Abstract
The prohormone convertases (PCs) 1/3 and 2 accomplish the major proteolytic cleavage events in neuroendocrine tissues; each of these convertases has a small associated binding protein that inhibits convertase action in the secretory pathway. The proSAAS protein binds to PC1/3, whereas the 7B2 protein binds to PC2. However, both convertase-binding proteins are more widely expressed than their cognate enzymes, suggesting that they may perform other functions as well. All known mammalian proSAASs are over 85% conserved; thus, identifying functionally important segments has been impossible. Here, we report the first identification of nonmammalian proSAAS molecules, from Xenopus and zebrafish (Danio rerio). Although these two proteins show an overall amino acid sequence identity of only 29 and 30% with mouse proSAAS, two 14–16 residue hydrophobic segments (predicted to form α-helices) and two, nine through 11 residue sequences containing basic convertase cleavage sites are highly conserved; therefore, these sequences may be of functional importance. Confidence that these nonmammalian molecules represent authentic proSAAS is supported by the finding that both inhibit mouse PC1/3 with nanomolar inhibition constants; human furin was not inhibited. In vitro, the two proteins were cleaved by PC2 and furin to three or more peptide products. Both zebrafish and Xenopus proSAAS exhibited neural and endocrine distributions, as assessed by in situ and PCR experiments, respectively. In summary, the identification of proSAAS molecules in lower vertebrates provides clues as to functional regions within this widely expressed neuroendocrine protein.
The identification of two proSAAS molecules in lower vertebrates provides clues as to functional regions within this widely-expressed neuroendocrine protein.
The biochemical mechanism of prohormone processing at pairs of basic residues is known to be conserved from yeast to man. The proteolytic convertases that accomplish this paired basic processing are also related: the catalytic domain of kex2, responsible for pro-α mating factor processing in yeast, exhibits 50% homology with proprotein convertase 1 [prohormone convertase (PC) 1/3], responsible for proinsulin processing in vertebrates (1). The convertase family has seven members with specificity for pairs of basic residues; among these, PC1/3 and PC2 are thought to accomplish the majority of proteolytic processing within neural and endocrine tissues. PC2 may be evolutionarily older than PC1/3 because PC2 homologs can be found in insects (2) and worms (3,4), whereas PC1/3 homologs do not appear to be present in these species.
Proprotein convertases also require binding proteins; the first of these to be discovered was the small neuroendocrine protein 7B2, first identified in 1983 (5) but only recognized as a PC2 binding protein in 1994 (6). Work by our group showed that this protein functions as an interesting bifunctional regulator of PC2 action. It is absolutely required for the production of an activatable zymogen (7), most likely by preventing proPC2 aggregation within the secretory pathway (8), and it also contains an inhibitory segment that is thought to prevent premature action of the enzyme before the granular compartment (9,10,11). 7B2 homologs are found in all organisms that express PC2, from worms (3,4) to flies (2), to higher vertebrates (reviewed in Refs. 14 and 15).
PC1/3 also has a specific binding protein, known as proSAAS (16). proSAAS is structurally organized similarly to 7B2 in that it also contains a potent PC1/3 inhibitory sequence at its C terminus; this peptide follows a furin cleavage site (17,18). proSAAS has inhibited the production of newly synthesized proopiomelanocortin (16), is involved in weight homeostasis (19), and has recently been implicated in control of the circadian clock (20). Despite the fact that PC1/3 has been identified in fish, amphibians, and even a protochordate (21), only mammalian proSAAS molecules have been identified to date (human, mouse, rat, cow, and dog). We speculated that this could be due to the conservation of only small segments of the proSAAS molecule, similarly to Caenorhabditis elegans 7B2, which exhibits only 30% overall homology with mouse 7B2 but very high conservation in two functional regions, such as the 36-residue functional domain (22) and the PC2 inhibitory region (9,10). By searching Expressed Sequence Tag databases for an analogously placed short sequence within proSAAS, we succeeded in identifying proSAAS molecules in fish and frogs. As detailed in Results, these molecules are indeed not well conserved overall but are highly conserved in short segments.
Materials and Methods
Construction of prokaryotic vectors
A full-length Xenopus laevis (Xenopus) proSAAS cDNA clone was purchased from Open Biosystems (Huntsville, AL) (clone identification no. 694770, accession no. CD253499). The GenBank sequence CD301470 contains only carboxy-terminal sequence, whereas the GenBank sequence CD253499, the source of the Open Biosystems clone, appears to contain only amino-terminal sequence, but actually encodes the entire coding sequence. The Danio rerio (zebrafish) proSAAS cDNA (GenBank EH441497) was a kind gift of Dr. C. Wei (Genome Institute of Singapore, Singapore) (clone FDR103-P00028-DEPE-F_C24). Xenopus and zebrafish proSAASs were amplified by PCR and then subcloned into the His-tag pQE30 vector at the SphI and HindIII sites to yield a construct lacking the signal peptide and with an amino-terminal Histag (amino-terminal sequence MRGSHHHHHHGSAC-KPLS for fish proSAAS, and MRGSHHHHHHGSAC-KPLG for Xenopus proSAAS). The PCRs used the following primers: 5′-AAC CGC ATG CAA GCC CCT GGG AAG TGT C-3′, directed toward the 5′-end of Xenopus proSAAS; 5′-CGA TAA AAG CTT TTA GTC TGG GAG GTA CTT CA-3′, directed toward the 3′-end of Xenopus proSAAS; 5′-AAC CGC ATG CAA GCC TCT TTC CGC CATG-3′, directed to the 5′-end of zebrafish proSAAS; and 5′-ATC GCA AAG CTT TTA GGC TGG GAG GTA CTG CAG-3′, directed to the 3′-end of zebrafish proSAAS. The annealing temperature for both PCRs was 58 C, and 30 cycles were used. The final concentration of each deoxynucleotide triphosphate was 200 μm, of primers was 2 μm, and of Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA) was 0.02 U. All cDNAs generated by PCR were verified by DNA sequencing.
