Abstract
We have studied sporadic Parkinson’s disease (sPD) from expression of patient mitochondrial DNA (mtDNA) in neural cells devoid of their own mtDNA, the “cybrid” model. In spite of reproducing several properties of sPD brain, it remains unclear whether sPD cybrid cells reflect more complex sPD brain bioenergetic pathophysiology. We characterized and correlated respiration of intact sPD cybrid cells with electron transport chain (ETC) protein assembly, complex I ETC gene expression and ETC protein levels in sPD brain. We also assayed expression for multiple ETC genes coded by mtDNA and nuclear DNA (nDNA) in sPD cybrid cells and brain. sPD cybrid cells have reduced levels of mtDNA genes, variable compensatory normalization of mitochondrial gene expression and show robust correlations with mitochondrial ETC gene expression in sPD brains. Relationships among ETC protein levels predict impaired complex I-mediated respiration in sPD brain. That sPD cybrid cells and sPD brain samples show very correlated regulation of nDNA and mtDNA ETC transcriptomes suggests similar bioenergetic physiologies. We propose that further insights into sPD pathogenesis will follow elucidation of mechanisms leading to reduced mtDNA gene levels in spD cybrids. This will require characterization of the abnormalities and dynamics of mtDNA changes propagated through sPD cybrids over time.
Keywords: Parkinson’s disease, mitochondria, cybrid, respiration, complex I, gene expression
Introduction
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder of adults, afflicts 1–2% of adults over 60 and is a brain-wide disorder that begins preclinically with 〈-synuclein (+) aggregates (“Lewy neurites”) in the dorsal medulla and olfactory bulb years before the advent of similar pathology in substantia nigra [1–5]. The subsequent advance of pathology into substantia nigra and death of nigral dopaminergic neurons produce the bradykinetic movement disorder that is the basis for the presenting clinical phenotype. Most but not all of PD symptomatic motor progression results from ongoing death of these nigral neurons, with later neuropsychiatric deficits likely arising from further rostral spread of Lewy neuritic changes and Lewy body formation [6, 7].
Individual pigmented nigral neurons isolated with laser capture microdissection demonstrate substantial bioenergetic deficiencies that increase with aging [8, 9] and appear worsened by PD [8]. The percentage of nigral dopamine neurons with negative histochemical staining for cytochrome oxidase (CO) activity is increased ~3-fold in PD brains [8], and these CO(−) nigral neurons have increased levels of mtDNA carrying deletions several kilobases in size [9, 10]. Although these mtDNA deletions occurred in the same general “common” area of the mitochondrial genome, each was unique in its boundaries, which tended to have direct repeat sequence homologies [9, 10]. These genetic findings implicate progressive mtDNA damage as contributing to the accelerated appearance of significant bioenergetic deficiency in PD neurons compared to that of normal aging. However, it is not yet clear whether similar mtDNA deletions will be found in other neuronal groups not as susceptible to degeneration as those in nigra.
We and others have explored the hypothesis that PD is a systemic mitochondrial disorder that is expressed clinically in postmitotic, energy intensive tissues like brain [11–13]. Mitochondria from multiple PD tissues demonstrate bioenergetic deficiencies, with a common finding being a reduction of activity of complex I in the electron transport chain [14, 15]. The complex I deficiency of catalytic activity is partially transferable through cytoplasmic fusion with mitochondrial transfer and mtDNA expression in cybrid cell models [16–18].
Multiple other abnormalities uncovered in PD cybrids, including spontaneous formation of cytoplasmic inclusions with all discernible characteristics of Lewy bodies [19], support the utility of the cybrid model for studying sporadic PD. In the present study we characterized sPD cybrid nuclear and mitochondrial gene expression profiles compared to that of sPD frontal cortex. We characterized with “high-resolution respirometry” [20] the respiration properties of sPD cybrids and examined the relationships among their respiration properties, mitochondrial ETC gene expression and complex I assembly. We also characterized the molecular genetic responses of sPD cybrids and sPD brain. Our findings define similarities among sPD cybrid lines and sPD brains, suggest future directions for investigations to increase understanding of sPD bioenergetics and support the use of sPD cybrids as translational models for elucidating sPD pathogenesis.
Materials and Methods
Detailed methods for cybrid creation, culture, differentiation into neurons, RNA isolation and analyses, microarray hybridization and analyses, real-time, quantitative PCR, Western blots and intact cell respiration measurements are provided in Supplemental Detailed Methods. Summary Methods are as follows:
Cybrid creation and neuronal differentiation
Cybrid cell lines were created by fusing platelets from individuals with idiopathic Parkinson’s disease or disease-free controls with rho0 cells created in SH-SY5Y neuroblastoma cells by long-term exposure to ethidium bromide to deplete selectively mitochondrial DNA (mtDNA). Blood samples were collected under an IRB-approved protocol, and details of cybrid creation are provided elsewhere [16, 19, 21]. Cybrid cell lines were routinely cultured in growth medium (GM), consisting of high glucose DMEM with 10% FBS, 100 µg/ml pyruvate, 50 µg/ml uridine, and antibiotic-antimycotic: 100 Units/ml penicillin G, 100µg/ml streptomycin, 0.25 µg/ml amphotericin β. Cells were fed every 2–3 days and passed once a week. Gradient-purified mitochondria were isolated from cybrid lines grown in bulk (20–26 T150 flasks) and purified on Ficoll gradients. Cybrid cells were differentiated into neurons by culturing in Neurobasal medium and adding staurosporine at 6–10 nM.
