Protein kinases are major signaling enzymes governing cellular physiology and are of intense interest in biomedical research.1 Understanding the regulation of protein kinase function has been an important aspect of elucidating signaling in phosphorylation networks.2 Autophosphorylation represents one critical control switch for modulating protein kinase action. In general, autophosphorylation reactions are thought to be intermolecular in which one kinase molecule phosphorylates a neighbor.3 In several instances, however, intramolecular kinase autophosphorylation has been proposed.3–5 One such proposal has been made for the Ser/Thr kinase, protein kinase A (PKA, cyclic AMP-dependent protein kinase).5 PKA is the best characterized member of the kinase superfamily and was the first to have its X-ray crystal structure solved.5–8 The PKA catalytic C-subunit has two regulatory phosphorylation sites, one in its activation loop targeted by PDK1 and a second near its C-terminus, thought to be self-catalyzed (Figure 1).5–8 Elegant studies by Taylor and colleagues examining concentration dependence and inactive mutants have suggested that the PKA C-terminal autophosphorylation event at Ser338 may be intramolecular.5 However, questions can still be raised as to whether the conformational flexibility of PKA exists to facilitate this intramolecular event or whether intermolecular autophosphorylation really occurs (see Figure 1). In this study, we use protein semisynthesis and computational modeling to obtain novel evidence for an intramolecular autophosphorylation reaction.
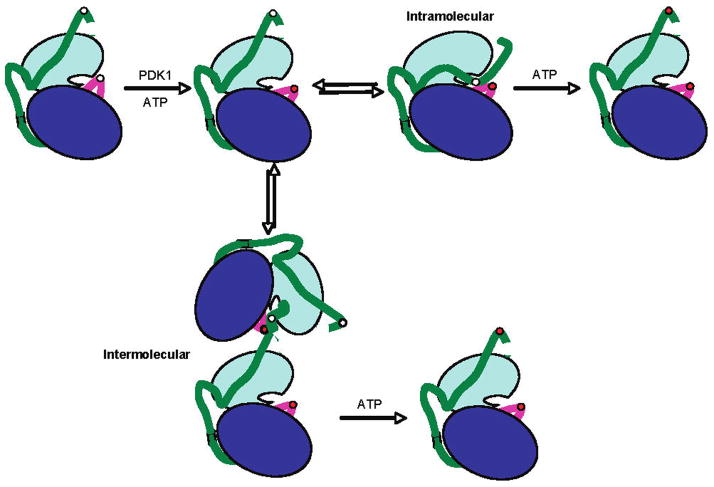
Figure 1.
Inter- versus intramolecular autophosphorylation models for PKA. Activating phosphorylation of PKA occurs at two sites, Thr197 and Ser338. PKA is initially phosphorylated, in vivo, on its activation loop (magenta) at Thr197 by PDK1. This is followed by autophosphorylation of Ser338 on its C-terminal tail (green). The N-terminal lobe is shown in cyan, and the C-terminal lobe in blue, and the active site is at their interface. Phosphorylation sites Thr197 and Ser338 are white circles when not phosphorylated and red circles when phosphorylated.