Purification of recombinant fish and Xenopus proSAAS
The pQE30 plasmids encoding fish and Xenopus proSAAS were expressed in XL1Blue, induced with isopropylthiogalactoside, and extracted with guanidinium chloride as previously described for mouse proSAAS (23). His-tagged proteins were purified by chromatography over a 5-ml nickel bead column (Amersham Pharmacia Biotech, Uppsala, Sweden); the column was washed with 30 ml each of: 1) 8 m urea, 100 mm sodium phosphate monobasic, 10 mm Tris (pH 8.0); 2) 8 m urea, 100 mm sodium phosphate, 10 mm Tris (pH 6.5); and 3) 8 m urea, 100 mm sodium phosphate, 10 mm Tris, 10 mm imidazole (pH 6.5), before elution with 8 m urea in 100 mm sodium phosphate (pH 4.0). The proteins were further purified by reverse-phase chromatography on a 10- × 250-mm Vydac C4 column (Grace Davison Discovery Sciences, Deerfield, IL). A 5-min wash of 20% acetonitrile in 0.1% trifluoroacetic acid, followed by a 75-min gradient from 20–80% acetonitrile in 0.1% trifluoroacetic acid, was used. Proteins were lyophilized and resuspended in 5 mm acetic acid for inhibition assays. Mass spectroscopy was performed on a Shimadzu/Kratos matrix-assisted laser desorption/ionization time-of-flight instrument (Shimadzu Biotech, Columbia, MD) using a sinapinic acid matrix by the Proteomics Core of the University of Maryland. His-tagged full-length mouse proSAAS was prepared as previously described (23). Protein concentration was measured with the Bradford method.
Enzyme assay
The assay for PC1/3 was performed in 96-well clear bottom black microtiter plates (BD Biosciences, Franklin Lakes, NJ) in a final volume of 100 μl, containing 100 mm sodium acetate (pH 5.5), 2 mm CaCl2, 0.2% octyl glucoside, 0.1% NaN3, and 0.1 mg/ml BSA. The substrate, pyr-Glu-Arg-Thr-Lys-Arg-methylcoumarinamide (Peptide Institute, Lexington, KY) was used at a final concentration of 50 μm. Recombinant mouse PC (mPC) 1/3 (24) was used at 8 nm final concentration, which generated 0.2 fluorescence U/min, in which one fluorescence unit corresponds to 3.8 pmol aminomethylcoumarin. All assays were performed in triplicate for 1 h at 37 C, and were quantitated using a Fluoroscan Ascent fluorometer (LabSystems, Waltham, MA) with excitation/emission wavelengths of 380/460 nm. Data were analyzed using GraphPad Prism 4 (GraphPad Software Inc., San Diego, CA). The Xenopus proSAAS inhibition curve was performed using final concentrations of 0.003–7.2 μm recombinant purified proSAAS, and the zebrafish proSAAS inhibition curve was performed between 0.3 nm and 2.4 μm fish proSAAS.
The assay for human furin (hfurin) (25) was performed using 1.5 nm furin, in 100 mm HEPES (pH 7.0), 2 mm CaCl2, 0.2% octyl glucoside, 0.1% NaN3, and 0.1 mg/ml BSA. The IC50 was calculated using nonlinear regression. The inhibition constants were calculated using the equation Ki = IC50/[1 + (substrate)/Michaelis-Menten constant], where Michaelis-Menten constant values for PC1/3 and furin are 11 and 8 μm, respectively, and Ki represents the inhibition constant.
Convertase digestion
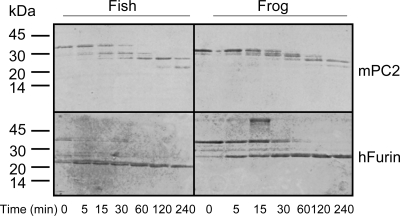
mPC2 and hfurin were purified from conditioned media taken from Chinese hamster ovary-overexpressing cells using procedures described previously (25,26). Each proSAAS digestion was performed with 2 μg proSAAS, 2 U mPC2 and hfurin in a 50-μl reaction. The reactions for PC2 contained 100 mm sodium acetate (pH 5.0), 5 mm CaCl2, and 0.1% Brij-35. For furin, 100 mm HEPES (pH 7.0), 5 mm CaCl2, and 0.1% Brij-35 was used. Reactions were incubated at 37 C for the times indicated in Fig. 5. One unit of enzyme activity is equal to the amount of the enzyme that is required to cleave 1 pmol/min of the pRTKR-aminomethyl coumarin fluorogenic substrate (Peptides Intl., Lexington, KY). After the reaction, products were separated on 18% Tris-HCl gels and stained with Coomassie blue.
