Abstract
The interest in exploring the role of epigenetics in the aging process has grown tremendously in recent years as demonstrated, in part, by the steadily increasing number of papers that have been published in the area. In addition, there has been and continues to be rapid improvement in the technologies needed to do the work. However, significant challenges remain, not the least of which is inherent to the aging process itself, that is, that even given a uniform genetic background and external environment, aging is a “heterogeneous” phenomenon with variation in the expression of the aging phenotype evident both between and within individuals. Thus, there is a pressing need to find experimental approaches that recognize this reality and deal with it effectively whether it is in the choice of animal model, cell, or tissue sampling or the use of techniques capable of analyzing small samples, ideally in situ and in a longitudinal fashion. Undoubtedly, because of the complexity of the situation and what are likely to be very large data sets, bioinformatics and systems biology are also going to be needed, something discussed in detail elsewhere in the report of the meeting.
Keywords: Epigenetics, Aging
THE impact of the genome on aging is well established. It clearly sets the broad boundaries of life expectancy for a species, and it has been repeatedly shown in both animal models and humans that even mutations in single genes can markedly affect the aging process and longevity. However, genomic DNA is by no means the whole story. For example, although it is clear that longevity at the individual level has a heritable component, that component seems to account for only about 25% of an individual’s life span. Consequently, other nongenomic mechanisms must play the predominant role in determining longevity. Moreover, even given an identical genetic background, aging is a heterogeneous process with individual-to-individual (even intraindividual) variation in the onset and extent of expression of the common features of aging (eg, cataracts, graying of hair, osteopenia, sarcopenia). Thus, not only is longevity strongly affected by nongenomic mechanisms but also is the aging process itself.
In general, two kinds of nongenomic mechanisms have been identified as playing major roles in aging and longevity. One of these is the stochastic behavior observed in all biologic processes (and in nonbiologic phenomena as well). This behavior refers to the spontaneous and unpredictable variations in events that occur at all levels of biologic organization from molecular to the whole organism. For example, at the subcellular level, a stochastic event might mean small, chance differences in protein folding leading to altered protein–ligand, protein–protein, or protein–nucleic acid interaction with structural and functional consequences downstream of the altered interaction. At the cellular level, chance may result in one cell receiving greater or lesser exposure to a cytokine or growth factor than a genetically identical cell located nearby, resulting in a difference in the two cells in subsequent proliferative activity and the pathways along which they differentiate or function. Finally, in utero or postpartum, one fetus or newborn may happen to receive more complete and regular nutrition than his or her identical twin leading to differences, in the adult, in metabolic activity, and in the risk in developing obesity and type II diabetes.
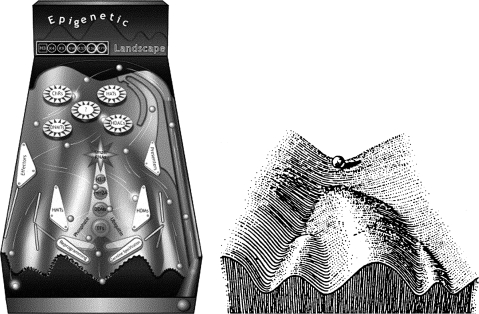
The second kind of nongenomic mechanism that almost certainly strongly affects both the aging process and the longevity is epigenesis. The concept of epigenesis has it origins in embryology where Waddington (1) represented it, graphically, as a landscape where a single channel or groove divides into several branches that descend from top to bottom of the slope (Figure 1). A multipotential cell (eg, a stem or progenitor cell) located initially at the top of the landscape differentiates along a particular developmental pathway depending upon which groove it happens to follow, the “linings” of the grooves corresponding to epigenetic signals, and events that dictate the future developmental fate of the cell. Another way of thinking about epigenesis is as a developmental cascade where earlier or concurrent events act sequentially during cell proliferation and maturation to influence the developmental destiny of the cell. At the time this original concept was developed, few epigenetic mechanisms were understood. Now, of course, many possible mechanisms have been identified resulting in a more detailed and complex rendering of the landscape with the various mechanisms shown as interacting with the “ball” (cell) to determine its fate (Figure 1) (2).
Figure 1.
This illustration shows the original (left) and a much more contemporary graphic representation (right) of the epigenetic landscape. In both, the essential idea is to depict how a cell, without changing its genome, can undergo differentiation along highly diverse pathways. The current vision of the Waddington’s epigenetic landscape proposed by Goldberg and colleagues uses the movement of pinballs to illustrate the complex and bidirectional epigenetic control of cell development and differentiation. The movement (representing different developmental stages) of the ball (the cell) in the machine depends on many epigenetic effectors (seen as flippers, obstacles, etc.) including DNA methyltransferases; histone modifiers such as acetyltranferases, deacetylases, methylases, and demethylases; and chromatin-remodeling factors. (Figures with permission of the authors (2)).