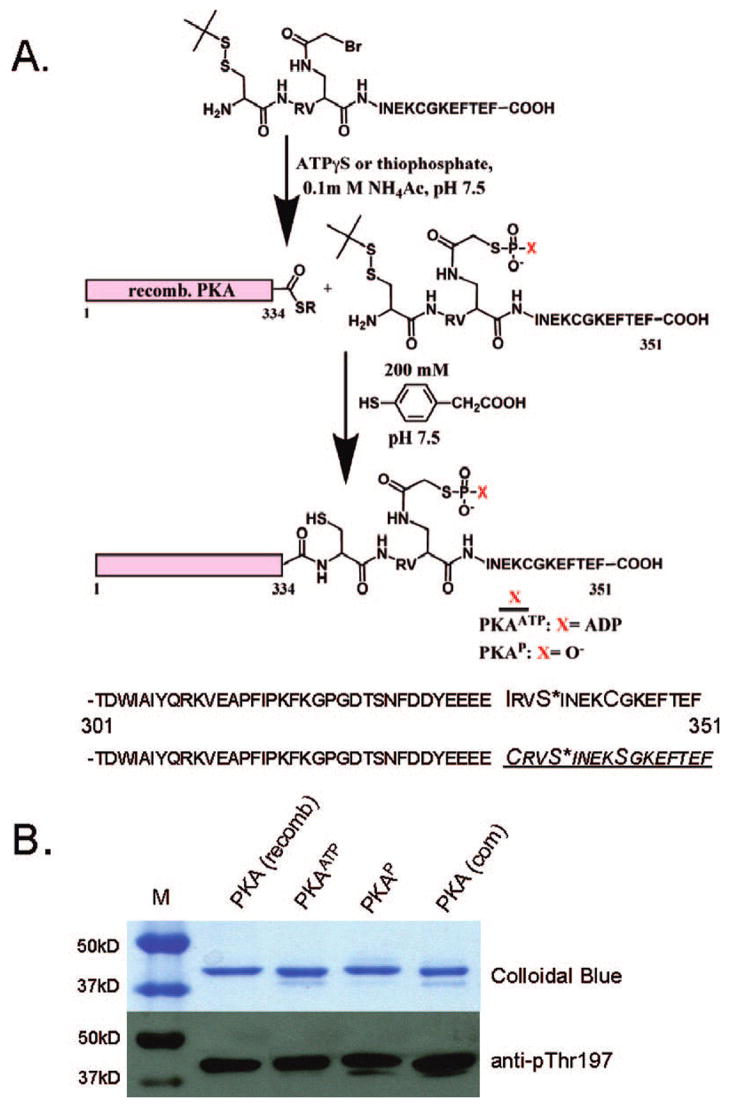
It has previously been shown that ATP–peptide bisubstrate analogues designed based on a dissociative transition state can show potent and selective inhibition of protein kinases.9 We considered that this ATP tethering strategy could be employed with PKA where the targeted tail phosphorylation site is replaced by an ATP-linked residue used for bisubstrate analogue inhibitors (Figure 2A). This would generate a protein–ATP conjugate that may exist in a conformation corresponding to an intramolecular kinase reaction coordinate arrangement. To execute this plan, we assembled semisynthetic PKA using an expressed protein ligation (EPL) strategy where the C-terminal segment of PKA carrying the ATP linkage was generated by chemical synthesis and the larger N-terminal section of PKA was prepared using recombinant methods (Figure 2A).9c,10 By exploiting the native chemical ligation reaction, the two segments were chemoselectively joined to produce the target PKAATP (Figure 2A). As a control, we prepared the corresponding phosphate containing semisynthetic PKAP, which would not be expected to exist in a conformation mimicking a kinase reaction (Figure 2A).
Figure 2.
Semisynthetic PKA. (A) Route to semisynthetic PKA linked to ATP (PKAATP) or phosphate (PKAP) at the C-terminal phosphorylation site Ser338. Natural (top) and semisynthetic (bottom) PKA sequences from 301 to 351 with synthetic peptide region underlined and Ser338 has an asterisk. (B) SDSPAGE and Western blot analysis of semisynthetic PKAs compared to commercially available [PKA(com)] and full length recombinant PKA [PKA(recomb)]. Western blot with anti-phosphoThr197 (T500P) showed comparable activation loop phosphorylation.