Brain samples
Slow-frozen frontal cortical ribbon samples from neuropathologically confirmed control and Parkinson’s Disease cases were obtained from the University of Virginia Brain Resource Facility that collects postmortem specimens under an IRB approved protocol. Tissue was stored in air-tight containers at −70°C from autopsy until time-of-use. The samples used were part of a larger study involving characterization of multiple aspects of mitochondrial function and ETC protein biochemistry [22]. The ages at death and postmortem intervals were not significantly different among CTL and PD cases [22].
DNA and RNA extraction and analyses
DNA and RNA were extracted using Qiagen kits and manufacturer’s instructions. RNA samples were analyzed for overall quality using automated electrophoresis with an Experion system (BioRad). All brain samples showed prominent 18S and 28S peaks and variable degrees of degradation that was represented by several small peaks. The mean +/− SD 28S/18S ratio for the 5 CTL brain samples used was 0.90 +/− 0.30 (range 0.50–1.29) and for the ten PD samples was 0.96 +/− 0.42 (range 0.52–1.35). For comparison, a commercial RNA sample (Stratagene human RNA) analyzed on our Experion system had a 28S/18S ratio of 1.31, and freshly isolated SH-SY5Y RNA had a ratio of 1.27. DNA and RNA levels were assayed with QuantIT kits.
Real-time, quantitative PCR (RT-qPCR)
Primers and TaqMan probes for mtDNA and nDNA-encoded ETC genes were designed with Beacon Designer software. All qPCR assays were carried out in an iQ5 instrument (BioRad) and used either primer pairs with SyberGreen detection or multiplex PCR with TaqMan probes. Relative gene expression was calculated with the method of Pfaffl [23].
Respiration measurements
Cybrid cells were harvested with trypsin, counted and placed into one of two chambers of an Oroboros Oxygraph II respirometer. Respiration in intact cells metabolizing glucose in DMEM media was assayed using the “high-resolution oximetry” approach [20]. All respiration experiments used the same numbers of Trypan blue-excluding live cells (typically 4.5 million cells/ml).
Cardiolipin assay
Cardiolipin is an abundant phospholipid in the inner mitochondrial membrane and was assayed as a marker for mitochondrial mass. Cell or mitochondrial samples were sonicated and mixed with Mito Tracker green and analyzed on a fluorescent plate reader. Cardiolipin external standards provided the standard curve.
Graphics and statistics
Data were graphed using Synergy KaleidaGraph and GraphPad Prism, both for Macintosh. Statistical analyses used GraphPad InStat for Macintosh. Correlation coefficients were derived from linear correlation curve fitting. Populations were compared using either t-tests for normally distributed data, or Mann-Whitney test for non-normally distributed data, as determined by InStat.
Results
PD cybrids have reduced levels of mitochondrial gene DNA’s and variable normalization of mitochondrial genes’ expression
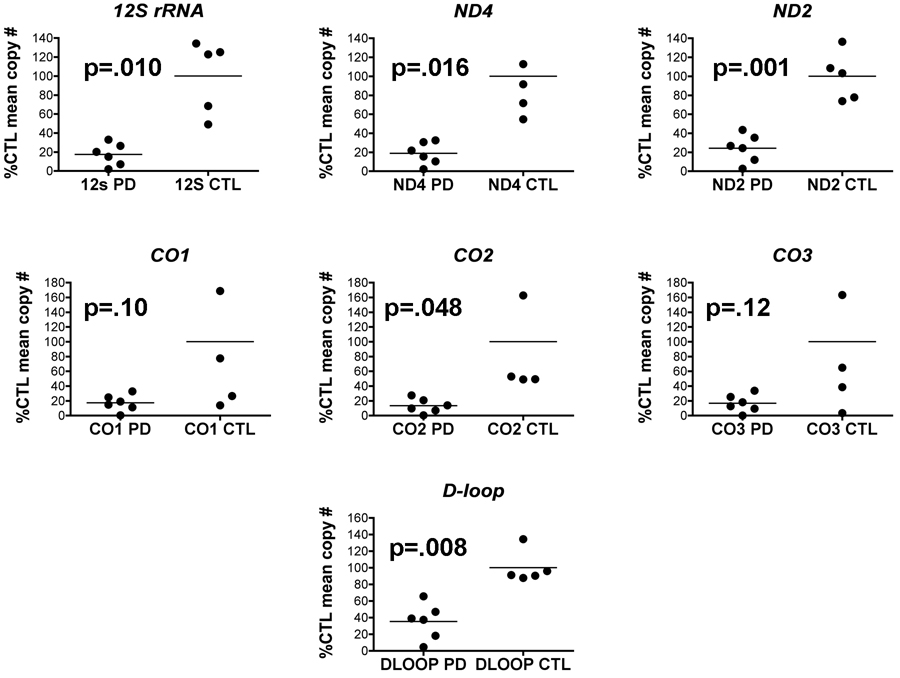
We extracted DNA from gradient purified mitochondria isolated from PD and CTL cybrids grown in bulk and assayed equal amounts of these mtDNA’s for levels of several mtDNA genes and D-loop using qPCR. When expressed as percentages of mean CTL cybrid levels, mtDNA genes (as DNA) from sPD cybrid mitochondria showed consistent several fold reductions (Figure 1). Overall the proportion of reduction was fairly constant across different genes’ DNA levels.
Figure 1.
Relative levels of mtDNA genes in sPD and CTL cybrid gradient-purified mitochondria. Mitochondria from cybrid lines were purified on Ficoll gradients, DNA extracted and analyzed with qPCR for mtDNA gene copy numbers. Data are expressed as percentages of mean CTL copy numbers for each gene. Statistical significances were determined by t-test.