Whatever the ultimate details, epigenesis accounts for the spectrum of cell types that emerge from the zygote, and for the differentiation of stem and progenitor cells in the growing or renewing tissues of juvenile and adult organisms (eg, bone marrow, gut, and surface epithelium). On this basis, it is not surprising that alterations or deviations in the epigenetic control of the fate of proliferating cells have been linked to cancer development (where there is extensive evidence), and in aging animals, it has been proposed to account for the changing composition of blood cell populations. What is important to remember here is that epigenesis is an ongoing, lifelong process where the epigenetic state can change either as a consequence of some stochastic event or by virtue of interaction with changes in either or both the internal (eg, hormones, growth factors) or external (eg, environmental stressors, diet, oxidative agents) environment. The result is a change in gene expression and in the future form and function of the cell, tissue, or organism, or all. A few notable examples of the latter include the control of the expression of the agouti gene by methionine in the diet; the profound impact, both positive and negative, of dietary restriction on the physiology; health and risk of mortality in animal models and humans; and the effect of maternal grooming of young rat pups on the subsequent behavior of the latter as adult animals, an effect apparently mediated by changes in the DNA methylation of neurons in the central nervous system (3).
What are then the mechanisms that account for epigenesis? To date, by far most work has focused on two circumstances that can dramatically alter gene expression: DNA methylation and posttranslational modifications (eg, acetylation, methylation, phosphorylation) of proteins that are part of chromatin structure, notably histones. DNA methylation is generally associated with the silencing of gene expression. Histone modification, however, can be associated with both gene activation and repression. Indeed, the combination of modifications within the histone tails, which was defined as the histone code, determines the open–close chromatin status and thus the degree of gene activity in a certain DNA region. Globally, but with important exceptions, the level of DNA methylation goes down with age. The pioneering work by Berdyshev and colleagues (4) showed that genomic global DNA methylation decreases with age in spawning humpbacked salmon. Subsequently, others also detected a global loss of cytosine methylation during aging in mice, rats, and humans. In contrast, a number of specific loci, including ribosomal DNA and tumor suppressor genes such as the estrogen receptor and myogenic differentiation antigen 1 (MYOD1), become hypermethylated during aging.
In addition to these most extensively epigenetic mechanisms, we now recognize a number of other mechanisms that are also likely to play important roles in epigenetic regulation, especially if some latitude is given for a more direct involvement of the genome itself. These other mechanisms include telomere shortening, variation in gene copy number, the involvement of noncoding RNAs in regulating translational activity, and the positioning of transposable elements that results in changes in the transcriptional activity of adjacent genes.
MODEL SYSTEMS
Despite some evidence supporting the role of epigenetics in aging and the logic that seems to dictate that the former must influence the latter, in general there has been relatively little progress in the area. Why? It appears that there are a number of contributing factors. Part of the answer is to be found in the nature of aging itself. As previously noted, aging is a heterogeneous process characterized by variations in the expression of the a variety of phenotypes being evident both within (at the tissue level) and between individuals, even if the latter are genetically identical and are of the same age. Thus, one important question is whether there are likely to be model systems better suited for assessing the epigenetic changes associated with and perhaps responsible for at least some components of the aging process. There is also the question of which possible elements of epigenetic control should be measured and are most likely to be informative. Invertebrate multicellular organisms like Caenorhabditis elegans are appealing as models for reasons related to their relative structural and functional simplicity, short life span, and accessibility to experimental manipulation. However, as adults, nematodes are essentially postmitotic, and results obtained from them may or may not be informative about epigenetics and aging in animals (including humans) where many tissues show cell replicative behavior throughout life. In addition, C. elegans DNA does not get methylated, eliminating one of the most common epigenetic modifiers seen in higher organisms. Finally, C. elegans also exhibits marked intra- and interindividual heterogeneity as it ages, and therefore, electing to use this species does not eliminate the problem of which animals, tissues, or cells will be most representative and informative of the role of epigenesis in aging.
Arguably and unfortunately, none of the other models currently used for aging research are problem free. Consequently, there continues to be a search for better alternatives and, perhaps even more importantly, for increasing the number of species studied so that the benefits of comparative analysis can be applied. In particular, there remains considerable interest in identifying and using vertebrates with relatively short life spans, that are amenable to genetic and other kinds of experimental manipulation, and exhibit features of aging similar to those seen in other vertebrates, including humans. A possibility, already in use in a number of laboratories, is the use of some species of teleost fish can be raised and bred in aquaria and may have life spans of less than 1 year (5).
Cells in culture represent another possible option as an experimental model to explore the role of epigenesis in the aging process. Cultured cells represent a simpler situation than the whole organism or even a tissue. They can be obtained from humans and animals of different ages and genetic backgrounds, and can be examined and tested with a very large array of techniques. Most importantly, unless transformed, such cells undergo a now well-described process of replicative senescence as a function of the number of cell divisions or in response to oxidative and other stresses. However, placing cells in vitro may minimize but does not eliminate heterogeneity or the possibility that, with time in culture, the cells will change at the genomic level (eg, in ploidy). Moreover, there is always the question of whether events assessed in culture accurately represent or mimic their in vivo counterparts, although cellular senescence has been documented in vivo in some tissues. Nonetheless, even given these limitations, there are compelling arguments for focusing on in vitro models at least in the near term, perhaps especially those based on the use of cells from human donors and embryonic stem cells. For example, early-passage euploid fibroblasts from human subjects ranging in age from young to elderly adult are available from the Coriell Institute (http://ccr.coriell.org). These can be maintained in culture through multiple passages until a senescent state is achieved (or induced) and, in the process, be manipulated and assessed by a full spectrum of experimental techniques. Similarly, embryonic stem cells and committed progenitor cells can be derived from human as well as animal sources and can be stimulated to differentiate (or dedifferentiate) along a multiplicity of developmental pathways. Moreover, stem cells in particular have already been subject to a number of epigenetic studies, most directed toward understanding the nongenomic mechanisms associated with and perhaps responsible for cell-type specific differentiation, the stability of the differentiated state, and its reversibility under some circumstances.
