Abstract
The transcription factor Etsrp is required for vasculogenesis and primitive myelopoiesis in zebrafish. When ectopically expressed, etsrp is sufficient to induce the expression of many vascular and myeloid genes in zebrafish. The mammalian homolog of etsrp, ER71/Etv2, is also essential for vascular and hematopoietic development. To identify genes downstream of etsrp, gain-of-function experiments were performed for etsrp in zebrafish embryos followed by transcription profile analysis by microarray. Subsequent in vivo expression studies resulted in the identification of fourteen genes with blood and/or vascular expression, six of these being completely novel. Regulation of these genes by etsrp was confirmed by ectopic induction in etsrp overexpressing embryos and decreased expression in etsrp deficient embryos. Additional functional analysis of two newly discovered genes, hapln1b and sh3gl3, demonstrates their importance in embryonic vascular development. The results described here identify a group of genes downstream of etsrp likely to be critical for vascular and/or myeloid development.
Introduction
The cardiovascular system of vertebrates provides a means to transport nutrients to and waste away from cells throughout the organism. As such, this system is critical for the survival of the organism. The cardiovascular system is made up of the vasculature and the blood that flows through it. It has been proposed that a common cell type, known as hemangioblasts, contribute to the development of vascular and blood cells [1], [2], [3], [4], [5]. This idea is supported by experimental evidence from several different organisms, notably the derivation of both blood and vascular endothelial cells from cultured mouse embryonic stem cells [6], [7], [8], and through in vivo lineage tracing studies in zebrafish and chick/quail chimeras [9], [10]. The zebrafish mutant line cloche provides genetic evidence since homozygous mutants lack both hematopoietic and vascular endothelial cells but organogenesis is otherwise normal [11]. As bi-potential precursor cells, hemangioblasts produce cells with more restricted potentials, angioblasts and hematopoietic progenitors [3]. Angioblasts subsequently produce the endothelial cells that line the vasculature during both vasculogenesis and angiogenesis, while hematopoietic progenitors differentiate into distinct blood lineages through hematopoiesis. Although some of the molecular factors that orchestrate these processes are known, many remain to be identified and studied.
Microarray studies were previously used to identify multiple novel hematopoietic and vascular genes misexpressed in the cloche mutant [12], [13], [14]. A particularly important gene identified in these studies was the transcription factor, Ets1-related protein (Etsrp), which is both necessary and sufficient to direct the development of the vascular and primitive myeloid lineages [13], [15], [16]. In zebrafish, hematopoietic/vascular development is separated into two distinct anatomical locations, the anterior lateral mesoderm (ALM) and posterior lateral mesoderm (PLM). The ALM gives rise to both primitive myeloid cells as well as the endothelial cells of the head vasculature. The PLM gives rise to primitive erythroid cells and the endothelial cells of the trunk and tail. Etsrp is expressed in both the ALM and PLM. Loss of etsrp function by morpholino antisense or genetic mutation, termed y11, results in loss of primitive myeloid cells and disrupted vasculogenesis and angiogenesis in the head and trunk. The defective vasculature in etsrp knockdown or mutant fish appears to be due to altered gene expression and cell behavior and not simply a loss of cells as the number of fli:gfp transgene positive cells is similar to control animals [15]. In contrast, the complete loss of pu.1 staining in etsrp knockdown and mutants suggests that primitive myeloid cells are never specified [16], [17]. Interestingly, the primitive erythroid population in the PLM appears relatively normal when etsrp function is blocked. Thus, etsrp is critical for primitive myeloid and endothelial development from the ALM and endothelial development from the PLM but not the primitive erythroid cell population of the PLM.
A mammalian homolog of etsrp, ER71/Etv2, has recently been identified. ER71/Etv2 knockout mice exhibit loss of vasculature and primitive erythrocytes suggesting it functions in the developing hemangioblast [18]. Additionally, overexpression of ER71/Etv2 in zebrafish embryos causes the ectopic induction of the hemangioblast marker scl/tal1 and the flk1:gfp transgene, identical to the results of etsrp overexpression. Therefore, both etsrp and ER71/Etv2 play an evolutionarily conserved role in hematovascular development.
As members of the Ets transcription factor gene family, Etsrp and ER71/Etv2 have a conserved DNA binding domain and presumably act as transcriptional activators. Limited in situ hybridization analysis of etsrp morpholino gene knockdown or y11 mutant embryos demonstrate that etsrp is necessary for the expression of flk1, scl, fli1, pu.1 and a few other known genes specific to vascular and hematopoietic cells [15], [19]. Additionally, overexpression of etsrp in zebrafish embryos can ectopically induce vascular and myeloid gene expression [16]. We therefore decided to search for novel blood and vascular related genes downstream of etsrp by analyzing expression profiles of zebrafish embryos overexpressing etsrp. Here we report the identification of 14 genes with little or no prior investigations that are expressed in blood and/or vascular endothelial cells and demonstrate the function of two of these genes, hapln1b and sh3gl3, in vascular development using morpholino antisense in transgenic zebrafish embryos.
Results
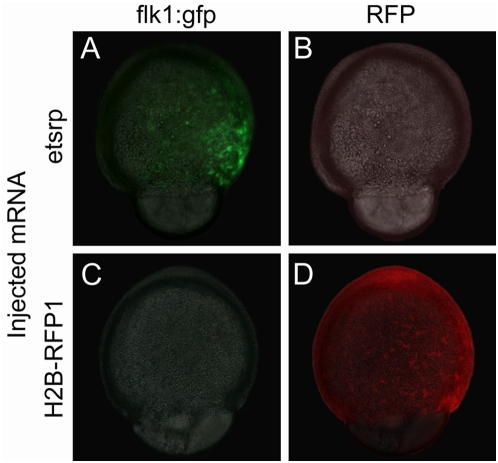
In order to identify novel blood or vascular related genes downstream of etsrp, the expression profiles of embryos ectopically expressing etsrp at late gastrulation stages, 80% epiboly to tail bud were compared to control embryos by microarray analysis. The flk1:gfp transgenic line was used to identify embryos successfully expressing etsrp since ectopic etsrp induces flk:gfp expression. Neither endogenous or transgenic flk1:gfp is normally expressed at 80% to tail-bud stage [20], [21], while early ectopic expression of flk1:gfp is evident in embryos injected with 75 pg of synthetic etsrp mRNA (Figure 1A). To ensure that the ectopic expression of flk1:gfp is induced by etsrp specifically, we injected up to 500 pg of synthetically transcribed mRNA encoding RFP tagged histone H2B, H2B-RFP1, as a control. Similar to uninjected controls, flk1:gfp expression is not induced in H2B-RFP1 injected embryos (Figure 1C and 1D).
Figure 1. Early ectopic expression of transgenic flk1:gfp is induced by etsrp over-expression.
(A) Injection of 75 pg of etsrp mRNA at one-cell stage results in the ectopic induction of flk1:gfp before the end of gastrulation. (B) No red fluorescence is observed in etsrp mRNA injected flk1:gfp transgenic embryos. (C) Injection of 500 pg of control H2B-RFP1 mRNA does not result in the early induction of flk1:gfp similar to uninjected embryos (not shown). (D) Uniform H2B-RFP1 expression in a H2B-RFP1 mRNA injected embryo. Panels A and C are composite images of light transmitted images merged with the green fluorescent channel, while panels B and D are composites of light transmitted images merged with the red fluorescent channel.
Prior to microarray analysis total RNA was extracted from pools of 70 embryos per treatment group and the change in expression of genes known to be induced by etsrp overexpression (OE) were assessed by quantitative RT-PCR. Both fli1a (11×) and scl (7.1×) were significantly induced by etsrp OE, while overexpression of H2B-RFP1 resulted in no difference from uninjected controls (Figure S1). Gene expression analysis was then performed using a microarray chip containing 34,647 oligos representing approximately 20,000 genes. 686 genes were identified as being induced greater than two-fold by ectopic etsrp expression. Although some of the genes affected are not restricted to the circulatory system (Figure S2), a great number of previously identified hematopoietic and vascular genes were induced, validating the microarray strategy used in this study. Table 1 lists the 59 genes with the greatest fold induction as measured on the microarray.