Figure 5.
His-tagged Xenopus and D. rerio proSAASs are cleaved by PC2 and furin. Two micrograms of each recombinant proSAAS were subjected to proteolysis using 2 U of either mPC2 or hfurin. Samples were separated on a 18% Tris-HCl gel and stained with Coomassie blue.
PCR analysis of mRNA expression in Xenopus tissues and developmental stages
Tissues were homogenized in TRIzol reagent (Invitrogen Corp., Carlsbad, CA), and 2 μg total RNA from each tissue or developmental stage was used for cDNA synthesis. Of the total pool of cDNA, 5% was used in the PCR. Primers used were: forward, 5′-AAGCCCCTGGGAAGTGTC-3′; and reverse, 5′-TTAGTCTGGGAGGTACTTC-3′. The annealing temperature was 54 C, and 40 cycles were run. Staging of embryos was performed according to Nieuwkoop and Faber (27).
D. rerio maintenance
Adult zebrafish were maintained and experimental embryos were produced essentially as described in The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio) (28). Embryos were staged according to hours post fertilization (hpf) at 28.5 C and morphological criteria (29).
Zebrafish proSAAS construction for probe and probe synthesis
The zebrafish proSAAS sequence originating from the pQE30/zebrafish proSAAS plasmid was subcloned into a pBluescript KS vector with BamHI and HindIII restriction enzyme sites. The new plasmid was digested with BamHI for synthesizing the antisense probe using T3 RNA polymerase and with HindIII for synthesizing the sense probe using T7 RNA polymerase. RNA probes were synthesized using standard procedures (30) without the alkaline hydrolysis step.
Whole mount in situ hybridization
Whole mount in situ hybridization with digoxigenin-labeled RNA probes was performed as described in Ref. (30) with the modifications described below. Following fixation with 4% paraformaldehyde in PBS, we peeled the head skin of embryos at 5 d post fertilization (dpf) and removed the brain from the upper jaw. After a rehydration step from storage in methanol at −20 C, a bleaching step with 6% hydrogen peroxide/PBS/0.1% Tween 20 was performed for 2–4 h, depending on the remaining pigmentation of embryos. The dissected brain samples were not digested with proteinase K before the hybridization step. After staining with nitro-blue tetrazolium chloride/5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (Roche Applied Science, Indianapolis, IN), we mounted the samples in 100% glycerol for photography using a Leica DM IRB inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany) and a DFC 480 charge-coupled device camera with Leica Application Suite software. We removed eyes for lateral-view pictures at 2 and 3-dpf stages before mounting.
Results
Zebrafish and Xenopus proSAASs are not highly conserved
We used a short peptide sequence from mouse preproSAAS (residues 191–218) and loose search parameters (expect 1000; PAM30; no filtering) to locate Xenopus proSAAS in the Xenopus Expressed Sequence Tags database; proSAAS sequences were identifiable as such because of the highly conserved downstream convertase inhibitory segment (LLR[V/I]KR[L/I]). The Xenopus sequence was then used to probe for the zebrafish sequence. Sequencing of cDNA clones identified through these Basic Alignment Search Tool searches resulted in the sequences shown in Fig. 1. Several other fish proSAAS sequences were also identified, such as: carp (Cyprinus carpio, GenBank CA967932.2); fathead minnow (Pimepheles promelas, DT250416.1); spined stickleback (Gasterosteus aculeatus, DW606844.1); and ricefish (Oryzias latipes, AM308793.1). Related Xenopus sequences were identified as well (Xenopus tropicalis, GenBank CX823517).
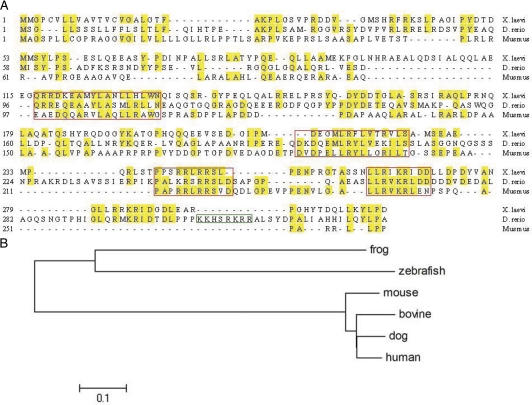
Figure 1.
Alignment of Xenopus, D. rerio, and Mus musculus proSAAS sequences. A, Sequence alignment. The red boxes correspond to highly conserved areas. Note the additional highly basic sequence within the fish CT peptide (green box). B, Phylogenetic tree.
The homology of Xenopus and zebrafish proSAAS with mouse proSAAS is shown in Fig. 1A. The optimal signal peptide cleavage site determined using the SignalP algorithm appears to vary by protein; it does not always precede the conserved A[R/K]P sequence present in the SAAS peptide. Overall conservation of the Xenopus and zebrafish sequences with mouse is only 29 and 30% identity, respectively; this low degree of conservation makes the identification of convertase binding protein homologs extremely difficult. However, there are four segments that exhibit over 60% identity; these are shown boxed in red in Fig. 1A. The first two segments exhibit 67 and 64% overall amino acid identity, whereas the second two segments display 76 and 85% identity with mouse proSAAS. The last segment represents the inhibitory CT peptide (17,18); this sequence is also well conserved in 7B2 from flies to man (3). Mouse proSAAS has an isoelectric point (pI) of 5.7, which is considerably lower than that of Xenopus or zebrafish proSAAS (6.1 and 7.4, respectively). No cysteines are present in any proSAAS sequences identified to date.
