Summary
In many bacteria expression of lateral gene transfer and of virulence factors is controlled by cell–cell signalling systems. Molecular interactions of microbial signal molecules with their cognate receptors are not well understood. For the Enterococcus faecalis conjugative plasmid pCF10, the PrgX protein serves as a molecular switch controlling expression of conjugation and virulence genes encoded by the plasmid. The induction state of a pCF10-carrying donor cell is determined by the ratio of two signalling peptides, cCF10 pheromone and iCF10 inhibitor. Recent analysis of PrgX/cCF10 interactions suggests a mechanism for conversion to the induced state. However, the means by which iCF10 peptide antagonizes cCF10 activity is unclear, and it has been suggested that inhibitor peptides block import of pheromone peptides. We now show that both of these peptides interact with the same binding pocket of PrgX, but they differentially alter the conformation of the protein and its oligomerization state, resulting in opposing biological activities.
Introduction
Intercellular communication mediated by small molecules controls a plethora of important microbial properties, including virulence, horizontal genetic transfer ability and antibiotic production (Dunny and Leonard, 1997; Lazazzera, 2001). In one important form of microbial cell–cell communication, quorum sensing (autoinduction), all members of a population comprised of a single cell type participate in both signal production and response (Nakayama et al., 2001). The genes for signal production and signal detection are frequently linked. In contrast, the enterococcal mating pheromone systems involve communication between two cell types (donors and recipients), where production of the peptide pheromone signal is encoded by a genetic element (the chromosome) that is distinct from the donor plasmid that codes for the pheromone response (Chandler and Dunny, 2004). The pheromone response results in increased expression of conjugation genes, leading to efficient lateral transfer of the plasmid to recipient cells in close proximity. In this situation, the donor cells use the pheromone peptide to monitor population density of recipients. As pheromone-responsive plasmids spread to new host cells via conjugation there is likely selective pressure to avoid wasteful self-induction by endogenous pheromone, while enabling the newly created donor cell to detect the same pheromone signal from an exogenous source. The plasmid encodes a membrane protein that reduces production of endogenous pheromone activity, as well as an inhibitor peptide that neutralizes the remaining endogenous pheromone activity (Nakayama et al., 1994; Chandler et al., 2005a). Thus the induction state of a donor cell is not determined by the absolute concentration of pheromone but by the ratio of pheromone to inhibitor in the growth medium. We have recently identified a mechanism by which this rather complex control system is used by plasmid-containing cells not only to detect recipients, but also to induce expression of virulence during growth in the mammalian bloodstream (Chandler et al., 2005b).
Some signalling peptides are detected by two-component signal transduction systems, and others, including the enterococcal pheromones, are imported into the responder cell where they interact with cytoplasmic effector molecules (Leonard et al., 1996; Nakayama et al., 1998). Until recently there were no published data describing the molecular mechanism by which the interaction of a bacterial signalling peptide with its cognate receptor triggers a response.
The tetracycline-resistance plasmid pCF10 enables its Enterococcus faecalis host cell to execute a mating response to a heptapeptide pheromone cCF10 (LVTLVFV) produced by recipient cells (Mori et al., 1988; Dunny et al., 2001). The pheromone is imported into donor cells by the concerted action of the pCF10-encoded PrgZ peptide binding protein (Ruhfel et al., 1993) and a chromosomal oligopeptide permease (Leonard et al., 1996). The key regulatory switch in pheromone induction of pCF10 transfer is the cytoplasmic pCF10 protein PrgX (Bae et al., 2000; Kozlowicz et al., 2004). PrgX represses initiation of transcription of the major conjugation operon of pCF10 in a pheromone-sensitive fashion (Bae and Dunny, 2001; Bae et al., 2004). In the absence of exogenous cCF10, PrgX partially represses transcription from the prgQ promoter (PQ) and the transcription that does occur only extends about 400 bp from the start site. In induced cells prgQ transcription increases and some transcripts extend several kb downstream, allowing for expression of multiple conjugation proteins (Chung and Dunny, 1995).