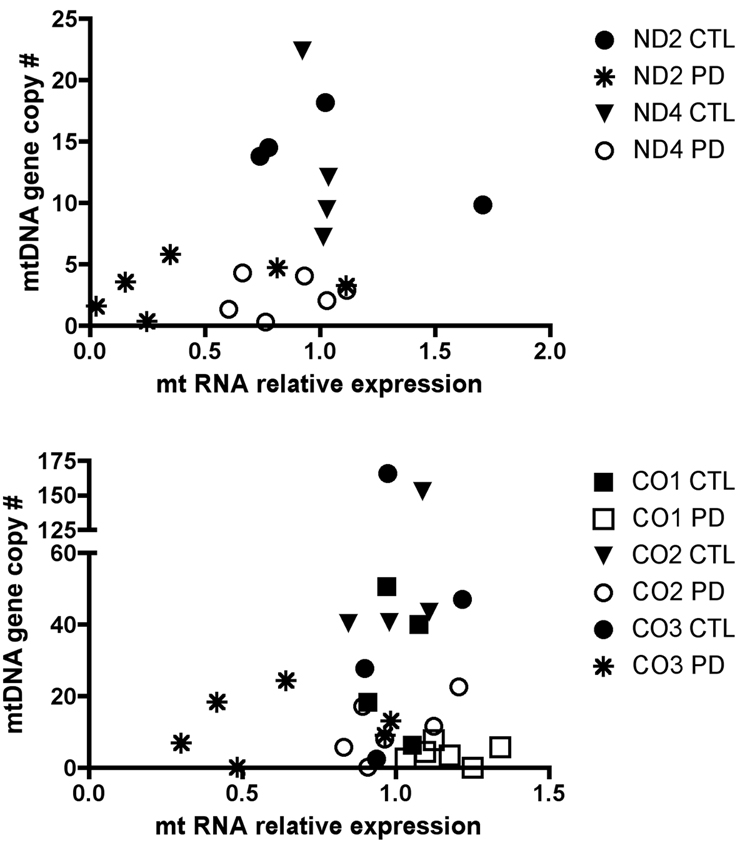
We also determined relative expression levels of these genes in cybrid cell lines’ RNA samples treated with reverse transcriptase. We followed the approach described by Pfaffl [23] to calculate gene expression relative to mean CTL cybrid levels. Figure 2 shows that for ND4, CO1 and CO2 genes, the RNA levels in sPD cybrids approximated those of CTL cybrids, even though there were substantial decreases in mtDNA levels for these genes. For ND2 and CO3, the relative RNA levels in four of the six sPD cybrids were lower than in CTL cybrids.
Figure 2.
Comparison of mtDNA copy numbers for ETC genes with relative expression levels of those mtDNA genes in sPD and CTL cybrid lines. mtDNA gene copy numbers were derived as in Figure 1 from DNA samples. Total RNA (1 ug) from each cybrid line was reversed transcribed into cDNA and analyzed with qPCR for levels of mtDNA-encoded genes. The method of Pfaffl [23] was used to calculate relative expression levels compared to the CTL cybrids.
Expression of mtDNA-encoded genes is very highly correlated in PD brain compared to PD cybrid neurons
Because the mtDNA 12S and 16S rRNA genes are independently transcribed from their own transcription initiation site and made in excess compared to the mtDNA-encoded ETC gene transcripts, we used the level of 12S rRNA expression as a normalizing factor across samples, which thus represents the averaged relative expression levels from 9 PD and 4 CTL cybrid neurons, and 10 PD and 5 CTL brain samples.
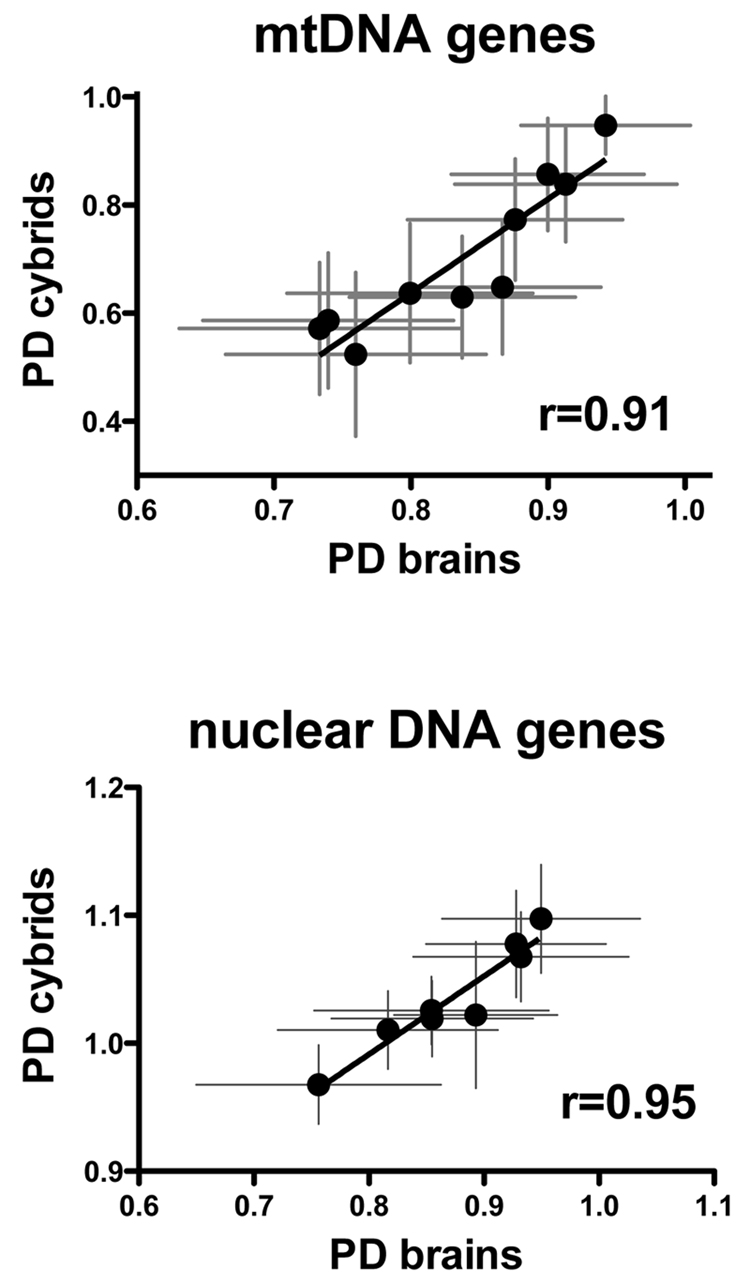
The relative expression levels of mtDNA encoded genes in PD brain and PD cybrid neurons are shown in Supplemental Spreadsheet File “ETC Expression.xls”. There were no statistically significant differences for individual mtDNA-encoded genes between PD and CTL in either group (brain or cybrid neurons). However, Figure 3 (top) shows that there was a very high correlation between averaged relative expression levels for 10 mtDNA-encoded genes in PD brain samples compared to PD cybrids, with a correlation coefficient (R) of 0.91.