Table 1. Top 59 genes induced by overexpression of etsrp.
| GB accession | Unigene ID | Location | Gene name | Fold induction |
| BI980779 | Dr.6315 | chr14 | Cdc42 guanine nucleotide exchange factor (GEF) 9 | 55.92 |
| AI965222 | Dr.45597 | chr14 | Cadherin 5 (cdh5) | 54.96 |
| DQ021472.1 | Dr.47591 | chr16 | Ets1b/etsrp Ψ | 47.01 |
| AW233044 | Dr.24158 | Serglycin | 39.62 | |
| NM_131626 | Dr.81423 | HAND2 | 34.52 | |
| AF045432 | Dr.75812 | chr22 | Tal1/SCL | 34.35 |
| NM_152964 | Dr.17548 | chr6 | Gastrulation brain homeo box 2 (Gbx2) | 27.73 |
| NM_131768 | Dr.3499 | chr3 | Claudin I | 26.36 |
| AW420632 | Dr.12726 | chr21 | Yes-relayed kinase | 25.02 |
| NM_131530 | Dr.5729 | chr11 | Homeo box C6b | 24.88 |
| AI601685 | Dr.5365 | chr22 | Dual specificity phosphatase 5 (dusp5) | 24.52 |
| BI673802 | Dr.113768 | chr11 | Fgd5 | 23.26 |
| AI477500 | Dr.4847 | chr11 | Wnt7A | 20.14 |
| BI880801 | Dr.83871 | chr17 | Rasgrp3 | 20.10 |
| AW115759 | Dr.23967 | chr17 | Complement receptor C1qR-like | 19.35 |
| AI477956 | Dr.76040 | chr8 | Nexilin f actin binding | 19.32 |
| AW595297 | Dr. 105109 | chr8 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 3 | 18.21 |
| Y14538 | Dr.5730 | chr19 | Homeo box A9a | 17.82 |
| BI842764 | Dr.14186 | chr17 | Si:ch211-214p16.1 | 17.50 |
| AI793554 | Dr.104443 | chr16 | Cellular retinoic acid binding protein 2a | 15.98 |
| BI882214 | Dr.13039 | chr15 | Solute carrier family 43, member 2 | 14.62 |
| BM181739 | Dr.8453 | chr2 | PLA2-like otoconin | 14.23 |
| AI959344 | Dr.28288 | chr7 | Matrix metalloproteinase precursor | 13.34 |
| BM036580 | Dr.84866 | chr16 | Similar to hemicentin | 12.93 |
| AB055670 | Dr.30464 | chr23 | Wnt1 | 12.77 |
| Y14532 | Dr.75789 | chr12 | Homeo box B6b | 12.71 |
| AI626374 | Dr.2644 | Dehydrogenase/reductase (SDR family) member 3 | 12.60 | |
| AI477237 | Dr.77024 | chr13 | Similar to Olig3 protein | 12.31 |
| AW279740 | Dr.22713 | chr23 | Zinc finger protein 385 | 12.18 |
| BI982208 | Dr.11052 | Ornithine decarboxylase antizyme 2, like | 11.42 | |
| BM103177 | Dr.24950 | Creatine kinase CKM3 | 11.27 | |
| AW076731 | Dr.23392 | chr25 | integrin alpha, VLA protein type | 10.63 |
| AW233713 | Dr.81043 | Dusp2 | 10.56 | |
| BG892475 | Dr.11064 | chr2 | amyloid precursor-like protein 2 (aplp2) | 10.31 |
| AA605884 | Dr.38472 | cDNA clone IMAGE:7137566 | 10.22 | |
| Y14527 | Dr.5721 | chr17 | Homeo box A10b | 10.15 |
| NM_131359 | Dr.75781 | Bone morphogenetic protein 2a | 9.90 | |
| NM_131419 | Dr.272 | NK2 transcription factor related 7 | 9.81 | |
| AI626636 | Dr.45596 | Adrenomedullin receptor (admr) | 9.76 | |
| NM_131070 | Dr.20373 | Midkine-related growth factor | 9.68 | |
| AW233616 | Dr.121119 | Calpain 8 | 9.52 | |
| AI522349 | Dr.44232 | Rhoub | 9.46 | |
| NM_131000 | Dr.20912 | Activated leukocyte cell adhesion molecule | 9.29 | |
| BG985561 | Dr.8617 | chr2 | Hyaluronan and proteoglycan link protein 1b | 9.21 |
| NM_131767 | Dr.26572 | Claudin h (cldnh) | 9.09 | |
| NM_131101 | Dr.5572 | Homeo box B5a | 8.81 | |
| AF071249 | Dr.28647 | chr17 | Homeo box A9b | 8.47 |
| BG727645 | Dr.11977 | chr16 | Leucine rich repeat 33 | 8.00 |
| BG985673 | Dr.24989 | chr10 | Myosin light chain 2 like | 7.80 |
| Y13948 | Dr.21032 | chr9 | Homeo box D3a | 7.71 |
| AI601355 | Dr.77902 | Similar to Fraser syndrome 1 isoform 1 | 7.61 | |
| NM_131419_1 | Dr.272 | NK2 transcription factor related 7 | 7.60 | |
| BM025541 | Dr.3373 | chr9 | Collagen type XVIII, alpha 1 | 7.56 |
| AF071568 | Dr.20962 | chr19 | Homeo box B2a | 7.43 |
| AY028584 | Dr.18294 | Prostaglandin-endoperoxide synthase 1 | 7.38 | |
| AI601770 | Dr.21376 | chr23 | Plexin precursor | 7.27 |
| AI721522 | Dr.8674 | Septin 9b | 6.78 | |
| BG728550 | Dr.81575 | chr15 | Cytochrome b5 reductase 4 | 6.58 |
| AI964237 | Dr.76050 | ATP synthase, H+ transporting, mitochondrial F1 complex, alpha subunit 1 | 5.68 |
Note: Genebank accession number and unigene ID correspond to the probes on the array. The fold change column is the average ratio of eight measurements. Ψ The large induction ratio of etsrp reflects array probe hybridization to both the exogenously injected RNA as well as the induced expression of endogenous etsrp, (See Supplementary Figure 1).
From the genes affected by etsrp OE, we set up four criteria to select candidates for further studies: 1. They have not been well studied in hematopoiesis and/or vasculogenesis; 2. They are induced by ectopic etsrp in embryos as validated by whole mount in situ hybridization (WISH); 3. They are expressed in hematopoietic and/or vascular tissues; and 4. Expression is reduced by etsrp deficiency.
To identify genes with few or no prior studies for further analysis, the list of genes induced by etsrp OE was compared to the data sets for cloche mutants [13], [14]. We reasoned that genes identified in both etsrp OE and cloche mutant microarrays were highly likely to be involved in blood and/or vascular development. From this group of overlapping genes we screened for genes that had not been well studied using the PubMed database at the NCBI website. This resulted in the selection of 31 genes (Table 2), which were cloned and analyzed further.
Table 2. Genes screened for blood and/or vascular expression.
| GB Accession | Unigene ID | Gene location | Gene name | Fold Induction | WISH for etsrp OE | down in cloche |
| BI980779 | Dr.6315 | chr14 | Cdc42 guanine nucleotide exchange factor GEF 9 (arhgef9) | 55.92 | X | X |
| AW171479 | Dr.2433 | chr12 | Carboxylesterase 2-like(cbe2) | 3.32 | X | X |
| AF109780 | Dr.81287 | Kreisler maf-related leucine zipper homolog 2 (krml2) | 4.82 | X | X | |
| AF071255 | Dr.117289 | chr12 | Hoxb8b | 3.57 | X | X |
| BI880801 | Dr.83871 | chr17 | RAS guanyl releasing protein 3 (rasrgrp3) | 20.51 | X | |
| BG727645 | Dr.11977 | chr16 | Leucine rich repeat containing 33 (lrrp33) | 8.00 | X | X |
| AW279740 | Dr.22713 | chr23 | Zinc finger protein 385 (znf385) | 12.18 | X | |
| BI673729 | Dr.14541 | chr6 | Similar to leucine rich repeat containing 51 (lrrp51)* | 6.08 | X | |
| AI544475 | Dr.33183 | chr13 | SPARC related modular calcium binding 2 (smoc2) | 6.68 | X | |
| BI867572 | Dr.9564 | chr21 | Similar to testis-expressed sequence 2(tex2) | 4.91 | X | |
| AW421939 | Dr.5562 | chr20 | Apolipoprotein B (apolipo b) | 3.07 | X | |
| BI430050 | Dr.418 | Protocadherin2 gamma20 (pcdhg) | 3.50 | |||
| BI672391 | Dr.14449 | weakly similar to XP_422395.1 (novel 2) | 5.61 | X | ||
| BC093143 | Dr.20125 | chr1 | hypothetical LOC550501 | 6.80 | ||
| BI864031 | Dr.13409 | chr12 | Anthrax toxin receptor 1/similar to tumor endothelial marker 8 (antxr1/tem8) | 7.29 | X | |
| BM095233 | Dr.17582 | chr23 | C1orf192 homolog | 4.47 | ||
| BI671403 | Dr.120608 | chr24 | PTPL1-associated RhoGAP 1 (arhgap29) | 5.65 | X | |
| BI673802 | Dr.113768 | chr11 | Fyve rhogef and ph domain containing zinc finger fyve domain containing protein 23 (Fgd5) | 23.26 | X | X |
| AY028584 | Dr.18294 | Prostaglandin-endoperoxide synthase 1 (ptgs1) | 7.38 | |||
| BG985561 | Dr.8617 | chr2 | Hyaluronan and proteoglycan link protein 1 precursor (hapln1B) | 9.21 | X | |
| BI842967 | Dr.76442 | chr8 | hypothetical LOC555375 | 3.68 | ||
| BI880784 | Dr.12941 | Similar to testican 3 | 3.81 | |||
| AW420632 | Dr.12726 | chr21 | Yes-relayed kinase (yrk) | 25.02 | X | X |
| BM036580 | Dr.84866 | chr16 | Similar to hemicentin | 12.93 | X | |
| BI983776 | Dr.12218 | chr25 | SH3-domain GRB2-like 3 (sh3gl3) * | 2.76 | X | |
| AI884043 | chr24 | Similar to immune costimulatory protein * | 3.34 | X | ||
| X60095 | Dr.510 | chr23 | Hoxc3a | 3.37 | ||
| BI428793 | Lim domain binding 2 (ldb2) | 4.21 | X | |||
| AI721944 | Dr.21605 | chr3 | Est- AI721944 | 2.92 | X | |
| AW019729 | chr8 | Est- AW019729 | 2.68 | X | ||
| AA542593 | Dr.75525 | chr23 | C20.orf.112 | 5.09 |
Note: Genebank accession number and unigene ID correspond to the probes on the array. The fold change column is the average ratio of eight measurements. (*) ectopic induction was observed for lrrp51 but no expression was detected between 24–30 hpf. Genes in bold indicate those that are expressed in blood or vascular endothelial cells at 24–30 hpf.