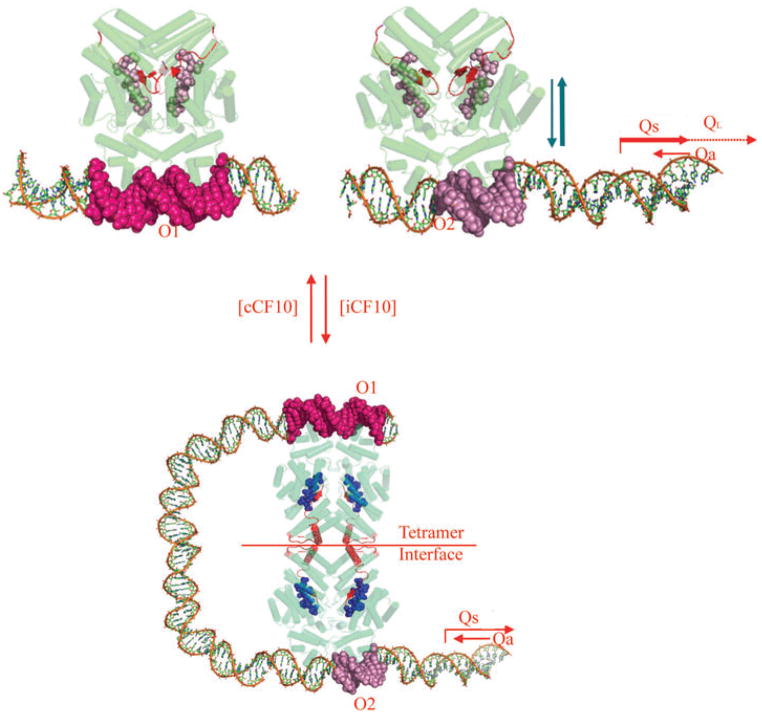
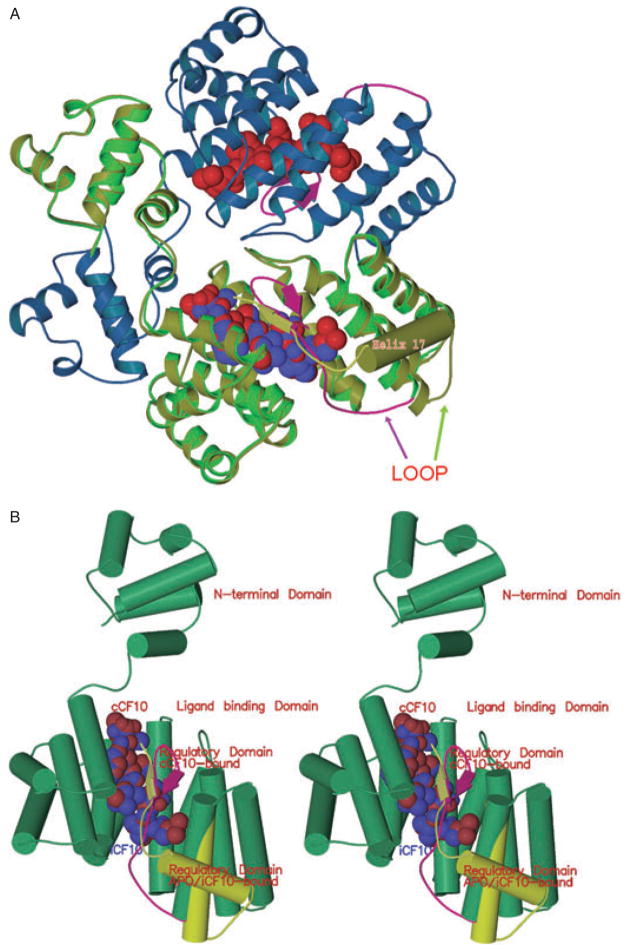
As shown in Fig. 1 (top), PrgX binds to two different DNA operators in the region between prgX and prgQ (Bae et al., 2002), one of which overlaps PQ. The current model for repression of prgQ by PrgX involves the interaction of the two binding sites via DNA looping as a result of PrgX oligomerization (Fig. 1, bottom). According to the model, the formation of a DNA loop stabilizes PrgX binding to both sites and reduces (but does not eliminate) transcription from PQ (Kozlowicz et al., 2004; Shi et al., 2005). The basal constitutive activity of PQ generates a 400 nt RNA named Qs (Fig. 1) (Chung and Dunny, 1995). Read through of prgQ transcription past a stem–loop structure (IRS1) at the 3′ end of Qs appears to be blocked by the interaction of Qs with an antisense RNA, Qa (Bae et al., 2004). In induced cells, the oligomerization state of PrgX is reduced and the DNA loop is proposed to open, ultimately destabilizing DNA binding and allowing for greater rates of transcription from PQ and more Qs RNA(Fig. 1, top). The increase in Qs RNA titrates the Qa RNA, resulting in extension of unbound prgQ transcripts into downstream regions (Bae et al., 2004). A second prgQ RNA, known as QL (Fig. 1; 530 nt), is formed after cCF10 induction (Chung and Dunny, 1995). Subsequent to QL production, downstream genes including prgB, which encodes the aggregation protein Asc10, are transcribed. Asc10 expression on the donor cell surface results in aggregation with recipients, leading to high rates of plasmid transfer (Dunny et al., 1978).
Fig. 1.
PrgX regulation of the prgQ conjugative transfer operon of plasmid pCF10. Predicted functional activity of intracellular PrgX bound to cCF10 or iCF10 are shown in the top and bottom panels respectively. PrgX binds two operator sites in pCF10 DNA. The O2 site overlaps the prgQ promoter and PrgX occupancy of this site inhibits prgQ transcription, but the binding interaction at this site is very weak unless the O1 site is also occupied.
Top. PrgX is shown in green with barrels indicating α-helices and the C-terminal region in red. Pheromone cCF10 is illustrated by red spheres. Target DNA is shown in multicolour. Pheromone binding alters the C-terminal region of PrgX such that the interaction of pairs of PrgX dimers to form tetramers is weakened. Pheromone-bound PrgX ineffectively represses PQ. The resulting high levels of the Qs transcript produced under these conditions titrates all of the Qa RNA produced from the complementary DNA strand, allowing QL production (right).
Bottom. The data presented in this paper indicate that PrgX/iCF10 complexes are comprised of the inhibitor peptide ligand (blue spheres) bound to the same PrgX cleft as cCF10, but that differences in interaction of the bound peptide with the C-terminus of PrgX actually stabilize a tetramer interface that is predicted to cause formation of a DNA loop in vivo. This loop increases occupancy of the operator sites, leading to reduced expression from PQ. The resulting lower level of Qs RNA allows it to be completely bound by the constitutively expressed Qa, resulting in termination of prgQ transcription. Even in the uninduced state, repression of PQ is not complete, and a significant basal level of Qs, which encodes iCF10, is produced.
