Abstract
Accurate modification of the 3 billion-base-pair human genome requires tools with exceptional sequence specificity. Here, we describe a general strategy for the design of enzymes that target a single site within the genome. We generated chimeric zinc finger recombinases with cooperative DNA-binding and catalytic specificities that integrate transgenes with >98% accuracy into the human genome. These modular recombinases can be reprogrammed: New combinations of zinc finger domains and serine recombinase catalytic domains generate novel enzymes with distinct substrate sequence specificities. Because of their accuracy and versatility, the recombinases/integrases reported in this work are suitable for a wide variety of applications in biological research, medicine, and biotechnology where accurate delivery of DNA is desired.
Keywords: recombinases, zinc finger, gene delivery, gene targeting, protein engineering
The postgenomic era of medicine will be defined by our ability to achieve biological control through genetic reprogramming. New tools are needed to accurately rewrite the genomic script and specifically alter genes, gene expression, and epigenetic state at any desired loci. To date, no enzyme—natural or synthetic—has been able to accurately modify only a single targeted site within the human genome (1). Scientists in biology, biotechnology, stem cell research, and gene therapy currently rely on naturally occurring enzymes to perform functions like DNA integration and excision. However, these enzymes recognize multiple sites within the human genome, often resulting in off-target DNA integration and chromosomal translocation (2–6). Our recent work with serine resolvases and invertases led us to hypothesize that we could use a modular approach that capitalizes on cooperative specificity to design synthetic enzymes that would uniquely recognize a single site within the 3 billion-base-pair human genome and allow us to deliver DNA specifically to this site (Fig. 1A) (7).
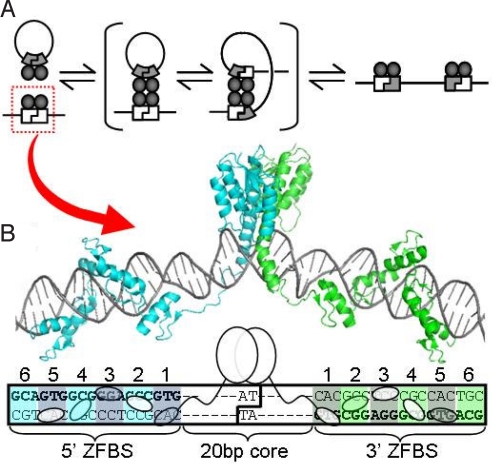
Fig. 1.
Structure and mechanism of RecZF-mediated integration. (A) Recombinase dimers bind to genomic and episomal target sites, form a tetrameric complex, cleave both target sites, exchange strands by a 180° rotation, and religate to generate the integrative product. (B) The DBDs of naturally occurring serine resolvases and invertases bind short and degenerate sequence motifs. When those domains are replaced with zinc finger proteins, recombination is retargeted to an extended sequence in which zinc finger binding sites (ZFBS) flank a 20-bp core recognized by the catalytic domain. Selective recognition of both of these elements affords RecZFs cooperative specificity. GinC5 was generated by fusing a hyperactive catalytic domain of the Gin invertase to the C5 zinc finger protein. The composite structure shown here is derived from those of DNA-bound γδ resolvase (12) and the Aart zinc finger protein (14).
In their native contexts, serine resolvases and invertases selectively recombine target sites within the same DNA molecule. This intramolecular specificity is assured by obligate assembly of large protein complexes, wherein accessory factors bound at neighboring sites impose topological and spatial constraints on the recombination reaction (8). Hyperactive mutants of several serine resolvases and invertases have been discovered that efficiently catalyze unrestricted recombination between minimal dimer-binding sites (Table S1) (9, 10). Furthermore, unlike other site-specific recombinases, serine resolvases and invertases are well suited to synthetic reengineering. These enzymes are modular in both structure and function, each comprised of a distinct catalytic domain flexibly tethered to a small helix–turn–helix DNA-binding domain (DBD). However, these DBDs are poorly suited for accurate genomic recombination (11) because the recognition motifs are short (4–6 bp) and degenerate (12, 13).
In contrast, zinc finger DNA-binding proteins recognize target sites of variable lengths with high specificity. These proteins are composed of a series of modular zinc finger domains that each bind specifically to 3 or 4 base pairs of DNA (14). Synthetic zinc finger DNA-binding proteins have been generated for many sequences, using several approaches (15–17). Capitalizing on this work, researchers have incorporated synthetic zinc finger proteins into a wide variety of molecular tools (7, 18–29).
The DBD of a hyperactive serine recombinase can be replaced with a zinc finger protein of higher affinity and specificity (7, 25). This substitution retargets recombination to sequences flanked by zinc finger binding sites (ZFBS) (Fig. 1B). However, these zinc finger-recombinases (RecZFs) retain a second, complementary specificity. The serine catalytic domain imposes its own sequence requirements on the interior of the RecZF target site (20-bp core, Fig. 1B) (7). Functional RecZF recombination sites must contain sequences compatible with both the zinc finger DNA-binding protein and recombinase catalytic domain.