The overall structure of the protein, an inhibitory peptide separated from the remainder of the protein by a canonical furin site, is similar in all species. It is interesting to note that the carboxy-terminal region of zebrafish proSAAS has an additional highly basic sequence, reminiscent of the acquisition of additional carboxy-terminal inhibition sequences in 7B2s from invertebrates such as Lymnaea (31). Both proSAAS proteins are considerably larger than the mouse protein.
The conservation of processed peptides between any of these vertebrate proSAASs is not strong, suggesting that proSAAS may represent an actual peptide precursor only in higher vertebrates. The eponymous SAAS sequence is not present in either the fish or frog precursors. With the exception of the furin consensus cleavage sequences, the location of the basic processing sites is also not conserved between fish and Xenopus, although the number of processing sites is roughly equivalent in all three species. Aside from the inhibitory sequences, the most highly conserved segments consist of two short hydrophobic sequences, shown in the first two red boxes in Fig. 1A. Using the NNPredict program, (www.cmpharm.ucsf.edu/∼nomi/nnpredict.html) these are both strongly predicted to assume a helical structure.
Figure 1B depicts the phylogenetic tree of the various proSAAS sequences available to date.
D. rerio proSAAS exhibits a predominantly neural distribution during development
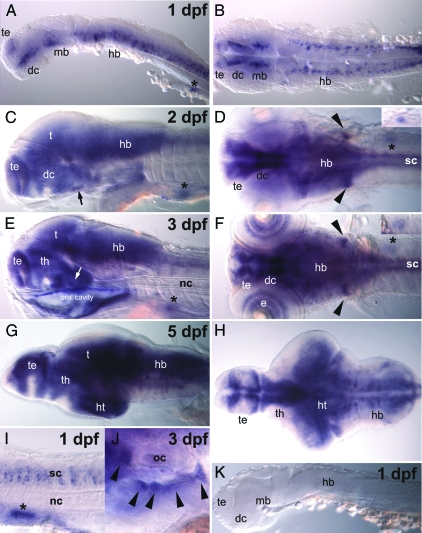
A strong hybridization signal for proSAAS was detectable at 24 hpf in the developing brain and spinal cord (Fig. 2, A, B, and I). Strong signals were seen in many parts of the central nervous system, including developing telencephalon, diencephalon, rhombencephalon, and spinal cord (Fig. 2, C and D) at 48 hpf. The vagal ganglia and pituitary gland displayed strong signals as well. Strong expression throughout the nervous system continued through 3–5 dpf (Fig. 2, E–H), the last day examined here. From 2 dpf onwards, proSAAS expression was also seen in developing pancreas and eyes (Fig. 2, C, E, and F). Nonneuronal and nonendocrine tissues did not show significant expression. Cranial nerve ganglia exhibit strong signals (Fig. 2J). Samples hybridized with the sense probe did not show a reaction (Fig. 2K).
Figure 2.
Distribution of D. rerio proSAAS. proSAAS gene expression was observed in neural tissues, which are telencephalon (te), diencephalon (dc), midbrain (mb), hindbrain (hb), spinal cord (sc), pituitary (arrows in C and E), and cranial nerves (arrowheads in D and F) at 1 dpf (A, B, and I), 2 dpf (C and D), 3 dpf (E and F), and 5 dpf (G and H). Observations made at high magnification around the left optic capsule revealed the proSAAS expression in cranial nerves of gIX and gX (arrowheads in J). In nonneural tissue the proSAAS gene was expressed in pancreatic cells and its primordium, shown as asterisks (A, C–F, and I) and superimposed in D and F. Samples hybridized with sense probe did not show any staining (K). The orientation is anterior at the left in all figures. A, C–E, G, and I–K are from a lateral view. B, D, and F are from a dorsal view, and H is from a ventral view. e, Eye; ht, hypothalamus; nc, notochord; oc, otic capsule; t, tegmentum; th, thalamus.
Xenopus proSAAS exhibits a neuroendocrine distribution
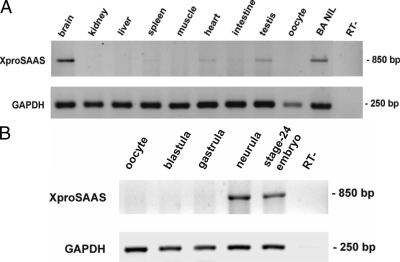
Figure 3A shows the distribution of Xenopus proSAAS mRNA in various tissues obtained from black-adapted Xenopus. The highest expression was observed in the brain; pituitary and testes also exhibited strong expression. Figure 3B confirms that proSAAS is not expressed in oocytes and demonstrates that expression begins in the neurula stage.
Figure 3.