The semisynthetic PKAATP and PKAP showed high purity by SDSPAGE gel (Figure 2B) and mass spectrometry, indicating that EPL proceeded to greater than 90% completion. Western blot analysis confirmed that the activation loop (Thr197) was phosphorylated to a similar extent to standard recombinant protein (Figure 2B), implying that intein fusion and C-terminal deletion did not prevent this autophosphorylation known to occur when recombinant PKA is generated in Escherichia coli.11
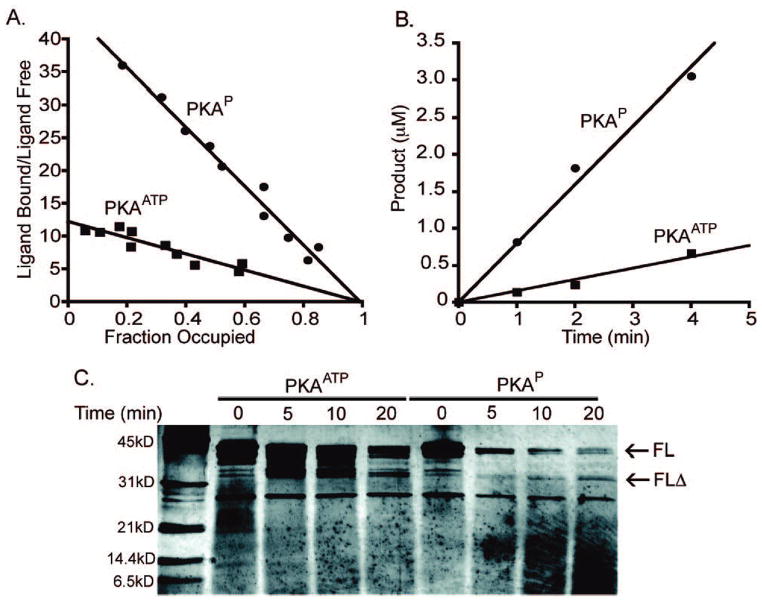
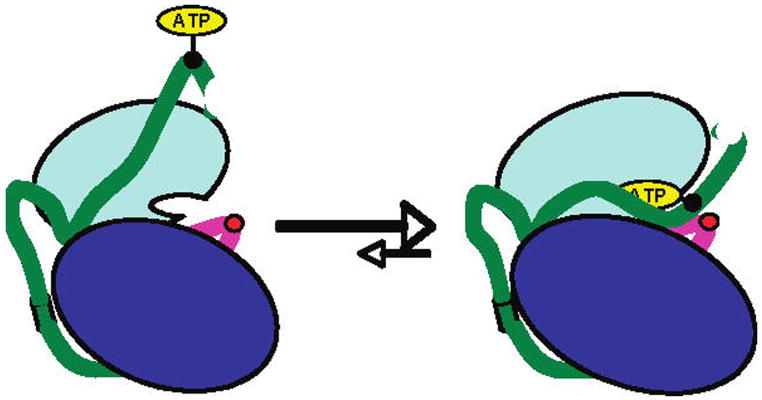
It is possible that ATP attachment could encourage the oligomerization of PKA molecules in which an ATP of one molecule interacts with the active site of another. Analysis of PKAATP by size exclusion chromatography indicated no difference in its elution profile compared with PKAP or standard recombinant PKA, indicating that PKAATP is monomeric (Figure S1). We next investigated the affinity of the semisynthetic PKAs for binding a fluorescent ADP analogue (Figure 3A).12 There was a striking 4-fold increase in the Kdapp for PKAATP compared with that of PKAP, suggesting that the nucleotide binding site of PKAATP was occluded by the covalently linked ATP. Consistent with this possibility, PKAATP showed diminished kinase activity compared with that of PKAP (Figure 3B). To further assess the conformation of PKAATP, we subjected the semisynthetic proteins to limited proteolysis with trypsin (Figure 3C). In comparison with PKAP, PKAATP showed a reduced rate of breakdown and the buildup of a metastable fragment (FLΔ) consistent with cleavage in the N-terminus. It is noteworthy that this proteolysis site is in a region where the C-terminal segment of PKA overlaps with the N lobe. This suggests that ATP occupancy in the PKA kinase active site of PKAATP (Figure 4) may induce a conformational change in this overlap region that modulates accessibility to trypsin.