The specificity of each RecZF is thus a product of modular site-specific DNA-binding and sequence-dependent catalysis. We hypothesized that this cooperative specificity would ensure accurately targeted recombination within a genomic context. To validate this design principle, we evaluated the ability of RecZFs to orchestrate plasmid integration into a single site in the human genome. We found that RecZFs catalyze targeted modification of the human genome with high accuracy (>98%). Moreover, additional RecZFs can be generated for novel target sequences. Because of this combination of precision and versatility, future development of RecZF technology should result in a powerful new set of tools for genetic studies, biotechnology, stem cell research, and gene therapies. More broadly, cooperative specificity may serve as a general strategy for designing the next generation of genome-modifying tools.
Results
To generate a highly specific zinc finger-recombinase (RecZF), we fused the catalytic domain of a hyperactive mutant of the Gin invertase, GinH106Y (9), to the zinc finger protein C5, yielding GinC5 (Fig. 1B and Table 1). A native Gin homodimer recognizes 3 elements within its substrate: 2 DBD-binding sites flanking an internal sequence required for catalysis (the 20-bp core). We replaced the natural DBD sites with those of C5 to generate a recombination site for GinC5, C.20G (Table 1). The RecZF site is thus the target of complementary zinc finger and catalytic domain specificities.
Table 1.
RecZF composition and DNA sequence specificity
| RecZF | Catalytic domain | Zinc finger | Zinc finger binding site |
|---|---|---|---|
| GinC1 | Gin(H107Y) | C1 | GTG |
| GinC2 | Gin(H107Y) | C2 | GGCGTG |
| GinC3 | Gin(H107Y) | C3 | GGAGGCGTG |
| GinC4 | Gin(H107Y) | C4 | GCGGGAGGCGTG |
| GinC5 | Gin(H107Y) | C5 | GTGGCGGGAGGCGTG |
| GinC6 | Gin(H107Y) | C6 | GCAGTGGCGGGAGGCGTG |
| Tn3C5 | Tn3(G70S, D102Y, E124Q) | C5 | GTGGCGGGAGGCGTG |
| Recombination site | RecZF target site |
||
|---|---|---|---|
| 5' ZFBS | 20 bp core | 3′ ZFBS | |
| C.20G | GCAGTGGCGGGAGGCGTGTCCAAAACCATGGTTTACAGCACGCCTCCCGCCACTGC | ||
| C.20T | GCAGTGGCGGGAGGCGTGTGATAATTTATAATATTTCGCACGCCTCCCGCCACTGC | ||
RecZFs were assembled by the fusion of hyperactive serine catalytic domains and zinc finger DNA-binding proteins (ZFPs). Each RecZF target site is a composite of sequences required for zinc finger DNA binding (ZFBS) and serine catalysis (20-bp core). RecZFs assembled from ZFPs C1 through C6 were designed to bind overlapping segments of the C6 target sequence and, therefore, are compatible with target sites C.20G and C.20T.
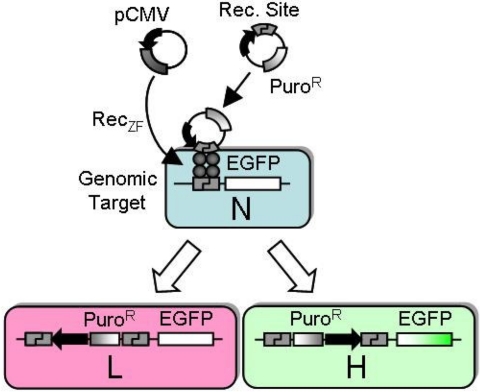
To evaluate the specificity of GinC5-mediated recombination, we created a reporter cell line (293-C.20G) by positioning a single copy of the GinC5 target site (C.20G) upstream of a promoterless EGFP transgene in the genome of human cells as described in Materials and Methods. The GinC5 target site was also introduced into the donor plasmid (C.20G-Puro) downstream of a CMV promoter. Because the puromycin resistance gene is constitutively expressed from the C.20G-Puro donor plasmid, integration anywhere in the genome confers puromycin resistance. Successful site-specific integration of donor plasmid into the genomic target site in the 293-C.20G cells should alter expression of EGFP. We anticipated that bidirectional integration would yield 2 phenotypically distinct products (Fig. 2). In one outcome, the CMV promoter would lie adjacent to EGFP, enhancing cellular fluorescence (EGFP-high). In the other, the CMV promoter and EGFP would be distant from each other and in opposite orientations (EGFP-low).
Fig. 2.
A model system for assaying RecZF-targeted integration. Donor plasmid and RecZF expression vector were cotransfected into a human cell line containing a single genomic target site (naïve, N). Bidirectional integration yielded 2 phenotypically distinct products. In one case, the CMV promoter lies adjacent to EGFP, enhancing cellular fluorescence (EGFP-high). In the other, CMV promoter and EGFP are distant and in opposite orientations (EGFP-low).
