Abstract
KRASis the most frequently mutatedrasfamily member in lung carcinomas1,2, whereasHRASmutations are common in tumours from stratified epithelia such as bladder or skin (www.sanger.ac.uk/genetics/CGP/cosmic/). Using a mouse model (HrasKI)3 in which theHrascoding sequence was inserted into theKraslocus, we demonstrate that specificity forKrasmutations in lung andHras mutations in skin tumours is determined by local regulatory elements in the targetrasgenes. We further show that, whileKras-4Ais dispensable for mouse development4,5, it is necessary both for lung carcinogenesisin vivoand for the previously reported6,7 inhibitory effect of wild-type (WT)Krason the transforming properties of the mutant allele. Kras-4A expression is detected in a sub-population of normal lung epithelial cells, but at very low levels in lung tumours, suggesting a role in tumour initiation rather than in tumour maintenance. The two Kras isoforms undergo different post-translational modifications8, therefore these findings can have important implications for the design of therapeutic strategies for inhibiting oncogenic Kras activity in the prevention and treatment of cancer.
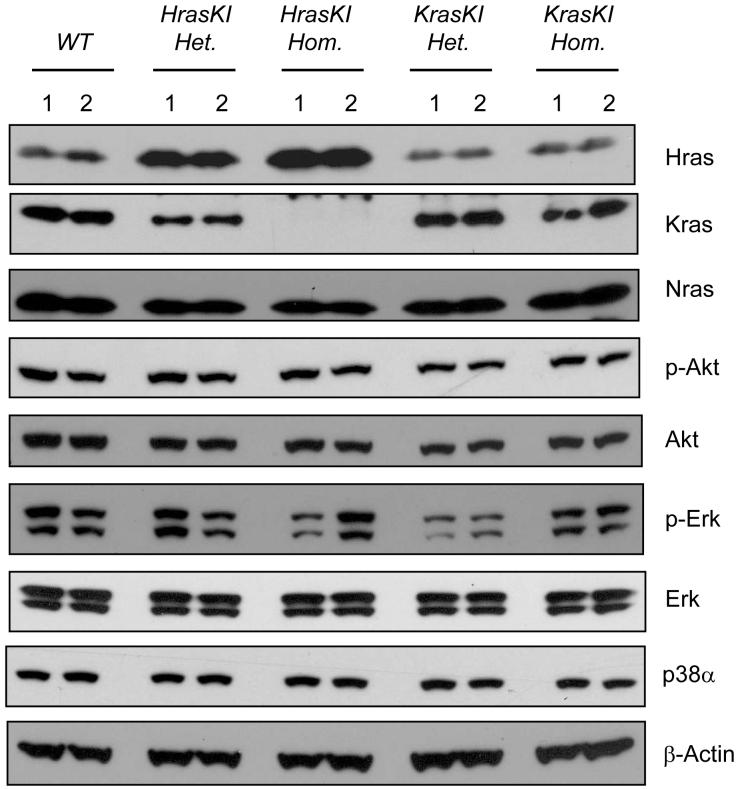
Kras deficiency in mice leads to embryonic lethality4,5, thus it has not been possible to determine whether the selection for Kras mutations in lung tumours reflects a specific oncogenic function that is required for lung carcinogenesis and cannot be compensated for by mutant Hras or Nras. The HrasKI mouse provides a viable model that completely lacks Kras expression3 (Fig. 1), but carries a “knock-in” Hras cDNA expressed under the control of Kras regulatory elements. We reasoned that HrasKI mice should be resistant to lung tumour development if Kras protein is essential for lung carcinogenesis. To control for the possibility of structural alterations affecting experimental outcomes, we used KrasKI mice in which the coding sequence corresponding to the Kras-4B isoform was knocked back into the Kras locus in the exact manner used to generate the HrasKI mouse3. HrasKI mice expressed elevated levels of Hras protein, consistent with the increase in Hras gene dosage from two copies to four in these animals (Fig. 1). KrasKI mice express similar levels of Kras protein as WT mice, as expected since Kras-4B is the major splice isoform9,10. Levels of Akt and Erk activation in these animals were similar to WT mice (Fig. 1). There was also no difference in levels of p38α, recently shown to affect ras mediated lung carcinogenesis11. The elevated level of Hras therefore does not affect the major downstream effector pathways relevant to cancer development.
