [DOI] [PubMed] [Google Scholar]
- 34.Potter GB, Zarach JM, Sisk JM, Thompson CC. The thyroid hormone-regulated corepressor hairless associates with histone deacetylases in neonatal rat brain. Mol Endocrinol. 2002;16:2547–2560. doi: 10.1210/me.2002-0115. [DOI] [PubMed] [Google Scholar]
- 35.Moraitis AN, Giguere V, Thompson CC. Novel mechanism of nuclear receptor corepressor interaction dictated by activation function 2 helix determinants. Mol Cell Biol. 2002;22:6831–6841. doi: 10.1128/MCB.22.19.6831-6841.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skorija K, Cox M, Sisk JM, Dowd DR, MacDonald PN, Thompson CC, Demay MB. Ligand-independent actions of the vitamin D receptor maintain hair follicle homeostasis. Mol Endocrinol. 2005;19:855–862. doi: 10.1210/me.2004-0415. [DOI] [PubMed] [Google Scholar]
- 37.Panteleyev AA, Botchkareva NV, Sundberg JP, Christiano AM, Paus R. The role of the hairless (hr) gene in the regulation of hair follicle catagen transformation. Am J Pathol. 1999;155:159–171. doi: 10.1016/S0002-9440(10)65110-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perakyla M, Malinen M, Herzig KH, Carlberg C. Gene regulatory potential of nonsteroidal vitamin D receptor ligands. Mol Endocrinol. 2005;19:2060–2073. doi: 10.1210/me.2004-0417. [DOI] [PubMed] [Google Scholar]
- 39.Boehm MF, Fitzgerald P, Zou A, Elgort MG, Bischoff ED, Mere L, Mais DE, Bissonnette RP, Heyman RA, Nadzan AM, Reichman M, Allegretto EA. Novel nonsecosteroidal vitamin D mimics exert VDR-modulating activities with less calcium mobilization than 1,25-dihydroxyvitamin D3. Chem Biol. 1999;6:265–275. doi: 10.1016/S1074-5521(99)80072-6. [DOI] [PubMed] [Google Scholar]
- 40.Moore JM, Galicia SJ, McReynolds AC, Nguyen NH, Scanlan TS, Guy RK. Quantitative proteomics of the thyroid hormone receptor-coregulator interactions. J Biol Chem. 2004;279:27584–27590. doi: 10.1074/jbc.M403453200. [DOI] [PubMed] [Google Scholar]
- 41.Estebanez-Perpina E, Moore JM, Mar E, Delgado-Rodrigues E, Nguyen P, Baxter JD, Buehrer BM, Webb P, Fletterick RJ, Guy RK. The molecular mechanisms of coactivator utilization in ligand-dependent transactivation by the androgen receptor. Journal of Biological Chemistry. 2005;280:8060–8068. doi: 10.1074/jbc.M407046200. [DOI] [PubMed] [Google Scholar]
- 42.Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 43.Rochel N, Wurtz JM, Mitschler A, Klaholz B, Moras D. The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol Cell. 2000;5:173–179. doi: 10.1016/s1097-2765(00)80413-x. [DOI] [PubMed] [Google Scholar]
- 44.Zamir I, Harding HP, Atkins GB, Horlein A, Glass CK, Rosenfeld MG, Lazar MA. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16:5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna N, Lanz R, O'Malley B. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 46.Jurutka PW, Hsieh JC, Nakajima S, Haussler CA, Whitfield GK, Haussler MR. Human vitamin D receptor phosphorylation by casein kinase II at Ser-208 potentiates transcriptional activation. Proc Natl Acad Sci U S A. 1996;93:3519–3524. doi: 10.1073/pnas.93.8.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bramlett KS, Wu Y, Burris TP. Ligands specify coactivator nuclear receptor (NR) box affinity for estrogen receptor subtypes. Mol Endocrinol. 2001;15:909–922. doi: 10.1210/mend.15.6.0649. [DOI] [PubMed] [Google Scholar]
- 48.Liao L, Kuang SQ, Yuan Y, Gonzalez SM, O'Malley BW, Xu J. Molecular structure and biological function of the cancer-amplified nuclear receptor coactivator SRC-3/AIB1. J Steroid Biochem Mol Biol. 2002;83:3–14. doi: 10.1016/s0960-0760(02)00254-6. [DOI] [PubMed] [Google Scholar]