Xenopus proSAAS tissue distribution and developmental expression. RNA was prepared from various tissues of black-adapted (BA) Xenopus (A) or from oocytes, blastula embryos (stage 5), gastrula (stage 10), neurula (stage 12.5), and stage-24 embryos (B). PCRs were performed according to the method described in Materials and Methods. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for RNA integrity. NIL, Neurointermediate lobe; RT-, negative control (absence of reverse transcriptase).
Xenopus and zebrafish proSAASs can both be cleaved by PC2 and furin
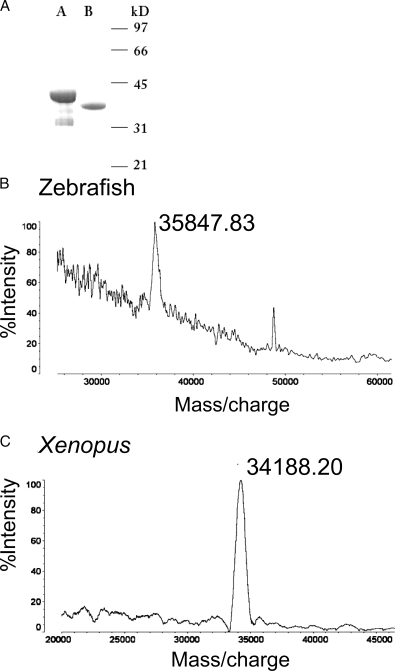
We generated recombinant amino-terminally His-tagged Xenopus and zebrafish proSAAS in bacteria. Purity was verified by SDS-PAGE (Fig. 4A) and mass spectroscopy, which yielded masses of 35848 and 34188 (Fig. 4B) (theoretical masses for zebrafish and Xenopus His-tagged proSAAS are 35887 and 34026, respectively). We examined the ability of these proteins to serve as substrates for furin and PC2 (Fig. 5). Both proteins were cleaved by both enzymes; stable 25-kDa products were generated from Xenopus proSAAS, whereas fish proSAAS was sequentially cleaved to a slightly smaller product by both enzymes. Four strong proteolytic products were observed upon digestion of fish and Xenopus proSAAS with PC2; five or six paired basic sites, including the furin consensus sequences, RXXR, are present within the Xenopus and fish sequences, respectively. However, the close proximity of the terminal processing sites renders separation of certain processing products impossible. Furin generated only three major proteolytic products for both proteins; there are three furin consensus sites in Xenopus proSAAS, but four in the zebrafish. Again, it is probable that cleaved peptides occurring at sites in close proximity in the carboxy-terminal tail cannot be readily distinguished. These results are in general agreement with the cleavage pattern previously determined for mouse proSAAS (23), with the exception of the production of a small peptide (most likely the SAAS peptide itself). These data show that both the zebrafish and Xenopus proteins generate about a 20-kDa central core of unprocessed protein that is likely to contain one or both of the internal hydrophobic segments mentioned previously.
Figure 4.
Purity of recombinant Xenopus and D. rerio proSAASs. A, Ten to 15 μg recombinant protein was electrophoresed on a 15% SDS-PAGE gel and stained with Coomassie blue. Lane A, Zebrafish. Lane B, Xenopus. Mass spectroscopic analysis of zebrafish (B) and Xenopus (C) proSAAS.
Xenopus and zebrafish proSAASs are both nanomolar inhibitors of PC1/3
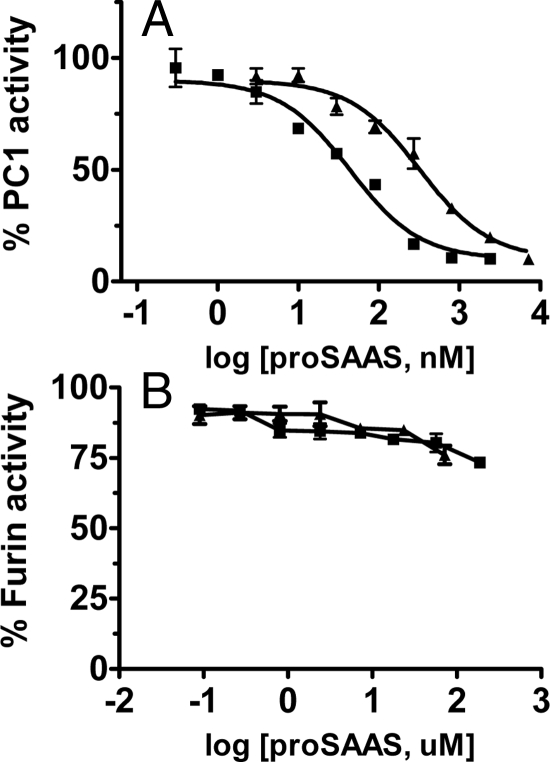
The carboxy-terminal portion of mammalian proSAAS is known to inhibit mPC1/3 (17,18); therefore, recombinant frog and fish proSAASs were used in an inhibition assay against mPC1/3. The IC50 values for Xenopus and zebrafish proSAASs were 320 and 44 nm, which correspond to Ki values of 58 and 7.8 nm, respectively (Fig. 6A). We believed that the second inhibitory sequence in zebrafish proSAAS located near the carboxyl terminus might represent a furin inhibitor, given the fact that peptides with multiple basic residues have been highly inhibitory to furin (32); however, neither protein inhibited hfurin. At the highest concentration of Xenopus proSAAS obtainable, 72 μm, hfurin retained 76% activity, whereas at 188 μm zebrafish proSAAS, hfurin retained 73% activity (Fig. 6B). Thus, the IC50 values for both proSAASs are greater than 0.1 and 0.2 mm, supporting the notion that these proteins will not inhibit furin in vivo. The second stretch of basic residues in the CT peptide may account for the much greater inhibitory potency of zebrafish as opposed to Xenopus proSAAS.