Pheromone treatment of donor cells alters the PrgX oligomerization state in vivo (Bae and Dunny, 2001), suggesting the possibility that PrgX is the functional cCF10 receptor for mating induction. This has recently been confirmed by the determination of high-resolution structures of both PrgX and PrgX/cCF10 complex (Shi et al., 2005). In addition to providing direct evidence for cCF10-PrgX interaction, these structures also indicated that PrgX could form pheromone-sensitive tetramers, with each pair of dimers bound to a DNA target, consistent with the looping model. As noted above, pheromone responsive plasmids encode two gene products that prohibit or antagonize self-induction. One of these control elements is a protein PrgY which reduces the level of endogenous pheromone activity produced by donor cells (Chandler et al., 2005a). To neutralize the remaining endogenous pheromone escaping PrgY, the plasmid encodes an inhibitor peptide known as iCF10 (AITLIFI). The iCF10 peptide is produced from the only functional open reading frame within the Qs transcript (Nakayama et al., 1994); this likely is the reason for the significant basal level of Qs production in uninduced cells. There exist no published data demonstrating how iCF10 mediates its activity, and previous studies of analogous inhibitor peptides encoded by other pheromone plasmids suggested that the inhibitor peptides might act at the cell surface to interfere with pheromone import rather than functioning internally (Fujimoto and Clewell, 1998).
Here we describe functional analysis of the mechanism by which the cCF10-mediated pheromone response is inhibited by iCF10. We demonstrate that PrgX is the only pCF10-encoded protein whose presence in an enterococcal responder cell is both sufficient and necessary for iCF10-mediated inhibition of cCF10 activity, and that PrgX mutants deficient in repression of conjugation can be rescued by exogenous iCF10. In addition, in vivo cross-linking studies suggest that iCF10 blocks cCF10-mediated reduction in the PrgX oligomerization state. A direct molecular interaction between PrgX and iCF10 was confirmed by determination of the crystal structure of the iCF10/PrgX complex. Comparison of this structure with that of cCF10/PrgX shows that both peptides bind to a cleft in the α-helical dimerization domain, and that specific amino acid residues in the PrgX C-terminus are positioned to interact closely with residues of the peptides lying in the cleft. However, when bound, the two peptides interact with different residues in the PrgX C-terminus, significantly affecting the secondary and tertiary structures of the C-terminal loop that stabilizes tetramer formation. While cCF10 binding causes a significant structural change in which helix 17 flips 120° and refolds into a β-duplex that covers cCF10, iCF10 binding results in ordering of the C-terminal 10 residues into a β-strand over iCF10. This ordering stabilizes the tetrameric interactions between dimers. These data provide a molecular basis for differential effects of antagonistic molecules in a bacterial signalling system.
Results
iCF10 functions internally to inhibit PrgX-dependent, pheromone-inducible lacZ activity
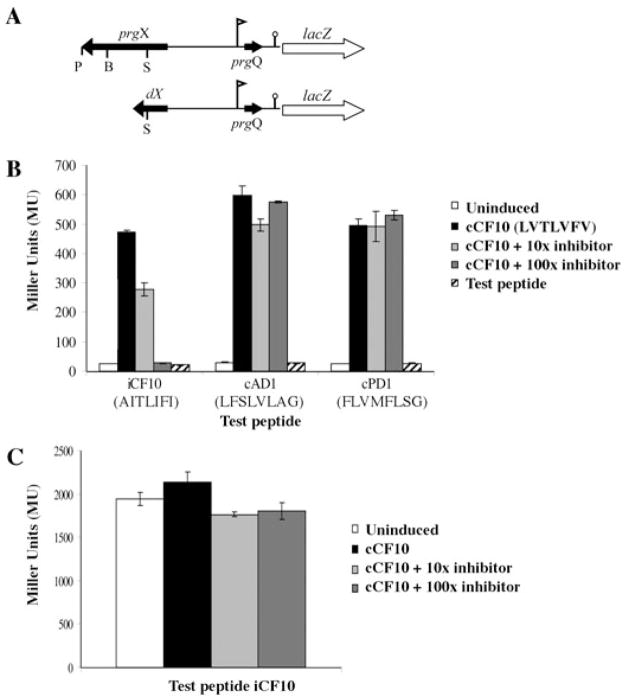
It has been demonstrated that PrgX can bind cCF10 using both affinity chromatography and crystallography techniques (Kozlowicz et al., 2004; Shi et al., 2005). In the affinity chromatography studies (Kozlowicz et al., 2004), PrgX also bound to its cognate inhibitor peptide iCF10, as well as to cAD1 and cPD1. The latter peptides are of similar size and physical properties to cCF10 and act as pheromones for other plasmids but not pCF10 (Dunny and Leonard, 1997). In order to establish whether interactions of PrgX in vivo with peptides other than cCF10 could have any biological effect that had not been detected previously, we first tested the ability of iCF10 and of the non-cognate peptides to compete with cCF10 using a strain carrying the plasmid p043lacZ (Fig. 2A). This construct contains the prgX-Q region of pCF10 with a lacZ reporter gene fused downstream of the IRS1 sequence. E. faecalis cells carrying this plasmid show PrgX-dependent, pheromone- inducible expression of lacZ (Fig. 2B) (Kozlowicz et al., 2004). Test peptides were added alone (at a concentration fivefold that used for cCF10) to test their induction capability, or along with cCF10 to test their ability to inhibit cCF10 (Fig. 2B). As seen in Fig. 2B, none of the non-cognate peptides had any inducing effect when added alone. It was also seen that cAD1 and cPD1 did not function as inhibitors when added in combination with cCF10 at 100-fold the concentration of cCF10 used. However, iCF10 partially inhibited the induction of β-galactosidase activity when added at a 10-fold excess relative to the concentration of cCF10. Induction of β-galactosidase activity by cCF10 was completely inhibited when a 100-fold molar excess of iCF10 was added to the cells immediately after cCF10 addition (Fig. 2B). The inhibitory activity of iCF10 on β-galactosidase activity from p043lacZ occurred in the absence of the cell surface cCF10 binding protein PrgZ, and therefore was unlikely due to competition between iCF10 and cCF10 for entry into the cell at the surface. Likewise, the fact that cAD1 and cPD1 showed no effect when added at the same high concentrations in combination with cCF10 indicates that the effect of iCF10 is due to an intracellular activity rather than just a blocking or clogging of the general oligopeptide import (Opp) system of the cell (Leonard et al., 1996). The control plasmid p043lacZdX has a deletion of two-thirds of the prgX sequence from the SpeI site to the 3′ end of the prgX gene (Fig. 2A), does not express PrgX protein, and confers constitutive lacZ expression (Fig. 2C) (Kozlowicz et al., 2004). As seen in Fig. 2C, inhibition by iCF10 was dependent upon PrgX; β-galactosidase activity from cells carrying p043lacZdX was not inhibited by addition of iCF10. Thus we propose that iCF10 antagonizes pheromone-inducible gene expression inside the cell via a previously undescribed interaction with PrgX. While these results did not directly indicate whether cAD1 or cPD1 can bind PrgX in vivo, they did show that there were no significant interactions between either of these peptides and PrgX that impacted prgQ regulation.
