Abstract
We carried out a genome-wide association study of genetic predictors of anti-cyclic citrullinated peptide antibody (anti-CCP) level in 531 self-reported non-Hispanic Caucasian Rheumatoid Arthritis (RA) patients enrolled in the Brigham Rheumatoid Arthritis Sequential Study (BRASS). For replication, we then analyzed 289 single nucleotide polymorphisms (SNPs) with P < 0.001 in BRASS in an independent population of 849 RA patients from the North American Rheumatoid Arthritis Consortium (NARAC). BRASS and NARAC samples were genotyped using the Affymetrix 100K and Illumina 550K platforms respectively. Association between SNPs and anti-CCP titer was tested using general linear models. The five most significant SNPs from BRASS all were within the major histocompatibility complex (MHC) region (P ≤ 3.5 × 10−6). After controlling for the human leukocyte antigen shared epitope (HLA-SE), the top SNPs still yielded P values < 0.0002. In NARAC, a single SNP from the MHC region near BTNL2 and HLA-DRA, rs1980493 (r2 = 0.85 with the top five SNPs from BRASS), was associated significantly with CCP titer (P = 6.1 × 10−5) even after adjustment for the HLA-SE (P = 0.0002). The top SNPs found in BRASS and NARAC had r2 = 0.46 and 0.64, respectively, to HLA-DRB1 DR3 alleles. These results confirm that the most significant genome region affecting anti-CCP titers in RA is the MHC region. We identified a SNP in moderate linkage disequilibrium (LD) with HLA-DR3, which may influence anti-CCP titer independently of the HLA-SE.
INTRODUCTION
Rheumatoid arthritis (RA) is one of the most common autoimmune diseases, affecting 0.5% to 1% of most populations (1). This systemic disease is marked by chronic inflammation that affects the synovial membrane of diarthrodal joints predominantly. RA is heterogeneous in terms of clinical presentation in disease aspects, progression, and severity, and in response to different clinical treatments. Its susceptibility is determined by a combination of multiple genetic and environmental factors. The most important genetic factor, which has been associated unequivocally with RA susceptibility, is variability in the major histocompatibility complex (MHC). Although the HLA associations with RA are complex (2,3), the majority of the genetic signal from the MHC is explained by alleles at the HLA-DRB1 locus (4), and accounts for approximately 30% of the genetic risk of RA (5). In individuals of European ancestry, the associated HLA-DRB1 alleles share a region of sequence similarity or “shared epitope” (SE) at amino acid positions 70–74 in the third hypervariable region of the HLA-DRB1 molecule (HLA-SE) (5).
Anti-cyclic citrullinated peptide (anti-CCP) antibody is an important bio-marker for RA. It is as sensitive as, but more specific than, rheumatoid factor for diagnosing RA (6). Multiple studies have shown that the presence of anti-CCP antibodies in RA correlates with radiographic progression (6–11). Known genetic risk factors for RA are associated more strongly with CCP positive (CCP+) RA than with CCP negative (CCP−) RA (12–14); the most recent study by Ding et al. (14) definitively showed that the HLA region is associated specifically with CCP+ subset of RA. Previous studies of genetic determinants of anti-CCP have been limited to the HLA region (7,10,11, 15–18). Almost all of the studies published on HLA-SE alleles and anti-CCP have the consistent finding that HLA-SE alleles are associated with CCP positivity, while some of these found that the HLA-SE is specifically associated with higher anti-CCP titer. Irigoyen et al. reported that HLA-DR3 is associated with lower anti-CCP titer, in contrast with HLA-SE alleles (16), using the NARAC collection.
Anti-CCP is relatively stable over time, making it a good quantitative trait to investigate. In this study, we treat anti-CCP antibody titer among RA subjects as a quantitative outcome and proxy for progressive RA in a genome-wide association study (GWAS) to elucidate the genetic basis of anti-CCP titer. Analyses were performed both with and without adjusting for HLA-SE status and HLA-DR3 to identify the additional contribution of genes other than HLA-DRB1 to variability in anti-CCP titer and severity of RA.
METHODS
Study Population
RA patients were recruited at the Robert Breck Brigham Arthritis Center at Brigham and Women’s Hospital as described previously (19). In brief, patients were eligible to join the study if they had a diagnosis of RA, were over 18 years old, and did not have a diagnosis of pso-riatic arthritis or systemic lupus erythe-matosus. All study protocols were approved by the Brigham and Women’s Hospital Institutional Review Board and informed consent was obtained from all subjects. A total of 575 RA samples were genotyped in this study, including all CCP+ patients enrolled in the BRASS cohort at that time, and 142 CCP− patients. The cohort is predominantly female (82.3%) with a mean age of 58.0 (±13.6) years old and average disease duration of 16.1 (±12.5) years. Anti-CCP antibodies were measured using a second generation Enzyme-Linked ImmunoSorbent Assay (ELISA) from Inova (Inova Diagnostics Inc., San Diego, CA, USA).