Figure 6.
Recombinant His-tagged Xenopus and D. rerio proSAASs are nanomolar inhibitors of PC1/3 but do not inhibit furin. A, PC1 inhibition. Various concentrations of Xenopus proSAAS (3, 10, 30, 90, 270, 800, 2400, and 7200 nm) and zebrafish proSAAS (0.3, 1, 3, 10, 30, 90, 270, 800, and 2400 nm) were assayed for the inhibition against PC1/3. Squares show recombinant zebrafish proSAAS. Triangles indicate recombinant Xenopus proSAAS. The values shown represent triplicate determinations of the percent (%) control activity (mean ± sd). B, Furin inhibition. Various concentrations of Xenopus (0.09, 0.27, 0.8, 2.4, 7.2, 24, and 72 μm) and zebrafish (0.09, 0.27, 0.8, 2.4, 7.2, 17.8, 57, and 188 μm) His-tagged proSAASs were assayed for furin inhibition. Squares show recombinant zebrafish proSAAS. Triangles indicate recombinant Xenopus proSAAS. The values shown represent triplicate determinations of the percent (%) control activity (mean ± sd).
Discussion
7B2 and proSAAS are interesting convertase-binding proteins that are widely distributed in early embryonic tissues but restricted to neuroendocrine tissues in the adult (33,34,35). ProSAAS has been shown to bind to the convertase PC1/3 (16); however, unlike the binding of 7B2 to proPC2, proSAAS is not required for production of an active PC1/3 enzyme (36). The proSAAS carboxy-terminal peptide is a potent inhibitor of PC1/3, analogous to the inhibition of PC2 by the 7B2 carboxy-terminal peptide (17,18). However, the function of the large domain preceding the furin cleavage site is presently unclear. Because vertebrate proSAAS molecules are nearly completely conserved, little information is available on functional areas within this 20-kDa domain.
Our data show that only small portions of proSAAS are conserved between mammals and other vertebrates. In 7B2 a short, reasonably well-conserved sequence precedes the furin cleavage site; this sequence contains the distinctive diagnostic motif PPNPCP (14). In proSAAS a similarly conserved hydrophobic sequence appears in this general position. The function of this conserved sequence is not known; in 7B2, the conserved 36-residue segment that contains the PPNPCP motif acts to facilitate proPC2 maturation (22), most likely by blocking unproductive aggregation (8). It is interesting to note that this region was one of three proSAAS sequences identified by mass spectroscopic analysis of τ-inclusions in Pick’s disease, initially by cross-reaction with diacylglycerol kinase antibodies (37).
If proSAAS functions physiologically to inhibit PC1/3 within the secretory pathway (11), proSAAS molecules should be identifiable in any species containing PC1/3. PC1/3-like sequences have been identified in mammals, fish, amphibians, and platypus, though not in any birds or reptiles as yet. The protochordate Amphioxus (21), the sea slug Aplysia (38,39), and Hydra (40) also contain PC1/3-like sequences, suggesting that this enzyme evolved early in metazoan evolution. The PC1/3-like enzyme described in Hydra shows almost equal similarity to furin in its catalytic domain (40). The Aplysia PC1/3 sequence is also quite different from any vertebrate PC1/3s, with only 51% overall homology to mPC1/3 in the catalytic domain. Indeed, this homology is similar to the divergence of the yeast convertase kex2 and PC1/3 (1), suggesting that vertebrate and invertebrate PC1s could operate quite differently. We were unable to find proSAAS-like sequences in the Aplysia genome. Because we were similarly unsuccessful in locating chicken proSAAS using chicken databases, this failure may reflect the quality of sequence information currently available rather than the presence or absence of proSAAS. It will be extremely interesting to examine the structure of PC1/3 binding proteins in invertebrate species, if they are found to exist.
The observed tissue distribution of proSAAS in Xenopus and zebrafish is consistent with that found in mammals. In zebrafish, expression on 1–5 dpf was limited to neural and endocrine organs. proSAAS expression was widespread in all parts of the zebrafish brain, similarly to observations made in rodents (33, 35, 42). In Xenopus, expression was detected in the neurula stage, similar to the expression of Xenopus PC1/3 (13) and consistent with the development of the nervous system. The lack of expression of proSAAS mRNA in the oocyte contrasts with the early expression of 7B2 and secretogranins II and III (13).
In conclusion, here, we have identified two nonmammalian proSAAS molecules that exhibit neuroendocrine localization and are functionally active in inhibiting PC1/3. The conservation of two PC1/3-inhibitory sequences along with two hydrophobic sequences suggests that these segments are likely to contribute to proSAAS function. Further studies are needed to determine the physiological role of the two hydrophobic helices.