Fig. 2. Inhibition of pheromone-inducible transcription from the prgQ promoter by iCF10 requires PrgX.
A. Schematic drawing of pheromone-inducible p043lacZ reporter construct (top) and the p043dX derivative lacking a functional prgX gene (bottom). Flag indicates the direction and location of PQ, lollipop indicates IRS1 sequence where prgQ transcription terminates in uninduced cells, genes are indicated by arrows showing approximate size and direction of transcription. P = PstI, B = BglII, S = SpeI restriction enzyme cleavage sites.
B. Effects of addition of various combinations of cCF10 and other peptides on prgQ expression in OG1RF(p043lacZ). Cells were exposed to various combinations of cCF10 and either iCF10, cAD1 or cPD1 at 10-fold and 100-fold molar excess. iCF10, cAD1 and cPD1 were also tested in the absence of cCF10 addition for their ability to act as inducers in this system; the amino acid sequence of each peptide is shown. cCF10 = cCF10 at 5 ng ml−1, 10X inhibitor = cCF10 at 5 ng ml−1 + test peptide at 50 ng ml−1, 100X inhibitor = cCF10 at 5 ng ml−1 + test peptide at 500 ng ml−1, Test peptide = test peptide at 25 ng ml−1, no cCF10.
C. Effects of various combinations of iCF10 and cCF10 on prgQ transcription in OG1RF containing p043lacZ versus p043dX. Same amounts of cCF10 and iCF10 as in (B); cAD1 and cPD1 not used as test peptides in this experiment.
Addition of iCF10 to culture medium rescues prgX regulatory mutations
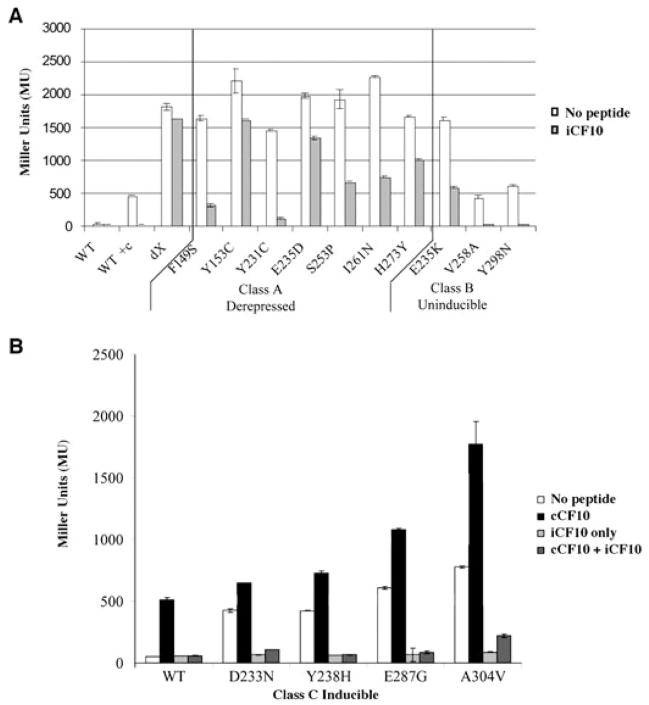
Several prgX mutations were previously isolated that resulted in defective repression of the prgQ promoter in p043lacZ due to single amino acid changes in PrgX (Kozlowicz et al., 2004). These mutant PrgX had various repression phenotypes (Class A, fully derepressed; Class B, partly derepressed and uninducible; Class C, partly derepressed and inducible). Because iCF10 appeared to interact with PrgX and function as a corepressor, we wanted to determine whether addition of iCF10 to the growth medium could rescue the defects caused by these mutations. All of the mutant strains responded to the addition of exogenous iCF10 (Fig. 3). Several mutant strains displaying fully derepressed or uninducible (Class A and B respectively) phenotypes were not completely repressed after addition of exogenous iCF10; however, seven of the 11 prgX alleles in Class A and B were repressed at least twofold by addition of iCF10 (Fig. 3A). As depicted in Fig. 3B each of the Class C, or inducible, mutants displayed an increased level of β-galactosidase expression relative to wild type when no exogenous peptides were added to the cultures. Pheromone addition increased expression to different extents for each mutant, and simultaneous addition of iCF10 resulted in complete repression of lacZ expression. When iCF10 alone was added, repression was equal to that of isogenic cells carrying wild-type prgX. These results show that iCF10 rescues the repression phenotypes of a variety of PrgX variants. Conceivably, iCF10 binding to PrgX may alter or stabilize the protein structure, thereby allowing it to function better as a repressor even in the presence of detrimental sequence changes in residues of the central dimerization domain or the C-terminal regulatory domain (CTD) important for structural integrity.
