Abstract
Logistic regression is often used to help make medical decisions with binary outcomes. Here we evaluate the use of several methods for selection of variables in logistic regression. We use a large dataset to predict the diagnosis of myocardial infarction in patients reporting to an emergency room with chest pain. Our results indicate that some of the examined methods are well suited for variable selection in logistic regression and that our model, and our myocardial infarction risk calculator, can be an additional tool to aid physicians in myocardial infarction diagnosis.
Keywords: logistic regression, diagnostic markers, variable selection methods, myocardial infarction
Background
Logistic regression is a statistical technique for predicting the probability of an event, given a set of predictor variables. Medical sciences use the technique commonly and, in particular, logistic regression has been applied for diagnosis in cardiology [1,2]. Variable selection is an important consideration when creating logistic regression models. Variables must be selected carefully so that the model makes accurate predictions, but without over‐fitting the data. Selecting variables by hand is a laborious task and can overlook important parameters. Thus, it is important that variable selection be automatic. In principle, given a set of variables, all possible models can be exhaustively enumerated and evaluated. The number of possible models quickly grows too large for this approach to be feasible, however. Given ’n‘ predictor variables, there are (2^n)‐1 possible models. For example, when there are above 20 variables, there are more than 1,000,000 models to check. The problem of variable selection is often addressed by sequential methods that start with a set of variables and attempt to grow or shrink the set by selecting which parameter should be added or removed from the set. This hill climbing approach has been traditionally called forward, backward and stepwise (or composite) selection [3]. The chief weakness of these techniques is that they examine only a very small sample of variable sets and stop iteration after discovering a local maximum. The vastness of the search space for variable selection has lead to heuristic approaches in addition to genetic algorithms [2], Markov Chain Monte Carlo random searches (MCMC) [4], and shrinkage techniques (e.g. LASSO) [5]. For a survey of variable selection (for linear regression, but same techniques apply to logistic regression), see [6].
Optimization techniques are often used to facilitate solution finding. Two of these methods, Particle Swarm Optimization (PSO) [7] and Simulated Annealing (SA) [8] have not been applied to variable selection for logistic regression. PSO is inspired by social psychology principles. PSO has been successfully used in a wide range of problems, some of which have been previously tackled by evolutionary techniques such as genetic algorithms. In medicine and chemistry, PSO has been used in combination with support vector machines and in chemistry also in multiple linear regression models. SA is a generic probabilistic meta‐algorithm used for locating a good approximation to the global optimum often when the search space is discrete. The SA algorithm is inspired by annealing in metallurgy, which is a technique of reducing the defects of a material by heating and controlling its cooling process.
Methodology
In this study, we implemented and evaluated the performance of these variable selection methods: binary version of PSO (BPSO) [13], the Bit Change Mutation enhancement to PSO (VBCM) [9], and finally, the SA algorithm. Additionally, we implemented a stochastic global search for variable selection (labeled ’Random‘ in Figure 1), that repeatedly generates random subsets of variables from the search space and evaluates the logistic regression models constructed with those subsets. When the terminating criterion is reached (as described below) the technique reports the best sample found so far.
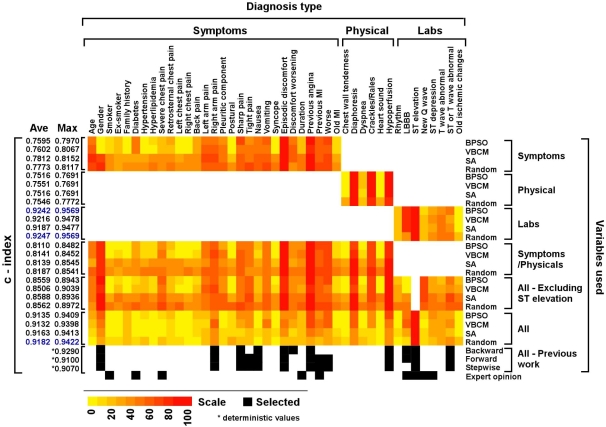
Figure 1.