- 49.McInerney EM, Rose DW, Flynn SE, Westin S, Mullen TM, Krones A, Inostroza J, Torchia J, Nolte RT, Assa-Munt N, Milburn MV, Glass CK, Rosenfeld MG. Determinants of coactivator LXXLL motif specificity in nuclear receptor transcriptional activation. Genes & Development. 1998;12:3357–3368. doi: 10.1101/gad.12.21.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang C, Norris JD, Gron H, Paige LA, Hamilton PT, Kenan DJ, Fowlkes D, McDonnell DP. Dissection of the LXXLL nuclear receptor-coactivator interaction motif using combinatorial peptide libraries: discovery of peptide antagonists of estrogen receptors alpha and beta. Mol Cell Biol. 1999;19:8226–8239. doi: 10.1128/mcb.19.12.8226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nolte RT, Wisely GB, Westin S, Cobb JE, Lambert MH, Kurokawa R, Rosenfeld MG, Willson TM, Glass CK, Milburn MV. Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-gamma. Nature. 1998;395:137–143. doi: 10.1038/25931. [DOI] [PubMed] [Google Scholar]
- 52.Zella LA, Chang CY, McDonnell DP, Wesley Pike J. The vitamin D receptor interacts preferentially with DRIP205-like LxxLL motifs. Arch Biochem Biophys. 2007;460:206–212. doi: 10.1016/j.abb.2006.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanhooke JL, Tadi BP, Benning MM, Plum LA, DeLuca HF. New analogs of 2-methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3 with conformationally restricted side chains: evaluation of biological activity and structural determination of VDR-bound conformations. Arch Biochem Biophys. 2007;460:161–165. doi: 10.1016/j.abb.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 54.Rachez C, Gamble M, Chang C, Atkins G, Lazar M, Freedman L. The DRIP complex and SRC-1/p160 coactivators share similar nuclear receptor binding determinants but constitute functionally distinct complexes. Molecular & Cellular Biology. 2000;20:2718–2726. doi: 10.1128/mcb.20.8.2718-2726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tagami T, Lutz WH, Kumar R, Jameson JL. The interaction of the vitamin D receptor with nuclear receptor corepressors and coactivators. Biochem Biophys Res Commun. 1998;253:358–363. doi: 10.1006/bbrc.1998.9799. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Martinez R, Zambrano A, Castillo AI, Aranda A. Vitamin D-dependent recruitment of corepressors to vitamin D/retinoid X receptor heterodimers. Mol Cell Biol. 2008;28:3817–3829. doi: 10.1128/MCB.01909-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ciesielski F, Rochel N, Moras D. Adaptability of the Vitamin D nuclear receptor to the synthetic ligand Gemini: remodelling the LBP with one side chain rotation. J Steroid Biochem Mol Biol. 2007;103:235–242. doi: 10.1016/j.jsbmb.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 58.Shimizu M, Miyamoto Y, Takaku H, Matsuo M, Nakabayashi M, Masuno H, Udagawa N, Deluca HF, Ikura T, Ito N. 2-Substituted-16-ene-22-thia-1alpha,25-dihydroxy-26,27-dimethyl-19-norvita min D(3) analogs: Synthesis, biological evaluation, and crystal structure. Bioorg Med Chem. 2008 doi: 10.1016/j.bmc.2008.05.043. [DOI] [PubMed] [Google Scholar]
- 59.Geistlinger TR, Guy RK. An inhibitor of the interaction of thyroid hormone receptor beta and glucocorticoid interacting protein 1. J Am Chem Soc. 2001;123:1525–1526. doi: 10.1021/ja005549c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.