Fig. 3. Exogenous iCF10 rescues the ability of PrgX mutants to repress prgQ transcription in p043lacZ. Beta-galactosidase activity was measured in extracts from OG1RF cells carrying p043lacZ derivatives containing mutations in prgX conferring single amino acid substitutions.
A. Fully derepressed (Class A) or partially depressed, uninducible (Class B) PrgX mutants in p043lacZ were grown in medium with or without 250 ng ml−1 iCF10. Controls: WT = p043lacZ; WT + c = WT + 5 ng ml−1 cCF10, dX = p043lacZdX.
B. Partially derepressed, inducible (Class C) mutations of PrgX were grown in medium with various combinations of iCF10 and cCF10. None = no peptide addition; cCF10 = 5 ng ml−1 cCF10; iCF10 only = 250 ng ml−1 iCF10; cCF10 + iCF10 = 5 ng ml−1 cCF10 + 250 ng ml−1 iCF10.
Reduction of PrgX oligomerization state in vivo by cCF10 is inhibited by iCF10
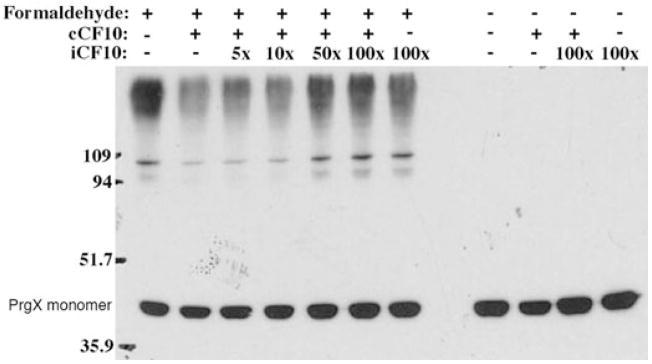
Previously, it was shown that cCF10, while having little effect on DNA binding in vitro, can reduce the oligomeric state of PrgX (Bae and Dunny, 2001). In order to determine whether iCF10 also had an effect on PrgX oligomerization, we used in vivo formaldehyde cross-linking to monitor the PrgX oligomerization state. As seen in Fig. 4, increasing amounts of excess iCF10 restored the higher molecular weight forms of PrgX that were initially lost when cells were treated with cCF10. This indicates that iCF10 directly inhibits cCF10-induced reduction of PrgX oligomeric state. These data are consistent with the possibility that in the presence of high concentrations of iCF10, most PrgX molecules in the cell are no longer bound to cCF10 and, as indicated in Fig. 2B, iCF10-bound PrgX can form productive repressors.
Fig. 4.
Dimerization of PrgX is restored upon addition of excess iCF10. OG1RF/p043lacZ cells were treated with 1% formaldehyde, lysed, and the resulting lysates were analysed by Western blotting for PrgX. During the growth of cells used for protein extraction, cCF10 was added at 25 ng ml−1 and excess quantities of iCF10 (5X = 125 ng ml−1; 10X = 250 ng ml−1; 50X = 1.25 μg ml−1; 100X = 2.5 μg ml−1) were added to aliquots of this culture immediately following pheromone addition. Prior to cell lysis, each culture was split, and one portion was treated with formaldehyde to cross-link PrgX, whereas the other was not treated with formaldehyde.
Both iCF10 and cCF10 ligands bind to the same PrgX pocket
The structures of apo-PrgX and PrgX/cCF10 were recently described in Shi et al. (2005). Briefly, each PrgX monomer consists of three domains (see also Fig. S1 and Table S1). The large central domain (residues 70–288) of PrgX forms a pocket into which cCF10 is bound (illustrated here in Fig. 5). Upon cCF10 binding, there is a reorganization of the CTD (residues 289–317) from an α-helix and disordered terminus to a loop and β-duplex that covers the cCF10-containing pocket (Fig. 5). The N-terminal DNA-binding domain is not significantly altered by pheromone binding. To examine the interaction of iCF10 with PrgX at the atomic level, we determined the structure of the PrgX/iCF10 complex. We were able to determine two PrgX/iCF10 complex crystal structures: a C-terminal deletion of residues 288–317 of PrgX (ΔCT)/iCF10 and Y231C/iCF10. Our previous study showed that the overall structure of the ΔCT/cCF10 complex is similar to the wild-type protein/cCF10 complex, except that the lack of the CTD destabilizes the formation of tetramers required for prgQ repression. The Y231C is also defective in repression, but can be rescued by addition of exogenous iCF10 (Fig. 3A) indicating that this protein retains ability to bind iCF10. The crystal data and refinement statistics for the PrgX/iCF10 complexes are presented in Table 1. There are four ΔCT/iCF10 complex molecules in the crystallographic asymmetric unit forming a PrgX tetramer. There are only three Y231C/iCF10 monomers (A,B,C) in an asymmetric unit. To eliminate the possibility of model bias the iCF10 and surrounding residues were deleted from the model and the result refined. The resulting 2Fo−Fc and Fo−Fc omit maps supported the final model. From these three monomers and their symmetry-related molecules, two crystallographically distinct tetramers (AAAA, BBCC) are formed in the Y231C/iCF10 crystal lattice. The two tetramers in the Y231C/iCF10 crystal are very similar (RMSD: 0.43 Å). The Y231C/iCF10 (tetramer AAAA of Y231C/iCF10 was used for RMSD calculations hereafter) also has very similar structure to the apo-PrgX tetramer with RMSD of 2.00 Å. However, the tetramers of Y231C/iCF10 and ΔCT/iCF10 are significantly different with RMSD of 7 Å because the deletion of CTD allows the two dimers in the ΔCT/iCF10 tetramer to rotate 18.9° as compared with the Y231C/iCF10 and apo-PrgX tetramers.