Genotyping and Quality Control
Genotyping of the samples was performed at the Broad Institute using the Affymetrix GeneChip 100K Mapping Array containing 116,204 SNPs (Affymetrix, Santa Clara, CA, USA). Samples were processed in a 96-well plate format using Biomek FX robotics according to the manufacturer’s protocol. Genotypes were called using the BRLMM (Bayesian Robust Linear Modeling using Mahalanobis distance) algorithm.
To ensure high quality data and to minimize false positive results due to technical artifact, filtering criteria were set as follows: genotype call rate ≥ 0.95 per SNP and 0.9 per individual, minor allele frequency (MAF) ≥ 0.01, and P value for Hardy-Weinberg equilibrium test ≥ 0.0001. There were 97,248 autosomal SNPs from 546 self-reported Caucasians (82.3% female) that met these criteria. The average genotyping rate in the remaining individuals was 98.0%, and the SNP-wise genotyping rate was 98.9%. Samples were genotyped for HLA-SE alleles by low-resolution genotyping.
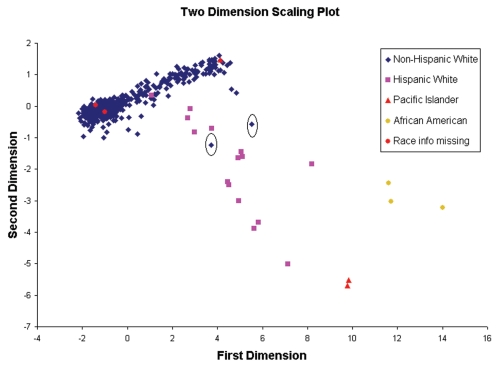
Individuals in BRASS were self-identified as Hispanic or non-Hispanic and as either Caucasian, American Native Indian, Pacific Islander/Asian, or African American. The entire study sample was utilized to screen for evidence of subtle population stratification through evaluation of pairwise identity by state (IBS) sharing distance across all samples using the software package PLINK (20). The hypothesis was that a more homogeneous population was more similar among individuals as reflected in small IBS distances. Clustering in terms of IBS-sharing distance among all possible pairs in the sample was explored through PLINK. A two-dimensional scaling plot from PLINK was used to visualize sample similarity. Standard classical multidimensional scaling was used to reduce representation of the data into two dimensions, enabling visualization on a two-axis scatter plot. Two dimensional scales (Figure 1) visualized four well-separated subgroups that correlated with self-reported ethnic groups. The IBS distance between African Americans, Asians, and Caucasians was distinguished. Also, the plot suggested Hispanic Caucasians were clearly genetically different from the majority of North American Caucasians. We further checked any potential outliers among the remaining self-reported non-Hispanic Caucasians using a Cook D test for the two-dimension scale. The Cook D value for each subject is the measurement of the parameter estimate change in analysis with that subject compared with the estimate without that subject. A larger value indicates that the subject is more different from the remaining subjects (for example, an outlier). We removed 19 individuals based on IBS analysis who also self-reported Hispanic and/or non-Caucasian race. Two more individuals who self-reported as non-Hispanic Caucasian, with Cook D value > 0.5 (1.07 and 0.62) were removed from analysis; the remaining subjects had a Cook D value no higher than 0.18. After quality control and removing outliers, 531 individuals were included in the genome-wide association testing.
Figure 1.
Population stratification assessments using a two-dimension plot of identity by state (IBS) distances for four self-reported ethnic groups. First and second dimension scales are shown on abscissa and ordinate respectively. Colors indicate different ethnic groups. The dimension scale results were generated from PLINK. Two individuals shown in the circles were considered outliers and excluded from further analysis.
Statistical Model
Anti-CCP titer was used as a continuous outcome variable. For the genome-wide analysis, each SNP was tested for association with anti-CCP titer using general linear regression (GLM) assuming an additive model for all SNPs in PLINK. Secondary analyses were performed in which the HLA-SE, the well-established risk locus for anti-CCP RA, was controlled to determine whether the association was independent from the HLA-SE for SNPs on Chromosome 6. Analyses using only CCP+ (anti-CCP titer > 20) subjects (N = 401) also were performed. We additionally controlled for HLA-DR3 to investigate whether the signal was independent of HLA-DR3.
Because CCP titer was not normally distributed, simulation was used to validate the use of GLM. Ten million random genotypes were generated based on different minor allele frequencies (MAF) (0.01, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5 respectively) for each individual. The empirical P value distribution was obtained by analyzing observed anti-CCP measures against the simulated genotypes using GLM. A pure null distribution was simulated, in which the type I error should follow the nominal level: for example, when we use 0.05 significance level, we should expect 1/20th of the results to be significant; if we use 0.001, we should get 1 in 1000 significant results; and so on. If no excess type I error was observed, this would support the validity of the GLM test.
