Abstract
Multidrug resistance continues to be a major impediment to successful chemotherapy in cancer patients. One cause of multidrug resistance is enhanced expression of the mdr1 gene, but the precise factors and physiological conditions controlling mdr1 expression are not entirely known. To gain a better understanding of mdr1 transcriptional regulation, we created a unique mouse model that allows noninvasive bioimaging of mdr1 gene expression in vivo and in real time. The model uses a firefly luciferase (fLUC) gene inserted by homologous recombination into the murine mdr1a genetic locus. The inserted fLUC gene is preceded by a neo expression cassette flanked by loxP sites, so that Cre-mediated recombination is required to configure the fLUC gene directly under the control of the endogenous mdr1a promoter. We now demonstrate that the mdr1a.fLUC knock-in is a faithful reporter for mdr1a expression in naive animals, in which fLUC mRNA levels and luminescence intensities accurately parallel endogenous mdr1a mRNA expression. We also demonstrate xenobiotic-inducible regulation of mdr1a.fLUC expression in real time, in parallel with endogenous mdr1a expression, resulting in a more detailed understanding of the kinetics of mdr1a gene induction. This mouse model demonstrates the feasibility of using bioimaging coupled with Cre/loxP conditional knock-in to monitor regulated gene expression in vivo. It represents a unique tool with which to study the magnitude and kinetics of mdr1a induction under a variety of physiologic, pharmacologic, genetic, and environmental conditions.
Keywords: bioimaging, conditional knock-in, gene regulation, MDR1, multidrug resistance
The human MDR1 gene encodes P-glycoprotein (Pgp), which functions as a transmembrane drug transporter and mediates the efflux of drugs from cells, thus conferring multidrug resistance on cancer cells and tumors that over-express it (1, 2). In addition, basal expression of MDR1 in organs such as liver, kidney, and colon, because of their involvement in drug excretion and absorption, affects the pharmacokinetics of drug uptake and excretion for agents that are substrates for Pgp transport (3–6). The mechanism of MDR1 regulation in tumors or in normal, nonmalignant tissue, particularly in response to xenobiotics and other environmental and physiologic stimuli, is not fully understood. The tumor-suppressor p53 has been implicated as a possible transactivator of MDR1 expression in tumors (7–9), although this association remains controversial (see ref. 10). In addition, nuclear receptors such as SXR and constitutive androstane receptor (CAR) have been suggested as possible master regulators of xenobiotic- and drug-inducible expression of MDR1, and of other genes involved in drug metabolism in organs such as the liver, kidney, and colon (11, 12). One recent report implicates the transcription factor FOXO3a in doxorubicin-mediated induction of MDR1 in the K562 human leukemia cell line (13).
Our limited understanding of MDR1 gene regulation in vivo is partly a result of the difficulty in conducting well-controlled clinical studies that require pretreatment and repeated posttreatment tissue sampling, and partly because of lack of adequate animal models for tracking MDR1/Pgp expression. A possible model system is the mouse, which has 2 MDR1 homologues, mdr1a and mdr1b, both of which can transport chemotherapy drugs and confer drug resistance (14, 15). Based on nucleotide and amino acid alignments in their coding regions, human MDR1 is more closely related to mdr1a than it is to mdr1b (16–18). In addition, the promoter regions of human MDR1 and murine mdr1a share ≈70% nucleotide sequence identity and they contain several cis-regulatory elements in common (17). Both genes are subject to similar regulation by SXR (11), or its mouse homolog PXR (19), and p53 (8, 20). Thus, mouse mdr1a is a reasonable surrogate with which to study MDR1 gene regulation in the in vivo setting. To gain a better understanding of mdr1 expression, we created a mouse model that allows noninvasive bioimaging of mdr1 gene expression in vivo and in real time by inserting a firefly luciferase (fLUC) gene into the murine mdr1a genetic locus by homologous recombination. We now report that the mdr1a.fLUC knock-in is a faithful reporter for basal mdr1a expression and induced expression under conditions of xenobiotic treatment. This model is a unique tool for noninvasively studying the magnitude and kinetics of mdr1a induction under a variety of physiologic, pharmacologic, and genetic conditions.
Results
Creation of a Mouse Model for Noninvasive Study of mdr1a Gene Expression in Vivo.
We created a gene replacement model that has the fLUC gene inserted by homologous recombination into the murine mdr1a chromosomal locus [supporting information (SI) Methods and Fig S1a]. In the targeting vector, the fLUC gene is preceded by a neo expression cassette that contains a synthetic polyadenylation sequence to stop transcription. The neo cassette is flanked by loxP sites, the targets for Cre-mediated recombination. We first established mouse embryonic stem cells and mice (Figs. S1 b and c) with the mdr1a+/flox genotype: 1 nontargeted mdr1a allele and 1 targeted mdr1a.fLUC allele before Cre-mediated removal of the floxed neo sequences. We mated these mice with Hprt-Cre transgenic mice to delete the floxed neo cassette in all tissues of the F1 offspring (21). The resulting progeny had the mdr1a+/fLUC genotype: 1 nontargeted mdr1a allele and 1 mdr1a.fLUC allele in the recombined state, with the fLUC gene directly under the control of the endogenous mdr1a promoter, in frame and in place of the Pgp ORF (see Fig. S1 a and c).
The mdr1a.fLUC Reporter Gene Faithfully Reflects Basal mdr1a Expression in Naive Animals.