Predicting risk for MI using different selection methods applied on different sets of diagnostic parameters. Parameters were subdivided into Symptoms (including medical history), physical signs (Physical) and laboratory tests (Labs) and then the c‐index was calculated using four methods; BPSO, VBCM, SA and Stochastic Search (Random) (see text). Previous modeled data [2] and experts opinion [12] are added for comparison. The c‐index for previous data/work is the result of a deterministic run (marked with an asterisk) rather than an average (Ave) of 100 runs as for the techniques we tested. Also maximum (Max) values for our runs are presented. The heatmap was created using Heatmap Builder: http://quertermous.stanford.edu/heatmap.htm. We use the heatmap color‐coded scale to range from yellow to red, which represents the weight of each parameter, meaning the number of times the parameter is selected in the winning set, out of the 100 runs. The black and white colored boxes represent the inclusion and exclusion of the parameters, respectively. The top three c‐indexes in the list are colored blue.
Discussion
Myocardial infarction (MI) is a medical condition that occurs when the blood supply to a part of the heart is interrupted. MI is a leading cause of mortality in the Western world. Severe chest pain is a frequently encountered symptom in adult health care and warrants immediate assessment due to the range of possible diagnoses including MI. We performed our experiments using data from 500 patients presented at an emergency room with chest pain in Sheffield, England [1] and then we evaluated it on an independent data set of 1253 patients from Edinburgh, Scotland [1]. The prevalence of MI in these datasets was 30% and 22%, respectively. The datasets contain only admitted patients and no misdiagnoses, and MI evaluation here might differ from other sets of evaluations. The datasets contain 43 variables (40 binary and 3 continuous) and an outcome that indicates whether the patient had MI. The datasets may not contain all laboratory test results that are available for the diagnosis of MI, for example enzyme tests. Adding such lab tests would likely improve the results. We performed split‐sample modeling using the Sheffield data, divided randomly into a training set of 335 patients and a test set of 165 patients. For each of the methods implemented in this study (BPSO, VBCM, SA and stochastic search ‐ Random), the logistic regression coefficients were calculated using the training set. Then, the (fitness) performance of the model was measured by calculating the c‐index, or the area under the Receiver Operating Characteristic (ROC) curve on the testing set [10]. To lower the risk of over‐fitting, the fitness score included a reward for parsimonious models, similarly to [2]. We ran BPSO and VBCM for 77 iterations, which resulted in 20 particles times 77 iterations = 1540 evaluations of the fitness function. We selected this iteration number to compare fairly with [2], who used 1549 evaluations. In the SA algorithm, we started with 50 subsets of variables chosen at random (with probability 0.5 of each variable included in the set) and ran each subset for 30 iterations, which resulted in 1500 evaluations. In the random search, we evaluated 1549 random subsets of the variables. In each subset, each variable was selected with probability of 0.5. The examined methods are non‐deterministic. Therefore, to reach a complete understanding of each method's performance, we repeated each experiment 100 times (each time split‐sample modeling was performed with a different random split). For each method, we collected the mean c‐index calculated on the evaluation (Edinburgh) dataset and the frequency with which each variable was selected to the final model by each of the examined selection methods.
Cardiologists diagnose MI using a combination of symptoms, medical history, physical examination signs and laboratory results. Lab results are often the easiest to obtain, especially in the emergency room, whereas extracting information about patient symptoms and the physical exam are more difficult to ascertain. Patients cannot always communicate the type of chest discomfort they are experiencing, the nature/quality of the pain, and may not relay all associated symptoms. Therefore, we saw the need to evaluate the contribution of different types of diagnosis parameters. We divided the parameters into symptoms (including medical history), physical examination results and laboratory tests, and generated models using only variables in the selected category. Additionally, we created models for a combination of symptoms (including medical history) and physical exam results (i.e., all variables other than lab results).