TECHNOLOGY
Technological innovations such as gene expression microarrays and ultrahigh-throughput sequencing are transforming current research on epigenetics. These new technologies permit global assessment of the epigenome, representing the totality of epigenetic “marks” in a given sample. The importance of these developments is highlighted in a recent article that describes the establishment of an international effort to expedite the epigenomics decoding effort (6). The initial work will focus on the diagnosis, prevention, and treatment of disease. However, it is easy to anticipate that human aging will sooner rather than later be incorporated into the effort.
In contrast, given that epigenesis is a heterogeneous process (and one that ideally needs to be tracked longitudinally), the development or adaptation of techniques that allow testing at the level of an individual cell or homogeneous group of cells seems imperative. One approach, already commonly used, is to isolate cells from donors that as a class are relatively homogenous and can be fractionated to further minimize heterogeneity, for example, peripheral white blood cells. However, assessing the epigenetic status of one cell type may be informative only of that cell type and not reflect important changes that are ongoing in other tissues or the aging organism as a whole. Thus, a much needed addition to this ex vivo approach is sampling cells from a second tissue from the same donor; something that to date appears to be rarely done. However, there are possibilities for obtaining this second sample, even most importantly, from human donors. These possibilities include cheek squamous cell epithelium (easiest to obtain and the least invasive), and biopsies of skin fibroblasts and skeletal muscle. The latter may be particularly informative because relative to the other cell types, muscle cells are essentially nonreplicative postdevelopment and show well-described and dramatic changes in phenotype with age.
Another experimental strategy that has received, thus far, very little attention in epigenetics in aging research is the use of imaging techniques of living cells and whole organisms; techniques that might be adapted to provide information on the epigenetic status in situations where cell or organism manipulation is also possible. For example, Yamagata (7) very recently reviewed the use of fluorescence-labeled probes in live cell imaging to detail the epigenetic dynamics, including DNA methylation, of very early mammalian development. Earlier, Neumann and associates (8) used histone–green fluorescence protein to assess the effects of specific siRNAs (small interfering RNAs) on the expression of genes associated with chromatin structure in living HeLa cells. Chromatin structure is one of the important epigenetic determinants of the availability of a gene for transcriptional activity. Finally, there is the possibility of adapting magnetic resonance imaging and positron emission tomography scans to visualize epigenesis, at least indirectly, in small animal models of aging. Whole-animal bioluminescence imaging using luciferase activity as an indicator of sites of epigenetic change is also an exciting prospect (9).
Acknowledgments
The ideas discussed in this article are the result of discussions held during the Biology of Aging Summit held in September 2008, and the authors are indebted to the participants in the discussions, Kenneth Dorshkind, John Greally, Gerald Schatten, and Felipe Sierra. M.F.F.’s research is supported in part by the Foundation of the Spanish Association Against Cancer. A.K. is funded by National Institutes of Health-National Institute on Aging grant U19 AG032122.
References
- 1.Waddington CH. The Strategy of the Genes; a Discussion of Some Aspects of Theoretical Biology. London, UK: Allen & Unwin; 1957. [Google Scholar]
- 2.Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell. 2007;128:635–638. doi: 10.1016/j.cell.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Fraga MF, Esteller M. Epigenetics and aging: the targets and the marks. Trends Genet. 2007;23:413–418. doi: 10.1016/j.tig.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Berdyshev GD, Korotaev GK, Boiarskikh GV, Vaniushin BF. Nucleotide composition of DNA and RNA from somatic tissues of humpback salmon and its changes during spawning. Biokhimiia. 1967;32:988–993. [PubMed] [Google Scholar]
- 5.Gerhard GS. Small laboratory fish as models for aging research. Ageing Res Rev. 2007;6:64–72. doi: 10.1016/j.arr.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 6.American Association for Cancer Research Human Epigenome Task Force; European Union, Network of Excellence, Scientific Advisory Board. Moving AHEAD with an international human epigenome project. Nature. 2008;454(7205):711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yamagata K. Capturing epigenetic dynamics during pre-implantation development using live cell imaging. J. Biochem. 2008;143:279–286. doi: 10.1093/jb/mvn001. [DOI] [PubMed] [Google Scholar]
- 8.Neumann B, Held M, Liebel U, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006;3(5):385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 9.Zinn KR, Chaudhuri TR, Szafran AA, et al. Noninvasive bioluminescence imaging in small animals. ILAR J. 2008;49(1):103–115. doi: 10.1093/ilar.49.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]