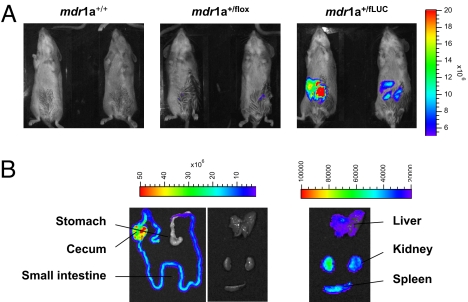
We injected luciferin into mdr1a+/fLUC mice heterozygous for the recombined mdr1a.fLUC allele and observed a strong luminescence signal in the abdominal area (Fig. 1A), consistent with the known pattern of mdr1a expression in several abdominal organs, such as liver, kidney, and intestine, with highest expression in intestine (14). To quantify the luminescence emitted by individual organs, we dissected organs from multiple animals after they had been injected with luciferin and measured the luminescence intensity in these isolated organs (Fig. 1B). As expected, luminescence signals were detected in the liver, kidney, spleen, and intestine, all of which have been reported to express mdr1a, with at least 100-fold stronger luminescence signals in the intestine relative to the other organs (Figs. 1B and 2A). To determine if the luminescence intensities visualized by the camera detection system accurately reflected basal expression of the intact endogenous mdr1a gene and the knock-in mdr1a.fLUC reporter gene, we extracted RNA from these organs and used real-time RT-PCR to measure mdr1a mRNA transcribed from the nontargeted allele and fLUC mRNA transcribed from the targeted allele. The fLUC mRNA levels and luminescence intensities paralleled mdr1a mRNA expression in all tissues analyzed, demonstrating that the mdr1a.fLUC reporter gene faithfully reported basal mdr1a expression in mdr1a+/fLUC mice (see Fig. 2A).
Fig. 1.
Luminescence imaging of mdr1a+/fLUC and control mice. (A) In vivo luciferase imaging of wild type mdr1a+/+ mice, mdr1a+/flox mice heterozygous for the targeted allele, and mdr1a+/fLUC mice heterozygous for the Cre-recombined mdr1a.fLUC allele. Imaging was performed as described in Methods and the same color scale is used to display the images of all mice shown. (B) Imaging of organs harvested from a representative mdr1a+/fLUC mouse heterozygous for the mdr1a.fLUC allele. Images in the first 2 panels are displayed on the same color scale (2–50 × 106 relative light units), thus illustrating the majority of luminescence emanating from the intestines. The images in the third column are displayed on a color scale representing 2–10 × 104 relative light units, thus illustrating the ≈100-fold lower expression of fLUC in those organs.
Fig. 2.
Measurement of basal luminescence and mRNA levels. (A) (Left) Bar graph showing the luminescence emitted by organs harvested from 3 mdr1a+/fLUC mice. The luminescence intensity of each organ was individually normalized to the luminescence intensity of the liver from the same mouse. Shown are the averages of the normalized values (± SE). (Right) Bar graph showing the fLUC and mdr1a mRNAs expressed in the same set of organs. Total RNA extracted from each organ was subjected to real-time RT-PCR (see Methods) and gene-specific levels in each organ were normalized to the respective levels in the liver. Shown are the averages of the normalized levels (± SE) for fLUC (filled bars) and mdr1a (hatched bars). (B) Total RNA was extracted from a separate cohort of six mdr1a+/+ (wild-type) mice and six mdr1a+/fLUC (knock-in) mice and subjected to real-time RT-PCR (see Methods). Mdr1a values in each mouse/organ were adjusted to GAPDH values in the same mouse/organ. (Left) Results for intestinal mRNAs. (Right) Results for liver and kidney mRNAs (note the 40-fold difference between the 2 scales). Statistical significance was determined by two-sided t test. (C) Regression analysis showing the relationship between intestinal mdr1a levels in individual mice (taken from B) and abdominal-area luminescence intensities measured just before killing of those same mice.
To determine if mdr1a.fLUC is also an accurate model for mdr1a expression in wild-type mice, we measured mdr1a mRNA in the organs of six mdr1a+/+ (wild-type) mice lacking the targeting construct and compared these with mdr1a and fLUC levels in 6 mdr1a+/fLUC mice heterozygous for the recombined knock-in allele. Mdr1a (Fig. 2B) and fLUC (not shown) mRNA expression patterns in mdr1a+/fLUC mice paralleled the mdr1a expression pattern in mdr1a+/+ mice. As expected, the knock-in mice, on average, expressed about half as much mdr1a mRNA in each organ as did wild-type mice. Finally, we plotted the live-animal luminescence intensities of the 6 mdr1a+/fLUC mice in this experiment against their own mdr1a mRNA levels in the intestine (the site of highest expression). There was a near-absolute correlation between intestinal mdr1a levels and luminescence (r2 = 0.9396) (Fig. 2C). We observed similar results for the correlation between intestinal mdr1a mRNA and fLUC mRNA levels within each of the 6 mice (r2 = 0.9582, not shown). Taken together, these results indicate that the mdr1a.fLUC locus is not dysregulated in our knock-in mice, in terms of tissue specificity, and that there is no compensatory dysregulation of mdr1a or mdr1a.fLUC expression levels because of knock-out of one mdr1a gene copy. Moreover, live-animal luminescence can reasonably be used as a reporter for mdr1a mRNA in the intestine, the site of highest mdr1a expression in mice.
