Abstract

Two novel spiroaminals, marineosins A and B (1, 2), containing two pyrrole functionalities, were isolated from cultures of a marine sediment-derived actinomycete related to the genus Streptomyces. The marineosins, which appear to be derived from unknown modifications of prodigiosin-like pigment pathways, showed significant inhibition of human colon carcinoma (HCT-116) in an in vitro assay (IC50 = 0.5 μM for marineosin A) and selective activities in diverse cancer cell types.
Marine actinomycetes have become an important source of structurally unique secondary metabolites1 including many with potent biological acitvities. Among the actinomycetes frequently recovered from marine samples are members assignable to the genus Streptomyces, a diverse group of soil bacteria that account for the majority of microbially derived antibiotics discovered as of 2002.2 Although the genus Streptomyces is best known from terrestrial soils, phylogenetically-related groups have been reported from marine samples including some that require seawater for growth3 suggesting a high level of marine adaptation.
Here, we report the isolation, structure determination, and biological activity of two unique spiroaminals, marineosins A and B (1, 2), from a marine-derived Streptomyces-related actinomycete (our strain CNQ-617).
Marineosin A (1)4 analyzed for the molecular formula C25H35N3O2 ([M]+ m/z 409.2726) by EI high-resolution mass spectrometry. The LRESIMS also showed ions that analyzed for [M+H]+ at m/z 410. This molecular formula, which contained ten degrees of unsaturation, was further indicated by analysis of 1H and 13C NMR spectral data (Table 1). Initial interpretation of DEPT and 13C NMR spectal data showed that the majority of the proton signals were from methylene protons. The 1H NMR spectrum of 1, measured in acetone-d6, also contained five aromatic protons at δ 5.44 (d, J = 3 Hz), 5.68 (d, J = 3 Hz), 6.11 (dd, J = 2.5, 3.5 Hz), 6.37 (dd, J = 1.5, 3.5 Hz), and 6.97 (dd, J = 1.5, 2.5 Hz). NMR analysis using both COSY and HMBC methods (Figure 1) showed correlations which illustrated that these proton signals were components of two pyrrole rings.
Table 1.
NMR spectral data for marineosin A (1) in acetone-d6
| C/H | δHa | mult (J in Hz) | δCb | HMBC data | |
|---|---|---|---|---|---|
| 1 | 6.97 | dd (1.5, 2.5) | 122.5 | CH | 2, 3, 4 |
| 2 | 6.11 | dd (2.5, 3.5) | 109.9 | CH | 1, 3, 4 |
| 3 | 6.37 | dd (1.5, 3.5) | 113.5 | CH | 1, 2, 4 |
| 4 | 129.1 | C | |||
| 5 | 164.4 | C | |||
| 6a | 1.88 | dd (8.5, 16.0) | 39.0 | CH2 | 5, 7 |
| 6b | 2.88 | dd (8.5, 16.0) | 39.0 | CH2 | 5, 7, 8 |