Acknowledgments
We thank J. Miceli for the expert purification of frog proSAASs. We are grateful to C. L. Wei for sending the D. rerio.
Footnotes
This work was supported by National Institutes of Health Grant DK49703 (to I.L.) and grants from the Academy of Finland and Sigrid Juselius Foundation (to P.P. and H.K.).
Disclosure Statement: The authors have nothing to disclose.
First Published Online October 23, 2008
Abbreviations: dpf, Days post fertilization; hfurin, human furin; hpf, hours post fertilization; mPC, mouse prohormone convertase; PC, prohormone convertase.
References
- Steiner DF, Smeekens SP, Ohagi S, Chan SJ 1992 The new enzymology of precursor processing endoproteases. J Biol Chem 267:23435–23438 [PubMed] [Google Scholar]
- Hwang JR, Siekhaus DE, Fuller RS, Taghert PH, Lindberg I 2000 Interaction of Drosophila melanogaster prohormone convertase 2 and 7B2. Insect cell-specific processing and secretion. J Biol Chem 275:17886–17893 [DOI] [PubMed] [Google Scholar]
- Lindberg I, Tu B, Muller L, Dickerson IM 1998 Cloning and functional analysis of C. elegans 7B2. DNA Cell Biol 17:727–734 [DOI] [PubMed] [Google Scholar]
- Kass J, Jacob TC, Kim P, Kaplan JM 2001 The EGL-3 proprotein convertase regulates mechanosensory responses of Caenorhabditis elegans. J Neurosci 21:9265–9272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidah NG, Hsi KL, De Serres G, Rochemont J, Hamelin J, Antakly T, Cantin M, Chretien M 1983 Isolation and NH2-terminal sequence of a highly conserved human and porcine pituitary protein belonging to a new superfamily. Immunocytochemical localization in pars distalis and pars nervosa of the pituitary and in the supraoptic nucleus of the hypothalamus. Arch Biochem Biophys 225:525–534 [DOI] [PubMed] [Google Scholar]
- Braks JA, Martens GJ 1994 7B2 is a neuroendocrine chaperone that transiently interacts with prohormone convertase PC2 in the secretory pathway. Cell 78:263–273 [DOI] [PubMed] [Google Scholar]
- Zhu X, Lindberg I 1995 7B2 facilitates the maturation of proPC2 in neuroendocrine cells and is required for the expression of enzymatic activity. J Cell Biol 129:1641–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SN, Lindberg I 2008 7B2 prevents unfolding and aggregation of prohormone convertase 2. Endocrinology 149:4116–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindberg I, van den Hurk WH, Bui C, Batie CJ 1995 Enzymatic characterization of immunopurified prohormone convertase 2: potent inhibition by a 7B2 peptide fragment. Biochemistry [Erratum (1995) 34:7780] 34:5486–5493 [DOI] [PubMed] [Google Scholar]
- Van Horssen AM, Van den Hurk WH, Bailyes EM, Hutton JC, Martens GJM, Lindberg I 1995 Identification of the region within the neuroendocrine polypeptide 7B2 responsible for the inhibition of prohormone convertase PC2. J Biol Chem 270:14292–14296 [DOI] [PubMed] [Google Scholar]
- Fortenberry Y, Liu J, Lindberg I 1999 The role of the 7B2 CT peptide in the inhibition of prohormone convertase 2 in endocrine cell lines. J Neurochem 73:994–1003 [DOI] [PubMed] [Google Scholar]
- Feng Y, Reznik SE, Fricker LD 2002 ProSAAS and prohormone convertase 1 are broadly expressed during mouse development. Brain Res Gene Expr Patterns 1:135–140 [DOI] [PubMed] [Google Scholar]
- Holling TM, van Herp F, Durston AJ, Martens GJ 2000 Differential onset of expression of mRNAs encoding proopiomelanocortin, prohormone convertases 1 and 2, and granin family members during Xenopus laevis development. Brain Res Mol Brain Res 75:70–75 [DOI] [PubMed] [Google Scholar]
- Muller L, Lindberg I 1999 The cell biology of the prohormone convertases PC1 and PC2. Prog Nucleic Acid Res Mol Biol 63:69–108 [DOI] [PubMed] [Google Scholar]
- Mbikay M, Seidah NG, Chretien M 2001 Neuroendocrine secretory protein 7B2: structure, expression and functions. Biochem J 357(Pt 2):329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricker LD, McKinzie AA, Sun J, Curran E, Qian Y, Yan L, Patterson SD, Courchesne PL, Richards B, Levin N, Mzhavia N, Devi LA, Douglass J 2000 Identification and characterization of proSAAS, a granin-like neuroendocrine peptide precursor that inhibits prohormone processing. J Neurosci 20:639–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron A, Fortenberry Y, Lindberg I 2000 The SAAS granin exhibits structural and functional homology to 7B2 and contains a highly potent hexapeptide inhibitor of PC1. FEBS Lett 473:135–138 [DOI] [PubMed] [Google Scholar]
- Qian Y, Devi LA, Mzhavia N, Munzer S, Seidah NG, Fricker LD 2000 The C-terminal region of proSAAS is a potent inhibitor of prohormone convertase 1. J Biol Chem 275:23596–23601 [DOI] [PubMed] [Google Scholar]