Figure 3.
(top) Correlation between mean relative expression levels of 10 mtDNA-encoded genes determined by qPCR in samples from PD brain and PD cybrid neurons. (bottom) Correlation between mean relative expression levels of 8 nDNA-encoded mitochondrial ETC genes determined by qPCR in samples from PD brain and PD cybrid neurons.
Supplemental Table 1 shows that in the PD brain and cybrid neuron samples there was a high degree of cross correlation of relative expression levels of the mtDNA-encoded genes. This finding is consistent with current views about mtDNA transcription, in that the individual genes are transcribed as part of a large polycistron that is then processed into individual rRNA’s, tRNA’s or coding mRNA’s for gene translation [24]. All coding genes are believed to arise from transcription of the mtDNA heavy strand, with the exception of ND6 that is transcribed off the mtDNA light strand.
Expression of nDNA-encoded mitochondrial ETC genes is also very highly correlated in PD brain compared to PD cybrid neurons
Approximately 74 out of 87 mammalian electron transport chain (ETC) proteins are coded for by the nuclear genome, with the protein products imported into mitochondria and assembled with the 13 mtDNA-encoded ETC proteins to form a functional respiratory chain. In light of our observation above that showed such a high correlation between PD brain and PD cybrid neurons among mtDNA-encoded genes’ expression, we next examined expression levels of eight nDNA-encoded complex I and complex IV ETC genes (data in “ETC Expression.xls”). As shown in Figure 3 (bottom), there was a very high correlation (R=0.95) among averaged relative expression levels for eight nDNA-encoded ETC genes between PD cybrid neurons and PD brain samples.
Complex I-mediated respiration in cybrids is related to expression of complex I genes and levels of functionally assembled complex I
Using an Oroboros Oxygraph II respirometer and the “high-resolution respirometry” approach devised by Gnaiger [20] we studied respiration in intact sPD and CTL cybrid cells metabolizing glucose. Cybrid cells at 80–90% confluence were harvested using trypsin, and equal numbers of trypan blue-excluding live cells were suspended in glucose-DMEM, placed into the calibrated Oxygraph chambers and saturated with room air. Basal respiration rate was then measured, followed by ATP synthase inhibition with oligomycin, stepwise uncoupling with multiple FCCP injections, inhibition of maximum uncoupled respiration with rotenone to determine the complex I-mediated component, followed by additions of antimycin A/myxothiazole to determine the component mediated by complexes II/III. With this approach, the mitochondrial contribution to the complex cellular control of respiration can be evaluated with all metabolic feedback systems in place in neural cells metabolizing glucose, the usual substrate for neuronal metabolism.
We developed an assay for mitochondrial cardiolipin, an abundant phospholipid in the inner mitochondrial membrane, as a marker for relative mitochondrial mass. In comparing our sPD to CTL cybrid cells, we found no difference between the ratios of µg mitochondrial cardiolipin content to mg cell protein (CTL=57.8 +/− 13.5 (SD); sPD=60.6 +/− 11.6 (SD)), indicating that relative mitochondrial mass was the same in the sPD and CTL cybrid cells.
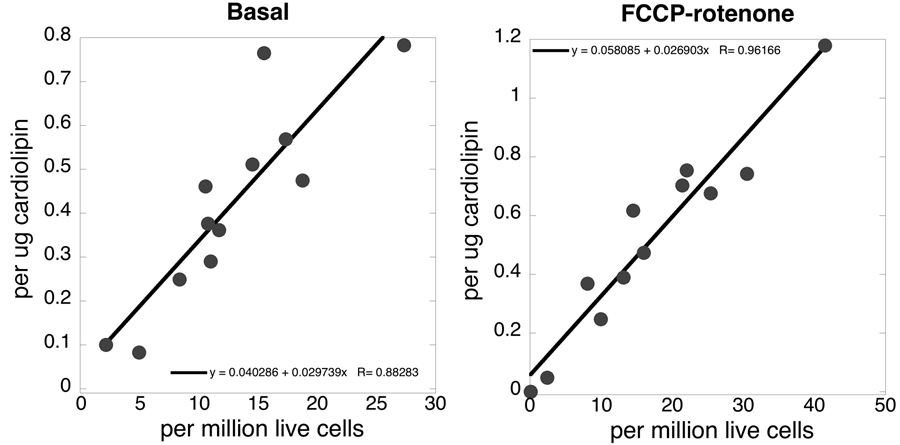
A comparison between basal and maximal complex I-mediated respiration rates normalized to either numbers of live cells or mitochondrial cardiolipin content in both sPD and CTL cybrids is shown in Figure 4. Both relationships were highly linear. The respiratory control ratios (maximum FCCP uncoupled respiration/basal respiration in presence of oligomycin) were 4.23 +/– 1.45 (SD) in CTL cybrid lines (n=5) and 3.57 +/– 1.67 in PD cybrid lines (n=7). The two groups were not significantly different from each other (P=0.55, two-tailed t test), indicating similar degrees of ETC coupling.