Permutation testing was utilized to evaluate the significance of association analysis results. Based on individual’s genome typing data, we permuted anti-CCP titer 1000 times. Then we randomly assigned genotype data to each CCP titer and ran pseudo-GWAS 1000 times to get a whole genome “association statistic.” Such permutation creates the null distribution (that is, no real associations) and hence the number of significant associations observed in the permutation analysis is a measure of the expected false positive level across the genome.
Replication Population
Samples from the NARAC population were used for replication. NARAC samples were collected by four different study designs. NARAC cases were from multiplex families (primarily affected sibling pairs) in which at least one sibling had documented erosions. The other collections that were used for replication included samples from the National Data Bank of Rheumatic Diseases, the National Inception Cohort of Rheumatoid Arthritis, and the Study of New Onset Rheumatoid Arthritis. A more detailed description is given elsewhere (21). We genotyped 908 RA cases by SNP assay with Infinium Human Hap 550 version 1.0 (Illumina, San Diego, CA, USA) and applied the same quality control (QC) criteria as in BRASS. Among all NARAC RA patients, 849 individuals who passed QC filters, had CCP measurements, and were determined to have at least 90% European ancestry using STRUCTURE (22) were utilized in the replication analysis. For this 849-member cohort, 74.1% were female, with a mean age of 56.0 ± 12.3 years and average disease duration of 10.6 ± 11.1 years; they were all CCP+. Low resolution HLA-DRB1 typing for the allele groups DRB1*01 through DRB1*18, and high-resolution DRB1*04 typing were performed by initial PCR amplification, followed by hybridization to immobilized sequence-specific oligonucleotide probes (16). CCP was measured using the same assay as in BRASS. Different sub-cohorts had different upper ranges of the titer; in particular some titers in the NARAC family collection were truncated at 413 (4%) or 210 (1%). In the primary analysis, CCP was transformed to a normal distribution by taking the inverse normal of the rank. GLM models for the 289 550K SNPs as predictors of ordinal CCP titer were run using PLINK (20), first unadjusted and then controlling for HLA-SE status for SNPs on Chromosome 6. As sensitivity analyses for the CCP range indicated differences among the sub-cohorts, logistic models also were run with CCP categorized as high (> 210) versus low (< 100) and the primary GLM models were rerun with cohort and disease duration as covariates.
RESULTS
Initial GWAS in BRASS
Demographics and clinical characteristics of the final set of 531 subjects in the BRASS cohort used in the genome-wide association analysis are summarized in Table 1.
Table 1.
Demographics of the BRASS cohort participants with 100K genotype data meeting quality control criteria (N = 531)
| Mean age (SD) | 58.0 ± 13.5 |
| Mean age at RA diagnosis (SD) | 41.6 ± 14.7 |
| Mean y disease duration (SD) | 16.4 ± 12.4 |
| Female, % | 82.7 |
| New onset RA,a % | 12.8 |
| Treated with medications at baseline | |
| Methotrexate alone, % | 31.1 |
| Anti-TNF alone, % | 22.2 |
| Methotrexate/anti-TNF, % | 17.1 |
| CCP+, % | 75.7 |
| Anti-CCP titer, Mean (SD) | 142.9 ± 121.4 |
| RF+, % | 76.4 |
| HLA-DRB1 Shared Epitope copies, % | |
| 0 | 31.4 |
| 1 | 40.7 |
| 2 | 27.9 |
New onset = RA onset within two years of enrollment.
The mean age at diagnosis was 41.6 years old. Patients had well-established disease; average disease duration was 16.4 years. Frequency of 0, 1, and 2 copies of the HLA-DRB1 SE among patients were 31.4%, 40.7%, and 27.9%, respectively. Seventy-six percent were clinically classified as CCP+.
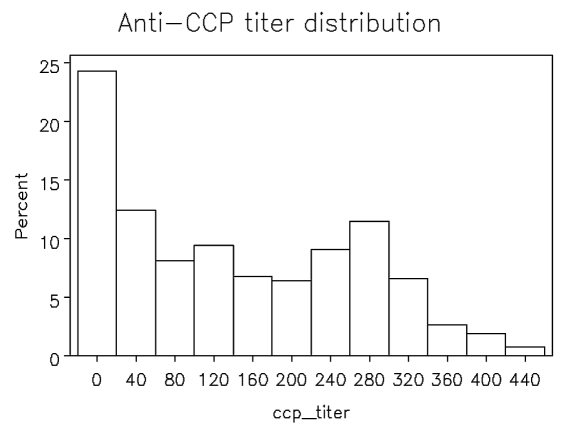
The anti-CCP titer in the BRASS patients demonstrated a wide spectrum (from 3 to 449 units), but did not follow a normal distribution (Figure 2). Skewness and kurtosis were 0.42 and −1.09 respectively, and suggest a relatively flat and long right tailed distribution. Simulation was done to evaluate whether a general linear model (GLM) would handle type I error properly. Considering minor allele frequency (MAF) values of 0.01, 0.05, 0.1, 0.2, 0.3, 0.4, and 0.5, 10 million genotypes were simulated. We observed the expected distribution of results at all α level and MAF thresholds (Table 2); no excess type I error was observed, supporting the validity of the GLM test in this analysis.