The mdr1a.fLUC Allele Is a Faithful Reporter for mdr1a Gene Induction.
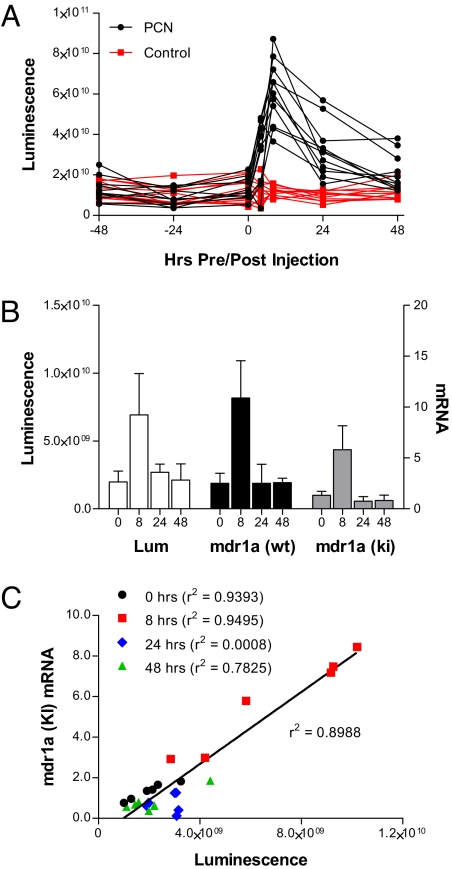
Mdr1a transcription is regulated by certain xenobiotics. For example, pregnenolone-16α-carbonitrile (PCN), a steroidal compound, induces mdr1a expression in rodents (22) by activating the nuclear receptor PXR (19). To determine if the fLUC gene could faithfully report changes in mdr1a expression induced by chemical signals, we first needed to determine the day-to-day variability in luminescence in individual mice. Thus, we performed live imaging on 22 heterozygous mdr1a+/fLUC mice on 3 consecutive days at 24-h intervals, without any prior injection of inducing agent (Fig. 3A). There was about a 5-fold mouse-to-mouse range between the least intense and most intense luminescences on the first day (–48-h time point in Fig. 3A), but less than 2-fold day-to-day fluctuation in luminescence, on average, for any given mouse over the 3-day sampling period.
Fig. 3.
Mdr1a gene induction by PCN. Heterozygous mdr1a+/fLUC mice were given 200 mg/kg of PCN by i.p. injection at time 0. Mice that were injected with corn oil were used as control. (A) Luciferase expression was quantified by measuring luminescence intensity in the abdominal area at various time points before and after PCN or vehicle injection. Shown are the luminescence intensities at each time point in each individual mouse. (B) Eighteen mdr1a+/fLUC mice and 18 mdr1a+/+ mice were given PCN (200 mg/kg by i.p. injection) and then groups of 6 mice of each genotype were killed at 8, 24, and 48 h after injection. Mice from Fig. 2B were used as the preinjection (0 h) cohort. Luminescence intensities in the abdominal area were measured immediately before sacrifice and various organs were dissected and frozen immediately after killing. Total RNAs were extracted from the intestinal tissues of each mouse and processed as in Fig. 2. Shown are the average luminescence intensities (Lum) measured in each group of mdr1a+/fLUC mice and the average mdr1a mRNA levels in the intestinal tissues of each group of mdr1a+/fLUC (ki) and mdr1a+/+ (wt) mice. Error bars indicate standard deviations. (C) Regression analysis showing the overall relationship between intestinal mdr1a levels in individual mice and abdominal-area luminescence intensities measured just before killing of those same mice. The correlation coefficient for all data points is shown next to the regression line. The correlation coefficient for data associated with each individual time point is shown next to the respective figure legend. The data for 0 h are the same as those shown in Fig. 2C.
We next treated the same cohort of mice with 200 mg/kg of PCN or vehicle control and quantified fLUC expression by performing live imaging at various time points after PCN delivery. In mice injected with PCN, luminescence intensity increased by an average of 5.2 ± 1.9-fold at the peak of induction (8 h), relative to each mouse's own 0-h baseline (see Fig. 3A). Luminescence in control mice treated with vehicle alone changed by an average of 1.3 ± 0.7-fold in that same time period (see Fig. S2 for images of representative PCN- and vehicle-treated mice). The difference in fold-change between the PCN-treated mice and the vehicle-treated mice was statistically significant (P < 0.0001). These results suggest that for future induction studies, assuming 8 mice per group and a pooled standard deviation of 1.5, we would be able to detect a difference in mean fold-change between 2 groups (drug vs. control) as small as 2.2-fold, with 80% power and a 2-tailed 0.05 level of significance.