Figure 4.
Correlations between sPD and CTL cybrid cell oxygen consumption rates normalized to numbers of live cells compared to ug of cardiolipin present. The graph on the left shows basal respiration of intact cells metabolizing glucose (R=0.88), and that on the right shows rotenone-sensitive maximal FCCP-uncoupled respiration (R=0.96).
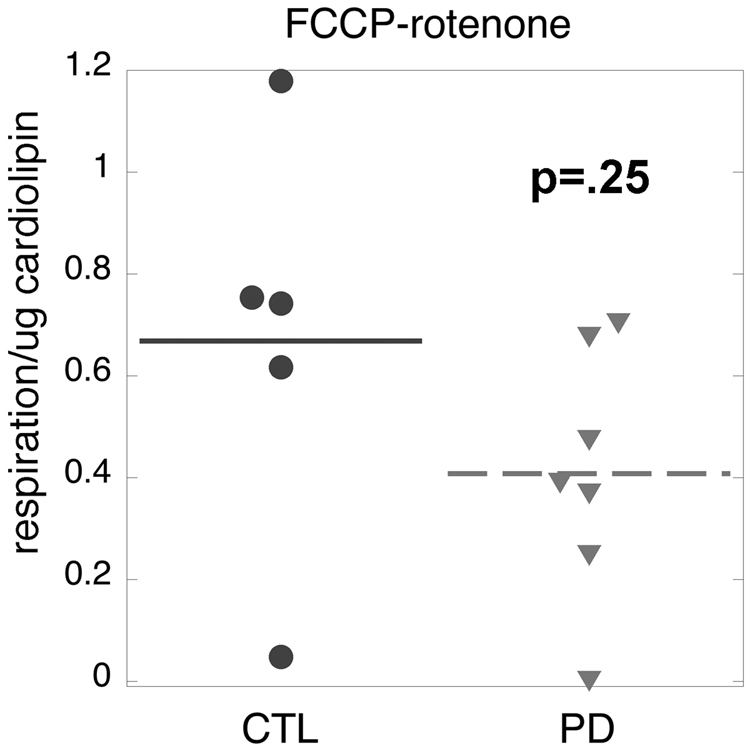
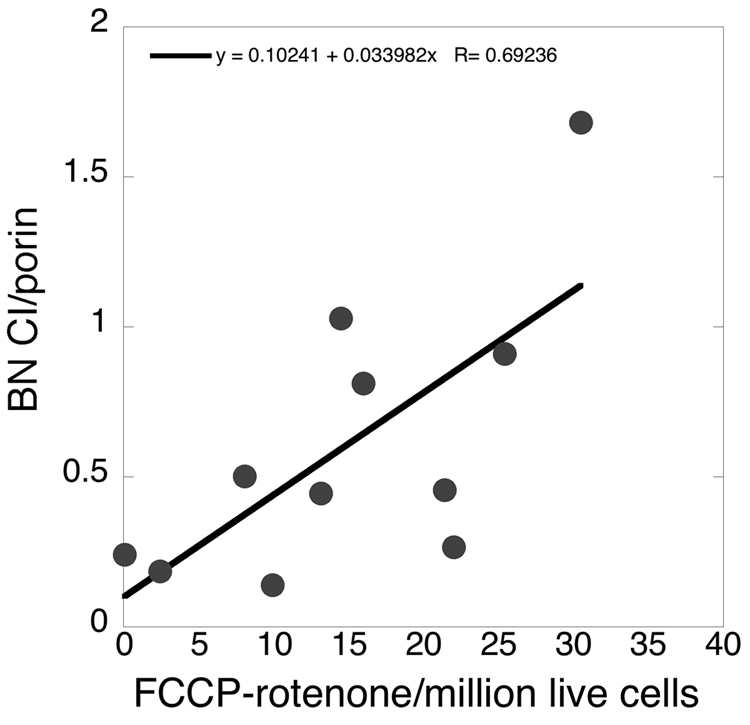
Figure 5 shows that there was a non-significant trend (p=0.24) for reduction of maximal complex I-mediated respiration in sPD compared to CTL cybrids. We then examined the relationship between maximal complex I-mediated respiration rate in intact sPD and CTL cybrid cells and the levels of fully assembled ~900 kDa complex I as assayed with blue-native (B-N) gel electrophoresis. We normalized the relative levels of B-N complex I to those of porin (a.k.a voltage dependent anion channel, VDAC), a marker for mitochondrial outer membrane. As shown in Figure 6 there was a moderately strong correlation (R=0.69) between maximal complex I-mediated respiration rates in eleven cybrid cell lines metabolizing glucose and their relative levels of fully assembled complex I.
Figure 5.
Distributions of maximal uncoupled complex I-mediated respiration in CTL and sPD cybrids. See text for details.
Figure 6.
Relationship in CTL and sPD cybrid cells between levels of the ~900 kDa complex I macroassembly assayed with blue-native gel electrophoresis and normalized to levels of the outer mitochondrial membrane protein porin, compared to maximal complex I-mediated uncoupled respiration. R=0.69.