Figure 1. Ras protein levels and effects on downstream signaling effectors in HrasKI and KrasKI mice.
Analysis of Ras protein levels in lungs of untreated mice showed elevated levels of Hras protein and complete absence of Kras protein in HrasKI homozygous mice. Mice homozygous and heterozygous for the KrasKI allele express similar Kras protein levels to WT mice. No apparent differences in activation of Akt and Erk, as measured by protein phosphorylation, and in level of p38α were observed among mice of the indicated genotypes.
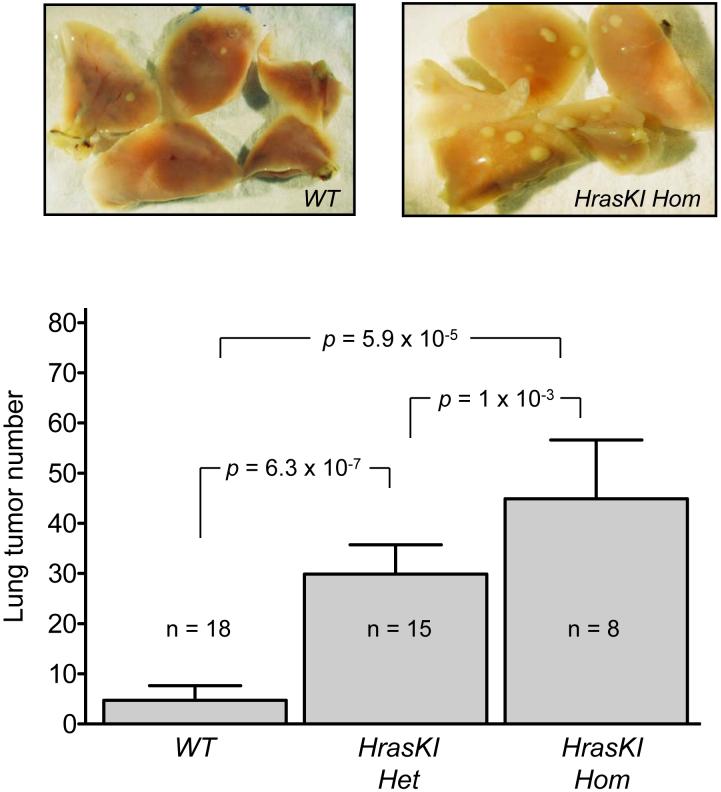
We treated HrasKI homozygous and heterozygous mice and WT littermates with urethane and sacrificed the animals 20 weeks later for analysis. Surprisingly, HrasKI homozygous mice developed ~10 fold more lung tumours than WT littermate controls (44.8 ± 11.7 vs. 4.7 ± 2.9 (Fig. 2)). The number of lung tumours in HrasKI heterozygous mice (29.9 ± 5.8) was intermediate between those of WT and HrasKI homozygous animals. While previous studies showed that activating mutations in Kras are important in lung cancer development2,7,12,13, our findings demonstrate that Kras is clearly dispensable for lung carcinogenesis.
Figure 2. HrasKI mice are highly susceptible to urethane induced lung tumours.
HrasKI homozygous mice developed significantly more lung tumours than WT littermates. The increase in lung tumour number correlated with the number of HrasKI alleles. Error bars indicate standard deviations, and statistics were performed using the Wilcoxon Mann-Whitney test.
The observation that lung tumour multiplicity correlates with the number of HrasKI alleles suggests that this allele is the target for mutations in HrasKI mice. The majority of lung tumours (21 of 27) from WT mice contain activating mutations in Kras, frequently at codon 61 but also at 12 or 13, as previously described2,7 (Supplementary Table 1). Of 35 lung tumours from HrasKI homozygous mice analyzed, 32 (91%) have mutations in the HrasKI allele. In heterozygous HrasKI mice, the vast majority of lung tumours (>90%) have a mutation in the HrasKI allele, with mutations in the endogenous Kras gene occurring in less than 10% of the tumours (p = 1.6 × 10-22, Fisher's exact test). In all cases, mutation in the HrasKI allele involves a CAA>CTA transversion at codon 61, which is the signature mutation in the Hras gene in mouse skin tumours induced by the two stage chemical carcinogenesis protocol14. However, we never found the endogenous Hras gene to be mutated in urethane induced lung tumours. The occurrence of Hras mutations in lung tumours therefore is completely dependent on Hras being expressed from the Kras locus.