We wanted to confirm the relationship between luminescence and mdr1a mRNA expression under inducing conditions, in both knock-in and wild-type mice. Because it is not possible to measure mRNA levels at different time points within a single mouse, we analyzed intestinal mdr1a expression in 24 mdr1a+/fLUC mice and 24 mdr1a+/+ mice, as follows: 6 mice of each genotype were killed without PCN injection (time 0, the same mice used in the basal expression analysis above) and 6 mice of each genotype were killed at 8, 24, and 48 h after PCN injection. For the mdr1a+/fLUC mice, live imaging was also performed immediately before killing. At every time point, average luminescence levels in the cohort of mdr1a+/fLUC mice paralleled average intestinal mdr1a expression levels in the cohorts of both mdr1a+/fLUC and mdr1a+/+ mice (Fig. 3B). Notably, the fold-induction levels (8-h cohorts vs. 0-h cohorts) for luminescence (3.5 ± 0.6-fold), mdr1a mRNA (4.4 ± 0.7-fold) and fLUC mRNA (3.9 ± 0.6-fold) in mdr1a+/fLUC mice and for mdr1a mRNA (4.3 ± 0.6-fold) in mdr1a+/+ mice were all very similar (no statistically significant differences by 2-tailed t test). Likewise, for the cohort of mdr1a+/fLUC mice killed at 8 h, the fold-induction of luminescence over each animal's own 0-h baseline was 3.5 ± 0.5-fold, suggesting that individual mice can reasonably be used to study gene expression behavior (and inter-individual differences) that could previously only be accomplished through cohort analysis. Moreover, the mdr1a mRNA levels in individual mdr1a+/fLUC mice correlated strongly with luminescence (r2 = 0.8988) (Fig. 3C) and fLUC mRNA levels (r2 = 0.9586, not shown) in those same mice at the 0-h (before injection) and 8-h time points. There was no correlation between mdr1a expression and luminescence at 24 h, however, and there was a moderate correlation at 48 h (see Fig. 3C). These data suggest that luminescence is an accurate reporter for mdr1a expression at steady state (before injection and 48 h after injection) and a faithful reporter of transcriptional induction of gene expression (8 h after injection), but luminescence does not appear to reflect the kinetics of mdr1a mRNA decay back to the steady state.
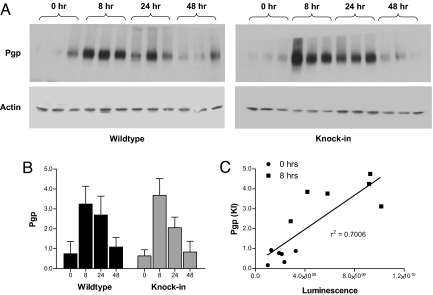
To look at the relationship between luminescence and Pgp protein, we analyzed Pgp expression in the intestines of each mouse in this experiment. Fig. 4a shows Western analysis of 3 mice at each time point and Fig. 4B shows the quantification of results from all 6 mice at each time point. Although Western analyses are not as quantitative as mRNA or luminescence measurements, there was general agreement between the timing and magnitude of Pgp induction and the timing and magnitude of luminescence and mRNA induction. On average, mdr1a+/fLUC mice had a 5.8 ± 0.5-fold induction of Pgp, compared with the 4.4 ± 0.7-fold induction of mRNA and 3.5 ± 0.6-fold induction of luminescence mentioned above. There was no significant difference between mdr1a+/fLUC and mdr1a+/+ mice in this regard (4.3 ± 0.5-fold Pgp induction; 4.3 ± 0.6-fold mdr1a mRNA induction in mdr1a+/+ mice). Moreover, Pgp levels in individual mdr1a+/fLUC mice correlated well with luminescence in those same mice, at least at the 0-h and 8-h time points (see Fig. 4C). As previously noted, the exact kinetics of Pgp decay did not correlate with the decay of luminescence in mdr1a+/fLUC mice (data not shown), but it is notable that Pgp levels returned to preinjection levels by 48 h in both mdr1a+/fLUC and mdr1a+/+ mice, as did mdr1a mRNA and luminescence (see Figs. 3 and 4). Taken together, these data show that live imaging can be used as a faithful reporter for the kinetics and magnitude of mdr1a gene induction, both at the mRNA and protein level. Moreover, there does not appear to be any obvious compensatory gene expression in mdr1a+/fLUC as a result of knocking out 1 copy of the mdr1a gene.
Fig. 4.
Western analysis of Pgp expression. (A) Proteins were extracted from the intestines of groups of 6 mdr1a+/+ and six mdr1a+/fLUC mice killed at time 0 (before injection) and at 8, 24, and 48 h after injection of PCN. Proteins were subjected to Western analysis as described in Methods. Results are shown for 3 mice of each genotype at each time point. (B) Pgp protein (for all 6 mice in each group) was quantified by measuring the density of each Pgp band and normalizing to actin in the same lane. Shown are the average Pgp expression levels (± SD). (C) Regression analysis showing the relationship between intestinal Pgp protein levels in each individual mouse and abdominal-area luminescence intensity measured in that same mouse just before killing (the 0-h and 8-h mice).
Common Chemotherapeutic Drugs Induce mdr1a Expression in Vivo.