Fig. 5. Comparison of PrgX conformation when bound to cCF10 and iCF10.
A. Half of a PrgX tetramer is depicted. The blue and green ribbons show the structure of PrgX when complexed with cCF10. The CTD of cCF10-bound PrgX is shown in magenta: the arrows depict a small beta sheet formed from the same residues that comprise helix 17 (barrel) of the apo-protein (Shi et al., 2005); cCF10 is represented by the red balls. The yellow/green ribbon superimposed on the bright green PrgX monomer delineates the structure of iCF10-bound PrgX. iCF10 is represented by the bright blue balls superimposed on the red cCF10 structure. The structure of iCF10-bound PrgX is shown in yellow-green with helix 17 represented by a barrel: this complex retains the apo-PrgX conformation.
B. Comparison of monomers of PrgX when bound to cCF10 or iCF10 (stereo image). The yellow-green cylinders and ribbon indicate the structure of the C-terminus when iCF10 is bound. The magenta ribbon shows the structure of the C-terminus when cCF10 is bound.
Table 1.
Data collection and refinement statistics.
| Y153C/iCF10 | ΔCT/iCF10 | |
|---|---|---|
| Space group | C222 | P21,21,21 |
| a (Å) | 94.32 | 83.75 |
| b (Å) | 134.75 | 110.12 |
| c (Å) | 192.48 | 188.60 |
| Resolution (Å) | 3.0 | 3.0 |
| Rmerge (%) | 8.2 (38.2)a | 10.4 (35.9)a |
| <I/σ(I)> | 18.7 (5.7)a | 12.8 (5.1)a |
| Redundancy | 8.9 (8.5)a | 7.1 (6.4)a |
| % completeness | 98.2 (95.3)a | 99.7 (97.8)a |
| Unique reflections | 24 231 | 35 826 |
| Rwork/Rfree (%) | 23.8/28.5 | 23.8/27.1 |
| #mol in AU | 3 | 4 |
| Ramachandran | ||
| Most favoured (%) | 86.0 | 88.8 |
| Allowed (%) | 13.0 | 10.4 |
| Generously allowed (%) | 0.7 | 0.7 |
| Disallowed (%) | 0.3 | 0.0 |
| RMSD | ||
| Bond length (Å) | 0.07 | 0.07 |
| Bond angle (°) | 1.12 | 1.11 |
| Dihedral (°) | 18.9 | 18.1 |
| Improper (°) | 0.81 | 0.78 |
Values in parentheses are for highest resolution shell.
As shown in Fig. 5, iCF10 occupies the same ligand-binding pocket in the PrgX central domain as cCF10. Although both peptides bind in the same pocket of the protein and in similar orientations, the repercussions of peptide binding on monomer and tetramer conformations vary. As shown in Fig. 5, iCF10 binding does not create the conformational change in the PrgX CTD that was seen when cCF10 bound. Binding of iCF10 instead appears to preferentially stabilize an altered PrgX CTD. Residues 306–317, which were disordered and not seen in any of the previous PrgX structures (Shi et al., 2005), are clearly present in the PrgX/iCF10 structure (Fig. 6 and Fig. S2). In this new conformation in the iCF10 complex residues 311–315 lie over the bound ligand and form main chain hydrogen bonds with it in the same space occupied by residues 295–299 in the cCF10 complex. This change allows residues 293–304 to fold into a new α-helix (helix 17).
Fig. 6.
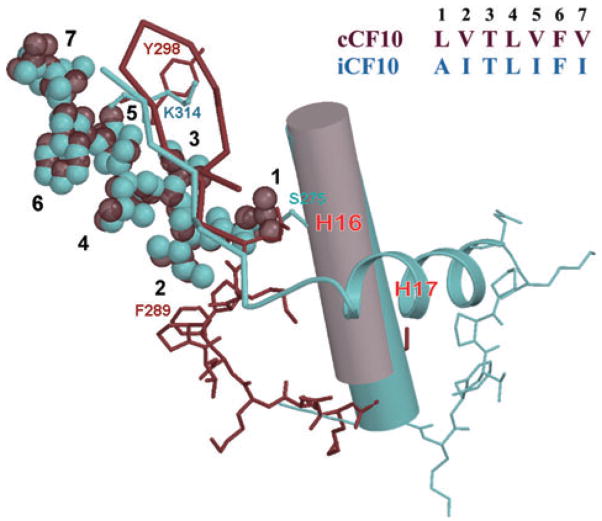
Comparison of the atomic interactions between amino acid residues of bound iCF10 and cCF10 with the PrgX C-terminus. cCF10 is shown as dark red van der Waals spheres; iCF10 is shown as light blue spheres. Positions of the side-chains of iCF10 and cCF10 are numbered 1–7. Dark red ribbon and sticks indicate the PrgX C-terminus in the cCF10 complex; light blue ribbon and sticks indicate the conformation in the iCF10 complex. Sequence of iCF10 and cCF10 are given in the upper right. The conserved helix 16 is shown as a cylinder.