Complex I-mediated respiration in PD brain may be reduced based on relationships among complex I gene expression and protein levels in PD brains and PD cybrids
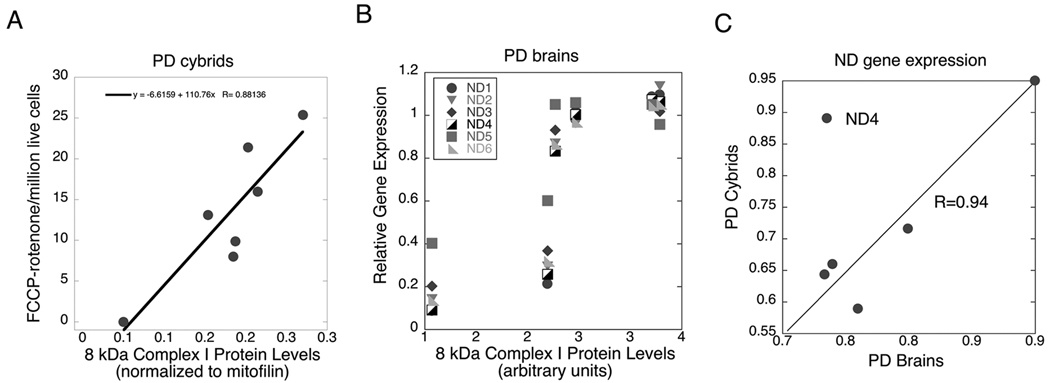
Because it is not possible to reliably measure coupled mitochondrial respiration in frozen postmortem brain specimens, we sought to develop a syllogistic argument relating complex I-mediated respiration in PD cybrids to respiration that could be predicted to occur in PD brain. The data used for this argument are shown in Figure 7. In part 7A we show that maximum complex I-mediated respiration in PD cybrids correlates well (R=0.88) with levels of the 8 kDa complex I protein that we showed previously were reduced in gradient-purified PD brain mitochondria and correlated in those mitochondria with reduced rates of NADH-driven electron flow through complex I [22]. In part 7B we show that the levels of this 8 kDa complex I protein in those sPD brain mitochondria correlate with levels of sPD brain complex I gene expression determined in the present study. The correlation coefficients range from a low of 0.78 for ND5 to a high of 0.88 for ND2. In part 7C we show that the mean relative expression levels of five out of the six mitochondrial complex I genes in sPD brains assayed in the present study correlate strongly with those found in sPD cybrids.
Figure 7.
Inter-relationships among complex I-mediated sPD cybrid respiration, complex I gene expression in sPD cybrids and sPD brains and levels of the 8 kDa complex I protein detected with the MS109 antibody from Mitosciences (see Supplemental Figure 1 and [22]). 7A shows the relationship in sPD cybrids between maximal complex I-mediated respiration and 8 kDa protein levels (normalized to mitofilin) in gradient-purified mitochondria from these cells; R=0.88). 7B shows the relationships among 8 kDa complex I protein levels in gradient-purified mitochondria from sPD brains and mtDNA-encoded complex I gene expression in the sPD brain samples; R varies from 0.78 to 0.88. 7C shows the relationship between mtDNA-encoded complex I gene relative expression in sPD cybrid cells compared to sPD brain. The outlier ND4 was not included in the regression analysis.
Discussion
In the present study we have developed the argument that cybrid cells expressing platelet mtDNA from subjects with sporadic PD are meaningful models of some molecular events in sPD brains and predict reduced complex I-mediated respiration in sPD brain. This argument is based on strong correlations among relative expression levels of genes for mitochondrial ETC proteins coded for by both the nuclear and mitochondrial genomes and inter-relationships among cybrid complex I-mediated respiration, levels of an 8 kDa complex I protein that discriminates sPD from CTL brain mitochondria and expression levels for multiple mtDNA-derived complex I genes. Our findings complement those recently described in sPD cybrids created in an NT2 host cell background [25]. In that study the authors found that sPD cybrids had reduced complex I catalytic activity, less basal ATP and greater progression into mitochondrial cell death.
In an earlier study we showed that the complex I macroassembly immunocaptured from mitochondria that had been gradient-purified from the same PD brain cases used in the present study showed increased oxidative damage and reduced rates of NADH-driven electron flow [22]. We also showed how some of the protein oxidative damage could originate within complex I. These findings are consistent with the concept that PD brain complex I proteins become oxidatively damaged more than age-matched controls through unclear mechanisms, with the end result of reduced complex I electron passage capacity that could itself contribute to additional oxidative damage.
Because coupled mitochondrial respiration cannot be reliably assayed in frozen postmortem tissue, we examined this physiological process in intact sPD cybrid cells metabolizing glucose. The “high-resolution respirometry” approach studies respiration under regulated conditions and provides understanding of respiratory coupling and control when all metabolic feedback systems are operative in intact cells [20]. This is a different situation compared to isolated mitochondria or permeabilized cells, where specific electron transport complex substrates are provided and maximal respiration under conditions of minimal (state 4) or maximal (state 3) ATP synthesis is measured. We have recently completed and will be reporting separately a study in which intact cybrid cell respiration, including its oxygen concentration-response [26], was compared to that of gradient-purified mitochondria isolated from the same cybrid cells. We observed similarities and some significant differences, including in isolated mitochondria the ability to stimulate substantial respiration through complex II that is minimally present in intact cells where the vast majority of respiration is mediated through complex I.