Figure 2.
Anti-CCP titer distribution among 531 RA cases in BRASS. Anti-CCP titer groups are shown on the abscissa. Percentage of subjects in each group is shown on the ordinate.
Table 2.
Simulation study of observed type I error (%) for general linear models using anti-CCP titer as outcome (Tests N = 1,000,000 random genotypes)
| Expected α level
| |||||
|---|---|---|---|---|---|
| MAF | 0.01 | 0.001 | 0.0001 | 0.00001 | 0.000001 |
| 0.01 | 0.0086 | 0.00006 | 0.00007 | 0.0000160 | 0.0000008 |
| 0.05 | 0.0097 | 0.00009 | 0.00009 | 0.0000104 | 0 |
| 0.1 | 0.0098 | 0.00106 | 0.00012 | 0.0000160 | 0.0000040 |
| 0.2 | 0.0010 | 0.00096 | 0.00010 | 0.0000098 | 0.0000020 |
| 0.3 | 0.0101 | 0.00101 | 0.00011 | 0.0000093 | 0.0000021 |
| 0.4 | 0.0099 | 0.0010 | 0.00011 | 0.0000134 | 0 |
| 0.5 | 0.0100 | 0.0010 | 0.00011 | 0.000014 | 0 |
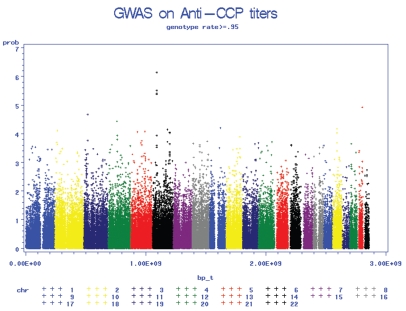
Genome-wide association analysis was conducted for anti-CCP titer using 97,248 SNPs in 531 BRASS samples with GLM (Figure 3). The top 25 SNPs with the smallest P values are shown in Table 3. The five most significant SNPs are in the MHC region, the well-known susceptibility region for RA, and the minor alleles all are associated with lower anti-CCP titers. After adjusting for HLA-SE status, coded as 0, 1, or 2 copies of SE allele, the association in these five SNPs remained significant (P < 10−5) suggesting that there may be other factors in this region, besides the HLA-SE locus, that affect anti-CCP titer.
Figure 3.
Genome-wide association P value plots showing the association of single nucleotide polymorphism (SNPs) with Anti-CCP titers. Chromosomal location is shown on the abscissa. −10 log(p) is shown on the ordinate.
Table 3.
Top 25 most significant associations between SNPs and anti-CCP titer using general linear models for BRASS and NARAC
| BRASS
|
NARAC
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chr | 100K RS number | Physical location | Gene | P valuea from GLM | P valueb | MAF | Call Rate | 550K RS numberc | LD R2 | P |
| 6 | rs2001097 | 32491836 | HLA-DRA | 3.88E-07 | 0.0036 | 0.132 | 0.970 | rs1980493 | 0.85 | 6.1E-05 |
| 6 | rs2395167 | 32496286 | BTNL2 | 5.95E-07 | 0.0034 | 0.131 | 0.987 | rs1980493 | 0.85 | 6.1E-05 |
| 6 | rs2213580 | 32496552 | HLA-DRA | 9.04E-07 | 0.0038 | 0.132 | 1 | rs1980493 | 0.85 | 6.1E-05 |
| 6 | rs2001099 | 32491611 | BTNL2 | 9.78E-07 | 0.005 | 0.134 | 0.998 | rs1980493 | 0.85 | 6.1E-05 |
| 6 | rs1041885 | 32520787 | HLA-DRA | 1.07E-06 | 0.0027 | 0.134 | 0.996 | rs1980493 | 0.85 | 6.1E-05 |
| 21 | rs2837108 | 39953480 | C21orf8 | 9.72E-06 | 0.0017 | 0.473 | 0.992 | rs2837108 | 1 | 0.8693 |
| 18 | rs1147760 | 32654700 | C18orf1 | 2.1E-05 | 0.0026 | 0.136 | 0.996 | rs13313604 | 0.908 | 0.9207 |
| 18 | rs10502668 | 32756590 | KIAA132 | 2.55E-05 | 0.0025 | 0.13 | 0.996 | rs10502668 | 1 | 0.9406 |