| 7 | 3.85 | t (8.5) | 89.9 | CH | 5, 6, 8, 25 |
| 8 | 106.1 | C | |||
| 9 | 2.91 | d (12.0) | 45.9 | CH | 7, 8, 10, 11, 20, 21, 22 |
| 10 | 129.9 | C | |||
| 11 | 5.68 | d (3.0) | 110.1 | CH | 10, 12, 13 |
| 12 | 5.44 | d (3.0) | 104.7 | CH | 10, 11, 13 |
| 13 | 131.6 | C | |||
| 14 | 2.23 | m | 28.6 | CH2 | 12, 13, 15, 16 |
| 15a | 1.37 | m | 25.5 | CH2 | |
| 15b | 1.64 | m | 25.5 | CH2 | |
| 16a | 1.28 | m | 28.0 | CH2 | 14, 18 |
| 16b | 1.34 | m | 28.0 | CH2 | 14, 18 |
| 17a | 0.52 | m | 25.5 | CH2 | |
| 17b | 0.69 | m | 25.5 | CH2 | |
| 18a | 1.02 | m | 25.7 | CH2 | 16, 17, 20 |
| 18b | 1.52 | m | 25.7 | CH2 | |
| 19a | 0.88 | m | 25.4 | CH2 | |
| 19b | 1.18 | m | 25.4 | CH2 | |
| 20a | 0.88 | m | 32.0 | CH2 | |
| 20b | 1.35 | m | 32.0 | CH2 | |
| 21 | 2.32 | m | 29.6 | CH | |
| 22a | 1.61 | td (6.5, 12.0) | 39.6 | CH2 | 9, 20, 21, 23, 24 |
| 22b | 1.77 | m (12.0) | 39.6 | CH2 | 9, 21 |
| 23 | 4.23 | m (6.5) | 70.4 | CH | 8, 21, 22, 24 |
| 24 | 1.51 | d (6.5) | 22.8 | CH3 | 22, 23 |
| 25 | 3.40 | s | 58.5 | CH3 | 7 |
| NHa | 10.93 | br, s | |||
| NHb | 8.23 | br, s |
Assignment by gHSQC methods;
500 MHz,
300 MHz
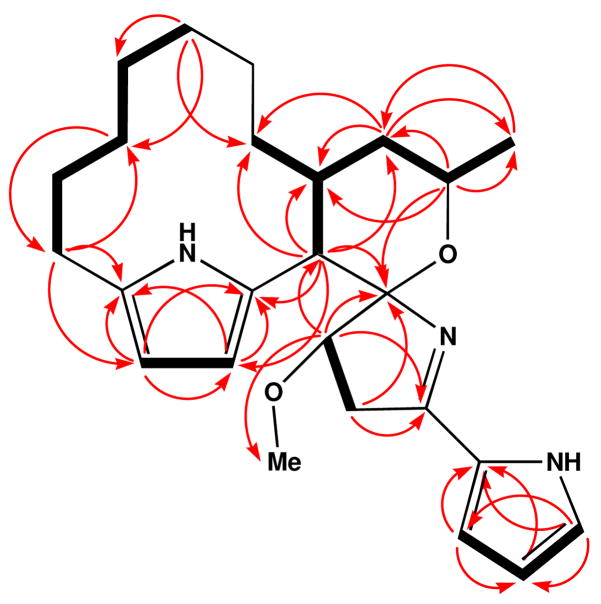
Figure 1.
Selected COSY (bold lines) and HMBC NMR correlations (red arrows) observed for marineosin A (1).
COSY NMR correlations between H-11 (δ 5.68) and H-12 (δ 5.44), and HMBC correlations from H-11 to C-10, C-12 and C-13, H-12 to C-10, C-11 and C-13, suggested the presence of a 2, 5-disubstituted pyrrole ring. The proton coupling constants of H-11 and H-12 (d, J = 3 Hz) also supported this substitution pattern. The remaining three aromatic proton signals showed COSY correlations that allowed the connectivity of C-1/C-2/C-3 to be established. Observed HMBC correlations from H-1 to C-3 and C-4, H-2 to C-4, and H-3 to C-1 and C-4 indicated that these olefinic protons could be assigned to a pyrrole ring with mono-substitution at C-4. Additional evidence, which confirmed the presence of this pyrrole ring, were the expected carbon and proton chemical shifts and proton coupling constants that matched with other known pyrrole derivatives.5
The major portion of the macrocyclic skeleton framework, which is composed of mostly methylene carbons, could be assembled through the interpretation of COSY, HMBC, TOCSY and HSQC-TOCSY NMR correlations. Interpretation of these data allowed C-14 to C-21 to be constructed. This carbon framework was connected to the pyrrole ring at C-13 on the basis of HMBC correlations from H-14 to C-12 and C-13. On the other side of the aliphatic chain, H-21 showed a COSY correlation to the methine proton at C-9. Lastly, HMBC correlations from H-9 to the pyrrole carbons C-10 and C-11 established the presence of the 12-membered macrocyclic ring. This part of the structure is reminiscent of the cycloalkyl structures of the prodigiosins, a series of bacterial pigments known for their bright colors and significant biological activites.6