In the present study we related sPD cybrid maximal complex I-mediated uncoupled respiration in intact cells to levels of fully assembled complex I and to levels of 8 kDa complex I subunit in the cybrids, to PD brain through the intermediaries of levels of RNA’s (cDNA’s) for mtDNA-encoded complex I genes and their relationship to levels of the same 8 kDa complex I subunit in PD brain mitochondria. While our approach is necessarily correlational and is consistent with but cannot prove that PD brain complex I-mediated respiration is reduced, we feel the results are supportive of the argument.
Although maximal complex I catalytic activity is reduced in many PD tissues including brain, it is unclear if such reductions in catalytic activity necessarily reduce respiratory capacity or instead serve as markers for inefficient electron flow that yields increased oxidative stress. Our findings suggest that this reduced complex I catalytic activity in sPD brain likely produces respiratory compromise, at least in terms of ability to engage in oxidative phosphorylation, in addition to whatever oxidative stress also results.
We also observed that sPD cybrid cells have several-fold reductions in multiple mtDNA genes. It is unclear why this is the case, but it suggests that sPD mtDNA is not propagated as well as CTL mtDNA in genetically identical host cells. There are many potential causes for this problem that can include oxidative damage to the specific mtDNA replicating DNA polymerase-gamma, defective replication due to deletions in mtDNA and stalling of replication forks, increased oxidative damage to mtDNA, or combinations of these and other processes. An important starting point is analysis of characteristics of sPD cybrid mtDNA itself. We have begun such analyses in terms of quantifying deletions present in the entire mitochondrial genome and presence of heteroplasmic mutations and will be reporting those findings separately (Quigley, et al, unpublished data).
In spite of reductions in mtDNA gene levels, sPD cybrids were able to normalize (to CTL levels) many but not all of their RNA levels for these mtDNA genes. Of the five mtDNA genes we examined, three (ND4, CO1, CO2) were normalized and two (ND2, CO3) were not normalized in four of the six sPD cybrid lines studied. Of these four sPD cybrid lines with the non-normalized ND2 and CO3 gene expression, three had the lowest respiration levels. Thus, reduced respiration in sPD cybrids relates to both overall loss of mtDNA genes and failure to normalize mitochondrial gene expression at the RNA level, at least for some mitochondrial genes. The mechanisms for how mtDNA gene expression is differentially regulated in these sPD cybrid lines are unclear.
In summary, we have conducted extensive analyses of the molecular genetic properties of sPD cybrids and compared those to sPD brains. We find substantial correlations of gene expression, particularly for both nuclear and mitochondrial genome-encoded electron transport chain genes, which indicate that sPD cybrids and sPD brains are regulating gene expression for mitochondrial respiratory function in very similar ways. Based on correlations with mitochondrial gene expression and complex I protein levels, respiration through complex I in intact sPD cybrid cells may relate to that of sPD brain. If true, sPD cybrids can serve as unique genetic models of sPD to assist both in unraveling how abnormalities in mtDNA originate and progress, and to screen therapeutics for their ability to enhance respiration through complex I. However, to consider properly the inherent heterogeneity of a sporadic disease, several different cell lines would need to be studied.
It is also important to note that most sPD postmortem brain tissues are derived from individuals who suffered from PD for many years and typically have advanced disease clinically and pathologically. In contrast, sPD cybrids are made from platelet mtDNA of patients typically at much earlier disease state. While the relationships between platelet and brain mtDNA’s are not yet known, at the minimum sPD cybrids represent an earlier disease stage compared to most postmortem sPD brain tissues. Given this situation, our findings that sPD cybrids and sPD brain tissues so closely co-regulate their mitochondrial bioenergetic systems is all the more remarkable.
Supplementary Material
Supplemental Figure 1. Western blot assay of complex I proteins from sPD and CTL cybrid mitochondria. The complex I proteins are identified according to the MitoSciences antibodies used. The lowest band shows SOD2. The code for samples: A-human heart mitochondria; B-SH-SY5Y; C-SH-SY5Y ρ0; D-C91; E-PD67; F-PD66; G-C68; H-C56; I-PD65; J-PD63; K-C64; L-PD61; M-PD60; N-C62; O-PD59.