Figure 3.
Comparative properties of PKAATP and PKAP. (A) Fluorescence binding analysis of semisynthetic PKA. PKAATP or PKAP (1 μM) was incubated with and without various concentrations of Mant-ADP (5–125 μM) in the presence of 1 M free Mn. Scatchard analysis revealed Mant-ADP showed a Kdapp of 77 ± 20 μM for PKAATP and 20 ± 2 μM for PKAP. (B) Kinase activities of semisynthetic PKAs. Radiometric assays were carried out using 5 nM enzyme, 30 μM biotinylated kemptide substrate, 10 μM ATP, and 10 mM MgCl2. Turnover numbers were found to be 36 ± 8 min−1 for PKAATP and 160 ± 10 min−1 for PKAP. (C) Limited proteolysis (trypsin) of PKAATP and PKAP using 1:5 trypsin/PKA at 4 °C for the period of time indicated. Silver-stained SDSPAGE revealed a metastable fragment (FLΔ) from PKAATP and N-terminal sequence revealed cleavage after Lys83 for FLΔ.
Figure 4.
Conformational model for PKAATP. Color scheme as in Figure 1.
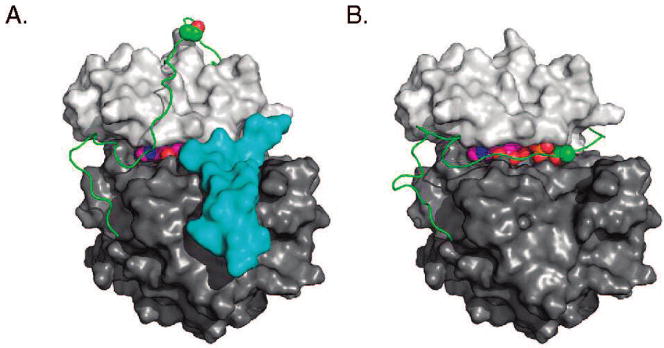
While the size exclusion chromatography, fluorescence, enzymatic, and protease studies are consistent with a conformation reflecting intramolecular PKA autophosphosphorylation, it was of interest to analyze the detailed structural implications of such a model. As can be seen in the crystal structure of the ternary complex of PKA bound to ATP and peptide substrate analogue PKI13 (Figure 5A, Protein Data Bank code: 1atp), Ser338 is more than 25 Å from the active site, and it is unclear whether the C-terminal tail containing Ser338 is capable of adopting a conformation that would put it in the active site. The diversity of C-terminal tail conformations in multiple crystallized states of PKA13,14 and the relatively high B-factors along most of this loop suggest that it is relatively unconstrained and free to adopt alternate conformations. In order to assess whether a physically plausible low-energy C-terminal tail conformation exists that places Ser338 in the active site in an appropriate orientation for autophosphorylation, we utilized a loop modeling protocol within the Rosetta software suite.15
Figure 5.
PKA structure. (A) Crystal structure of the ternary complex. (B) Proposed structural model for an autophosphorylation state. The N-terminal lobe is in light gray, the C-terminal lobe is in dark gray, PKI is in cyan, the C-terminal tail is in green, ATP is in magenta, and ATP and Ser338 are shown as spheres.
Starting from the crystal structure of the PKA ternary complex (Figure 5A), we defined residues 314–346 of the C-terminal tail as a flexible loop based on both B-factor data and a comparison of other crystallized forms of PKA, and we held the rest of the protein backbone rigid. We placed Ser338 in the same position as Ala21 of PKI of the PKA ternary complex to serve as an approximation for the substrate position within the active site for autophosphorylation. Finally, we anchored the end of the C-terminal tail (residues 347–350) to its position in the ternary complex, making the a priori assumption that, in a low-energy autophosphorylation state, the buried Phe347 and Phe350 would remain in the same position as has been observed in all major PKA crystal structures. We then carried out loop modeling of the N-terminal side of the loop (residues 314–338) and C-terminal side of the loop (residues 338–346) separately. We combined the lowest energy model from each simulation and refined along the middle stretch of the loop (residues 336–342), allowing minor movement of Ser338. Finally, we placed the ATP ligand into the active site and carried out one additional round of refinement to generate a final model, illustrated in Figure 5B (see Supporting Information for further details).