We further pursued the specificity of ras gene mutations in lung and skin tumours by inducing both tumour types in the same HrasKI homozygous mice. A single dose of urethane was injected intraperitoneally to induce lung tumours, followed by a single topical treatment on the skin with urethane and subsequent biweekly application of tetradecanoyl-phorbol acetate (TPA) to induce papilloma development. All lung tumours (n = 28) analyzed have the CAA>CTA mutation in the HrasKI allele. Of the papillomas (n = 20) developed in these same mice, 15 (75%) carried the CAA>CTA mutation in the endogenous Hras gene on chromosome 7, consistent with previous studies14,15. Of the remaining 5 papillomas, 4 (20%) contained the CAA>CTA mutation in the HrasKI allele. Mutations in the endogenous Kras gene are rarely seen in WT mice, but the frequency is increased in Hras-/- mice where no endogenous Hras target is available16. Regulatory elements in Kras are therefore capable of directing expression to at least a subset of target cells within the epidermis, but the overall preference is strongly in favour of Hras. These results indicate that the mechanisms underlying Kras mutation selection in lung cancer and Hras in skin cancer involve cis-acting regulatory elements specific to each gene rather than functional differences between the encoded proteins. Previous studies have demonstrated the existence of multiple sequence variants in the mouse Kras gene that may potentially affect regulation of expression17, and further studies would be required to identify the specific element(s) involved.
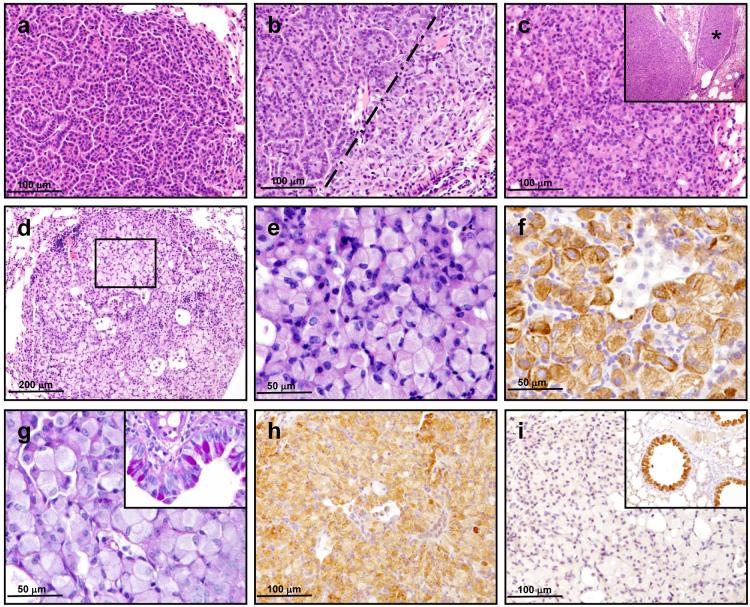
As with other Kras models of lung tumour development12,13, HrasKI mice developed papillary adenomas (Fig. 3a), but also significantly more mixed adenomas (Fig. 3b) and solid adenomas (Fig. 3c), sometimes with intrabronchiolar extension (Fig. 3c, inset). A significant percentage of the solid adenomas contain cells with epithelioid morphology (Fig. 3d, e) - large, plump cells with eccentrically located nuclei and abundant cytoplasm ranging from finely granular and eosinophilic to microvesicular and amphophilic. Positive cytokeratin 8/18 staining (Fig. 3f) and negative PAS staining (Fig. 3g) demonstrated that the vast majority of these cells were epithelioid, non-mucinous tumour cells. These solid epithelioid tumours stained positively for SP-C (Fig. 3h) and negatively for CCA/CC10 (Fig. 3i), like tumours from urethane treated WT mice and other mouse models of lung cancer12,13,18.
Figure 3. Lung tumours from HrasKI mice display papillary, solid and mixed growth patterns, and contain cells with an epithelioid morphology.