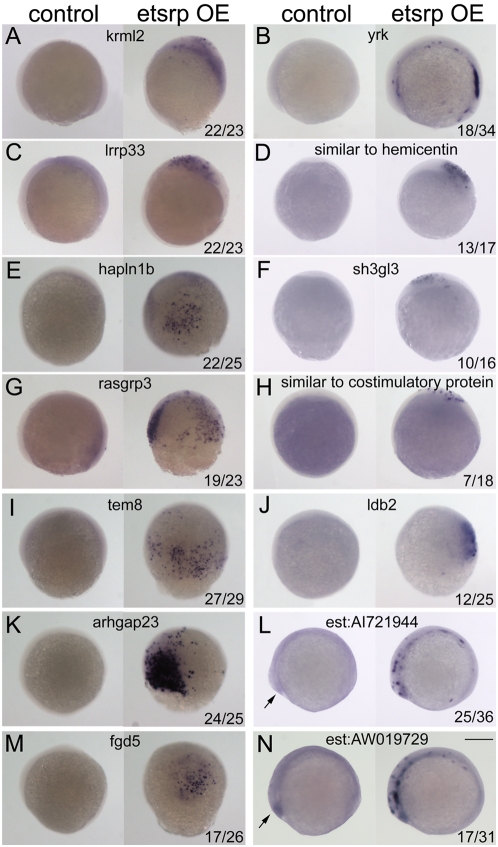
To validate the results obtained with the etsrp OE microarray, WISH was performed on embryos at the same developmental stage as was done for the microarray, 80% epiboly to tailbud. To ensure that the OE samples examined were indeed ectopically expressing etsrp, embryos were injected with plasmid DNA encoding an Etsrp-mcherry fusion protein. This fusion protein was functionally active since it efficiently induced ectopic expression of flk:gfp when injected into transgenic embryos (data not shown). Embryos expressing Etsrp-mcherry and flk:gfp were selected to test for ectopic induction for 23 of the 31 genes identified in Table 2. 21 of 23 genes were clearly induced ectopically by etsrp OE. Figure 2 shows the ectopic induction of the 14 genes identified below to be relevant to blood and/or vascular expression. There was ubiquitous endogenous expression in 2/23 genes, arhgef9 and znf385, prohibiting any qualitative discernment of their changes in expression by etsrp OE (data not shown). Overall, this demonstrates that the microarray data is low in false positives and reliable.
Figure 2. Ectopic induction of microarray identified genes by etsrp overexpression.
Ectopic gene expression was examined at 80% epiboly to tailbud stages in flk1:gfp transgenic embryos injected with 30 pg etsrp-mcherry DNA at the one cell stage. Embryos exhibiting both red and green fluorescence were selected for analysis. Representative control embryos are on the left and etsrp overexpressing (OE) embryos are on the right side of each panel. Represented genes: (A) krml2; (B) yrk; (C) lrrp33; (D) similar to hemicentin; (E) hapln1b; (F) sh3gl3; (G) rasgrp3; (H) similar to costimulatory protein; (I) tem8; (J) ldb2; (K) arhgap23; (L) est:AI721944; (M) fgd5; and (N) est:AW019729. Note that there is a low level of endogenous expression in the control embryos for the two EST's, L and N, at the polster (arrows). Ratios in bottom right hand corner in panels represent the number of embryos with ectopic induction of total embryos processed and scored in the injected groups; control embryos never displayed ectopic induction. All embryos are in lateral view, and those at tail bud stage are oriented with anterior to the left. Scale bar: 250 µm.
To screen the 31 selected genes for blood and/or vascular expression, their expression patterns were examined in wild type embryos by WISH at 24–30 hours post fertilization (hpf). Of the genes examined, two are expressed in blood lineages (Figure 3), and twelve in vasculature (Figure 4), while sixteen other genes demonstrate tissue specificity but are irrelevant to blood or vessels (Figure S2).
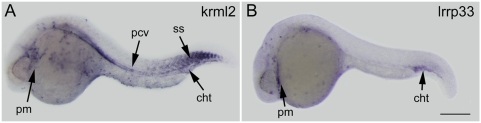
Figure 3. Expression of genes identified in primitive myeloid blood cells.
Gene expression was examined at 24–28 hours post fertilization by whole mount in situ hybridization in wildtype embryos. (A) krml2 is expressed in primitive myeloid cells (dispersed cells labeled throughout yolk and head) in the posterior cardinal vein, caudal hematopoietic tail region, and somites. (B) Lrrp33 is expressed in primitive myeloid cells and in the caudal hematopoietic tissue Abbreviations: pm, primitive myeloid; pcv, posterior cardinal vein; cht, caudal hematopoietic tail region; ss, somites. Scale bar: 250 µm.
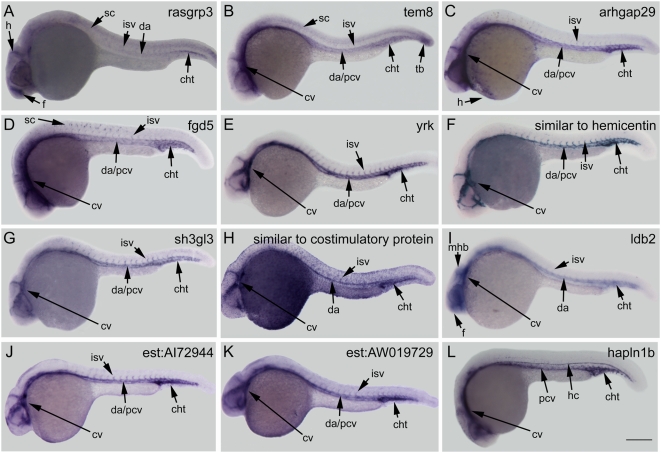
Figure 4. Expression of genes identified in vascular endothelial cells.
Gene expression was examined at 24–28 hours post fertilization by whole mount in situ hybridization in wildtype embryos. (A) rasgrp3 is expressed weakly in the forebrain, hindbrain, spinal cord, intersomitic vessels, dorsal aorta, and caudal hematopoietic tail region. (B) tem8 is expressed highly in the cranial vasculature, the dorsal aorta and posterior cardinal vein, tailbud and weakly in the intersomitic vessels and spinal cord. (C) arhgap29 is expressed in the cranial vessels, dorsal aorta, posterior cardinal vein, intersomitic vessels, caudal hematopoetic tail region, and hatching gland. (D) fgd5 is expressed in the cranial vasculature, dorsal aorta, posterior cardinal vein, intersomitic vessels, and caudal hematopoetic tail region. (E, F, and G) yrk, similar to hemicentin, and sh3gl3 are all expressed in the cranial vasculature, dorsal aorta, posterior cardinal vein, intersegmental vessels, and caudal vascular hematopoietic tail region. (H) similar to costimulatory protein is expressed in the cranial vasculature, dorsal aorta, intersomitic vessels, and caudal hematopoietic tail region; additional expression is present in the ectoderm layer throughout the embryo. (I) ldb2 is expressed in the forebrain, midbrain-hindbrain boundary, dorsal aorta, intersomitic vessels, and caudal hematopoietic tail region. (J and K) Both est:AI721944 and est:AW019729 are expressed in the cranial vasculature, dorsal aorta, posterior cardinal vein, intersomitic vessels, and caudal hematopoietic tail region. (L) hapln1b is expressed in the cranial vasculature, posterior cardinal vein, caudal hematopoietic tail region, and hypochord. Abbreviations: f, forebrain; h, hindbrain; sc, spinal cord; cv, cranial vasculature; da, dorsal aorta; pcv, posterior cardinal vein; isv, intersomitic vessels; cht, caudal hematopoietic tail region; tb, tail bud; h, hatching gland; and hc, hypochord. Scale bar: 250 µm.