| 8 | rs2945913 | 8235635 | — | 4.11E-05 | 0.0013 | 0.151 | 0.981 | rs2945913 | 1 | 0.1276 |
| 5 | rs2112342 | 116261135 | — | 4.34E-05 | 0.0026 | 0.368 | 0.998 | rs2112342 | 1 | 0.2793 |
| 18 | rs1445527 | 32769877 | KIAA132 | 4.93E-05 | 0.002 | 0.132 | 0.983 | rs13313604 | 1 | 0.9207 |
| 18 | rs2017027 | 33059192 | KIAA132 | 5.07E-05 | 0.004 | 0.13 | 0.996 | rs13313604 | 0.81 | 0.9207 |
| 9 | rs4877406 | 87990359 | SPIN | 6.32E-05 | 0.0005 | 0.276 | 0.964 | rs4877406 | 1 | 0.9854 |
| 4 | rs842873 | 73061500 | GPR74 | 7.78E-05 | 0.0007 | 0.44 | 1 | —c | ||
| 6 | rs10484713 | 140662577 | — | 0.000104 | 0.0146 | 0.255 | 0.977 | rs10457702 | 1 | 0.6854 |
| 18 | rs10502669 | 32758505 | KIAA132 | 0.000106 | 0.0052 | 0.132 | 0.994 | rs13313604 | 1 | 0.9207 |
| 5 | rs10514299 | 87699366 | MGC3321 | 0.000116 | 0.021 | 0.239 | 1 | rs10514299 | 1 | 0.4791 |
| 6 | rs5000563 | 32512113 | HLA-DRA | 0.000128 | 0.028 | 0.254 | 0.994 | rs2395174 | 1 | 0.883 |
| 6 | rs1357056 | 122405433 | GJA1 | 0.00014 | 0.0044 | 0.063 | 0.998 | rs2684265 | 1 | 0.5603 |
| 11 | rs313877 | 102830195 | DDI1 | 0.000141 | 0.0132 | 0.194 | 0.985 | rs1481980 | 0.958 | 0.1932 |
| 5 | rs2056403 | 52399256 | ITGA2 | 0.000156 | 0.0026 | 0.257 | 0.983 | rs1421933 | 1 | 0.7621 |
| 5 | rs390856 | 87583315 | MGC3321 | 0.000165 | 0.0215 | 0.241 | 0.998 | rs10514299 | 1 | 0.4791 |
| 3 | rs10510617 | 28989642 | RBMS3 | 0.000174 | 0.0017 | 0.48 | 1 | rs6768946 | 0.869 | 0.07224 |
| 4 | rs1246590 | 75463744 | —- | 0.000182 | 0.0084 | 0.252 | 0.981 | rs1265327 | 1 | 0.6153 |
| 6 | rs6922541 | 135524512 | HBS1L | 0.000189 | 0.0034 | 0.404 | 0.981 | rs6922541 | 1 | 0.4497 |
P value from GLM using all BRASS subjects, N = 531.
P value from GLM using only CCP+ subjects, N = 401.
550K entries did not have a tagging SNP on 550K genotyping platform.
The primary results suggest that five SNPs in the HLA region are associated with CCP titer; however, this could be interpreted as an HLA association with CCP positivity rather than titer since CCP− subjects were included in the analysis. To test whether these findings are robust, we limited the analysis to 401 CCP+ RA subjects in a secondary analysis. The associations still held with slightly weaker P values, suggesting that these SNPs are associated with CCP level among those who are CCP+, not just with CCP positivity (Table 3).
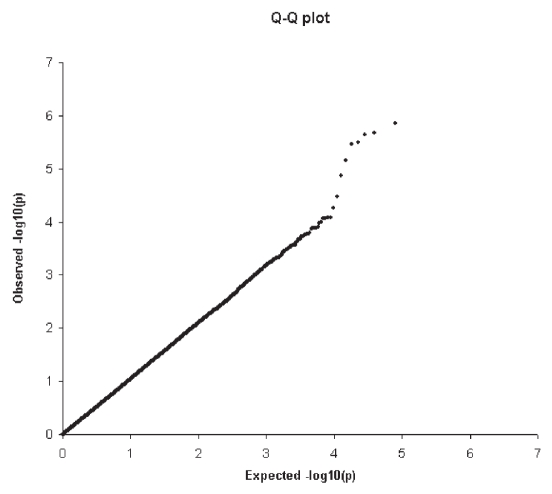
Permutation analyses to examine the expected number of false positive findings with observed associations are summarized in Table 4 for two genotype success rates. For SNPs with a genotype rate greater than 95%, we observed 134 tests that reached significance at the level of < 0.001; by random chance we would expect 93 significant tests. Furthermore, a Q-Q plot (Figure 4) demonstrated no excess signals that might be due to bias based on a genotype call rate of 95%.
Table 4.