Acknowledgements
This research was supported by NIH NS39788, the D. Loy Stewart Research Fund and fellowship support to K.P.M. from the Government of India.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Braak E, Sandmann-Keil D, Rub U, Gai WP, de Vos RA, Steur EN, Arai K, Braak H. alpha-synuclein immunopositive Parkinson's disease-related inclusion bodies in lower brain stem nuclei. Acta Neuropathol (Berl) 2001;101:195–201. doi: 10.1007/s004010000247. [DOI] [PubMed] [Google Scholar]
- 2.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging. 2003;24:197–211. doi: 10.1016/s0197-4580(02)00065-9. [DOI] [PubMed] [Google Scholar]
- 3.Braak H, Ghebremedhin E, Rub U, Bratzke H, Del Tredici K. Stages in the development of Parkinson's disease-related pathology. Cell Tissue Res. 2004;318:121–134. doi: 10.1007/s00441-004-0956-9. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, Sandmann-Keil D, Gai W, Braak E. Extensive axonal Lewy neurites in Parkinson's disease: a novel pathological feature revealed by alpha-synuclein immunocytochemistry. Neurosci Lett. 1999;265:67–69. doi: 10.1016/s0304-3940(99)00208-6. [DOI] [PubMed] [Google Scholar]
- 5.Del Tredici K, Rub U, De Vos RA, Bohl JR, Braak H. Where does parkinson disease pathology begin in the brain? J Neuropathol Exp Neurol. 2002;61:413–426. doi: 10.1093/jnen/61.5.413. [DOI] [PubMed] [Google Scholar]
- 6.Aarsland D, Perry R, Brown A, Larsen JP, Ballard C. Neuropathology of dementia in Parkinson's disease: a prospective, community-based study. Ann Neurol. 2005;58:773–776. doi: 10.1002/ana.20635. [DOI] [PubMed] [Google Scholar]
- 7.Bertrand E, Lechowicz W, Szpak GM, Lewandowska E, Dymecki J, Wierzba-Bobrowicz T. Limbic neuropathology in idiopathic Parkinson's disease with concomitant dementia. Folia Neuropathol. 2004;42:141–150. [PubMed] [Google Scholar]
- 8.Bender A, Krishnan KJ, Morris CM, Taylor GA, Reeve AK, Perry RH, Jaros E, Hersheson JS, Betts J, Klopstock T, Taylor RW, Turnbull DM. High levels of mitochondrial DNA deletions in substantia nigra neurons in aging and Parkinson disease. Nat Genet. 2006;38:515–517. doi: 10.1038/ng1769. [DOI] [PubMed] [Google Scholar]
- 9.Kraytsberg Y, Kudryavtseva E, McKee AC, Geula C, Kowall NW, Khrapko K. Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat Genet. 2006;38:518–520. doi: 10.1038/ng1778. [DOI] [PubMed] [Google Scholar]
- 10.Reeve AK, Krishnan KJ, Elson JL, Morris CM, Bender A, Lightowlers RN, Turnbull DM. Nature of mitochondrial DNA deletions in substantia nigra neurons. Am J Hum Genet. 2008;82:228–235. doi: 10.1016/j.ajhg.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bogaerts V, Theuns J, van Broeckhoven C. Genetic findings in Parkinson's disease and translation into treatment: a leading role for mitochondria? Genes, brain, and behavior. 2008;7:129–151. doi: 10.1111/j.1601-183X.2007.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schapira AH. Mitochondria in the aetiology and pathogenesis of Parkinson#x00027;s disease. Lancet neurology. 2008;7:97–109. doi: 10.1016/S1474-4422(07)70327-7. [DOI] [PubMed] [Google Scholar]
- 13.Thomas B, Beal MF. Parkinson#x00027;s disease. Human molecular genetics. 2007;16:R183–R194. doi: 10.1093/hmg/ddm159. Spec No. 2. [DOI] [PubMed] [Google Scholar]
- 14.Parker WD, Jr, Swerdlow RH. Mitochondrial dysfunction in idiopathic Parkinson disease. Am J Hum Genet. 1998;62:758–762. doi: 10.1086/301812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shults CW. Mitochondrial dysfunction and possible treatments in Parkinson's disease--a review. Mitochondrion. 2004;4:641–648. doi: 10.1016/j.mito.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 16.Swerdlow RH, Parks JK, Miller SW, Tuttle JB, Trimmer PA, Sheehan JP, Bennett JP, Jr, Davis RE, Parker WD., Jr Origin and functional consequences of the complex I defect in Parkinson's disease. Ann Neurol. 1996;40:663–671. doi: 10.1002/ana.410400417. [DOI] [PubMed] [Google Scholar]
- 17.Gu M, Cooper JM, Taanman JW, Schapira AH. Mitochondrial DNA transmission of the mitochondrial defect in Parkinson's disease. Ann Neurol. 1998;44:177–186. doi: 10.1002/ana.410440207. [DOI] [PubMed] [Google Scholar]
- 18.Swerdlow RH. Mitochondria in cybrids containing mtDNA from persons with mitochondriopathies. J Neurosci Res. 2007;85:3416–3428. doi: 10.1002/jnr.21167. [DOI] [PubMed] [Google Scholar]
- 19.Trimmer PA, Borland MK, Keeney PM, Bennett JP, Jr, Parker WD., Jr Parkinson's disease transgenic mitochondrial cybrids generate Lewy inclusion bodies. J Neurochem. 2004;88:800–812. doi: 10.1046/j.1471-4159.2003.02168.x. [DOI] [PubMed] [Google Scholar]
- 20.Hutter E, Unterluggauer H, Garedew A, Jansen-Durr P, Gnaiger E. High-resolution respirometry--a modern tool in aging research. Experimental gerontology. 2006;41:103–109. doi: 10.1016/j.exger.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 21.Swerdlow RH, Parks JK, Davis JN, 2nd, Cassarino DS, Trimmer PA, Currie LJ, Dougherty J, Bridges WS, Bennett JP, Jr, Wooten GF, Parker WD. Matrilineal inheritance of complex I dysfunction in a multigenerational Parkinson's disease family. Ann Neurol. 1998;44:873–881. doi: 10.1002/ana.410440605. [DOI] [PubMed] [Google Scholar]
- 22.Keeney PM, Xie J, Capaldi RA, Bennett JP., Jr Parkinson's disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Esteves AR, Domingues AF, Ferreira IL, Januario C, Swerdlow RH, Oliveira CR, Cardoso SM. Mitochondrial function in Parkinson's disease cybrids containing an nt2 neuron-like nuclear background. Mitochondrion. 2008;8:219–228. doi: 10.1016/j.mito.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Gnaiger E. Oxygen conformance of cellular respiration. A perspective of mitochondrial physiology. Adv Exp Med Biol. 2003;543:39–55. doi: 10.1007/978-1-4419-8997-0_4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Western blot assay of complex I proteins from sPD and CTL cybrid mitochondria. The complex I proteins are identified according to the MitoSciences antibodies used. The lowest band shows SOD2. The code for samples: A-human heart mitochondria; B-SH-SY5Y; C-SH-SY5Y ρ0; D-C91; E-PD67; F-PD66; G-C68; H-C56; I-PD65; J-PD63; K-C64; L-PD61; M-PD60; N-C62; O-PD59.