- Wei S, Feng Y, Che FY, Pan H, Mzhavia N, Devi LA, McKinzie AA, Levin N, Richards WG, Fricker LD 2004 Obesity and diabetes in transgenic mice expressing proSAAS. J Endocrinol 180:357–368 [DOI] [PubMed] [Google Scholar]
- Hatcher NG, Atkins Jr N, Annangudi SP, Forbes AJ, Kelleher NL, Gillette MU, Sweedler JV 2008 Mass spectrometry-based discovery of circadian peptides. Proc Natl Acad Sci USA 105:12527–12532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva AA, Chan SJ, Steiner DF 2000 Evolution of the prohormone convertases: identification of a homologue of PC6 in the protochordate amphioxus. Biochim Biophys Acta 1477:338–348 [DOI] [PubMed] [Google Scholar]
- Muller L, Zhu P, Juliano MA, Juliano L, Lindberg I 1999 A 36-residue peptide contains all of the information required for 7B2-mediated activation of prohormone convertase 2. J Biol Chem 274:21471–21477 [DOI] [PubMed] [Google Scholar]
- Sayah M, Fortenberry Y, Cameron A, Lindberg I 2001 Tissue distribution and processing of proSAAS by proprotein convertases. J Neurochem 76:1833–1841 [DOI] [PubMed] [Google Scholar]
- Zhou Y, Lindberg I 1993 Purification and characterization of the prohormone convertase PC1(PC3). J Biol Chem 268:5615–5623 [PubMed] [Google Scholar]
- Cameron A, Appel J, Houghten RA, Lindberg I 2000 Polyarginines are potent furin inhibitors. J Biol Chem 275:36741–36749 [DOI] [PubMed] [Google Scholar]
- Lamango NS, Zhu X, Lindberg I 1996 Purification and enzymatic characterization of recombinant prohormone convertase 2: stabilization of activity by 21 kDa 7B2. Arch Biochem Biophys 330:238–250 [DOI] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J 1967 Normal table of Xenopus laevis (Daudin). 2nd ed. Amsterdam: Elsevier/North Holland Publishing Co. [Google Scholar]
- Westerfield M 1995 The zebrafish book: a guide for the laboratory use of zebrafish (Brachydanio rerio). 3rd ed. Eugene, OR: Institute of Neuroscience, University of Oregon [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995 Stages of embryonic development of the zebrafish. Dev Dyn 203:253–310 [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH 1993 Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119:1203–1215 [DOI] [PubMed] [Google Scholar]
- Spijker S, Smit AB, Martens GJ, Geraerts WP 1997 Identification of a molluscan homologue of the neuroendocrine polypeptide 7B2. J Biol Chem 272:4116–4120 [DOI] [PubMed] [Google Scholar]
- Kacprzak MM, Peinado JR, Than ME, Appel J, Henrich S, Lipkind G, Houghten RA, Bode W, Lindberg I 2004 Inhibition of furin by polyarginine-containing peptides: nanomolar inhibition by nona-D-arginine. J Biol Chem 279:36788–36794 [DOI] [PubMed] [Google Scholar]
- Lanoue E, Day R 2001 Coexpression of proprotein convertase SPC3 and the neuroendocrine precursor proSAAS. Endocrinology 142:4141–4149 [DOI] [PubMed] [Google Scholar]
- Feng Y, Reznik SE, Fricker LD 2001 Distribution of proSAAS-derived peptides in rat neuroendocrine tissues. Neuroscience 105:469–478 [DOI] [PubMed] [Google Scholar]
- Morgan DJ, Mzhavia N, Peng B, Pan H, Devi LA, Pintar JE 2005 Embryonic gene expression and pro-protein processing of proSAAS during rodent development. J Neurochem 93:1454–1462 [DOI] [PubMed] [Google Scholar]
- Fortenberry Y, Hwang JR, Apletalina EV, Lindberg I 2002 Functional characterization of ProSAAS: similarities and differences with 7B2. J Biol Chem 277:5175–5186 [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Arawaka S, Koyama S, Kimura H, Ren CH, Wada M, Kawanami T, Kurita K, Daimon M, Kawakatsu S, Kadoya T, Goto K, Kato T 2003 An N-terminal fragment of ProSAAS (a granin-like neuroendocrine peptide precursor) is associated with τ inclusions in Pick’s disease. Biochem Biophys Res Commun 308:646–654 [DOI] [PubMed] [Google Scholar]
- Korner J, Chun J, Harter D, Axel R 1991 Isolation and functional expression of a mammalian prohormone processing enzyme, murine prohormone convertase 1. Proc Natl Acad Sci USA 88:6834–6838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorham EL, Nagle GT, Smith JS, Shen H, Kurosky A 1996 Molecular cloning of prohormone convertase 1 from the atrial gland of Aplysia. DNA Cell Biol 15:339–345 [DOI] [PubMed] [Google Scholar]
- Chan SJ, Oliva AAJ, LaMendola J, Grens A, Bode H, Steiner DF 1992 Conservation of the prohormone convertase gene family in Metazoa: analysis of cDNAs encoding a PC3-like protein from Hydra. Proc Natl Acad Sci USA 89:6678–6682 [DOI] [PMC free article] [PubMed] [Google Scholar]