RecZFs Mediate Accurately Targeted Integration into the Human Genome.
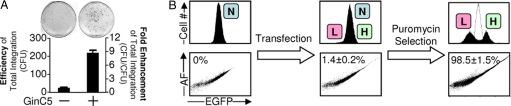
In the absence of recombinase, transfection of 293-C.20G cells with the C.20G-Puro donor plasmid yielded very few puromycin-resistant colonies [GinC5(−), Fig. 3A] and there was no difference in EGFP fluorescence between transfected cells and controls. By contrast, cotransfection with donor and the GinC5 expression vector substantially enhanced the efficiency of stable transgene integration (8.7 ± 1.6-fold more colonies relative to transfection with donor plasmid alone) (Fig. 3A). Additionally, a significant fraction of the cotransfected cell population (1.4 ± 0.2%) showed modified EGFP fluorescence (Fig. 3B). This percentage is equal to the efficiency of targeted integration. After puromycin selection, the vast majority of resistant cells exhibited either increased or decreased EGFP fluorescence (Fig. 3B). In 2 separate experiments, clonal cell populations were isolated and characterized. The frequency of targeted integration among clones from each experiment (23/23 and 32/33) indicated that the overall specificity of integration was 98.5 ± 1.5%. Genomic PCR analysis and sequencing confirmed that the 2 fluorescent phenotypes matched the expected genotypes at the C.20G locus (Fig. 4). These data indicate that GinC5 accurately targeted plasmid integration into the human genome. The level of nonrecombinase-mediated integration [GinC5(−), Fig. 3A], suggests that off-target integration catalyzed by GinC5 is below the detection limit of our assay.
Fig. 3.
GinC5 accurately targets plasmid integration into the human genome. (A) The donor plasmid constitutively expressed the puromycin resistance gene. After cotransfection of donor plasmid and RecZF expression vector, puromycin selection enriched for cells containing plasmid integrated anywhere in the genome. The number of puromycin resistant colonies was enhanced 8.7-fold by GinC5 expression. Samples were prepared and analyzed in triplicate and standard errors are shown. (B) Flow cytometry revealed the efficiency and specificity of targeted plasmid integration. Plasmid integrated at the target site in 1.4% of transfected cells, resulting in EGFP up- and down-regulation. Analysis of all integrative events (those that conferred puromycin resistance) indicated that the vast majority of plasmid integration occurred at the GinC5 target site. The specificity of GinC5-mediated integration (98.5 ± 1.5%) was determined by clonal analysis (Fig. 4). AF signifies cellular autofluorescence.
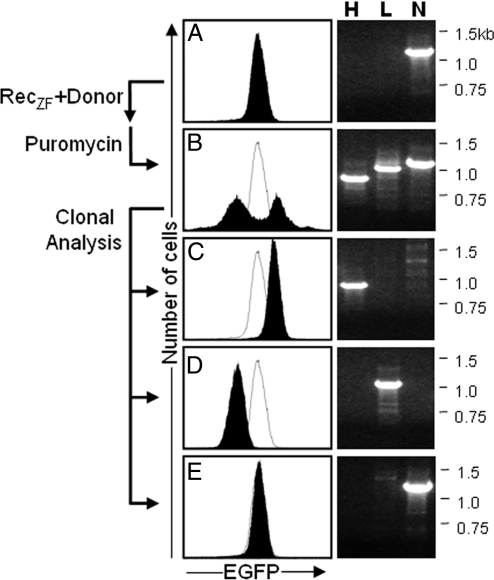
Fig. 4.
Clonal analysis of cellular fluorescence and genotype after plasmid integration. (A) The basal level of fluorescence in cells containing the naïve GinC5 genomic target locus (N). (B) EGFP expression was up- and down-regulated in the bulk population of puromycin-resistant cells containing stably integrated plasmid. (C–E) Clones isolated from this bulk population showed higher (C), lower (D), or no (E) change relative to levels of EGFP fluorescence in naïve cells. Genomic PCR confirmed that each phenotype matched the expected genotype at the genomic target locus: EGFP-high (H) (0.9 kb), EGFP-low (L) (1.1 kb), and naïve (N) (1.2 kb).