WT and HrasKI mice developed a spectrum of papillary (a), mixed (b) and solid adenomas (c). Whereas WT animals predominantly developed papillary adenomas with occasional mixed and solid tumours, the HrasKI mice tended to develop more mixed and solid adenomas, sometimes with prominent intrabronchiolar extension (c, inset, marked by asterisk). The dashed line in (b) marks the boundary between the papillary (left) and solid (right) portions of the mixed adenoma. Several solid adenomas from HrasKI mice contained epithelioid cells (d, inset enlarged in e) which stained positive for (f) cytokeratin 8/18 and negative for PAS (g). Occasional macrophage clusters (f, keratin-negative cells) were also present in the epithelioid adenomas. Goblet cell metaplasia served as a positive control for PAS/mucin (g, inset). These adenomas displayed features of alveolar type II pneumocytes like their papillary counterparts (not shown) as they stain positively for SPC (h) and negatively for CCA/CC10 (i). The adjacent bronchiolar epithelium stained positive for CCA/CC10 as expected (i, inset).
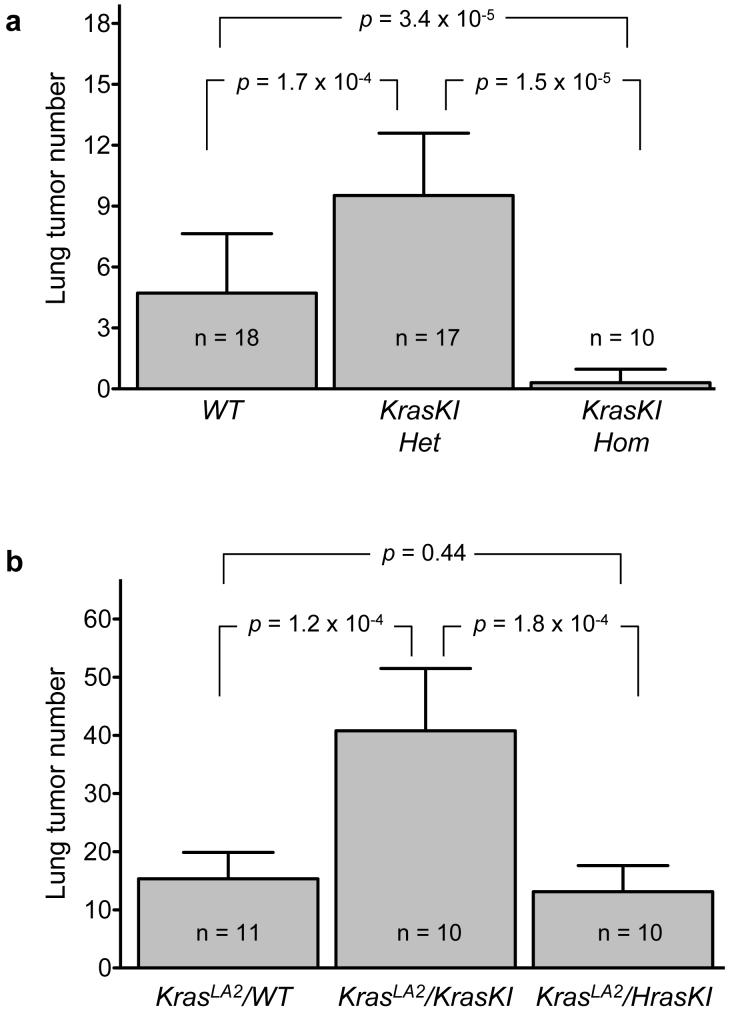
Both Kras-4A and Kras-4B splice variants are present in normal lungs, but Kras-4B is clearly the more abundant isoform9,10. However, KrasKI homozygous mice that express only Kras-4B proved to be highly resistant to urethane-induced lung tumour formation (Fig. 4a). Of 10 KrasKI homozygous mice examined, 8 did not develop any surface lung tumours. Two mice developed 1 and 2 small tumours each, but none of the tumours contained a mutation in the KrasKI allele, nor in the endogenous Hras or Nras genes. Consistent with our findings, mice with a genomic knockout of exon 4A and therefore able to express only Kras-4B displayed a dramatic decrease in lung tumor number following treatment with N-methyl-N-nitrosourea19.
Figure 4. The KrasKI allele renders mice resistant to urethane induced lung carcinogenesis, and is deficient in suppression of lung tumour development in KrasLA2 animals.