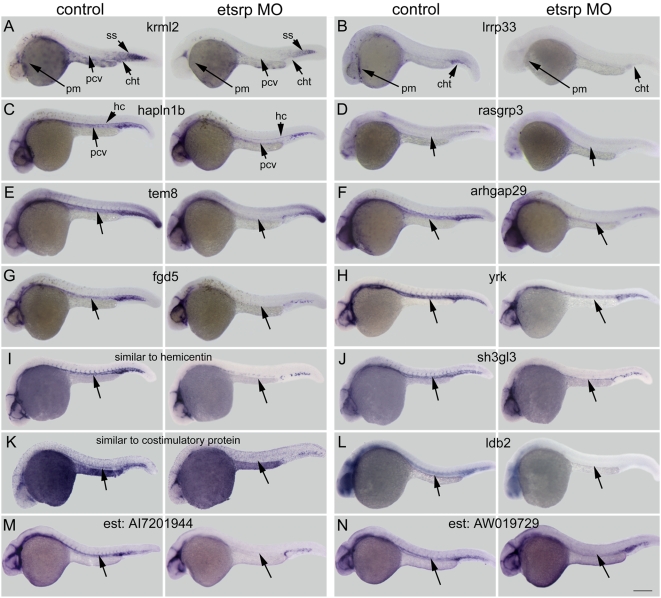
The dependence of the new blood and vascular specific genes on etsrp was then examined by comparing their expression in etsrp morphants to uninjected control embryos (Figure 5). In etsrp morphants, the expression of krml2 (Figure 5A) and lrrp33 (Figure 5B) was abolished in myeloid cells, while the remaining 12 vascular genes were also significantly reduced in expression, most noticeably in the trunk region containing the dorsal aorta, posterior cardinal vein, and intersomitic vessels (Figure 5, C–N). Note that gene expression in the cranial vasculature was less affected in etsrp morphants but still reduced. Gene expression in tissues outside of blood and vessels such as somites for krml2 (Figure 5A), neurons for rasgrp3 (Figure 5D) and ldb2 (Figure 5L), and tailbud for tem8 (Figure 5E) were not affected by etsrp knockdown. This demonstrates that etsrp is not only sufficient but also required for normal expression in primitive myeloid and/or vascular cells in the 14 genes identified here.
Figure 5. Morpholino knockdown of etsrp reduces expression of genes identified by etsrp overexpression microarray analysis.
Embryos were injected with a combination of two etsrp targeting morpholinos (4 ng each) and gene expression was examined by in situ hybridization at 24–26 hours post fertilization. Representative control embryos are on the left of each panel and etsrp morpholino (MO) injected embryos are on the right. (A) krml2 is reduced in primitive myeloid cells, posterior cardinal vein, and caudal hematopoietic tail region by etsrp knockdown. (B) lrrp33 expression is absent in myeloid cells and reduced in the caudal hematopoietic tail region. (C) hapln1b expression is lost in the posterior cardinal vein when etsrp is knocked down. (D–N) Etsrp knockdown causes a significant decrease of gene expression, most pronounced in the trunk (unlabeled arrows). (D) rasgfp3; (E) tem8; (F) arhgap29; (G) fgd5; (H) yrk; (I) similar to hemicentin; (J) sh3gl3; (K) similar to costimulatory protein; (L) ldb2; (M) est:AI7201944; and (N) est:AW019729. Note that non-hematovascular tissues such as somites in (A) and hypochord in (C) are not affected by etsrp knockdown. Abbreviations: pm, primitive myeloid; pcv, posterior cardinal vein; cht, caudal hematopoietic tail region; ss, somites; and hc, hypochord. Scale bar: 250 µm.
Genes downstream of etsrp with endogenous expression in blood
Krml2
Krml2 is a member of the Maf family of basic leucine zipper transcription factors that have high evolutionary conservation with orthologs existing in chicken, Xenopus, mice, and humans [22], [23]. The expression of krml2 in zebrafish was previously found in somites, reticulospinal oculomotor neurons, and lens [24]. In this study we have also detected its expression in primitive myeloid cells and the vasculature (Figure 3A). The mouse and avian orthologs of krml2 have also been detected in macrophages [25], and it has been demonstrated to regulate hematopoiesis by repressing erythropoiesis in favor of the myeloid fate through an interaction with Ets1 [26].
LRRP33
Leucine rich repeat containing protein 33 is a member of the leucine-rich repeat protein family. Lrrp33 is a single pass transmembrane protein that contains 21 leucine-rich repeats, and has orthologs in mice, rats, and humans, which remain to be characterized. The leucine-rich repeat family of proteins consists of over 60,000 members with diverse biological activities, but a common feature shared by proteins with LRR motifs is their propensity to associate with other proteins through LRRs [27]. Prominent members of this family with characterized roles in inflammatory and innate immunity are the toll-like receptors, which are comprised of ten members in humans, with homologs for all encoded in the zebrafish genome [28], [29]. Lrrp33 is excluded from this subfamily because it lacks the cytosolic toll-interleukin (TIR) receptor domain through which TLRs transduce intracellular signals [30], [31]. In this study we have detected lrrp33 expression in primitive myeloid cells and the caudal hematopoietic tail region (Figure 3B).
Genes downstream of etsrp with endogenous expression in vasculature
RAS guanyl releasing protein 3 (RASgrp3)
RASgrp3 is a member of the RAS family of GTPases which link cell surface receptor activation and RAS activation by switching its state of activity between inactive GDP and active GTP bound states [32]. With orthologs in mice and humans, zebrafish RASgrp3 encodes a protein with 708 residues containing an N-terminal and catalytic RhoGEF domains, followed by a pair of calcium binding EF-hand domains and a zinc dependent DAG/phorbol-ester binding domain. RASgrp3 is expressed at low levels in both axial and cranial vasculature, and the caudal vascular plexus region, as well as the neural tube and hindbrain (Figure 4A). A role for RASgrp3 in endothelial cells was initially identified in an embryonic stem cell gene trap assay [33]. In endothelial cells RASgrp3 is up-regulated by VEGF signaling, and functions as a phorbol ester receptor in both B-cells and endothelial cells, where it has been ascribed with forming a disorganized angiogenic vasculature in disease states [33], [34]. There are currently four RASgrp genes encoded in mammals, RASgrp-1, -2, -3, and -4, while zebrafish only have three, RASgrp-1, -2, and -3. The mouse ortholog is expressed in the vasculature during developmental stages, but the knockout is viable [33].
Similar to tumor endothelial marker 8 (tem8)/Anthrax Receptor 1 (antxr1)
Tem8 was initially identified as a gene up-regulated in tumors, and is one of two anthrax toxin receptors, the other being capillary morphogenesis gene 2 (CMG2)/ANTXR2 which together recognize and internalize anthrax toxins [35], [36], [37]. Zebrafish tem8 encodes a 552 amino acid transmembrane protein with a von Willebrand factor type A domain, an extracellular Ig anthrax toxin receptor domain and the cytoplasmic c-terminal anthrax receptor domain. It is expressed at low levels in neurons with higher expression levels in the tailbud as well as the axial and cranial vascular endothelial cells (Figure 4B). Two cmg2 orthologs have been identified in zebrafish, cmg2a and cmg2b, but their expression remains to be examined [38]. Tem8 regulates cell adhesion and migration by coupling extracellular stimuli to changes in the actin cytoskeleton [39], [40]. Regulation of cell adhesion and migration by tem8 plays a role in angiogenesis and is relevant to tumor angiogenesis, anthrax toxin responses, and potentially circulatory system development.
RhoGTPase-activating protein 29 (arhgap29)
Arhgap29 was identified in a screen for proteins that interact with the protein-tyrosine phosphatase PTPL1 (which is expressed in many tissues), and has the alternative name PTPL1-Associated RHOGAP1 (PARG1) [41]. Zebrafish arhgap29 is a guanine activating protein of RhoGTPases that is 1365 amino acids long with a protein kinase C domain and a RhoGAP domain which are conserved in orthologs in multiple species including humans. The expression of ARHGAP29 has been detected in the hearts of mouse embryos among other tissues [42], and in zebrafish it is expressed in the cranial vasculature, dorsal aorta, cardinal vein, intersomitic vessels, caudal vascular plexus, and hatching gland at 24 hpf (Figure 4C).
FYVE, rhoGEF and PH domain containing protein 5 (fgd5)
Fgd5 is a member of the Dbl homology (DH) protein family that functions as guanine nucleotide exchange factors for the RhoGTPases Rho, Rac and Cdc42 [43]. There are six paralogs of this gene in mammals. The family's name is derived from the founding member, fgd1, which causes faciogenital dysplasia in humans when mutated [44]. Fgd5 has been widely conserved through evolution and, in zebrafish, Fgd5 is 1603 amino acids long with the common domains of all DH family members, which includes a DH domain followed by two plextrin homology PH domains that flank a FYVE containing zinc finger domain. Fgd5 is expressed in the endocardium of mice during development [45], while in zebrafish it is expressed throughout the vascular system, including the heart (Figure 4D).
Yes-related kinase (yrk)
Yes-related kinase (yrk) is a homolog of the yes oncogene, and is a member of the src non-receptor tyrosine kinase family. It is distinct from other src members including fyn and yes, and is expressed in the cerebellum, spleen, lung, and skin in adult chicken tissues [46]. The protein is composed of 528 amino acids with single SH2, SH3 and tyrosine kinase domains. Unlike its ortholog in chicken, the zebrafish yrk is expressed exclusively in vascular endothelial cells where its function remains to be investigated (Figure 4E).