Having established that mdr1a.fLUC is a faithful reporter for basal and induced mdr1a gene expression, we wanted to evaluate the effects of common chemotherapeutic agents that are known substrates for the mdr1a gene product. Studies with cell lines have demonstrated that the human MDR1 gene is inducible by taxanes (11). To determine if the same is true in vivo, we treated mdr1a+/fLUC mice with paclitaxel or docetaxel as described in Methods, and quantified fLUC expression by live imaging at various time points before and after drug delivery. Luminescence intensities were induced by an average of 3.1 ± 1.3-fold above individual basal (0-h) levels in mice treated with docetaxel and 3.0 ± 2.8-fold above basal levels in mice treated with paclitaxel (Fig. 5). The average peak induction time was 8 h for both drugs, with return to baseline by 48 h. In mice treated with vehicle alone, luminescence showed a 1.0 ± 0.4-fold change at 8 h relative to baseline (see Fig. 5). The difference in mean fold-change between docetaxel and control was significant (P < 0.002), whereas the difference in mean fold-change between paclitaxel and control was borderline (P = 0.076) because of the higher standard deviation in the paclitaxel group. Finally, there was no difference in mean fold-change between the paclitaxel and docetaxel groups (P = 0.98). Power analysis of this experiment indicated that with a pooled standard deviation of 1.8 and a sample of 8 mice per group, we would have been able to detect a fold-change between taxane and control groups as small as 2.7-fold with 80% power and a 2-tailed 0.05 level of significance.
Fig. 5.
Mdr1a.fLUC induction by taxanes. Heterozygous mdr1a+/fLUC mice were given 10 mg/kg paclitaxel (circles) or 10 mg/kg docetaxel (triangles), as described in Methods. Mice that were injected with saline-diluted Cremophor EL/alcohol (squares) were used as controls. fLUC expression was quantified by measuring luminescence intensity in the abdominal area at various time points before and after drug or vehicle injection. The luminescence intensities recorded from 8 mice were individually normalized to their own luminescence intensities at time 0, before drug or vehicle treatment. Shown are the averages of the normalized values (± SE) at each time point.
Discussion
Results in this article demonstrate that the mdr1a.fLUC allele is a faithful reporter for basal mdr1a expression in naive animals and for real-time induction of mdr1a gene expression by xenobiotics. The changes in luminescence intensities induced by PCN and taxanes likely reflect expression of mdr1a.fLUC in the intestine, because the basal luminescence signals in this organ were 100-fold higher than in other organs, making changes in luminescence signals in other organs less detectable. The strong correlation between luminescence and intestinal mdr1a expression shown in Fig. 3 is consistent with this conclusion.
A significant feature of our model is the ability to monitor gene expression over time within individual animals. Notably, we observed significant mouse-to-mouse heterogeneity in both the basal expression of mdr1a and the magnitude of induction in response to xenobiotics. This heterogeneity in expression as determined by imaging is real, because whenever we were able to measure luminscence and mdr1a mRNA in the same animal and at the same time point, we observed near-absolute correlations between those measurements, suggesting that differences in luminescence were accurately reporting parallel differences in mdr1a gene expression. Such heterogeneity is consistent with known inter-individual heterogeneity in rodent and human intestinal mdr1 expression (19, 22–24). Likewise, considerable heterogeneity in mdr1 gene induction has been observed in rodents and in human tissues exposed to xenobiotics; both the presence or absence of induction and the magnitude of induction are variable between individuals (19, 22, 25, 26). Despite the heterogeneity, we were able to detect statistically significant induction of mdr1a.fLUC expression in drug-treated animals, relative to controls, with relatively good power.
We might be able to use our model to determine the underlying causes of heterogeneity in induction as well. In other model systems and humans, this type of heterogeneity has been attributed to genetic differences, differences in basal expression that could affect the calculated fold-change in expression compared to baseline, and differences in sensitivity to the inducing agents because of undetermined physiological or interacting environmental factors (25–27). The heterogeneity seen with our model is not likely to be the result of genetic differences between animals, because we used inbred mice in our experiments, but future studies could be designed to determine whether specific genetic backgrounds affect the extent and magnitude of heterogeneity in the induction response. In terms of the fold-change calculations, we used linear regression analysis and found that 0-h luminenscence was not predictive of 8-h luminescence when we examined either the PCN- (P = 0.36) or the taxane- (P = 0.49) treated mice. Thus, the variable baseline luminescence levels do not account for the variability in calculated fold changes relative to those baselines. The causes of heterogeneous induction in the intestine, therefore, are probably the result of a combination of environmental, physiological, and molecular factors that affect both drug pharmacokinetics (28) (unpublished observations) and the response to the drug once it reaches its target tissues. The exact identities of these factors are not entirely known and most likely are themselves quite heterogeneous between individual mice, but we believe that our model system will help us understand the parameters (kinetics and magnitude of induction) and mechanisms (environmental conditions, physiological factors, and specific molecular components) involved in mdr1 expression in response to xenobiotics.
In the current study, the kinetics of induction were such that gene expression peaked at about 8 h, on average, for all of the agents tested. It will be interesting to determine if other chemotherapeutic agents exhibit different induction kinetics, and whether such differences might reflect different pharmacologic properties in terms of tissue distribution or clearance. In addition, any observed differences in the kinetics of mdr1a induction by these or other agents might reflect different molecular mechanisms of induction. Although it has been shown that PCN and taxanes induce mdr1 by activating the nuclear receptor SXR/PXR (11, 19, 22), another nuclear receptor, CAR, has also been implicated as a regulator of transporter expression (29). A recent study suggests that another therapeutic, doxorubicin, might induce mdr1 expression by activating the transcription factor FOXO3a (13). Further studies crossing mdr1a+/fLUC mice with mice that are knocked out for PXR, CAR, or other genes of interest should help elucidate the trans-acting factors that are required for mdr1a gene regulation by different agents. Indeed, the heterogeneous expression or activity of such factors could account for some of the inter-individual heterogeneity in gene induction that was observed in our experiments, and this would be revealed by experiments with the respective knockout mice as well.