(a) KrasKI homozygous mice were almost free of lung tumours, while KrasKI heterozygous mice remained susceptible to lung carcinogenesis, and developed significantly more lung tumours than WT mice. Mutations in these tumours were found exclusively in the endogenous Kras gene and not the KrasKI allele. (b) KrasLA2 mice harboring the KrasKI allele, which generates only the 4B isoform, developed almost 3-fold more lung tumours than those carrying an intact Kras gene capable of generating both 4A and 4B isoforms. The HrasKI allele in KrasLA2 displayed an inhibitory effect on tumor number similar to WT mice. Error bars indicate standard deviations, and statistics were performed using the Wilcoxon Mann-Whitney test.
Tumours from mice heterozygous for the KrasKI allele were also positive for SPC and negative for CCA/CC10 (Supplementary Fig. 1). Kras mutations were present in 24 of 33 tumours (73%) from KrasKI heterozygous mice, but in all 24 cases the mutation occurred in the endogenous Kras gene (which retains both Kras-4A and Kras-4B) and not in the KrasKI allele (which expresses exclusively Kras-4B) (p = 6.2 × 10-14, Fisher's exact test). These findings are consistent with the resistant phenotype of KrasKI homozygous mice, and indicate that oncogenic mutations involving only Kras-4B do not provide the necessary advantage required for tumourigenesis. Since both Kras-4A and Kras-4B transcripts are present in lungs of mice containing the intact Kras gene9,10, our results suggest that the transforming effect of Kras mutations during lung carcinogenesis in vivo are mediated primarily through the activity of the Kras-4A protein.
Kras-4A and Kras-4B isoforms differ only at their carboxyl termini, resulting in differences in post-translational modifications8. Kras-4A and Hras proteins undergo similar post-translational modifications and therefore are localized within similar microdomains in the membrane, distinct from those involving Kras-4B8,20. The similarities of Kras-4A and Hras in this respect correlate with their ability to induce lung tumours in WT and HrasKI mice, respectively. In vitro studies also demonstrated that mutant Kras-4A and Hras are more potent oncogenes than mutant Kras-4B21,22.
The WT ras alleles in tumours that contain ras mutations are frequently deleted, and/or the mutant allele is amplified23-25. The possibility of a tumour suppressor function for WT Ras proteins was supported by in vitro functional assays26 and in vivo genetic studies involving genetically engineered mice6,7. These studies demonstrated that WT Kras can potently suppress the activity of oncogenic Kras to regulate lung tumour development6,7. We found that KrasKI heterozygous mice surprisingly developed ~2-fold more lung tumours than WT mice (Fig. 4a). The observation, together with results from our mutational analysis, suggested that the KrasKI allele may be less efficient in this suppressor activity. To further test this idea, KrasKI heterozygous mice were bred with KrasLA2 mice13, which undergo spontaneous recombination to activate Kras and do not require carcinogen treatment. The genotypes tested included animals with one KrasLA2 allele and either a WT Kras gene or a WTKrasKI allele. KrasLA2 mice carrying a KrasKI allele developed significantly more lung tumours than those that are able to express Kras-4A isoform from the WT Kras gene (Fig. 4b). Interestingly, the HrasKI allele is also more effective than the KrasKI allele in suppressing tumor multiplicity in KrasLA2 mice, indicating that it can substitute for Kras-4A in suppressor activity in vivo. While in vitro work showed that overexpressed WT Kras-4B has inhibitory effects, our collective in vivo findings suggest that efficient suppression of oncogenic Kras activity requires an intact WT Kras locus that is capable of generating the Kras-4A isoform.