Our results (Figure 1) show that the mean c‐index ranged from 0.75 to 0.92 indicating that all the methods tested perform well and can accurately predict MI in this dataset (for comparison see [11]). The bit change heuristic improved BPSO only in some cases (when symptoms and/or physical signs were involved) and only marginally. Regarding the variables selected, there is substantial overlap between the methods. For example, ’Previous angina‘ and ’Previous MI‘ are selected almost every time by all methods. However, there is little overlap with the expert opinion. For example, gender is an indicator that most methods consistently select, while the expert cardiologist did not indicate this variable as a factor in MI diagnosis. In contrast, smoking is a risk factor according to the cardiologist but not to the logistic regression models. One variable, ST elevation, stands out as being selected by all the methods, every time. The c‐index of a model computed for just this variable is 0.77, suggesting that the variable is, by itself, a good predictor of the outcome. We evaluated models created without the ST elevation variable and the results show a notable decrease in performance (from around 0.91 to 0.85 c‐indexes). When performing the variable selection methods using limited inputs, we observed compensatory shifts between the different parameters. For example, in the absence of ’ST elevation‘ (a dominant parameter) the weight of ’New Q wave‘ increases substantially in all selection methods. We observe that models created without lab results and only including symptoms (including medical history) and physical examination results perform worse than when lab results are available. However, mean c‐index performance of around 0.81 for those models indicates that a predictive diagnosis can be made in the absence of laboratory results. Another interesting observation is that models created with only laboratory results perform even better than those created by considering all variables. We are aware that our model does not confound a perfect diagnostic tool for MI as the data set does not take into account clinical variations such as patients with myocarditis, aortic dissection or pericarditis, which are rare conditions that mimic aspects of MI (and sometimes even co‐occur with it).
For a complete and accurate evaluation of MI, regardless of the c‐index, symptoms, medical history, physical examination and labs are required. Based on absolute predictive values obtained in a mathematical model/technical analysis, the conclusion reached from our analyses might be that fairly accurate diagnosis can be made without lab results. However, this might not easily translate to a clinical setting. It should be noted that patients symptoms and physical examination data add value to the analysis for consistency of diagnosis. We believe our diagnosis parameter evaluator could be of use for other researchers, and thus, we set a web‐accessible MI risk calculator based on the best models we found, with and without lab results. The MI risk calculator can be found at URL http://groups.csail.mit.edu/pag/AMICalculator.
Conclusion
We evaluate the use of Binary Particle Swarm Optimization (BPSO) and the Bit Change Mutation (VBCM) heuristic, Simulated Annealing (SA), and a stochastic model (Random) for selection of variables in logistic regression. We use data from more than 1700 patients to predict the diagnosis of MI in patients reporting to an emergency room with chest pain. Our results indicate that the examined methods are well suited for variable selection in logistic regression and that our model/risk calculator can be an additional tool to aid the physician in coming to a final diagnosis of MI and could be used in conjunction with other studies and laboratory data when available.
Acknowledgments
We thank Drs. Ronen Durst and Rajesh Swaminathan for helpful suggestions and comments and Drs. Vijaya Kolachalama, Sahil Parikh and Rami Tzafriri for commenting on the manuscript.
Footnotes
Citation:Kiezunet al., Bioinformation 3(7): 311-313 (2009)
References
- 1.Kennedy R, et al. European heart journal. 1996;17:118. doi: 10.1093/oxfordjournals.eurheartj.a015035. [DOI] [PubMed] [Google Scholar]
- 2.Vinterbo S, Ohno‐Machado L. J American Medical Informatics Assoc. 1999;6:984. [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen R. Long‐Linear models and logistic regression. New York, USA: Springer; 1997. [Google Scholar]
- 4.Qian Q, Field C. Proc Conf MCMC Methods; China. 2002. [Google Scholar]
- 5.Tibshirani R. J of the Royal Statistical Society. 1996;58:267. [Google Scholar]
- 6.Miller A. Subset selection in regression. CRC Press; 2002. [Google Scholar]
- 7.Kennedy J, Eberhart R. Proc IEEE Inter Conf Neutral Networks; 1995. p. 4. [Google Scholar]
- 8.Kirkpatrick S, et al. Science. 1983;220:671. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- 9.Lee S, et al. Communications and Computer Science. 2007;90:2255. [Google Scholar]
- 10.Harrell FE, et al. JAMA. 1982;247:2543. [PubMed] [Google Scholar]
- 11.Bulgiba AM, Razaz M. Int J Cardiol. 2005;102:87. doi: 10.1016/j.ijcard.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 12.Dreiseitl S, et al. Proc AMIA Symp; 1999. p. 246. [PMC free article] [PubMed] [Google Scholar]
- 13.Kennedy J, Eberhart R. IEEE International Conference on Computational Cybernetics and Simulation; 1999. p. 246. [Google Scholar]