Observed number of significant associations compared with expected associations by permutation testing
| Observed N
|
|||||
|---|---|---|---|---|---|
| Genotype rate | SNP N | <0.01 | <0.001 | <0.0001 | <0.00001 |
| 95% | 93709 | 1114 | 134 | 14 | 6 |
| 99% | 59885 | 690 | 78 | 9 | 4 |
| Corresponded expected N
|
|||||
| 95% | 93709 | 932.5 | 93.2 | 8.1 | 0.9 |
| 99% | 59885 | 596.0 | 59.7 | 5.8 | 0.6 |
Figure 4.
Quantile-Quantile plot (Q-Q) showing the comparison between whole genome association observed −log10(p) and expected –log10(p). Expected –log10(p) assumes there is no association between SNPs with anti-CCP and shown on the abscissa. The observed –log10(p) is shown on the ordinate.
Replication Analysis in NARAC
We selected 289 SNPs that were associated with anti-CCP titer in all BRASS cohort or CCP+ cohort at P ≤ 0.001 to assess replication in the NARAC Illumina 550K data set. We compared the two genotyping platforms using HapMap linkage disequilibrium (LD) information (23) to define proxy SNPs for replication. There were 289 tagging SNPs in the 550K panel that passed quality control (QC) and tagged one or more candidate SNPs with r2 > 0.8 in the Hap Map CEU population (24). Among the SNPs analyzed in the NARAC sample, the most significant was rs1980493, which is in LD with the top five SNPs from BRASS (R2 = 0.85) (unadjusted GLM P = 6.1 × 10−5 [Table 3], and SE-adjusted P = 0.00018). The minor allele was associated with lower anti-CCP levels. In sensitivity analyses to test for differences in subcohort CCP ranges (see Methods), rs1980493 remained the top SNP (GLM adjusted for HLA-SE, cohort, and disease duration, P = 0.0003; logistic regression of high/ low CCP subsets, P = 0.003). This SNP is located between HLA-DRA and the gene BTNL2. Two more candidate SNPs, rs12478948 and rs2046159, achieved a P value of < 0.01 in the NARAC study (data not shown).
Additional exploration of the HLA region was performed using BRASS data. Because the HLA-SE is actually a group of alleles in the HLA-DRB1 region, when we checked LD between the five identified SNPs with individual SE alleles DR1 and DR4, the highest LD r2 with SE risk alleles observed was 0.09. We also examined LD defining HLA-SE as any copy of SE alleles and found an r2 = 0.14 (Table 5).
Table 5.
Association between HLA-DRB1 alleles and anti-CCP titer, and LD with the most strongly associated SNP
| DRB1 allele | Association with anti-CCP, P value | LD with SNP rs1041885 (R2) |
|---|---|---|
| DR1 | 0.3531 | 0.0296 |
| DR2 | 0.1025 | 0.0019 |
| DR3 | 0.0003 | 0.4621 |
| DR4 | 4 × 10−7 | 0.0935 |
| DR7 | 0.5409 | 0.0119 |
| DR11_DR13 | 0.0184 | 0.0353 |
| SEa | 2 × 10−7 | 0.1447 |
DR1 and DR4 are known HLA-SE for RA.
A previous study demonstrated an inverse association between HLA-DR3 and anti-CCP titer (16) in NARAC subjects. The five top SNPs in BRASS had an r2 = 0.46 with DR3. We repeated the analysis adjusting for HLA-DR3 and the P value for the top five SNPs in BRASS were 0.0002 (data not shown). Among the CCP + BRASS subjects, the P values were 0.01. The P value for the single top SNP in NARAC was 0.6 after adjusting for DR3. The P value for a heterogeneity test between BRASS and NARAC was 0.23 for the top SNP. We then pooled the raw data and performed a pooled meta-analysis across the BRASS and NARAC studies, and the SNP rs1980493 was associated significantly with anti-CCP titer at 1.8 × 10−9, 5.3 × 10−7, 0.0014, and 0.0022 for unadjusted model, adjusted for HLA-SE, adjusted for HLA-DR3, and adjusted for both HLA-SE and DR3 in the same model respectively. When limiting BRASS to CCP+ only (N = 401) and pooling with NARAC for the same four analyses, results were 2.9 × 10−7, 1.0 × 10−6, 0.02, 0.01, respectively.
DISCUSSION
We performed GWAS on the quantitative RA trait of anti-CCP titer and confirmed that HLA is the most important region for this RA phenotype. However, the HLA-SE does not explain all the risk in the region, as there is a significant signal in the region that is independent of HLA-SE status. The minor alleles of the top SNPs in both cohorts are associated inversely with anti-CCP level. This signal, near BTNL2 and HLA-DRA, is in LD with HLA-DR3; therefore, the precise causal location of this effect remains to be determined.