Specific interactions between residues of the ligands and the PrgX backbone determine tetramer conformation
In both the PrgX/iCF10 and apo-PrgX structures, the helix 16-loop-helix 17 segment forms a triangle with the loop functioning as the tetramer interface as mentioned above (Fig. 5A). The PrgX/cCF10 structures show that the loophelix 17 segment pulls away from the tetramer interface (Fig. 5A and B) and reorganizes itself into a β-duplex that covers cCF10 in the ligand-binding pocket, thus disrupting the tetramer conformation. Both iCF10 and cCF10 form very similar β-sheet-like interactions with the amino acid side-chains at the bottom of the binding pocket (Table 2, Fig. 5 and Fig. S2). While iCF10 interacts with E312 and K314, residues that are C-terminal to the helix 16-loop-helix 17 segment (Fig. 6), cCF10 interacts with T296 and Y298 within the now unfolded helix 17. The number and character of local interactions are similar in both complexes (Table 2); however, by interacting preferentially with residues within unfolded helix 17, cCF10 promotes the reorganization of the CTD that destabilizes the tetrameric interaction. In contrast, the interaction of bound iCF10 with residues at the extreme C-terminus anchors this domain such that the positions of helix 17 and of the preceding tetramer interfacial loop are stabilized, as depicted in Fig. 6B.
Table 2.
Comparison of PrgX/iCF10 and PrgX/cCF10 interactions.
| PrgX/iCF10 interactions |
PrgX/cCF10 interactions |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Residue | Atom | Residue | Atom | Distance Å | Residue | Atom | Residue | Atom | Distance (Å) |
| A1 | N | E235 | OE2 | 2.65 | L1 | N | T199 | OG1 | 2.85 |
| L1 | N | E279 | OE2 | 2.77 | |||||
| A1 | O | N196 | OD1 | 3.29 | L1 | O | T199 | OG1 | 3.09 |
| A1 | O | K195 | NZ | 2.78 | L1 | O | K195 | NZ | 2.89 |
| T3 | N | N196 | OD1 | 2.73 | T3 | N | N196 | OD1 | 2.61 |
| T3 | O | N196 | ND2 | 2.83 | T3 | O | N196 | ND2 | 2.90 |
| T3 | OG1 | E312 | OE1 | 2.35 | T3 | OG1 | T296 | OG1 | 3.06 |
| T3 | T3 | OG1 | Y302 | OH | 3.09 | ||||
| T3 | OG1 | K195 | NZ | 2.42 | T3 | OG1 | K195 | NZ | 2.89 |
| L4 | N | E312 | O | 2.73 | L4 | N | T296 | O | 3.26 |
| L4 | O | K314 | N | 2.79 | L4 | O | Y298 | O | 2.82 |
| 15 | N | N158 | OD1 | 3.27 | V5 | N | N158 | OD1 | 2.95 |
| 15 | O | N158 | ND2 | 3.45 | V5 | O | N158 | ND2 | 2.98 |
| F6 | N | K314 | O | 2.81 | F6 | N | Y298 | O | 3.16 |
| 17 | N | E154 | OE2 | 3.27 | V7 | N | E154 | OE2 | 3.35 |
| 17 | O | K79 | NZ | 2.74 | V7 | O | K79 | NZ | 2.84 |
Bold letters show the interactions between iCF10 and cCF10 with the CTD of PrgX.
Discussion
Our results establish that iCF10 acts as an intracellular effector peptide that antagonizes cCF10 disruption of PrgX oligomerization by binding directly to PrgX. These data define this antagonistic activity at the molecular level by identifying the peptide and PrgX residues involved that determine the fate of the response. PrgX was shown to be both necessary and sufficient for iCF10 activity, and the regulatory phenotype of mutations in PrgX could be rescued by addition of iCF10 exogenously. The structural data elucidate the subtle, but critical differences in the molecular interactions between PrgX and its two peptide ligands, and also support previous genetic and biochemical findings.
These results reinforce the notion that the cognate peptides cCF10 and iCF10, but not other enterococcal pheromone peptides, have any biological impact on the pCF10 system. Using a reporter system in which only prgX and prgQ were present, we saw that iCF10 acts to inhibit cCF10 in a PrgX-dependent fashion (Fig. 2). We recently have shown that a prgX gene expressed from an ectopic location in the E. faecalis chromosome complements the defect in peptide response of cells carrying the p043dX reporter plasmid (Fig. 2C), providing further confirmation that PrgX is the critical peptide-responsive switch in this system (K.R. Fixen et al., submitted). Previous results have indicated that cPD1 has no inhibitory effects on transfer of pCF10 (Dunny et al., 2001). In this study we tested both cAD1 and cPD1 as inducers and inhibitors of prgQ expression (Fig. 2B). Our results are consistent with previous findings that non-cognate peptides have no effect on pheromone induction of pCF10, even when delivered in excess. Thus the previously reported ability of the non-cognate peptides to bind PrgX in vitro may not have biological significance (Kozlowicz et al., 2004). In our original analysis of the structure of PrgX crystallized in the absence of exogenously added peptides, we observed some diffuse electron density in the region of the peptide binding pocket (Shi et al., 2005; unpubl. data). Thus it is likely that this pocket can accommodate a variety of peptides. However, we suspect that only cCF10, iCF10 and perhaps other peptides with very similar amino acid sequences can participate in sufficient atomic interactions with both the peptide binding pocket and the C-terminus of PrgX to mediate a high-affinity binding, with the concomitant changes in the PrgX CTD that either stabilize or disrupt the tetrameric state of the protein in vivo. To further examine this question, we recently characterized the biological activities of a series of cCF10 variants carrying amino acid substitutions that were identified in a genetic screen (K.R. Fixen et al., submitted); the structures of complexes between PrgX and these peptides are being analysed currently. The comparative analysis of complexes between PrgX and either cCF10 or iCF10 presented here and previously (Shi et al., 2005) indicate that productive peptide binding by PrgX is governed by interactions that can vary dramatically between these peptides even though they share similar size, amino acid sequence and hydrophobicity.