Similar to hemicentin
Hemicentin is an extracellular matrix ECM protein of the immunoglobulin superfamily identified originally in c.elegans and found to facilitate tissue organization and cell migration [47]. The human ortholog has been associated with age related macular degeneration [48] and in mice its expression has been observed at the pericellular ECM of various epithelial cells including embryonic trophectoderm, skin, tongue, in addition to the ECM of some blood vessels [49]. Distinct from hemicentin, zebrafish similar to hemicentin differs in size, structure and possibly function. While hemicentin contains a von willberand A domain, 48 tandem Ig domains, multiple tandem epidermal growth factor repeats (EGFs), and a single fibulin-like C-terminal domain, zebrafish similar to hemicentin is 264 amino acids long, has a single Ig domain and a transmembrane domain. Its specific expression in the vascular system suggests a potential role for this gene in endothelial cells (Figure 4F).
SH3 domain Growth factor Receptor Bound protein 2-like 3 (sh3gl3)
Sh3gl3 encodes for a protein that is 386 amino acids long which contains a Bin-Amphiphysin-Rvs (BAR) domain implicated in synaptic vesicle internalization, actin regulation, differentiation, and cell survival, and a src homology 3 (SH3) domain [50]. The mammalian SH3gl3, also referred to as endophilin A3 and EEN-B2, is a member of a small family of SH3 containing proteins known as the SH3GL family that includes three members, EEN, EEN-B1 and EEN-B2 [51]. In adult mice, EEN-B2 is expressed in the brain, testis and spinal cord, while its embryonic expression is ubiquitous and there appears to be a marked transient expression in the vascular system [52]. In zebrafish, sh3gl3 expression is vascular specific at the stage examined here (Figure 4G). Besides sh3gl3, paralogs in zebrafish include sh3gl1a, sh3gl1b, sh3gl2, sh3gl2b, and zgc:158742.
Similar to immune costimulatory protein
Similar to immune costimulatory protein is a novel gene that encodes a 372 amino acid long transmembrane protein with two tandem Ig-like domains at the N-terminus followed by a transmembrane domain. It is expressed in the cranial vasculature, the caudal vascular plexus, and is restricted to the artery in the axial vessels. It is also expressed in a spotted pattern throughout the ectoderm (Figure 4H).
LIM domain binding protein 2 (ldb2)
LIM domain binding protein 2 (ldb2) is a LIM homeodomain co-activator that is one of four ldb genes found in zebrafish. Ldb2 has been reported to be induced in human vascular endothelial cells following stimulation with VEGF [53]. The expression of ldb2 in zebrafish has been described in the central nervous system and vasculature [54], which we confirmed in Figure 4I. We have also found that ldb2 is silenced in the axial vasculature of etsrp morphants (Figure 5L).
Novel Expressed Sequence Tags (ESTs)
The overexpression of etsrp resulted in the identification of two ESTs that could not be linked to any annotated genes but which are clearly induced ectopically (Figure 2L and 2N), demonstrate vascular specific expression, including cranial, axial, caudal and intersomitic vessels (Figure 4J and 4K), and whose expression is absent in the axial and intersomitic vessels in etsrp morphants (Figure 5M and 5N). Their accession numbers are: AI721944 and AW019729.
Hapln1b
Hyaluronan and proteoglycan link protein 1b is a member of the vertebrate hyaluronan and proteoglycan link protein family which consists of four members in mice and humans, HAPLN-1,-2,-3,-4 [55]. Hapln1, also known as cartilage link protein, is a secreted extracellular matrix protein that stabilizes aggrecan-hyaluronan complexes in cartilage and other tissues, with the knockout in mice resulting in dwarfism and craniofacial abnormalities [56]. Zebrafish also contain four hapln genes, hapln-1a, -1b, -2, and -3. During early stages of zebrafish development, hapln1b is expressed in the hypochord, cranial vasculature, posterior cardinal vein, and caudal hematopoietic tail region (Figure 4L and [57]).
Functional Analysis of hapln1b and sh3gl3
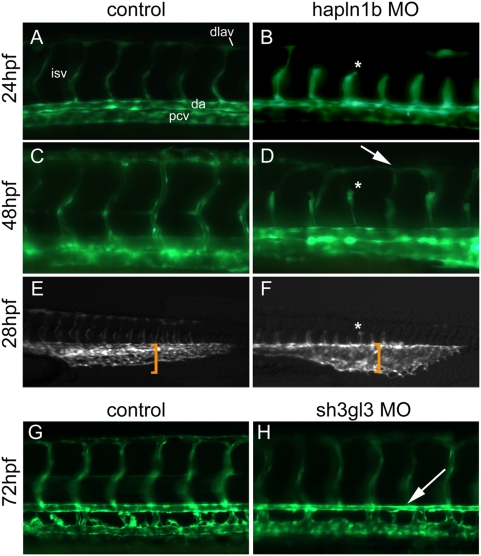
To determine if the genes highlighted here might play a functional role in vascular development we used antisense morpholino (MO) oligos to knockdown hapln1b and sh3gl3 in flk:gfp transgenic fish. Hapln1b knockdown using 2 ng of morpholino results in the arrested angiogenic sprouting of many intersomitic vessels (asterisks in Figure 6B, 6D and 6F), preventing them from forming stereotypical dorsolateral anastamosing vessels (dlavs) by 48 hpf (arrow in Figure 6D). Furthermore, the vascular remodeling that occurs at the caudal vascular plexus around 28–30 hpf is severely defective in hapln1b morphants (compare bracketed region in Figure 6E and 6F). These defects were observed in 87 out of 92 (92.6%) embryos injected with hapln1b-MO while standard control MO (5 ng) injected embryos never exhibited this phenotype (0 out of 70 embryos). Sh3gl3 morphants (3 ng) had a less severe phenotype than hapln1b morphants. Sh3gl3-MO treated embryos had relatively normal intersomitic vessel development, but displayed reduced circulation as visualized by red blood cell movement in the axial vasculature when compared to controls (data not shown). A closer examination of the axial vessels at 72 hpf revealed a significant reduction in the diameter of the dorsal aorta with a concomitant increase in the diameter of the posterior cardinal vein in morphants (arrow in Figure 6H compared to 6G). This constriction of the dorsal aorta was likely the cause of the observed decrease in circulation. 61 of 83 (73.5%) embryos injected with sh3gl3-MO displayed the shrunken dorsal aorta/reduced circulation phenotype while none of the standard control MO injected (5 ng, 0 out of 70 embryos) or uninjected control embryos did. The demonstration that hapln1b or sh3gl3 knockdown results in distinct vascular phenotypes suggest that other genes identified in this gene expression screen will be functionally important and should be studied in further detail.
Figure 6. Hapln1b and sh3gl3 are mediators of vascular development.
(A–F) Flk1:gfp transgenic embryos injected with 2 ng of a translation blocking morpholino (MO) targeting hapln1b arrests angiogenic sprouting of the intersomitic vessels (asterisks in panels B, D, F; compare to their counterparts in A, C, and E respectively), resulting in delayed and improper dorsal longitudinal anastomotic vessel formation (arrow in D). Furthermore, the caudal vascular plexus is dilated relative to wild-type controls at 28 hours post fertilization (hpf) (compare the identical sized bracket in panels E and F). (G and H) Injection of 3 ng of translation blocking MO targeting sh3gl3 results in a thinner dorsal aorta (arrow in H) relative to wild type controls (G) at 72 hpf. Abbreviations: dlav, dorsal longitudinal anastomotic vessels; isv, intersomitic vessels; da, dorsal aorta; and pcv, posterior cardinal vein.
Discussion
The transcription factor Etsrp is a critical mediator of vasculogenesis, angiogenesis, and primitive myelopoiesis in zebrafish. In this study we have used gene expression profiling to identify a group of genes that are induced by etsrp OE during early development. Among these genes are many previously identified vascular and myeloid genes, supporting the previous identified function of etsrp in vascular and myeloid development. The array data was validated by WISH in etsrp OE embryos. Gene expression was then examined at 24 hpf for expression in vascular or blood lineages. Genes that met these criteria were then assayed for expression in embryos with etsrp knocked down by morpholino antisense. Fourteen genes were found to be both ectopically induced by etsrp OE and down regulated in etsrp knockdown embryos. Of these genes two, krml2 and lrrp33, are found in myeloid cells and the remaining twelve are vascular. Any expression of these genes outside of hematopoietic or vascular tissue was unaffected by etsrp knockdown demonstrating the specificity of etsrp's function. Two newly identified vascular genes, hapln1b and sh3gl3, were then independently knocked down by morpholino antisense. Each knockdown resulted in distinct vascular phenotypes demonstrating the utility of the screening method used and the importance of the genes identified. Together, these studies have identified genes downstream of etsrp that are likely to mediate its observed role in primitive myeloid and early vascular development in zebrafish.
The ER71/Etv2 mammalian homolog of etsrp has been shown to mediate vascular development in mice. Given that etsrp has a similar function in zebrafish, it is likely that many of the genes identified in this microarray screen will have mammalian homologs functionally important for angiogenesis and/or vasculogenesis. Of the fourteen genes we focused on, ten have confirmed mammalian homologs. The remaining four cannot yet be determined due to uncertainty in the assembly of the zebrafish genomic sequence. However, it is expected that mammalian homologs for these genes will be found in the future.