Besides genetic susceptibility analysis in RA, there also are studies of genetic associations for anti-CCP level as a quantitative trait. Most of these studies focused on the HLA region (7,10,11, 15–18), and consistently found that the HLA-SE is associated with higher anti-CCP titer. Irigoyen et al. reported that in contrast to the SE, HLA-DR3 alleles were found to be associated with lower anti-CCP titers. The top SNPs from BRASS have moderate LD with DR3, also are associated with lower anti-CCP titers, and the association holds after adjustment for DR3. The proxy SNP from NARAC has higher LD with DR3 (r2 = 0.64, with D’≈ 1), and, after adjusting for DR3, the association is no longer significant. We do see significant association between the top SNP and anti-CCP titer after controlling for DR3 in the combined cohort. Although our results support independence of the SNP from the HLA-SE, further evidence will be needed to determine whether the SNP is associated with anti-CCP independent of HLA-DR3.
Aside from traditional linkage and candidate gene studies, the availability of cost effective high-throughput SNP genotyping platform technology has generated high enthusiasm for GWAS for a wide range of disorders. Among these, four were done within several large RA case-control studies looking for RA risk loci (13,21,25,26). Remmers et al. (25) reported that a STAT4 SNP, rs7574865,was associated significantly with RA risk in two large cohorts. There were two SNPs on the 100K SNP platform, rs3821236 and rs3024886, in LD with STAT4 rs7574865 with R2 of 0.74; however, we failed to find any association with anti-CCP titer. For non-MHC reported regions from WTCCC (26) that associated with RA risk, we did see signals from Chromosomes 6q, 21, and 22 in analysis of BRASS samples, but failed to replicate these results in the NARAC samples. Plenge et al.(21) reported that SNPs in LD with the TRAF1 and C5 genes were associated significantly with RA, and Raychaudhuri et al.(27) reported new risk loci for RA, CD40 and CCL21 from meta analysis. However, in the 100K SNP platform, none of the available SNPs are in high LD with the reported SNPs.
An important strength of this study is the inclusion of two large, well-characterized RA cohorts. Limitations include the risk of false positive findings due to the large number of tests inherent in genome-wide association testing. Therefore, it is crucial to confirm findings by replication studies. We included 130 CCP− subjects from BRASS in the primary analysis which raises the question of whether the significant associations in the HLA region are due to an HLA association with the CCP positive/negative phenotype. However, in a secondary analysis limited to CCP+ subjects only, our findings were similar, suggesting that the association with CCP titer is robust. Although we have a fairly homogeneous cohort with a sample size >500, we still lack power to detect loci with small effect sizes. The 100K SNP platform only covers ~20% of the common variants through the genome, so more comprehensive sets of SNPs are needed for a better understanding of the genetic basic of the quantitative trait anti-CCP titer.
After testing our top findings in NARAC sample, we identified a locus in the HLA-DRA/BTNL2 region in LD with DR3 with a strong signal in both samples. After adjustment for HLA-SE status, this signal remained significant at P = 5.3 × 10−7, suggesting a signal in this region that is independent of the HLA-SE.
ACKNOWLEDGMENTS
The BRASS Registry is supported by a grant from Millennium Pharmaceuticals, Biogen-Idec, and Crescendo Biosciences. The NARAC is supported by US National Institutes of Health grants R01-AR44422, M01 RR-00079, and NO1-AR-2-2263. J Cui is supported by NIH grants R01 AR49880, P60 AR047782, and the Alliance for Lupus Research. LA Criswell is supported by NIH grants K24 AR02175 and R01 AI065841. RM Plenge is supported by a NIH K08 AI55314-3, a private donation from the Fox Trot Fund, the Burroughs Wellcome Fund, and the William Ran-dolph Hearst Fund of Harvard University. EW Karlson is supported by NIH grants R01 AR49880, P60 AR047782, and K24 AR0524-01. The Broad Institute Center for Genotyping and Analysis is supported by grant U54 RR020278 from the National Center for Research Resources. We thank all participants and Investigators from BRASS and NARAC, especially the principle investigator Peter K Gregersen.
Footnotes
Online address: http://www.molmed.org
Epub (www.molmed.org) ahead of print March 6, 2009.
DISCLOSURE
The BRASS registry is supported by a research grant from Millenninum Pharmaceuticals, Biogen-Idec, and Crescendo Bioscience. E Izmailova and A Parker are employed at Millennium. R Roubenoff is employed at Biogen.