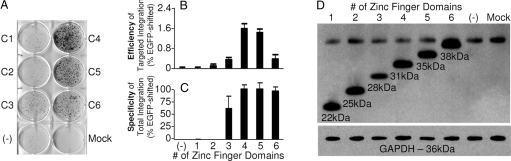
We also compared integration mediated by RecZFs containing different numbers of zinc finger domains (Table 1). The smallest RecZF, GinC1, is expected to recognize a 3-bp zinc finger binding site and the largest, GinC6, should specifically bind to an 18-bp site. We found that RecZFs with 4 or 5 zinc finger domains integrated donor plasmid into the genomic target with the highest integration efficiencies [1.6 ± 0.2% (Fig. 5 A and B) and 1.4 ± 0.2% (Fig. 3B), respectively] and specificities of total integration [98.7 ± 6.7% (Fig. 5C) and 98.5 ± 1.5% (Fig. 3B), respectively]. By contrast, recombinases with 1 or 2 fingers showed no activity above background suggesting that the binding activity of 1 and 2 finger proteins is insufficient. Western blot analysis confirmed that these differences were not caused by uneven protein expression levels (Fig. 5D).
Fig. 5.
Zinc finger dependence of RecZF-mediated integration. (A–C) The Gin catalytic domain was fused to zinc finger proteins of varied length. RecZFs containing 4 or 5 zinc finger domains afforded the largest increase in numbers of puromycin-resistant colonies (A), highest efficiency of targeted integration (B), and highest specificity of total integration (C). Standard error is shown for samples prepared in triplicate. (D) To ensure that these assays were not influenced by different levels of protein expression, Western blot analysis was performed. HA-tagged RecZFs were detected with HRP-conjugated anti-HA antibody. GAPDH served as an internal control to ensure uniform sample loading.
RecZF Target Sequence Specificity Is Programmable by Modular Design.
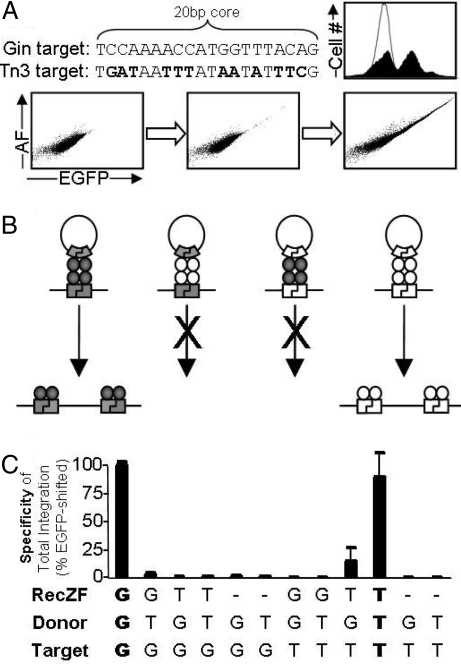
To demonstrate that additional highly specific RecZFs can be created by modular assembly, we fused the C5 zinc finger protein to a hyperactive mutant catalytic domain derived from Tn3 resolvase (S70G, D102Y, E124Q) (7). The resulting chimera, Tn3C5, was expected to recombine C.20T (Table 1), a RecZF site containing the central 20 bp of the Tn3 native substrate (Fig. 6A) (10). Tn3C5-mediated integration in the 293-C.20T cell line occurred with high specificity of total integration (88.3 ± 20.5%). Background fluorescence of unmodified cells prevented detection of the expected EGFP-low phenotype, but highly fluorescent cells (EGFP-high) were readily quantified (see Materials and Methods). Efficiency of targeted integration by Tn3C5 was 0.13 ± 0.01%, considerably lower than that of GinC5 (Fig. 6C).
Fig. 6.
Study of the contribution of catalytic domain specificity to RecZF target specificity. Novel RecZFs can be generated by combining hyperactive serine catalytic domains (Table S1) and zinc finger proteins. Tn3C5 was generated by fusing a hyperactive catalytic domain of the Tn3 resolvase to the C5 zinc finger protein. (A) Like GinC5, Tn3C5 catalyzed highly specific plasmid integration. (B) Illustration of RecZF target site selectivity. RecZF specificity is a product of both zinc finger recognition and catalytic domain sequence dependence. Because the Gin and Tn3 catalytic domains are specific for different core sequences (20G and 20T, respectively; Table 1), RecZFs derived from these 2 elements are functionally orthogonal. (C) Selectivity of the Gin and Tn3 catalytic domains was assayed under all possible combinations of RecZF (GinC5, G; Tn3C5, T), donor (C.20G-Puro, G; C.20T-Puro, T), and genomic target (293-C.20G, G; 293-C.20T, T). High levels of specific recombination were only observed when enzyme matched both substrates (shown in bold). Standard error is shown for samples prepared in triplicate.
We tested the selectivity of the Gin and Tn3 catalytic domains (Fig. 6B) by assaying integration with all possible combinations of RecZF (GinC5, Tn3C5), donor (C.20G-Puro, C.20T-Puro), and genomic target (293-C.20G, 293-C.20T) (Fig. 6C). High levels of specific recombination were only observed when enzyme matched both substrates. Notably, the genomic target sites were particularly stringent: Integration was only allowed when target and RecZF were properly paired. These results demonstrate that the cooperative specificities of the zinc finger protein and the recombinase catalytic domain are necessary to create a functional enzyme at a particular target site. This mechanism accounts for the stringent activity of these enzymes and exceptional accuracy of plasmid integration.