It is also interesting to note that intestinal mdr1a, Pgp, and luminescence all return to baseline levels by 48 h, despite differences in the details of their respective decay rates. It is perhaps not surprising that, in the absence of a selection pressure or an aberrant cellular environment (i.e., cancer), mdr1a mRNA and protein expression readily return to their pretreatment steady states. A future application of the mdr1a.fLUC model will be to determine the conditions under which mdr1a/Pgp expression (as reported by fLUC) becomes elevated, relative to baseline, in a sustained fashion, either as a result of repeated xenobiotic injections or during tumorigenesis. It is important to recognize, however, that mdr1a.fLUC is a transcriptional model that reports on steady state and newly induced gene expression. Further modifications to the targeting vector, incorporating elements that act in cis to influence mRNA stability or translation, might allow the model to be used to study those elements of gene regulation as well.
Our reporter gene model also demonstrates the feasibility of coupling the conditional Cre/loxP system with a bioimaging reporter gene to control the in vivo monitoring of regulated gene expression both temporally and spatially. Other luciferase knock-in reporter systems have been described (30–32), but they either lack the ability to control knock-in of the transgene via Cre/loxP recombination (30, 31) or they are not used for monitoring transcription from a regulated gene (32). With our vector design, which requires Cre-mediated recombination to achieve the fLUC knock-in, we can potentially control knock-in spatially by mating mdr1aflox mice with Cre-donor mice that express Cre in a tissue-restricted way, thus limiting the mdr1a.fLUC configuration to selected tissues and organs in any given animal. Although there appeared to be some leakiness in the nonrecombined configuration, which will need to be overcome by modifying the targeting construct, the tissue-specific knock-in approach should nevertheless allow us to study gene regulation in individual organs without the confounding effects of overwhelming luminescence emanating from the intestine. In addition to using this strategy to study tissue-specific mdr1a regulation, we anticipate that this approach can be generalized to any number of other genes, other physiologic or developmental settings, and other disease states.
Methods
Imaging and Gene Expression Assays.
For in vivo imaging, mice were given 0.2 ml of luciferin (15 mg/ml) by i.p. injection and then anesthetized by inhalation of isoflurane (4% isoflurane carried by a flow of 1.5 L/min oxygen). Live in vivo imaging was carried out on a Xenogen IVIS imaging system (Caliper Life Sciences) 8 minutes after luciferin injection. For image analysis, consistent regions of interest were drawn that covered the entire abdominal area. For ex vivo imaging of organs, mice were killed about 6 minutes after luciferin injection. Organs were isolated and imaged on the same Xenogen imaging system within 5 minutes after the death of the animal. After imaging, organs were frozen immediately in liquid nitrogen, pulverized, and total RNA was extracted using a Qiagen RNease Plus kit. For experiments in Figs. 2B and 3, total intestinal RNA was extracted from the entire tract below the stomach. Total RNA was reverse transcribed and the levels of mdr1a and fLUC mRNA were measured, in triplicate, by SYBR Green-based real time PCR (Applied Biosystems), using primers specific for mdr1a sequences present only in the nontargeted mdr1a allele (forward: 5′-GTCGTGATGGAACTTGAA; reverse: 5′-CACGTTCATTATAAATGTCGTTCGG) and primers specific for fLUC sequences (forward: 5′-GTCGTGATGGAACTTGAAG; reverse: 5′-CACACTGACTGCTGGTTTCTTT), respectively.
Protein Extraction and Western Analysis.
Membrane proteins were extracted from pulverized intestines according to published procedure (33), with minor modifications. Briefly, pulverized tissues were dissolved into ice-cold PBS containing a mixture of protease inhibitors, Complete (Roche Applied Science) and 2 mM PMSF, with 8 strokes of a tissue homogenizer. The homogenates were centrifuged at ≈1,300 × g to pellet the intact cellular materials. The pellets were suspended in extraction buffer (10 mM Tris-HCl, pH 8.6, 1.6 mM MgCl2, 0.14 mM NaCl, 1% Nonidet P-40, and 2 mM PMSF) and incubated on ice for 5 min, followed by centrifugation at ≈21,000 × g for 30 min. The supernatant containing the cell membranes and other cytoplasmic components (33) was analyzed using the RC DC Protein Assay kit purchased from Bio-Rad to determine total protein concentration. Crude protein extracts were mixed with sample loading buffer containing 1% SDS and 50 mM DTT and boiled for 5 min before being resolved by electrophoresis in a 4% to 15% linear gradient Tris-HCl-polyacrylamide gel. Proteins were transferred onto a nitrocellulose membrane and probed with the C219 monoclonal anti-p-glycoprotein antibody, purchased from Covancer. After incubation with a peroxidase-conjugated anti-mouse IgG secondary antibody, the Pgp was detected using an ECL kit purchased from GE HealthCare. Membranes were then reprobed with the AC-40 monoclonal anti-actin antibody (Sigma), followed by secondary antibody and ECL detection. Western images were digitized with a high-resolution scanner and the densities of individual bands were measured by ImageQuantMT 5.2.
Animal Husbandry and Xenobiotic Treatment.