Simulations of the N-terminal side of the loop resulted in a large number of low-energy conformations, and the lowest energy model was also found in the largest cluster of structurally similar models, suggesting convergence on this particular conformation. Simulations of the C-terminal side of the loop, however, resulted in closed loop conformations without chain breaks in a small number of cases (~1%), suggesting that a loop conformation that places the Ser338 in the appropriate location may be relatively strained. The final two stages of refinement allowed the loop to close completely without steric clashes, while placing the Ser338 within 1 Å of the Ala21 in PKI and keeping Phe347 and Phe350 buried in their appropriate locations, thus achieving a physically realistic model that satisfies all a priori assumptions (Figure 5B).
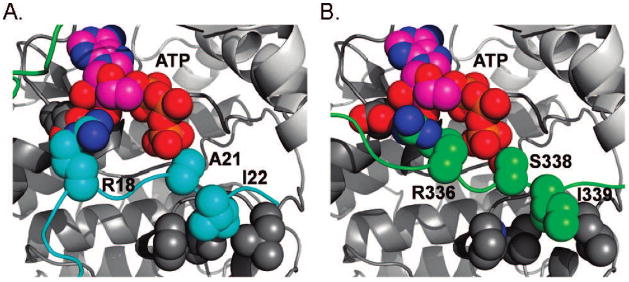
Beyond satisfying these constraints, there are a number of notable structural features in the model. First, the conformation of the long N-terminal side of the loop bears some resemblance to its corresponding conformation in the crystallized ternary complex, and many of the original residue–residue contacts remain intact. Second, there are a number of interactions between the middle and the C-terminal section of the loop and the substrate binding site that are similar to PKI–enzyme interactions. Specifically, Arg336 forms a hydrogen bond with Glu113 in the active site, similar to that involving Arg18 of PKI, and Ile339 makes similar contacts with a hydrophobic pocket formed by Leu198, Pro202, and Leu205 as Ile22 in PKI (Figure 6A,B). Taken together, this computational effort provides a plausible model for how intramolecular autophosphorylation may be achieved.
Figure 6.
Substrate binding site of PKA. (A) Crystal structure of the ternary complex. (B) Proposed structural model for an autophosphorylation state. Colors scheme as in Figure 5. ATP and the side chains of Ile198, Pro202, Leu205, Glu113, Arg336, and Ser338 of PKA and Arg18, Ala21, and Ile22 of PKI are shown as spheres.
The combination of biophysical studies described provides unique evidence for an intramolecular autophosphorylation reaction involving the C-terminus of PKA (Figure 1). The fact that the active site of PKAATP is still partially accessible to the fluorescent probe and catalytically active does suggest that there is a dynamic equilibrium between the conformation corresponding to ATP occupancy and the unengaged form (Figure 4). These results extend our understanding of the conformational plasticity that is known to be so critical to the regulation and function of protein kinases.2 Furthermore, by incorporation of the appropriate transition state analogue motif, the semisynthetic approach applied here may be useful in clarifying the molecularity and mechanisms of other autoprocessing proteins and enzymes.
Supplemental Material
Acknowledgments
We would like to thank Lawrence Szewczuk and Dirk Schwarzer for helpful discussions, and the NIH for financial support (CA74305 to P.A.C. and GM078221 to J.J.G.).