Discussion
In order to specifically alter genomes, researchers have developed a variety of tools for targeted genetic modification. Progress toward an accurate genomic integrase began with the characterization of naturally occurring enzymes such as Cre recombinase (30). More recently, bacteriophage serine integrases have gained prominence because they catalyze unidirectional recombination; this selectivity facilitates stable transgene integration (31). Additional contributions were made by synthetically coupling a highly specific DNA-binding domain with a nonspecific integrase (26) and transposase (27). Unfortunately, each of these enzymes suffers from a lack of specificity. Using these systems in human cells, only 1–14% of plasmid integration occurs at the targeted genomic site and sophisticated selection strategies are necessary to obtain pure populations (26, 27, 30, 31). Additionally, off-target recombination (2, 6) can result in deletion, chromosomal translocation, cytotoxicity, oncogenesis, and other adverse consequences that have all been documented (3–6, 32). Substantial off-target activity has also been observed with other classes of DNA-modifying proteins (1, 33, 34). Additional experiments are necessary to determine whether RecZFs can also cause these abnormalities, although the high level of specificity of plasmid integration suggests that this new class of enzymes will be less susceptible to these limitations.
Clearly, there remains an unmet need for tools with exceptional sequence specificity. To address this problem, we set out to assemble enzymes from highly specific DNA-binding and catalytic domains to provide 2 complementary layers of stringent regulation. Our experiments with GinC5 validated this design strategy: The combination of cooperative specificities—zinc finger binding and the sequence-dependent catalysis—ensured that plasmid integrated precisely into the desired genomic target site. Although integration anywhere in the genome would have conferred puromycin resistance, 55 of 56 resistant clones (98.5%) contained an integration event at the target locus. The single clone containing a nonspecific integration event is likely attributable to the background level of puromycin resistant colonies that arose in the absence of RecZF (Fig. 3A). It remains possible that RecZF-mediated nonspecific integration occurred at a level below the threshold of our assays.
Compared with naturally occurring enzymes, RecZFs not only integrate with high specificity, but high efficiency as well. RecZF efficiencies are comparable to other integration technologies, including phage integrases (31), adeno-associated virus vectors (35), and transposases (36). For example, the efficiency of targeted integration catalyzed by GinC5 (1.4%; Fig. 3B) is similar to that of the φC31 integrase (0.7%) (31). φC31 is a member of the family of large serine recombinases and has received substantial attention because of the advantage of this enzyme to mediate unidirectional integration (6). It remains to be determined why these 2 reactions, one irreversible (φC31) and the other kinetically disfavored (GinC5), yield similar integrative efficiencies.
Another approach to achieving genomic specificity relies on cellular DNA repair (37). Mediated by endogenous DNA-repair enzymes, homologous recombination (HR) accurately modifies the genetic sequence of the target locus. The chief drawback of this approach is its relative inefficiency: HR is orders of magnitude less frequent than random plasmid integration (1). Several catalysts have been developed that dramatically enhance the efficiency of HR, including triplex forming oligonucleotides and adeno-associated viral vectors (1). A very promising method relies on endonucleases (e.g., homing nucleases such as I-SceI and zinc finger-nuclease fusion proteins) that recognize rare sites (23, 29, 38). Although zinc finger-nuclease fusion proteins have been used to modify genomic DNA with high efficiency, this strategy currently suffers from fundamental limitations that hamper its overall safety and efficacy in gene therapy scenerios. In particular, aberrant cleavage of unknown off-target chromosomal sequences can result in significant toxicity in treated cells (33). Additionally, repeated cleavage at both the target site and off-target sites stimulates error-prone DNA repair mechanisms that lead to random mutations at these sites (23, 24, 29). These events are unpredictable and extremely difficult to comprehensively characterize experimentally and 1 recent study found that off-target modifications occurred with a frequency of >13% (23). In contrast, RecZF specificity can be readily quantified by clonal analysis as performed in this study (Fig. 4). More importantly, RecZFs eliminate the need to damage cellular DNA and do not rely on cellular mechanisms of DNA repair and homologous recombination. A more thorough comparison of RecZFs with other molecular tools will require further experiments that directly characterize their effect on cytotoxicity and genome stability (i.e., chromosome loss or translocation).