All experiments involving live animals were reviewed and approved by the City of Hope Institutional Animal Care and Use Committee. Mice were kept on a normal Chow diet in a specific-pathogen-free facility at City of Hope. PCN, paclitaxel, and docetaxel were purchased from Sigma. PCN was dissolved in corn oil and administered to mice by i.p. injection (200 mg/kg) at the start of the experiment. Animals injected with corn oil (vehicle) were used as control. Paclitaxel and docetaxel were dissolved in 50.3% Cremophor EL and 49.7% dehydrated alcohol (vol/vol) as concentrates (6 mg/ml) and then diluted in saline to a final concentration of 1.2 mg/ml, before being administrated to mice by i.p. injection (10 mg/kg). Mice injected with saline-diluted Cremophor EL/alcohol were used as controls.
Statistical Analysis.
To perform statistical analysis of xenobiotic treatment data, we first calculated fold changes at 8 h relative to basal expression at 0 h for each individual mouse. Comparisons of treatment groups were made using a 2-sample t-test implementing a 0.05 two-tailed significance level. Linear regression was used to assess if basal (0 h) luminescence was predictive of 8-h luminescence, implementing a Wald test to determine if the slope was significantly different from zero. In the experiments examining the relationship between luminescence and mdr1a mRNA or Pgp protein, the coefficient of determination (r2) was used to describe the fraction of the variance in the outcome variable (e.g., mdr1a mRNA or Pgp protein) explained by the predictor (e.g., luminescence) from a linear regression analysis. Statistical analyses were done using the software packages R (R Foundation for Statistical Computing) and graphPad Prism (version 5.0.1 for Windows, GraphPad Software).
Supplementary Material
Acknowledgments.
The authors thank Donna Isbell in the Animal Resource Center for her assistance, Brenda Aguilar for help with the bioimaging, and Jenny Glavin for her work on the initial targeting constructs. Purified human CG used to produce embryos for injection of ESC was supplied by the National Hormone and Peptide Program (University of California, Los Angeles). This work was partially supported by Grant DAMD 17-03-1-0461 from the Department of Defense (to S.E.K.), Grant CA91135 from the National Cancer Institute (to T.W.S.), and by City of Hope's Cancer Center Support Grant CA33572.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807343106/DCSupplemental.
References
- 1.Gottesman MM, Pastan I. Biochemistry of multidrug resistance mediated by the multidrug transporter. Annu Rev Biochem. 1993;62:385–427. doi: 10.1146/annurev.bi.62.070193.002125. [DOI] [PubMed] [Google Scholar]
- 2.Kane SE, Pastan I, Gottesman MM. Genetic basis of multidrug resistance of tumor cells. J Bionenerg Biomembr. 1990;22:593–618. doi: 10.1007/BF00762963. [DOI] [PubMed] [Google Scholar]
- 3.Chen CC, et al. Detection of in vivo P-glycoprotein inhibition by PSC 833 using Tc-99m sestamibi. Clin Cancer Res. 1997;3:545–552. [PubMed] [Google Scholar]
- 4.Luker GD, Fracasso PM, Dobkin J, Piwnica-Worms D. Modulation of the multidrug resistance P-glycoprotein: detection with technetium-99m-sestamibi in vivo. J Nucl Med. 1997;38:369–372. [PubMed] [Google Scholar]
- 5.Schinkel AH, Wagenaar E, van Deemter L, Mol CA, Borst P. Absence of the mdr1a P-Glycoprotein in mice affects tissue distribution and pharmacokinetics of dexamethasone, digoxin, and cyclosporin A. J Clin Invest. 1995;96:1698–1705. doi: 10.1172/JCI118214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sikic BI, et al. Modulation and prevention of multidrug resistance by inhibitors of P-glycoprotein. Cancer Chemother Pharmacol. 1997;40(Suppl):S13–S19. doi: 10.1007/s002800051055. [DOI] [PubMed] [Google Scholar]
- 7.Chin KV, Ueda K, Pastan I, Gottesman MM. Modulation of activity of the promoter of the human MDR1 gene by Ras and p53. Science. 1992;255:459–462. doi: 10.1126/science.1346476. [DOI] [PubMed] [Google Scholar]
- 8.Thottassery JV, Zambetti GP, Arimori K, Schuetz EG, Schuetz JD. p53-dependent regulation of MDR1 gene expression causes selective resistance to chemotherapeutic agents. Proc Natl Acad Sci USA. 1997;94:11037–11042. doi: 10.1073/pnas.94.20.11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zastawny RL, Salvino R, Chen J, Benchimol S, Ling V. The core promoter region of the P-glycoprotein gene is sufficient to confer differential responsiveness to wild-type and mutant p53. Oncogene. 1993;8:1529–1535. [PubMed] [Google Scholar]
- 10.Bush JA, Li G. Cancer chemoresistance: the relationship between p53 and multidrug transporters. Int J Cancer. 2002;98:323–330. doi: 10.1002/ijc.10226. [DOI] [PubMed] [Google Scholar]
- 11.Synold TW, Dussault I, Forman BM. The orphan nuclear receptor SXR coordinately regulates drug metabolism and efflux. Nat Med. 2001;7:584–590. doi: 10.1038/87912. [DOI] [PubMed] [Google Scholar]