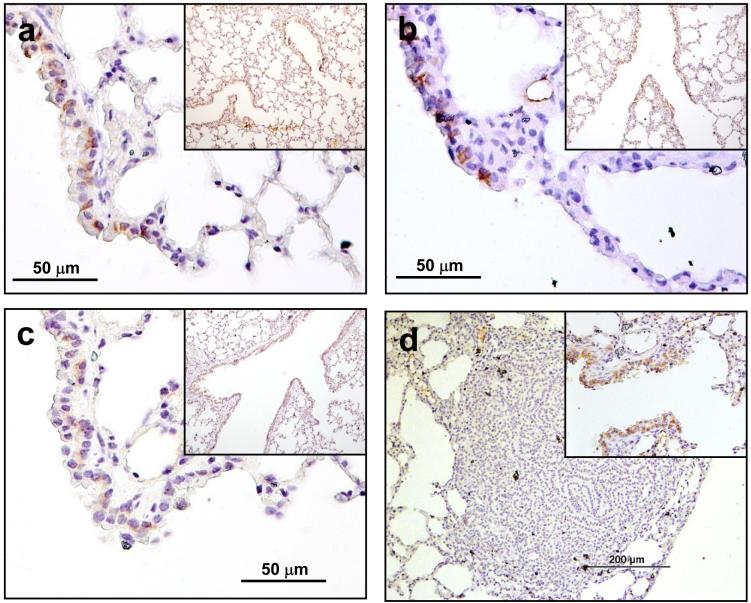
Studies of human tumours have shown that Kras-4B is the more abundant splice variant, and in fact that levels of Kras-4A may be even further reduced compared to Kras-4B in tumors carrying Kras mutations27. This is in apparent contradiction to our genetic evidence for a major role of the Kras-4A isoform in lung carcinogenesis. We therefore investigated the localization of Kras-4A expression in normal lung and lung tumours. Using an antibody specific for Kras-4A, expression was detected in a sub-population of Clara cells throughout the bronchial tree, including at the bronchio-alveolar junctions (Fig. 5a, b). Similar sections from mice homozygous for the KrasKI allele (Fig. 5c) or the HrasKI allele (not shown), both of which lack Kras-4A, were negative. Antibodies against Kras-4B yielded non-specific signals, as positive staining was observed even in sections from HrasKI homozygous mice that completely lack Kras expression (data not shown). The bronchio-alveolar junctions have been reported to contain the putative bronchio-alveolar stem cells (BASCs)28, but Kras-4A positive cells are certainly more abundant than BASCs. Sections of tumours from WT mice however showed almost no detectable signal for Kras-4A protein (Figure 5d), although low level expression was detected at the RNA level, as previously described for human cancers27. These data, taken together with the genetic evidence presented above, suggest that Kras-4A is required for tumor initiation, but may not be necessary for maintenance of tumour growth. It is possible that Kras-4A expression may affect the stem cell fate decision leading to the adenocarcinoma lineage, but further detailed studies will be required to elucidate the relationship between Kras isoform expression and lung stem cells. Interestingly, differential roles for Kras-4A and -4B have previously been proposed in embryonic stem cell differentiation9, but there are presently no data on adult stem cells.
Figure 5. Kras4A is expressed in normal lung epithelium but not significantly in tumours.
Kras-4A staining was observed in a sub-population of Clara cells in WT129/Sv (a) and FVB/N mice (b) but absent in homozygous KrasKI mice (expressing only Kras-4B) (c). The staining was observed throughout the bronchial tree, terminating at bronchio-alveolar duct junctions, locations of the BASCs. In contrast, papillary adenomas from 129/Sv mice do not show significant staining for Kras4A (d), although adjacent bronchial structure showed positive staining (d, inset).
Inhibitors targeting specific Ras post-translational events, in particular farnesylation, have not been successful in clinical trials, the reasons for which are unclear. Nevertheless, in vitro studies as well as mouse models strongly suggest that the activity of oncogenic Ras proteins can be effectively inhibited through manipulation of their carboxyl domains8,20. The identification of Kras-4A, and not Kras-4B as previously thought, as the main mediator of both the oncogenic activity of mutant Kras and the suppressor activity of WT Kras provides new opportunities for designing and testing novel targeted therapies.
Methods
Mouse breeding and Carcinogenesis
The HrasKI and KrasKI alleles were originally generated, and maintained in a 129/Sv background. Animals heterozygous for the HrasKI or KrasKI allele were intercrossed to generate WT, KI heterozygous or KI homozygous animals. Genotyping of mice was performed on DNA extracted from tail clippings (see Supplementary Table 2 for PCR primer sequences). Chemical carcinogenesis of the lungs and skin was performed as previously described2,7,14,15. KrasLA2 mice (on FVB/N background) were bred with KrasKI heterozygous mice, and all mice that inherited the KrasLA2 were selected for analysis. Animals were sacrificed at 4 months of age for analysis.
All mouse experiments were approved by the University of California, San Francisco Laboratory Animal Resource Center.