Because of the modular composition of the RecZFs, recombinases/integrases with distinct sequence specificities can be synthesized. We have demonstrated that RecZFs are fully programmable, with target sites defined by the cooperative specificities of both zinc finger protein (Fig. 5) (7) and serine recombinase catalytic domain (Fig. 6). Although it remains to be demonstrated that RecZFs can be readily prepared to act at any given DNA sequence, a large variety of RecZFs can be assembled by drawing on the growing number of hyperactive serine catalytic domains (Table S1) and the expansive pool of polydactyl zinc finger DNA-binding proteins of high affinity and specificity. In addition, we have shown that the specificities of naturally occurring serine recombinase catalytic domains can be modified by directed evolution (7). Indeed, large changes in catalytic domain specificity have already been achieved. Beyond vastly increasing the number of catalytic domains available for RecZF assembly, such evolutionary selection should enable the design of RecZFs tailored to endogenous genomic loci. We envision that an array of orthogonal recombinases/integrases will permit site-specific genomic manipulation comparable to that allowed by the restriction enzymes that presently facilitate genetic manipulations in vitro (39). Our results with GinC5-mediated integration suggest that cooperative specificity can be used to create a new family of highly specific genetic tools that might play an important role in biology and medicine by allowing scientists to rewrite the genomic script with exceptional accuracy.
Materials and Methods
Zinc Finger-Recombinase Expression Vectors.
Our group has selected zinc finger domains that selectively bind many of the 64 DNA triplets (40–42). A synthetic DNA-binding protein can be designed for a given target sequence by the modular assembly of corresponding fingers. A hyperactive Gin invertase catalytic domain (H106Y) (9) was PCR amplified using a 3′ primer encoding the F1 zinc finger domain (GTG). This fusion product (GinC1) was subsequently digested so that 3′ zinc finger domains F2 through F6 (GGC, GGA, GCG, GTG, GCA) could be iteratively inserted to generate a series of RecZFs of increasing length: GinC2, GinC3, GinC4, GinC5, and GinC6 (Table 1). These genes were then cloned into pcDNA3.1-Zeocin (Invitrogen). The same method was used to generate Tn3C5, a fusion of a hyperactive Tn3 resolvase catalytic domain (S70G, D102Y, E124Q) (7) and the zinc finger protein C5. Sequences of each RecZF are included in Table S2.
Recombination Substrates.
The CMV promoter was PCR amplified from pcDNA3.1-Zeocin (Invitrogen), using a long 3′ primer encoding recombination site C.20G (Table 1), and cloned into a derivative of pBabe-Puromycin (43) to generate C.20G-Puro. The 20G sequence differs from the natural substrate of the Gin invertase (Table S1) at the 2 central base pairs: TC→AT (7). Donor plasmid C.20T-Puro was generated in similar fashion using a 3′ primer encoding recombination site C.20T (Table 1).
Cell lines containing genomic target sites for GinC5 and Tn3C5 were generated using the Flp-In system (Invitrogen). The EGFP gene (Clontech) was PCR amplified with a 5′ primer encoding recombination site C.20G and cloned into a derivative of pcDNA5/FRT (Invitrogen) to generate EGFP-C.20G. EGFP-C.20T was generated in similar fashion, using a 5′ primer encoding recombination site C.20T. The Flp-In-293 cell line (Invitrogen) was cotransfected with EGFP-C.20G (or EGFP-C.20T) and the Flp expression plasmid (pOG44, Invitrogen) to create a human cell line containing a single target recombination site. Single colonies for each RecZF target site were isolated by hygromycin selection, characterized by flow cytometry and genomic PCR, and used as target cell lines (293-C.20G and 293-C.20T) in subsequent experiments. Cells were maintained in DMEM containing 10% (vol/vol) FBS, hygromycin (150 μg/mL), and 1% penicillin/streptomycin (Gibco/BRL, Invitrogen).
Targeted Integration Assays.
Target cells (293-C.20G, 293-C.20T) were seeded onto polylysine-coated 6-well plates at a density of 7.5 × 105 cells per well. After 24 h of incubation, these cells were cotransfected with RecZF expression vector (2 μg) and donor plasmid (C.20G-Puro or C.20T-Puro, 200 ng), using Lipofectamine 2000 (Invitrogen) according to the manufacturer's directions. Negative controls contained pcDNA3.1-Zeocin (2,000 ng) rather than the RecZF expression vector. At 24 h after transfection (day 1), cells were transferred at 1:10 dilution into new wells. On day 2, hygromycin (150 μg/mL) or hygromycin and puromycin (2 μg/mL) were added to the media. Cells were then passaged as necessary for 16 days. On day 18, cells were subjected to cytometric analysis with a FACScan dual laser flow cytometer (BD Biosciences). Calculations of efficiency and specificity were based on populations with high EGFP fluorescence (EGFP-high) (Figs. 2–5). The gating parameters used to analyze unselected populations captured 0.05% of background. A different set of gating parameters was used for puromycin-selected populations, capturing 3% of background. Clonal analysis (see below) indicated that GinC5-mediated integration of C.20G-Puro into 293-C.20G, followed by puromycin selection, yielded cells that were 98.5% EGFP modified (Fig. 3 and Fig. 4). All other values of efficiency and specificity were interpolated between the background populations (0% EGFP-modified) and this positive control.