Interestingly, ER71/Etv2 is critical for primitive erythropoiesis in mice, while etsrp knockdown or mutation in zebrafish does not affect primitive erythropoiesis. The data presented here supports the previous finding that etsrp regulates primitive myelopoiesis and vasculogenesis but not erythropoiesis since genes related to the former group are highly induced by etsrp OE while no erythroid genes are induced. So it appears that ER71/Etv2 and etsrp have evolutionarily divergent functions in the context of primitive erythropoiesis. One possible explanation for this difference is that zebrafish primitive erythropoiesis may not progress through an endothelial cell intermediate while mammalian primitive erythropoiesis in the yolk sac blood islands does. Alternatively, the evolutionary function of etsrp in primitive erythrocytes may have been co-opted by another ets transcription factor in zebrafish, possibly due to a historical genome duplication event [58]. Another possibility is that the observed difference is due to differential timing of primitive erythrocyte cell death in etsrp/ER71/Etv2 null fish and mammals. In etsrp mutant fish, primitive erythroid cells marked by gata1 expression are initially specified but then later lost through apoptosis [15]. In ER71 null mice, no primitive erythrocytes are observed but an increase in apoptosis was noted [18]. This suggests the possibility that primitive erythrocytes are initially specified in ER71 null mice, but quickly die due to compromised vasculature as happens in fish.
An issue arising from these studies is that etsrp OE results in the induction of many nonhematopoietic and nonvascular genes (Figure S2). It is likely that etsrp OE directly mis-targets genes normally regulated by other Ets family members. When overexpressed at abnormally high levels it might also overcome repressive mechanisms and thereby transactivate non-specific genes. Included in the list of genes ectopically induced are many hox genes that may represent a secondary response to etsrp OE. We have found that krml2 is expressed in myeloid cells and axial vasculature. It was reported that the expression of hoxb3 was induced by krml2 overexpression [59]. In this study, not only is krml2 induced, but hoxb3 is also induced about five-fold. Ectopic induction of nonvascular/nonmyeloid target genes may also be the result of secondary effects of etsrp overexpression. A possible mechanism is that etsrp induces premature differentiation of vascular and myeloid cells and these cells signal to adjacent cells to differentiate into additional nonvascular/nonmyeloid cell types. The strong ectopic induction of genes in the early embryo emphasizes that Etsrp is a very potent, early acting transactivator whose roles in gene regulation must be tightly controlled.
It should also be noted that the overexpression of etsrp alone is sufficient to induce many other transcription factors such as scl and fli1a. The overexpression of scl has also been demonstrated to induce other blood and vascular specific genes alone [60]. Recently, it was proposed that fli1a appears to be at the top of the transcriptional network driving blood and vascular development. Constitutively active fli1a, in which the artificial trans-activation domain of the viral VP16 protein was fused to full-length fli1a coding sequences, can ectopically induce blood and vascular genes [61]. However, overexpression of fli1a alone is not sufficient to induce blood or vascular markers as demonstrated here for etsrp. This argues that etsrp is more likely the top regulator of hemangioblast development.
The two etsrp target genes, hapln1b and sh3gl3, examined by morpholino gene knockdown each displayed interesting vascular phenotypes. Hapln1b is a member of a protein family that functions as extracellular linkers for hyaluronans and proteoglycans. Hapln1b MO treated embryos display truncated intersomitic vessels and disorganized caudal vascular plexus. This phenotype is potentially due to decreased angioblast cell migration or adhesion due to extracellular matrix problems. Future studies will be necessary to establish the exact cellular defect responsible for the vascular phenotype observed in hapln1b morphants. Sh3gl3 is an intracellular protein whose homologs have been suggested to regulate endocytosis and intracellular membrane trafficking [62]. The phenotype observed in the sh3gl3 morphant, reduced dorsal aorta and increased posterior cardinal vein diameter, is suggestive of disrupted notch signaling as notch signaling mediates artery/vein specification and relative size [63]. Indeed endocytotic mechanisms are critical for proper notch signaling [64]. We hypothesize that sh3gl3 may regulate artery/vein size and notch signaling through receptor endocytosis.
In summary, we have identified a set of genes induced by etsrp OE in zebrafish embryos. Within this set we have identified multiple genes that have restricted expression in the vascular or myeloid lineages and are reduced by morpholino knockdown of etsrp. These genes are likely to play important roles in the development of their respective tissues. Additionally, this data can be used in the future to clarify gene hierarchies necessary for hemangioblast differentiation by comparing with mammalian stem cell or zebrafish mutant, morphant, or overexpression gene array data, as we have already done with cloche. The results presented here provide a basis for future studies of the hemangioblast.
Materials and Methods
Microarray
flk1:gfp transgenic embryos were injected with 75 pg of synthetic etsrp mRNA, transcribed with Ambion's mMessage mMachine T3 polymerase kit from XbaI linearized etsrp-T3TS as previously described [19], and harvested at ∼80% epiboly in pools of 70 embryos per group. The transcription profile of uninjected transgenic embryos was used as controls. Total RNA was isolated with Trizol reagent (Invitrogen), according to manufacturer's instructions. The detailed protocol for microarray hybridization and normalization procedures were described by Horak et al [65] and are available upon request. First-strand cDNA probes were generated by aminoallyl dUTP incorporation and then coupled to either Cy3 or Cy5. The resulting cDNA probes were purified, concentrated, and then hybridized to arrays. Each chip contains 34,647 printed oligo elements (Compugen, Operon, and MWG) designed from zebrafish EST assemblies and representing approximately 20,000 genes, approximately 60% of the total predicted genes according to the public Ensembl database. After hybridization, the slides were washed, dried, and scanned using an Agilent DNA microarray scanner (Agilent Technologies) at 635 nm (Cy5) and then at 532 nm (Cy3). Fluorescence intensities were quantified using Agilent feature extraction software (Agilent Technologies), and changes in expression levels were determined by comparing the signal intensities between etsrp OE and uninjected control groups. Hybridizations were performed four times, switching the fluorescent labeling to eliminate biases caused by the labeling process. Samples were normalized using the Lowess calculations and cutoffs for significance were set at a two-fold change in either direction. Oligo sequences were mapped to multiple databases, including RefSeq, UniGene, Ensembl, TIGR, and genomic coordinates to maximally determine gene identity and function. Data were deposited into searchable FilemakerPro and Excel databases for analysis.
Quantitative RT-PCR
Total RNA was purified with Trizol reagent (Invitrogen) from equal numbers of embryos between experimental groups, and single strand cDNA was synthesized with Superscript II reverse transcriptase (Invitrogen). Real-time PCR was performed using the iCycler iQ Real-Time PCR Detection System (BioRad, Hercules, CA) with iQ SYBR Green Supermix (BioRad). Gene expression levels were measured by the ΔΔ Ct method, comparing etsrp OE or H2B-mRFP injected groups relative to uninjected controls, with β-actin used as the reference gene. Pre-microarray validation was performed from the same samples used for the microarray experiment, and biological triplicates were examined for the injection control experiment reported in Figure S1. Quantitative RT-PCR primers are listed in Table S1.
Gene cloning and RNA whole mount in situ hybridization (WISH)
Genes were cloned into the pCRII Topo vector using the TOPO TA cloning kit (dual promoter) according to manufacturer's instructions (Invitrogen), after amplifying genes of interest from a 24 hpf wild type embryo cDNA library. The primers used are noted in Table S2. Positive clones were analyzed by restriction digest and confirmed by sequencing. WISH was performed as described in [66]. DIG labeled RNA probes were generated by linearizing TOPO-cloned genes with restriction endonuclueases (New England Biolabs), and transcribing with SP6 or T7 RNA polymerase (Promega).
Embryo injections
Injections were performed with borosilicate glass microcapillaries (Harvard apparatus) that were pulled with a flaming/brown micropipette puller, model P-97 (Sutter Instrument Co) on a PLI-90 microinjector (Harvard apparatus). For microarray validation by WISH, an etsrp-mcherry construct was generated by sub-cloning mcherry from mcherry-tol2 (a gift from Hong Zhang) into pSPORT6 between XbaI and XhoI, and etsrp was subsequently subcloned into this vector at the 5′end of mcherry between BamHI and XbaI sites. To test the requirement of etsrp for the expression of the blood/vascular related genes a mixture of etsrp morpholinos, MO1 and MO2 (4 ng each) described in [19] was injected into embryos at 1–2 cell stage, and harvested for WISH at 24–28 hpf. Translation blocking morpholinos targeting the first codon of hapln1b and sh3gl3 were synthesized by Gene-Tools, LLC. Hapln1b MO sequence: 5′-GAGCAGGAAGGTCATCTTGATTTAT-3′; sh3gl3 MO sequence: 5′-TTAAACCCGGCCACTGACATCCTTC-3′; Standard control MO sequence: 5′-CCTCTTACCTCAGTTACAATTTATA-3′.
Image acquisition and processing
In situ stained embryos were further processed by a serial dehydration in ethanol, followed by rehydration into 1×PBS. Embryos were imaged either in 1×PBS on agarose coated dishes, or in 2% methylcellulose in depression microscope slides. Images were captured with a color digital CCD camera (Axiocam, Zeiss) mounted on a dissecting microscope (Stemi SV11, Zeiss) with Openlab 4.0.2 software (Improvision, Lexington, MA). Fluorescence images were obtained with the same equipment but on an Axioskop2 plus compound microscope with 10× and 20× objections. In several cases separate images were captured per embryo and areas in focus were compiled with Adobe Photoshop CS version 8.