The H-9 and H-23 methine protons both demonstrated HMBC correlations to a quaternary carbon at C-8, suggesting the presence of a 6-membered cyclic ether. Oxygen was placed between C-8 (δ 106.1) and C-23 (δ 70.4) initially on the basis of their appropriate carbon chemical shifts. HMBC correlations from the methylene protons at H-6 and the methine proton at H-7 to C-8 were observed. The methoxy group was connected to C-7 on the basis of HMBC correlation between H3-25 and C-7. Both H2-6 and H-7 also showed HMBC correlations to the quaternary carbon at C-5, which based on its chemical shift at δ 164.4, was initially thought to be a carbonyl group. However, analysis of IR spectral data indicated a lack of carbonyl groups in this molecule. Based upon the molecular formula and the partial structure defined above, C-5 was deduced to be an imine carbon attached to the mono-substituted pyrrole ring at C-4. The imine nitrogen was then connected to C-8. This completed construction of a five-membered ring with C-8 forming a spiro center. This part of the structure, which includes two five-member rings, is similar to structural features of the common 4-methoxy-2, 2-bipyrrole chromophores from the marine-derived tambjamines7 and the bacterial pigments which contain the prodiginine central core.6
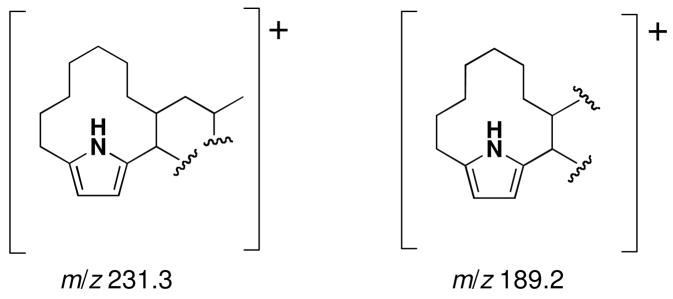
Additional evidence that confirmed the structure of 1 came from the EIMS experiment that illustrated prominent fragment ions in support of the structure. Two fragment ions were observed at m/z 189.2 and 231.3, supporting the partial structures in Figure 2. This result also indicated the presence of a pyrrole chromophore.
Figure 2.
Fragment ions observed in the EIMS experiment performed on marineosin A (1)
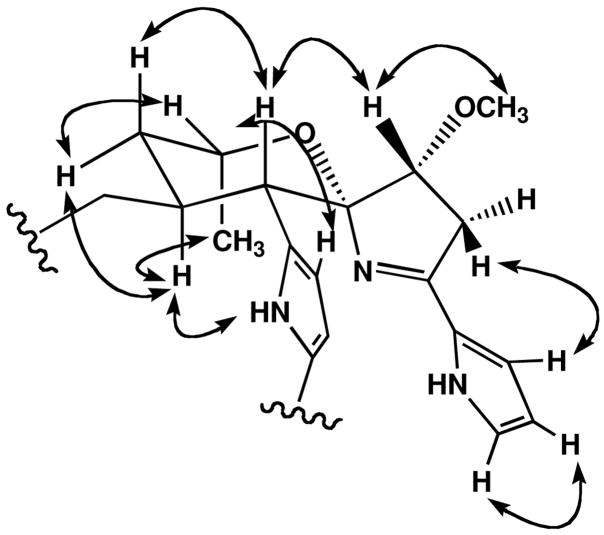
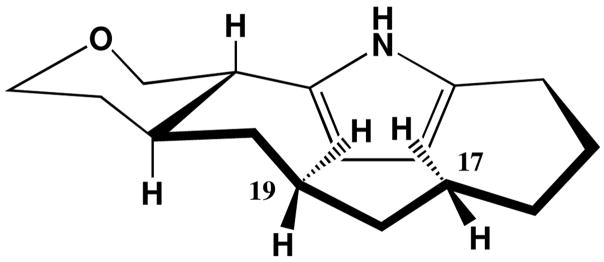
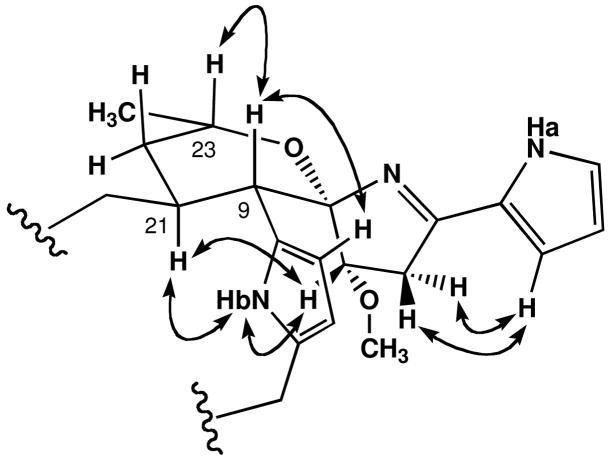
The assignment of the relative configuration of marineosin A (1), and confirmation of the overall structure, was achieved by interpretation of NOESY NMR spectral data (Figure 3) and by analysis of 1H NMR vicinal coupling constants. The methine protons H-9(δ 2.91) and H-21 (δ 2.32) were both placed in axial positions on a chair tetrahydropyran ring based upon their large vicinal coupling constant (J = 12 Hz). Me-24 (δ 1.51) was