For clonal analysis, cell populations from 2 independent experiments were seeded into 96-well plates at limiting dilution with media containing both hygromycin and puromycin. Each well was visually inspected to ensure single colony formation. EGFP expression by clonal populations was determined by flow cytometry. Genomic PCR was used to genotype the colonies and parental bulk populations. Each PCR contained 100 ng of genomic DNA isolated with the QIAamp DNA mini kit (QIAGEN). Three sets of PCR primer pairs were used, each pair corresponding to an expected genotype at the target locus (Fig. S1).
Colony Counting Assay.
To determine the efficiency of plasmid integration, transfected cells were transferred at 24 h after transfection (day 1) onto polylysine-coated 10-cm plates containing media with puromycin (2 μg/mL). On day 10, cells were stained with crystal violet solution for colony counting. Parallel incubation of diluted cell populations in the absence of puromycin indicated that 1.30 × 104 ± 7 × 102 CFUs were seeded in each 10-cm plate. The 6-well plates shown in Fig. 6A were generated in similar fashion.
Western Blot Analysis.
Western Blot Analysis is described in SI Methods.
Supplementary Material
Acknowledgments.
This work was supported by The Skaggs Institute for Chemical Biology and National Institutes of Health Grant R21CA126664. C.A.G. is a National Institutes of Health Postdoctoral Fellow and is supported by National Institutes of Health Grant F32 CA125910.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812502106/DCSupplemental.
References
- 1.Sorrell DA, Kolb AF. Targeted modification of mammalian genomes. Biotechnol Adv. 2005;23(7–8):431–469. doi: 10.1016/j.biotechadv.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Thyagarajan B, Guimaraes MJ, Groth AC, Calos MP. Mammalian genomes contain active recombinase recognition sites. Gene. 2000;244(1–2):47–54. doi: 10.1016/s0378-1119(00)00008-1. [DOI] [PubMed] [Google Scholar]
- 3.Schmidt EE, Taylor DS, Prigge JR, Barnett S, Capecchi MR. Illegitimate Cre-dependent chromosome rearrangements in transgenic mouse spermatids. Proc Natl Acad Sci USA. 2000;97:13702–13707. doi: 10.1073/pnas.240471297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loonstra A, et al. Growth inhibition and DNA damage induced by Cre recombinase in mammalian cells. Proc Natl Acad Sci USA. 2001;98:9209–9214. doi: 10.1073/pnas.161269798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu J, Jeppesen I, Nielsen K, Jensen TG. PhiC31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Therapy. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- 6.Chalberg TW, et al. Integration specificity of phage phiC31 integrase in the human genome. J Mol Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- 7.Gordley RM, Smith JD, Graslund T, Barbas CF., III Evolution of programmable zinc finger-recombinases with activity in human cells. J Mol Biol. 2007;367:802–813. doi: 10.1016/j.jmb.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 8.Grindley ND, Whiteson KL, Rice PA. Mechanisms of site-specific recombination. Ann Rev Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 9.Klippel A, Cloppenborg K, Kahmann R. Isolation and characterization of unusual gin mutants. EMBO J. 1988;7:3983–3989. doi: 10.1002/j.1460-2075.1988.tb03286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arnold PH, Blake DG, Grindley ND, Boocock MR, Stark WM. Mutants of Tn3 resolvase which do not require accessory binding sites for recombination activity. EMBO J. 1999;18:1407–1414. doi: 10.1093/emboj/18.5.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rozsa FW, Viollier P, Fussenegger M, Hiestand-Nauer R, Arber W. Cin-mediated recombination at secondary crossover sites on the Escherichia coli chromosome. J Bacteriol. 1995;177:1159–1168. doi: 10.1128/jb.177.5.1159-1168.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 13.Chiu TK, Sohn C, Dickerson RE, Johnson RC. Testing water-mediated DNA recognition by the Hin recombinase. EMBO J. 2002;21:801–814. doi: 10.1093/emboj/21.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Segal DJ, Crotty JW, Bhakta MS, Barbas CF, III, Horton NC. Structure of Aart, a designed six-finger zinc finger peptide, bound to DNA. J Mol Biol. 2006;363:405–421. doi: 10.1016/j.jmb.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 15.Pabo CO, Peisach E, Grant RA. Design and selection of novel Cys2His2 zinc finger proteins. Ann Rev Biochem. 2001;70:313–340. doi: 10.1146/annurev.biochem.70.1.313. [DOI] [PubMed] [Google Scholar]
- 16.Maeder ML, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beerli RR, Barbas CF., III Engineering polydactyl zinc-finger transcription factors. Nat Biotechnol. 2002;20:135–141. doi: 10.1038/nbt0202-135. [DOI] [PubMed] [Google Scholar]