- 12.Urquhart BL, Tirona RG, Kim RB. Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: Implications for interindividual variability in response to drugs. J Clin Pharmacol. 2007;47:566–578. doi: 10.1177/0091270007299930. [DOI] [PubMed] [Google Scholar]
- 13.Hui RC-Y, et al. Doxorubicin activates FOXO3a to induce the expression of multidrug resistance gene ABCB1 (MDR1) in K562 leukemic cells. Mol Cancer Ther. 2008;7:670–678. doi: 10.1158/1535-7163.MCT-07-0397. [DOI] [PubMed] [Google Scholar]
- 14.Croop JM, et al. The three mouse multidrug resistance (mdr) genes are expressed in a tissue-specific manner in normal mouse tissues. Mol Cell Biol. 1989;9:1346–1350. doi: 10.1128/mcb.9.3.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gros P, Raymond M, Bell J, Housman D. Cloning and characterization of a second member of the mouse mdr gene family. Mol Cell Biol. 1988;8:2770–2778. doi: 10.1128/mcb.8.7.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Devault A, Gros P. Two members of the mouse mdr gene family confer multidrug resistance with overlapping but distinct drug specificities. Mol Cell Biol. 1990;10:1652–1663. doi: 10.1128/mcb.10.4.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SI, et al. Structural analysis of the mouse mdr1a (P-glycoprotein) promoter reveals the basis for differential transcript heterogeneity in multidrug-resistant J774.2 cells. Mol Cell Biol. 1990;10:3596–3606. doi: 10.1128/mcb.10.7.3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang-Wai DF, et al. Human (MDR1) and mouse (mdr1, mdr3) P-glycoproteins can be distinguished by their respective drug resistance profiles and sensitivity to modulators. Biochemistry. 1995;34:32–39. doi: 10.1021/bi00001a005. [DOI] [PubMed] [Google Scholar]
- 19.Cheng X, Klaassen CD. Regulation of mRNA expression of xenobiotic transporters by the pregnane x receptor in mouse liver, kidney, and intestine. Drug Metab Dispos. 2006;34:1863–1867. doi: 10.1124/dmd.106.010520. [DOI] [PubMed] [Google Scholar]
- 20.Bush JA, Li G. Regulation of the Mdr1 isoforms in a p53-deficient mouse model. Carcinogenesis. 2002;23:1603–1607. doi: 10.1093/carcin/23.10.1603. [DOI] [PubMed] [Google Scholar]
- 21.Tang SH, Silva FJ, Tsark WM, Mann JR. A Cre/loxP-deleter transgenic line in mouse strain 129S1/SvImJ. Genesis. 2002;32:199–202. doi: 10.1002/gene.10030. [DOI] [PubMed] [Google Scholar]
- 22.Brady JM, et al. Tissue distribution and chemical induction of multiple drug resistance genes in rats. Drug Metab Dispos. 2002;30:838–844. doi: 10.1124/dmd.30.7.838. [DOI] [PubMed] [Google Scholar]
- 23.Lown KS, et al. Role of intestinal P-glycoprotein (mdr1) in interpatient variation in the oral bioavailability of cyclosporine. Clin Pharmacol Ther. 1997;62:248–260. doi: 10.1016/S0009-9236(97)90027-8. [DOI] [PubMed] [Google Scholar]
- 24.Hashida T, et al. Pharmacokinetic and prognostic significance of intestinal MDR1 expression in recipients of living-donor liver transplantation. Clin Pharmacol Ther. 2001;69:308–316. doi: 10.1067/mcp.2001.115142. [DOI] [PubMed] [Google Scholar]
- 25.Olinga P, et al. Coordinated induction of drug transporters and phase I and II metabolism in human liver slices. Eur J Pharm Sci. 2008;33:380–389. doi: 10.1016/j.ejps.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Schwarz UI, et al. Induction of intestinal P-glycoprotein by St John's wort reduces the oral bioavailability of talinolol. Clin Pharmacol Ther. 2007;81:669–678. doi: 10.1038/sj.clpt.6100191. [DOI] [PubMed] [Google Scholar]
- 27.Kerb R. Implications of genetic polymorphisms in drug transporters for pharmacotherapy. Cancer Lett. 2006;234:4–33. doi: 10.1016/j.canlet.2005.06.051. [DOI] [PubMed] [Google Scholar]
- 28.Eiseman JL, et al. Plasma pharmacokinetics and tissue distribution of paclitaxel in CD2F1 mice. Cancer Chemother Pharmacol. 1994;34:465–471. doi: 10.1007/BF00685656. [DOI] [PubMed] [Google Scholar]
- 29.Burk O, Arnold KA, Geick A, Tegude H, Eichelbaum M. A role for constitutive androstane receptor in the regulation of human intestinal MDR1 expression. Biol Chem. 2005;386:503–513. doi: 10.1515/BC.2005.060. [DOI] [PubMed] [Google Scholar]
- 30.Yoo S-H, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ishikawa T, Jain NK, Takeo MM, Herschman HR. Imaging cyclooxygenase-2 (Cox-2) gene expression in living animals with a luciferase knock-in reporter gene. Mol Imaging Biol. 2006;8:171–187. doi: 10.1007/s11307-006-0034-7. [DOI] [PubMed] [Google Scholar]
- 32.Safran M, et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2:297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- 33.Lankas GR, Cartwright ME, Umbenhauer D. P-glycoprotein deficiency in a subpopulation of CF-1 mice enhances avermectin-induced neurotoxicity. Toxicol Appl Pharmacol. 1997;143:357–365. doi: 10.1006/taap.1996.8086. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.