Mutational Analysis
Lung tumours were dissected from fixed lungs under a dissecting microscope, and genomic DNA was purified by phenol/chloroform extraction following proteinase K digestion. Mutations were detected by sequencing of PCR products corresponding to the endogenous Kras, HrasKI, KrasKI fragments containing the mutation hotspots at codon 12, 13 or 61 (Supplementary Table 2 for primer sequences). Since mutations in the HrasKI and Hras were found by sequencing to be exclusively the CAA-CTA alteration at codon 61, we took advantage of the XbaI digestion assay to detect this mutation29.
Immunoblotting
Protein lysates of lung tissues were prepared as previously described30. Immunoblots were probed with antibodies against Kras (F234 and C-19), Hras (C-20), Nras (C-20), and Erk1 (C-16) from Santa Cruz Biotechnology; phospho-p44/p42 Map Kinase, Akt, and αp38 from Cell Signaling; β-actin from Sigma; and phospho-Akt from Dr. David Stokoe (UCSF).
Immunohistochemistry
Sections were deparaffinized and antigen retrieval was performed in 10mM sodium citrate solution (pH 6). Endogenous peroxidase was quenched with a 3% H2O2 solution (Sigma) for 10 min at room temperature, and blocking was performed with 5% goat serum (Zymed) diluted in PBS containing 0.1% Tween-20 for 1h at room temperature. Overnight incubation with the primary antibodies against CCA/CC10 (a gift from Dr. A. Mukherjee, NICHD/NIH), SP-C (Chemicon), and Kras-4A (Santa Cruz Biotechnology) was performed at 4 °C. After washing in PBS containing 0.1% Tween-20 (PBS-T) for 3× 5min sections were incubated with a biotinylated secondary antibody anti rabbit (Vector) at a dilution of 1:200 for 1h at room temperature. Sections were washed with PBS-T, incubated in Vectastain ABC reagent (Vector) according to the manufacturer's instructions, and developed using DAB.
Supplementary Material
Acknowledgements
This work was supported by CA111834 and CA084244 (AB). MDT was supported in part by a Sandler Foundation Postdoctoral Research Fellowship. CEW was supported by the Swiss National Science Foundation. RDL was supported by a grant of Associazione Italiana per la ricerca sul cancro (AIRC). A.B. acknowledges support from the Bruce and Davina Isackson Foundation and from the Barbara Bass Bakar Chair of Cancer Genetics.
References
- 1.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–9. [PubMed] [Google Scholar]
- 2.You M, Candrian U, Maronpot RR, Stoner GD, Anderson MW. Activation of the Ki-ras protooncogene in spontaneously occurring and chemically induced lung tumors of the strain A mouse. Proc Natl Acad Sci U S A. 1989;86:3070–4. doi: 10.1073/pnas.86.9.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potenza N, et al. Replacement of K-Ras with H-Ras supports normal embryonic development despite inducing cardiovascular pathology in adult mice. EMBO Rep. 2005;6:432–7. doi: 10.1038/sj.embor.7400397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson L, et al. K-ras is an essential gene in the mouse with partial functional overlap with N-ras. Genes Dev. 1997;11:2468–81. doi: 10.1101/gad.11.19.2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plowman SJ, et al. While K-ras is essential for mouse development, expression of the K-ras 4A splice variant is dispensable. Mol Cell Biol. 2003;23:9245–50. doi: 10.1128/MCB.23.24.9245-9250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, et al. Wildtype Kras2 can inhibit lung carcinogenesis in mice. Nat Genet. 2001;29:25–33. doi: 10.1038/ng721. [DOI] [PubMed] [Google Scholar]
- 7.To MD, et al. A functional switch from lung cancer resistance to susceptibility at the Pas1 locus in Kras2LA2 mice. Nat Genet. 2006;38:926–30. doi: 10.1038/ng1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov. 2007;6:541–55. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- 9.Pells S, et al. Developmentally-regulated expression of murine K-ras isoforms. Oncogene. 1997;15:1781–6. doi: 10.1038/sj.onc.1201354. [DOI] [PubMed] [Google Scholar]
- 10.Wang Y, You M, Wang Y. Alternative splicing of the K-ras gene in mouse tissues and cell lines. Exp Lung Res. 2001;27:255–67. doi: 10.1080/019021401300054028. [DOI] [PubMed] [Google Scholar]