Supporting Information
Quantitative RT-PCR of etsrp overexpression and H2B-mRFP injection controls for selected genes. Relative gene expression levels were calculated for etsrp or H2B-mRFP1 expressing as compared to uninjected control embryos at 80 percent epiboly to gastrulation stages for the genes indicated on the x-axis. The induction of endogenous etsrp (3′UTR amplicon from injections encoding etsrp lacking the 3′UTR), fli1a, scl, yrk, and atxr1/tem8 are statistically significant (p<0.05, Student's t-test). Although slightly increased hapln1b and sh3gl3 are not significantly different from controls.
(2.50 MB TIF)
Genes induced by etsrp overexpression but not expressed in blood or vessels. (A) arhgef9; (B) pchdg; (C) novel2; (D) LOC555375 (expression is in ventral somites not vasculature); (E) cbe2; (F) smoc2; (G) LOC550501; (H) similar to testican; (I) hoxb8b; (J) tex2; (K) c1orf192; (L) c20.orf112; (M) znf385; (N) apolipoB; (O) ptgs1; and (P) hoxc3a. Scale bar: 250 um.
(6.74 MB TIF)
Quantitative RT-PCR primers
(0.03 MB DOC)
Primers used to clone in situ hybridization probes
(0.09 MB DOC)
Acknowledgments
We thank Hong Jiang and Anqi Liu for assistance with in situ hybridization and animal husbandry.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: HIN. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ciau-Uitz A, Walmsley M, Patient R. Distinct origins of adult and embryonic blood in Xenopus. Cell. 2000;102:787–796. doi: 10.1016/s0092-8674(00)00067-2. [DOI] [PubMed] [Google Scholar]
- 2.Eichmann A, Corbel C, Nataf V, Vaigot P, Breant C, et al. Ligand-dependent development of the endothelial and hemopoietic lineages from embryonic mesodermal cells expressing vascular endothelial growth factor receptor 2. Proc Natl Acad Sci U S A. 1997;94:5141–5146. doi: 10.1073/pnas.94.10.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmidt A, Brixius K, Bloch W. Endothelial precursor cell migration during vasculogenesis. Circ Res. 2007;101:125–136. doi: 10.1161/CIRCRESAHA.107.148932. [DOI] [PubMed] [Google Scholar]
- 4.Shalaby F, Rossant J, Yamaguchi TP, Gertsenstein M, Wu XF, et al. Failure of blood-island formation and vasculogenesis in Flk-1-deficient mice. Nature. 1995;376:62–66. doi: 10.1038/376062a0. [DOI] [PubMed] [Google Scholar]
- 5.Walmsley M, Ciau-Uitz A, Patient R. Adult and embryonic blood and endothelium derive from distinct precursor populations which are differentially programmed by BMP in Xenopus. Development. 2002;129:5683–5695. doi: 10.1242/dev.00169. [DOI] [PubMed] [Google Scholar]
- 6.Nishikawa SI, Nishikawa S, Hirashima M, Matsuyoshi N, Kodama H. Progressive lineage analysis by cell sorting and culture identifies FLK1+VE-cadherin+ cells at a diverging point of endothelial and hemopoietic lineages. Development. 1998;125:1747–1757. doi: 10.1242/dev.125.9.1747. [DOI] [PubMed] [Google Scholar]
- 7.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 8.Palis J, Robertson S, Kennedy M, Wall C, Keller G. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development. 1999;126:5073–5084. doi: 10.1242/dev.126.22.5073. [DOI] [PubMed] [Google Scholar]
- 9.Dieterlen-Lievre F, Le Douarin NM. From the hemangioblast to self-tolerance: a series of innovations gained from studies on the avian embryo. Mech Dev. 2004;121:1117–1128. doi: 10.1016/j.mod.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 10.Vogeli KM, Jin SW, Martin GR, Stainier DY. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature. 2006;443:337–339. doi: 10.1038/nature05045. [DOI] [PubMed] [Google Scholar]
- 11.Stainier DY, Weinstein BM, Detrich Z, Li F, Mc. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- 12.Qian F, Zhen F, Ong C, Jin SW, Meng Soo H, et al. Microarray analysis of zebrafish cloche mutant using amplified cDNA and identification of potential downstream target genes. Dev Dyn. 2005;233:1163–1172. doi: 10.1002/dvdy.20444. [DOI] [PubMed] [Google Scholar]
- 13.Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106:534–541. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber GJ, Choe SE, Dooley KA, Paffett-Lugassy NN, Zhou Y, et al. Mutant-specific gene programs in the zebrafish. Blood. 2005;106:521–530. doi: 10.1182/blood-2004-11-4541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pham VN, Lawson ND, Mugford JW, Dye L, Castranova D, et al. Combinatorial function of ETS transcription factors in the developing vasculature. Dev Biol. 2007;303:772–783. doi: 10.1016/j.ydbio.2006.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sumanas S, Gomez G, Zhao Y, Park C, Choi K, et al. Interplay between Etsrp/ER71, scl and alk8 signaling controls endothelial and myeloid cell formation. Blood. 2008;111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu F, Patient R. Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ Res. 2008;103:1147–1154. doi: 10.1161/CIRCRESAHA.108.179713. [DOI] [PubMed] [Google Scholar]
- 18.Lee D, Park C, Lee H, Lugus JJ, Kim SH, et al. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell. 2008;2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sumanas S, Lin S. Ets1-related protein is a key regulator of vasculogenesis in zebrafish. PLoS Biol. 2006;4:e10. doi: 10.1371/journal.pbio.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin SW, Beis D, Mitchell T, Chen JN, Stainier DY. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- 21.Liao W, Bisgrove BW, Sawyer H, Hug B, Bell B, et al. The zebrafish gene cloche acts upstream of a flk-1 homologue to regulate endothelial cell differentiation. Development. 1997;124:381–389. doi: 10.1242/dev.124.2.381. [DOI] [PubMed] [Google Scholar]
- 22.Coolen M, Sii-Felice K, Bronchain O, Mazabraud A, Bourrat F, et al. Phylogenomic analysis and expression patterns of large Maf genes in Xenopus tropicalis provide new insights into the functional evolution of the gene family in osteichthyans. Dev Genes Evol. 2005;215:327–339. doi: 10.1007/s00427-005-0476-y. [DOI] [PubMed] [Google Scholar]
- 23.Moens CB, Cordes SP, Giorgianni MW, Barsh GS, Kimmel CB. Equivalence in the genetic control of hindbrain segmentation in fish and mouse. Development. 1998;125:381–391. doi: 10.1242/dev.125.3.381. [DOI] [PubMed] [Google Scholar]
- 24.Schvarzstein M, Kirn A, Haffter P, Cordes SP. Expression of Zkrml2, a homologue of the Krml1/val segmentation gene, during embryonic patterning of the zebrafish (Danio rerio). Mech Dev. 1999;80:223–226. doi: 10.1016/s0925-4773(98)00220-2. [DOI] [PubMed] [Google Scholar]
- 25.Eichmann A, Grapin-Botton A, Kelly L, Graf T, Le Douarin NM, et al. The expression pattern of the mafB/kr gene in birds and mice reveals that the kreisler phenotype does not represent a null mutant. Mech Dev. 1997;65:111–122. doi: 10.1016/s0925-4773(97)00063-4. [DOI] [PubMed] [Google Scholar]
- 26.Sieweke MH, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 27.Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- 28.Meijer AH, Gabby Krens SF, Medina Rodriguez IA, He S, Bitter W, et al. Expression analysis of the Toll-like receptor and TIR domain adaptor families of zebrafish. Mol Immunol. 2004;40:773–783. doi: 10.1016/j.molimm.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 29.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 30.Kopp EB, Medzhitov R. The Toll-receptor family and control of innate immunity. Curr Opin Immunol. 1999;11:13–18. doi: 10.1016/s0952-7915(99)80003-x. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Tao X, Shen B, Horng T, Medzhitov R, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 32.Rebhun JF, Castro AF, Quilliam LA. Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. J Biol Chem. 2000;275:34901–34908. doi: 10.1074/jbc.M005327200. [DOI] [PubMed] [Google Scholar]
- 33.Roberts DM, Anderson AL, Hidaka M, Swetenburg RL, Patterson C, et al. A vascular gene trap screen defines RasGRP3 as an angiogenesis-regulated gene required for the endothelial response to phorbol esters. Mol Cell Biol. 2004;24:10515–10528. doi: 10.1128/MCB.24.24.10515-10528.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aiba Y, Oh-hora M, Kiyonaka S, Kimura Y, Hijikata A, et al. Activation of RasGRP3 by phosphorylation of Thr-133 is required for B cell receptor-mediated Ras activation. Proc Natl Acad Sci U S A. 2004;101:16612–16617. doi: 10.1073/pnas.0407468101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St Croix B, Rago C, Velculescu V, Traverso G, Romans KE, et al. Genes expressed in human tumor endothelium. Science. 2000;289:1197–1202. doi: 10.1126/science.289.5482.1197. [DOI] [PubMed] [Google Scholar]