also placed in an axial position on the same side of the ring as H-21 on the basis of a mutual NOESY correlation. This result confirmed that the tetrahydropyran ring was in the chair conformation. The methine signal at H-9 also showed a NOESY correlation to the pyrrole proton at H-11, and thus showed that both protons were on the same side of the molecule. NOESY correlations observed from H-7 to H-9 and from H-6 to H-3 and H-11 confirmed the assignment of the spiro center placing nitrogen on the opposite side of the molecule from the H-9 methine proton. The 2, 5-disubstitution pattern of the pyrrole ring was also supported by NOESY correlations observed between H-11 and H-12. In the large, pyrrole-containing ring unusual high field shifts were observed for H-17 and H-19. Inspection of molecular models illustrated a feasible conformation in which these protons are shielded by the transannular pyrrole ring (Figure 4).
Figure 3.
Assigned relative configuration of 1 based upon selected NOESY NMR correlations.
Figure 4.
Large-ring conformation of 1 suggested by the pyrrole-ring shielded proton chemical shifts of H-17 and H-19.
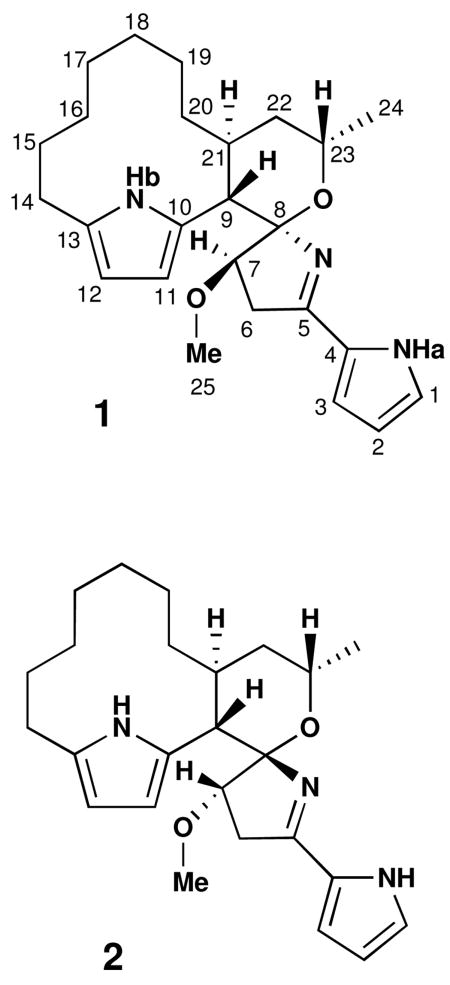
The molecular formula for marineosin B (2)8, a minor component of the mixture, was analyzed as C25H35N3O2, a formula isomeric with 1, by HRFABMS (m/z 410.2804, [M+H]+) and by analysis of NMR spectral data. Initial analysis of 1H NMR spectral data for 2 illustrated features very similar to those of 1, although most chemical shifts were slightly different. From this observation, it was deduced that these molecules were most likely related as diastereomers. As in 1, the major portion of marineosin B was assembled by the interpretation of COSY, TOCSY and HMBC NMR data (see supporting information). Comparison of all spectral data with those derived from compound 1 allowed the structure of 2 to be established. Several of the relevant carbon chemical shifts were, however, obtained only by interpretation of HMBC and HSQC NMR data (see supporting information).
The correlation patterns derived from interpretation of COSY and HMBC NMR spectroscopic data were found to be identical to those from 1, and consequently, the same planar structure could be assigned to 2. Despite the fact that marineosin B (2) possessed the same planar structure, differences in the proton and carbon chemical shifts suggested that this compound had significant stereochemical differences. Consequently, the relative stereochemistry of 2 was also established by interpretation of NOESY NMR spectroscopic data (Figure 5) and by analysis of vicinal coupling constants.