- 11.Ventura JJ, et al. p38alpha MAP kinase is essential in lung stem and progenitor cell proliferation and differentiation. Nat Genet. 2007;39:750–8. doi: 10.1038/ng2037. [DOI] [PubMed] [Google Scholar]
- 12.Jackson EL, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–8. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson L, et al. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–6. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 14.Balmain A, Pragnell IB. Mouse skin carcinomas induced in vivo by chemical carcinogens have a transforming Harvey-ras oncogene. Nature. 1983;303:72–4. doi: 10.1038/303072a0. [DOI] [PubMed] [Google Scholar]
- 15.Nelson MA, Futscher BW, Kinsella T, Wymer J, Bowden GT. Detection of mutant Ha-ras genes in chemically initiated mouse skin epidermis before the development of benign tumors. Proc Natl Acad Sci U S A. 1992;89:6398–402. doi: 10.1073/pnas.89.14.6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ise K, et al. Targeted deletion of the H-ras gene decreases tumor formation in mouse skin carcinogenesis. Oncogene. 2000;19:2951–6. doi: 10.1038/sj.onc.1203600. [DOI] [PubMed] [Google Scholar]
- 17.Wang M, Wang Y, You M. Identification of genetic polymorphisms through comparative DNA sequence analysis on the K-ras gene: implications for lung tumor susceptibility. Exp Lung Res. 2005;31:165–77. doi: 10.1080/01902140490495543. [DOI] [PubMed] [Google Scholar]
- 18.Dankort D, et al. A new mouse model to explore the initiation, progression, and therapy of BRAFV600E-induced lung tumors. Genes Dev. 2007;21:379–84. doi: 10.1101/gad.1516407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patek CE, et al. Mutationally activated K-ras 4A and 4B both mediate lung carcinogenesis. Exp Cell Res. 2008;314:1105–14. doi: 10.1016/j.yexcr.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Hancock JF. Ras proteins: different signals from different locations. Nat Rev Mol Cell Biol. 2003;4:373–84. doi: 10.1038/nrm1105. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Zhu T, Guan KL. Transformation potential of Ras isoforms correlates with activation of phosphatidylinositol 3-kinase but not ERK. J Biol Chem. 2004;279:37398–406. doi: 10.1074/jbc.M405730200. [DOI] [PubMed] [Google Scholar]
- 22.Voice JK, Klemke RL, Le A, Jackson JH. Four human ras homologs differ in their abilities to activate Raf-1, induce transformation, and stimulate cell motility. J Biol Chem. 1999;274:17164–70. doi: 10.1074/jbc.274.24.17164. [DOI] [PubMed] [Google Scholar]
- 23.Quintanilla M, Brown K, Ramsden M, Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 24.Li J, et al. LOH of chromosome 12p correlates with Kras2 mutation in non-small cell lung cancer. Oncogene. 2003;22:1243–6. doi: 10.1038/sj.onc.1206192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guerrero I, Villasante A, Corces V, Pellicer A. Loss of the normal N-ras allele in a mouse thymic lymphoma induced by a chemical carcinogen. Proc Natl Acad Sci U S A. 1985;82:7810–4. doi: 10.1073/pnas.82.23.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diaz R, et al. Inhibition of Ras oncogenic activity by Ras protooncogenes. Int J Cancer. 2005;113:241–8. doi: 10.1002/ijc.20563. [DOI] [PubMed] [Google Scholar]
- 27.Plowman SJ, et al. K-ras 4A and 4B are co-expressed widely in human tissues, and their ratio is altered in sporadic colorectal cancer. J Exp Clin Cancer Res. 2006;25:259–67. [PubMed] [Google Scholar]
- 28.Kim CF, et al. Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell. 2005;121:823–35. doi: 10.1016/j.cell.2005.03.032. [DOI] [PubMed] [Google Scholar]
- 29.Nagase H, Mao JH, Balmain A. Allele-specific Hras mutations and genetic alterations at tumor susceptibility loci in skin carcinomas from interspecific hybrid mice. Cancer Res. 2003;63:4849–53. [PubMed] [Google Scholar]
- 30.Mao JH, et al. Mutually exclusive mutations of the Pten and ras pathways in skin tumor progression. Genes Dev. 2004;18:1800–5. doi: 10.1101/gad.1213804. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.