REFERENCES
- 1.Gabriel SE, Crowson CS, O’Fallon WM. The epidemiology of rheumatoid arthritis in Rochester, Minnesota, 1955–1985. Arthritis Rheum. 1999;42:415–20. doi: 10.1002/1529-0131(199904)42:3<415::AID-ANR4>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 2.Jawaheer D, Gregersen PK. Rheumatoid arthritis. The genetic components. Rheum Dis Clin North Am. 2002;28:1–15. v. doi: 10.1016/s0889-857x(03)00066-8. [DOI] [PubMed] [Google Scholar]
- 3.Newton JL, Harney SM, Wordsworth BP, Brown MA. A review of the MHC genetics of rheumatoid arthritis. Genes Immun. 2004;5:151–7. doi: 10.1038/sj.gene.6364045. [DOI] [PubMed] [Google Scholar]
- 4.Jawaheer D, et al. Screening the genome for rheumatoid arthritis susceptibility genes: a replication study and combined analysis of 512 multicase families. Arthritis Rheum. 2003;48:906–16. doi: 10.1002/art.10989. [DOI] [PubMed] [Google Scholar]
- 5.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis. An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–13. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 6.Vallbracht I, et al. Diagnostic and clinical value of anti-cyclic citrullinated peptide antibodies compared with rheumatoid factor iso-types in rheumatoid arthritis. Ann Rheum Dis. 2004;63:1079–84. doi: 10.1136/ard.2003.019877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Gaalen FA, et al. Association between HLA class II genes and autoantibodies to cyclic citrullinated peptides (CCPs) influences the severity of rheumatoid arthritis. Arthritis Rheum. 2004;50:2113–21. doi: 10.1002/art.20316. [DOI] [PubMed] [Google Scholar]
- 8.Meyer O, et al. Serial determination of cyclic citrullinated peptide autoantibodies predicted five-year radiological outcomes in a prospective cohort of patients with early rheumatoid arthritis. Arthritis Res Ther. 2006;8:R40. doi: 10.1186/ar1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mierau R, Genth E. Diagnosis and prognosis of early rheumatoid arthritis, with special emphasis on laboratory analysis. Clin Chem Lab Med. 2006;44:138–43. doi: 10.1515/CCLM.2006.026. [DOI] [PubMed] [Google Scholar]
- 10.Berglin E, et al. Radiological outcome in rheumatoid arthritis is predicted by presence of antibodies against cyclic citrullinated peptide before and at disease onset, and by IgA-RF at disease onset. Ann Rheum Dis. 2006;65:453–8. doi: 10.1136/ard.2005.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huizinga TW, et al. Refining the complex rheumatoid arthritis phenotype based on specificity of the HLA-DRB1 shared epitope for antibodies to citrullinated proteins. Arthritis Rheum. 2005;52:3433–8. doi: 10.1002/art.21385. [DOI] [PubMed] [Google Scholar]
- 12.Karlson EW, et al. Associations between human leukocyte antigen, PTPN22, CTLA4 genotypes and rheumatoid arthritis phenotypes of autoantibody status, age at diagnosis and erosions in a large cohort study. Ann Rheum Dis. 2008;67:358–63. doi: 10.1136/ard.2007.071662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Plenge RM, et al. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477–82. doi: 10.1038/ng.2007.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding B, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–8. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee HS, et al. Interaction between smoking, the shared epitope, and anti-cyclic citrullinated peptide: a mixed picture in three large North American rheumatoid arthritis cohorts. Arthritis Rheum. 2007;56:1745–53. doi: 10.1002/art.22703. [DOI] [PubMed] [Google Scholar]
- 16.Irigoyen P, et al. Regulation of anti-cyclic citrullinated peptide antibodies in rheumatoid arthritis: contrasting effects of HLA-DR3 and the shared epitope alleles. Arthritis Rheum. 2005;52:3813–8. doi: 10.1002/art.21419. [DOI] [PubMed] [Google Scholar]
- 17.van der Helm-van Mil AH, et al. The HLA-DRB1 shared epitope alleles are primarily a risk factor for anti-cyclic citrullinated peptide antibodies and are not an independent risk factor for development of rheumatoid arthritis. Arthritis Rheum. 2006;54:1117–21. doi: 10.1002/art.21739. [DOI] [PubMed] [Google Scholar]
- 18.van der Helm-van Mil AH, et al. The HLA-DRB1 shared epitope alleles differ in the interaction with smoking and predisposition to antibodies to cyclic citrullinated peptide. Arthritis Rheum. 2007;56:425–32. doi: 10.1002/art.22373. [DOI] [PubMed] [Google Scholar]
- 19.Shadick NA, et al. Opposing effects of the D70 mutation and the shared epitope in HLA-DR4 on disease activity and certain disease phenotypes in rheumatoid arthritis. Ann Rheum Dis. 2007;66:1497–502. doi: 10.1136/ard.2006.067603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Plenge RM, et al. TRAF1-C5 as a risk locus for rheumatoid arthritis — a genomewide study. N Engl J Med. 2007;357:1199–209. doi: 10.1056/NEJMoa073491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian C, et al. Analysis and application of European genetic substructure using 300 K SNP information. PLoS Genetics. 2008;4:e4. doi: 10.1371/journal.pgen.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 24.Frazer KA, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–61. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remmers EF, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythe-matosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–78. doi: 10.1038/nature05911. et al. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raychaudhuri S, et al. Common variants at CD40 and other loci confer risk of rheumatoid arthritis. Nat Genet. 2008;40:1216–23. doi: 10.1038/ng.233. [DOI] [PMC free article] [PubMed] [Google Scholar]