Figure 5.
Assigned relative configuration of 2 based upon selected NOESY NMR correlations.
The methine protons H-9 (δ 2.71) and H-21 (δ 2.26) were both placed in axial positions due to their large vicinal coupling constants (J = 13 Hz). Proton H-21 was placed on the same side of the molecule as NHb (δ 9.39) based on observed mutual NOESY correlations. A NOESY correlation between H-9 and H-23 suggested that the tetrahydropyran ring may adopt a boat or twist boat conformation. The NOESY correlations from NHb to H-14b (δ 2.67), H-12 to H-14a (δ 2.44), and from H-9 to H-11 confirmed that the pyrrole embeded in the macrocyclic ring also possessed a 2, 5-disubstitution pattern.
The difference between marineosin A (1) and B (2) was also concluded to be the configuration of the spiro aminal carbon, C-8. NOESY correlations from 2 indicated that H-7 (δ 4.03) correlated to NHb and H-21, an observation which proved that these protons were aligned on the same side of the molecule. This assignment explains why the chemical shifts of NHb and H-7 were shifted downfield in 2, since this part of the molecule became more structurally constrained than the analogous region in 1.
The marineosins were isolated from a previously undescribed, sediment-derived actinomycete related under current taxonomic rules to the genus Streptomyces. The new strain, identified by analysis of 16S rDNA sequence data (genebank deposit # EU161093), requires seawater for growth, indicating that this may be a true marine actinomycete.
The marineosins possess 4-methoxy-2-pyrrolyl-azacyclopentene chromophores and a unique spiro aminal constellation formed with the adjacent tetrahydropyran ring. This is the first time this carbon skeleton has been observed. It is interesting that the spiro-tetrahydropyran-dihydropyrrole aminals in the marineosins adopt different configurations. An explanation possibly lies in the stabilizing anomeric effect expected in marineosin A (1).9 However, in marineosin B (2), in which the aminal nitrogen atom is inverted, this stabilization may be absent.
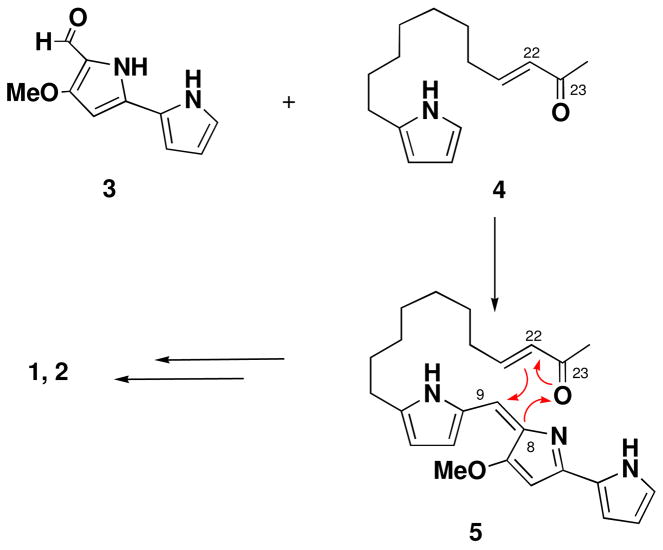
The marineosins are clearly related to the prodigiosin class of bacterial pigments.6 While possessing the fundamental components of the undecylprodigiosins, the origin of the tetrahydropyran ring-aminal system is intriguing and unprecedented. One possibility shown in Figure 5 is the condensation of 3, the known primary precursor bipyrrole aldehyde of the prodigiosins, which is formed from proline, serine and glycine6, with a novel 2-keto-undec-3-enyl pyrrole to lead to 5. The enone 5 then could undergo a hetero Diels Alder cyclization which forms the tetrahydropyran ring.10 and spiroaminal in a single step. An origin via hetero cyclization explains the diastereomeric mixture of 1 and 2 obtained by visualizing cyclization from above and below the eneone plane in 4.