- 18.Beerli RR, Dreier B, Barbas CF., III Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci USA. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan S, et al. Zinc-finger protein-targeted gene regulation: Genomewide single-gene specificity. Proc Natl Acad Sci USA. 2003;100:11997–12002. doi: 10.1073/pnas.2035056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papworth M, et al. Inhibition of herpes simplex virus 1 gene expression by designer zinc-finger transcription factors. Proc Natl Acad Sci USA. 2003;100:1621–1626. doi: 10.1073/pnas.252773399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberhardy SR, et al. Inhibition of human immunodeficiency virus type 1 replication with artificial transcription factors targeting the highly conserved primer-binding site. J Virol. 2006;80:2873–2883. doi: 10.1128/JVI.80.6.2873-2883.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urnov FD, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 23.Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26:808–816. doi: 10.1038/nbt1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Santiago Y, et al. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci USA. 2008;105:5809–5814. doi: 10.1073/pnas.0800940105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akopian A, He J, Boocock MR, Stark WM. Chimeric recombinases with designed DNA sequence recognition. Proc Natl Acad Sci USA. 2003;100:8688–8691. doi: 10.1073/pnas.1533177100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan W, Dong Z, Wilkinson TA, Barbas CF, III, Chow SA. Human immunodeficiency virus type 1 incorporated with fusion proteins consisting of integrase and the designed polydactyl zinc finger protein E2C can bias integration of viral DNA into a predetermined chromosomal region in human cells. J Virol. 2006;80:1939–1948. doi: 10.1128/JVI.80.4.1939-1948.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ivics Z, et al. Targeted Sleeping Beauty transposition in human cells. Mol Ther. 2007;15:1137–1144. doi: 10.1038/sj.mt.6300169. [DOI] [PubMed] [Google Scholar]
- 28.Nomura W, Barbas CF., III In vivo site-specific DNA methylation with a designed sequence-enabled DNA methylase. J Am Chem Soc. 2007;129:8676–8677. doi: 10.1021/ja0705588. [DOI] [PubMed] [Google Scholar]
- 29.Carroll D. Progress and prospects: Zinc-finger nucleases as gene therapy agents. Gene Therapy. 2008;15:1463–1468. doi: 10.1038/gt.2008.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fukushige S, Sauer B. Genomic targeting with a positive-selection lox integration vector allows highly reproducible gene expression in mammalian cells. Proc Natl Acad Sci USA. 1992;89:7905–7909. doi: 10.1073/pnas.89.17.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thyagarajan B, Olivares EC, Hollis RP, Ginsburg DS, Calos MP. Site-specific genomic integration in mammalian cells mediated by phage phiC31 integrase. Mol Cell Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hacein-Bey-Abina S, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 33.Cornu TI, et al. DNA-binding specificity is a major determinant of the activity and toxicity of zinc-finger nucleases. Mol Ther. 2008;16:352–358. doi: 10.1038/sj.mt.6300357. [DOI] [PubMed] [Google Scholar]
- 34.Smith AE, Hurd PJ, Bannister AJ, Kouzarides T, Ford KG. Heritable gene repression through the action of a directed DNA methyltransferase at a chromosomal locus. J Biol Chem. 2008;283:9878–9885. doi: 10.1074/jbc.M710393200. [DOI] [PubMed] [Google Scholar]
- 35.Hirata R, Chamberlain J, Dong R, Russell DW. Targeted transgene insertion into human chromosomes by adeno-associated virus vectors. Nat Biotechnol. 2002;20:735–738. doi: 10.1038/nbt0702-735. [DOI] [PubMed] [Google Scholar]
- 36.Wu SC, et al. piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and MosI in mammalian cells. Proc Natl Acad Sci USA. 2006;103:15008–15013. doi: 10.1073/pnas.0606979103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- 38.Paques F, Duchateau P. Meganucleases and DNA double-strand break-induced recombination: Perspectives for gene therapy. Curr Gene Therapy. 2007;7:49–66. doi: 10.2174/156652307779940216. [DOI] [PubMed] [Google Scholar]
- 39.Roberts RJ. How restriction enzymes became the workhorses of molecular biology. Proc Natl Acad Sci USA. 2005;102:5905–5908. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dreier B, Beerli RR, Segal DJ, Flippin JD, Barbas CF., III Development of zinc finger domains for recognition of the 5′-ANN-3′ family of DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2001;276:29466–29478. doi: 10.1074/jbc.M102604200. [DOI] [PubMed] [Google Scholar]
- 41.Dreier B, et al. Development of zinc finger domains for recognition of the 5′-CNN-3′ family DNA sequences and their use in the construction of artificial transcription factors. J Biol Chem. 2005;280:35588–35597. doi: 10.1074/jbc.M506654200. [DOI] [PubMed] [Google Scholar]
- 42.Dreier B, Segal DJ, Barbas CF., III Insights into the molecular recognition of the 5′-GNN-3′ family of DNA sequences by zinc finger domains. J Mol Biol. 2000;303:489–502. doi: 10.1006/jmbi.2000.4133. [DOI] [PubMed] [Google Scholar]
- 43.Morgenstern JP, Land H. Advanced mammalian gene transfer: High titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 1990;18:3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.