- 36.Scobie HM, Rainey GJ, Bradley KA, Young JA. Human capillary morphogenesis protein 2 functions as an anthrax toxin receptor. Proc Natl Acad Sci U S A. 2003;100:5170–5174. doi: 10.1073/pnas.0431098100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bradley KA, Mogridge J, Mourez M, Collier RJ, Young JA. Identification of the cellular receptor for anthrax toxin. Nature. 2001;414:225–229. doi: 10.1038/n35101999. [DOI] [PubMed] [Google Scholar]
- 38.Bolcome S, Se Z, R B ApC, Rj C. Anthrax lethal toxin induces cell death-independent permeability in zebrafish vasculature. Proc Natl Acad Sci U S A. 2008;105:2439–2444. doi: 10.1073/pnas.0712195105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Werner E, Kowalczyk AP, Faundez V. Anthrax toxin receptor 1/tumor endothelium marker 8 mediates cell spreading by coupling extracellular ligands to the actin cytoskeleton. J Biol Chem. 2006;281:23227–23236. doi: 10.1074/jbc.M603676200. [DOI] [PubMed] [Google Scholar]
- 40.Hotchkiss KA, Basile CM, Spring SC, Bonuccelli G, Lisanti MP, et al. TEM8 expression stimulates endothelial cell adhesion and migration by regulating cell-matrix interactions on collagen. Exp Cell Res. 2005;305:133–144. doi: 10.1016/j.yexcr.2004.12.025. [DOI] [PubMed] [Google Scholar]
- 41.Saras J, Franzen P, Aspenstrom P, Hellman U, Gonez LJ, et al. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. J Biol Chem. 1997;272:24333–24338. doi: 10.1074/jbc.272.39.24333. [DOI] [PubMed] [Google Scholar]
- 42.Miller RA, Christoforou N, Pevsner J, McCallion AS, Gearhart JD. Efficient array-based identification of novel cardiac genes through differentiation of mouse ESCs. PLoS ONE. 2008;3:e2176. doi: 10.1371/journal.pone.0002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall A. Rho GTPases and the control of cell behaviour. Biochem Soc Trans. 2005;33:891–895. doi: 10.1042/BST20050891. [DOI] [PubMed] [Google Scholar]
- 44.Zheng Y, Fischer DJ, Santos MF, Tigyi G, Pasteris NG, et al. The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J Biol Chem. 1996;271:33169–33172. doi: 10.1074/jbc.271.52.33169. [DOI] [PubMed] [Google Scholar]
- 45.Narumiya H, Hidaka K, Shirai M, Terami H, Aburatani H, et al. Endocardiogenesis in embryoid bodies: novel markers identified by gene expression profiling. Biochem Biophys Res Commun. 2007;357:896–902. doi: 10.1016/j.bbrc.2007.04.030. [DOI] [PubMed] [Google Scholar]
- 46.Martins-Green M, Bixby JL, Yamamoto T, Graf T, Sudol M. Tissue specific expression of Yrk kinase: implications for differentiation and inflammation. Int J Biochem Cell Biol. 2000;32:351–364. doi: 10.1016/s1357-2725(99)00118-1. [DOI] [PubMed] [Google Scholar]
- 47.Vogel BE, Hedgecock EM. Hemicentin, a conserved extracellular member of the immunoglobulin superfamily, organizes epithelial and other cell attachments into oriented line-shaped junctions. Development. 2001;128:883–894. doi: 10.1242/dev.128.6.883. [DOI] [PubMed] [Google Scholar]
- 48.Schultz DW, Klein ML, Humpert AJ, Luzier CW, Persun V, et al. Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet. 2003;12:3315–3323. doi: 10.1093/hmg/ddg348. [DOI] [PubMed] [Google Scholar]
- 49.Xu X, Dong C, Vogel BE. Hemicentins assemble on diverse epithelia in the mouse. J Histochem Cytochem. 2007;55:119–126. doi: 10.1369/jhc.6A6975.2006. [DOI] [PubMed] [Google Scholar]
- 50.Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, et al. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- 51.Giachino C, Lantelme E, Lanzetti L, Saccone S, Bella Valle G, et al. A novel SH3-containing human gene family preferentially expressed in the central nervous system. Genomics. 1997;41:427–434. doi: 10.1006/geno.1997.4645. [DOI] [PubMed] [Google Scholar]
- 52.So CW, Sham MH, Chew SL, Cheung N, So CK, et al. Expression and protein-binding studies of the EEN gene family, new interacting partners for dynamin, synaptojanin and huntingtin proteins. Biochem J. 2000;348 Pt 2:447–458. [PMC free article] [PubMed] [Google Scholar]
- 53.Yang S, Toy K, Ingle G, Zlot C, Williams PM, et al. Vascular endothelial growth factor-induced genes in human umbilical vein endothelial cells: relative roles of KDR and Flt-1 receptors. Arterioscler Thromb Vasc Biol. 2002;22:1797–1803. doi: 10.1161/01.atv.0000038995.31179.24. [DOI] [PubMed] [Google Scholar]
- 54.Toyama R, Kobayashi M, Tomita T, Dawid IB. Expression of LIM-domain binding protein (ldb) genes during zebrafish embryogenesis. Mech Dev. 1998;71:197–200. doi: 10.1016/s0925-4773(97)00202-5. [DOI] [PubMed] [Google Scholar]
- 55.Spicer AP, Joo A, Bowling A hyaluronan binding link protein gene family whose members are physically linked adjacent to chondroitin sulfate proteoglycan core protein genes: the missing links. J Biol Chem. 2003;278:21083–21091. doi: 10.1074/jbc.M213100200. [DOI] [PubMed] [Google Scholar]
- 56.Watanabe H, Yamada Y. Mice lacking link protein develop dwarfism and craniofacial abnormalities. Nat Genet. 1999;21:225–229. doi: 10.1038/6016. [DOI] [PubMed] [Google Scholar]
- 57.Kudoh T, Tsang M, Hukriede NA, Chen X, Dedekian M, et al. A gene expression screen in zebrafish embryogenesis. Genome Res. 2001;11:1979–1987. doi: 10.1101/gr.209601. [DOI] [PubMed] [Google Scholar]
- 58.Woods IG, Kelly PD, Chu F, Ngo-Hazelett P, Yan YL, et al. A comparative map of the zebrafish genome. Genome Res. 2000;10:1903–1914. doi: 10.1101/gr.10.12.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Manzanares M, Cordes S, Kwan CT, Sham MH, Barsh GS, et al. Segmental regulation of Hoxb-3 by kreisler. Nature. 1997;387:191–195. doi: 10.1038/387191a0. [DOI] [PubMed] [Google Scholar]
- 60.Gering M, Rodaway AR, Gottgens B, Patient RK, Green AR. The SCL gene specifies haemangioblast development from early mesoderm. EMBO J. 1998;17:4029–4045. doi: 10.1093/emboj/17.14.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu F, Walmsley M, Rodaway A, Patient R. Fli1 acts at the top of the transcriptional network driving blood and endothelial development. Curr Biol. 2008;18:1234–1240. doi: 10.1016/j.cub.2008.07.048. [DOI] [PubMed] [Google Scholar]
- 62.Ringstad N, Gad H, Low P, Di Paolo G, Brodin L, et al. Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron. 1999;24:143–154. doi: 10.1016/s0896-6273(00)80828-4. [DOI] [PubMed] [Google Scholar]
- 63.Kim YH, Hu H, Guevara-Gallardo S, Lam MT, Fong SY, et al. Artery and vein size is balanced by Notch and ephrin B2/EphB4 during angiogenesis. Development. 2008;135:3755–3764. doi: 10.1242/dev.022475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Le Borgne R. Regulation of Notch signalling by endocytosis and endosomal sorting. Curr Opin Cell Biol. 2006;18:213–222. doi: 10.1016/j.ceb.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 65.Horak CE, Lee JH, Elkahloun AG, Boissan M, Dumont S, et al. Nm23-H1 suppresses tumor cell motility by down-regulating the lysophosphatidic acid receptor EDG2. Cancer Res. 2007;67:7238–7246. doi: 10.1158/0008-5472.CAN-07-0962. [DOI] [PubMed] [Google Scholar]
- 66.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat Protoc. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Quantitative RT-PCR of etsrp overexpression and H2B-mRFP injection controls for selected genes. Relative gene expression levels were calculated for etsrp or H2B-mRFP1 expressing as compared to uninjected control embryos at 80 percent epiboly to gastrulation stages for the genes indicated on the x-axis. The induction of endogenous etsrp (3′UTR amplicon from injections encoding etsrp lacking the 3′UTR), fli1a, scl, yrk, and atxr1/tem8 are statistically significant (p<0.05, Student's t-test). Although slightly increased hapln1b and sh3gl3 are not significantly different from controls.
(2.50 MB TIF)
Genes induced by etsrp overexpression but not expressed in blood or vessels. (A) arhgef9; (B) pchdg; (C) novel2; (D) LOC555375 (expression is in ventral somites not vasculature); (E) cbe2; (F) smoc2; (G) LOC550501; (H) similar to testican; (I) hoxb8b; (J) tex2; (K) c1orf192; (L) c20.orf112; (M) znf385; (N) apolipoB; (O) ptgs1; and (P) hoxc3a. Scale bar: 250 um.
(6.74 MB TIF)
Quantitative RT-PCR primers
(0.03 MB DOC)
Primers used to clone in situ hybridization probes
(0.09 MB DOC)