Marineosins A (1) and B (2) were tested for their ability to arrest cell division in the HCT-116 human colon tumor cell line and for their antifungal effects against wild-type and amphotericin B-resistant strains of C. albicans. Antifungal activity against Candida albicans was shown to be extremely weak with MIC value over 100 μg/mL. Despite their structural similarities, 1 and 2 show rather different cytotoxic activities. Marineosin A (1) was more active in the HCT-116 cytotoxicity assay showing an IC50 value of 0.5 μM. In contrast, marineosin B exhibited considerably weaker cytotoxicity showing IC50 = 46 μM. The difference in configuration at the spiroaminal center, and in the tetrahydropyran conformation appears to significantly affect their bioactivities. Focused testing of marineosin A in the NCI 60 cell line panel showed broad cytotoxicity with considerable selectivity against the melanoma and leukemia cell lines. Further testing of these compounds in related cancer-relevant bioassays is in progress.
Supplementary Material
Experimental procedures, NMR spectral data tables, complete 2D spectra of 1–2, EIMS data, identification and cultivation of the strain CNQ-617, and the details of extraction and isolation of 1 and 2. This material is available free of charge via the Internet at http://pubs.acs.org.
Figure 6.
Possible biosyntheses of 1 and 2 via a hetero Diels Alder cyclization.
Acknowledgments
We thank Alejandra Prieto-Davó (SIO) for the phylogenetic analysis of strain CNQ-617. We also thank K. Freel and S. Kelly (SIO) for performing the HCT-116 cytotoxicity bioassays. This work is a result of generous financial support provided by the NIH, National Cancer Institute under grants CA44848 and CA 52955.
References
- 1.(a) Fenical W, Jensen PR. Nat Chem Biol. 2006;2:666–673. doi: 10.1038/nchembio841. [DOI] [PubMed] [Google Scholar]; (b) Ward AC, Bull AT, Goodfellow M. Int J Syst Evol Microbiol. 2005;55:1759. doi: 10.1099/ijs.0.63625-0. [DOI] [PubMed] [Google Scholar]
- 2.Bérdy J. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 3.Gontang EA, Fenical W, Jensen PR. Appl Env Micro. 2007;73:3272–3282. doi: 10.1128/AEM.02811-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marineosin A (1): colorless oil; [α]D −101.7 (c 0.06, MeOH); UV (MeOH) λmax (log ε) 285 nm (4.06), 227 nm (3.88); IR νmax (KBr) 2925, 2856, 1610, 1508, 1435, 1261, 763 cm−1; NMR spectral data, see Table 1 and SI; HREIMS [M]+ m/z 409.2726 (calcd for C25H35N3O2, 409.2729).
- 5.(a) Morales JJ, Rodriguez AD. J Nat Prod. 1991;54:629–631. doi: 10.1021/np50077a021. [DOI] [PubMed] [Google Scholar]; (b) Shen X, Perry TL, Dunbar CD, Kelly-Borges M, Hamann MT. J Nat Prod. 1998;61:1302–1303. doi: 10.1021/np980129a. [DOI] [PubMed] [Google Scholar]
- 6.Fürstner A. Angew Chem Int Ed. 2003;42:3582–3603. doi: 10.1002/anie.200300582. and references cited therein. [DOI] [PubMed] [Google Scholar]
- 7.(a) Carté B, Faulkner DJ. J Org Chem. 1983;48:2314–2318. [Google Scholar]; (b) Paul VJ, Lindquist N, Fenical W. Mar Ecol Prog Ser. 1990;59:109–118. [Google Scholar]; (c) Lindquist N, Fenical W. Experientia. 1991;47:504–506. [Google Scholar]
- 8.Marineosin B (2): colorless oil; [α]D +143.5 (c 0.09, MeOH); UV (MeOH) λmax (log ε) 287 nm (3.24); IR νmax (KBr) 2926, 2856, 1607, 1434, 1106, 743 cm−1; NMR data, see Table S2; HRFABMS [M+H]+ m/z 410.2804 (calcd for C25H36N3O2, 410.2807).
- 9.Cottier L, Descotes G, Grenier MF, Metras F. Tetrahedron. 1981;37:2515–2524. [Google Scholar]
- 10.We thank an anonymous reviewer for suggesting that a hetero Diels Alder cyclization pathway might be involved.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental procedures, NMR spectral data tables, complete 2D spectra of 1–2, EIMS data, identification and cultivation of the strain CNQ-617, and the details of extraction and isolation of 1 and 2. This material is available free of charge via the Internet at http://pubs.acs.org.