Footnotes
Supporting Information Available: Experimental details, supporting figure, and structural model coordinates are available. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Hunter T. Cell. 2000;100:113. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 2.Huse M, Kuriyan J. Cell. 2002;109:275. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 3.Wang ZX, Wu JW. Biochem J. 2002;368:947. doi: 10.1042/BJ20020557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Pango MA, Sarno S, Poletto G, Giogio C, Pinna LA, Meggio F. Mol Cell Biochem. 2005;274:23. doi: 10.1007/s11010-005-3116-y. [DOI] [PubMed] [Google Scholar]; (b) Cole A, Frame S, Cohen P. Biochem J. 2004;377:249. doi: 10.1042/BJ20031259. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Gao X, Harris TK. Bioorg Chem. 2006;34:200. doi: 10.1016/j.bioorg.2006.05.002. [DOI] [PubMed] [Google Scholar]; (d) Lochhead PA, Sibbet G, Morice N, Cleghon V. Cell. 2005;121:925. doi: 10.1016/j.cell.2005.03.034. [DOI] [PubMed] [Google Scholar]; (e) Lochhead PA, Kinstrie R, Sibbet G, Rawjee T, Morrice N, Cleghon V. Mol Cell. 2006;24:627. doi: 10.1016/j.molcel.2006.10.009. [DOI] [PubMed] [Google Scholar]; (f) Cann AD, Kohanski RA. Biochemistry. 1997;36:7681. doi: 10.1021/bi970170x. [DOI] [PubMed] [Google Scholar]
- 5.Iyer GH, Moore MJ, Taylor SS. J Biol Chem. 2005;280:8800. doi: 10.1074/jbc.M407586200. [DOI] [PubMed] [Google Scholar]
- 6.Yonemoto W, McGlone ML, Grant B, Taylor SS. Protein Eng. 1997;10:915. doi: 10.1093/protein/10.8.915. [DOI] [PubMed] [Google Scholar]
- 7.(a) Knighton DR, Zheng JH, Ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Science. 1991;253:407. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]; (b) Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Science. 1991;253:414. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 8.Toner-Webb J, van Patten S, Walsh DA, Taylor SS. J Biol Chem. 1992;267:25174. [PubMed] [Google Scholar]
- 9.(a) Parang K, Till JH, Ablooglu AJ, Kohanski RA, Hubbard SR, Cole PA. Nat Struct Biol. 2001;8:37. doi: 10.1038/83028. [DOI] [PubMed] [Google Scholar]; (b) Hines AC, Cole PA. Bioorg Med Chem Lett. 2004;14:2951. doi: 10.1016/j.bmcl.2004.03.039. [DOI] [PubMed] [Google Scholar]; (c) Shen K, Cole PA. J Am Chem Soc. 2003;125:16172. doi: 10.1021/ja0380401. [DOI] [PubMed] [Google Scholar]
- 10.Muir TW, Sondhi D, Cole PA. Proc Natl Acad Sci USA. 1998;95:6705. doi: 10.1073/pnas.95.12.6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herberg FW, Bell SM, Taylor SS. Protein Eng. 1993;6:771. doi: 10.1093/protein/6.7.771. [DOI] [PubMed] [Google Scholar]
- 12.Ni Q, Shaffer J, Adams JA. Protein Sci. 2000;9:1818. doi: 10.1110/ps.9.9.1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng J, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. Acta Crystallogr. 1993;49:362. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 14.(a) Wu J, Yang J, Kannan N, Madhusudan, Xuong NH, Ten Eyck LF, Taylor SS. Protein Sci. 2005;14:2871. doi: 10.1110/ps.051715205. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Kim C, Xuong NH, Taylor SS. Science. 2005;307:690. doi: 10.1126/science.1104607. [DOI] [PubMed] [Google Scholar]
- 15.(a) Rohl CA, Strauss CE, Chivian D, Baker D. Proteins. 2004;55:656. doi: 10.1002/prot.10629. [DOI] [PubMed] [Google Scholar]; (b) Wang C, Bradley P, Baker D. J Mol Biol. 2007;373:503. doi: 10.1016/j.jmb.2007.07.050. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.