Results of previous studies raised the possibility that the inhibitor peptides encoded by the various enterococcal pheromone-inducible plasmids might function at the donor cell surface by interfering with pheromone import (Fujimoto and Clewell, 1998; Dunny et al., 2001). The pCF10 pheromone binding protein PrgZ and the TraC homologues encoded by other pheromone plasmids increase sensitivity of donor cells the cognate exogenous pheromone at low concentrations (Weaver and Clewell, 1990; Leonard et al., 1996), and it is theoretically possible that pheromone-inhibitor competition for PrgZ/TraC binding might be the basis for inhibitor activity. However, previous studies (Leonard et al., 1996) and the work described here suggest that the one or more chromosomally encoded OppA-like peptide binding proteins and associated permease systems can mediate import of inhibitor and pheromone peptides at concentrations above 10−11 M. In the experiments reported here (Fig. 2), no PrgZ was present. It is unlikely that the inhibition of cCF10 by iCF10 resulted from interference with general peptide import, because the non-cognate peptides cAD1 and cCPD1, which are likely imported by the same peptide permease systems, had no effect on cCF10 activity. While it remains possible that iCF10 competes to some extent with cCF10 for binding to PrgZ, our results suggest that this interaction may not have much influence on the activities of these peptides under normal physiological conditions.
Phenotypic effects of mutations in prgX were rescued by excess iCF10 addition (Fig. 3). The original analysis of these mutations in prgX showed that many of the regulatory phenotypes were intermediate, and at that time it was suggested that the mutations were affecting structural stability or oligomerization. The screen used to isolate the mutant strains was specifically designed to isolate mutants in PrgX that were deficient in regulation of PQ but retained the ability to autoregulate PrgX production (Kozlowicz et al., 2004). Thus, if oligomerization was required for PrgX autoregulation, then it would be impossible to isolate mutants in which oligomerization was completely abolished. This argument is supported by the data provided from the solution of the PrgX and PrgX/peptide crystal structures (Fig. 5) (Shi et al., 2005). While space constraints preclude a detailed discussion of each of these mutations here, our cumulative structural data indicate that many of the mutations are adjacent to regions important for both dimerization and peptide binding. For example, in the wild-type PrgX molecule, residue E235 makes a critical contact with the N-terminus of bound iCF10, whereas E279 mediates a similar interaction with cCF10 (Fig. 6). The E235K substitution resulted in a partially derepressed phenotype that was unresponsive to pheromone (Kozlowicz et al., 2004), but could still be rescued by excess iCF10 (Fig. 3A). Another mutation in this region, D233N, is also partially derepressed but remains responsive to both peptides (Fig. 3B); the structure indicates that this residue contributes to dimerization (which in turn is required for tetramerization) by interacting with Y231 on the adjacent monomer. The cumulative structural and biochemical analyses of these mutations suggest that the effect of both mutations is to make the ‘induced’ conformation of PrgX favoured. It is conceivable that some of the derepressed mutants shown in Fig. 3 might show this phenotype only in the presence of endogenously produced pheromone, while others could assume the induced conformation in the absence of cCF10; the recent construction of a mutation in the chromosomal ccfA gene abolishing pheromone production (Chandler et al., 2005b) makes it possible to test this experimentally.
As shown in Fig. 4, cCF10 addition resulted in a decreased amount of detectable cross-linked PrgX oligomers. Addition of increasing amounts of iCF10 resulted in the reappearance of PrgX higher-order oligomers; this is consistent with a predominance of PrgX tetramers in these cells. We cannot make this conclusion with certainty from these data alone, because formaldehyde treatment of PrgX-containing cells could potentially produce different cross-linked species in terms of the number of inter- and intramolecular cross-links/molecule, which could affect the migration of the protein in SDS gels (attempts to use native gels for analysis of the products of these experiments were unsuccessful due to failure of the protein to enter the gels). However, this interpretation is supported strongly by the structural data showing that, the CTD of PrgX is stabilized upon interaction with iCF10, and thus the tetramer interface would be also stabilized, leading to an increase in PrgX oligomers detectable by formaldehyde cross-linking. This claim of C-terminal stabilization is also supported by the appearance of the last 13 amino acids in the PrgX/iCF10 structure that were not visible in any other PrgX crystal structure (Fig. 5) (Shi et al., 2005); while we cannot totally rule out the possibility that tight packing of PrgX molecules in crystals forces them into tetramers, considerable genetic and biochemical evidence also suggests that the model shown in Fig. 1 is operative in pCF10-containing cells (Kozlowicz et al., 2004). We suggest that iCF10 functions as a corepressor molecule when bound to PrgX and that apo-PrgX may only be partially functional in its absence. The fact that iCF10 has no function in the absence of PrgX (Fig. 2C) also satisfies the original definition of a corepressor molecule defined by Jacob and Monod (1961).
The structural data presented here also provide insights into the interactions between PrgX and its ligands. The two ligands form similar interactions with the pocket (Table 2 and Fig. 6); however, key differences were observed. Leu 1 in cCF10 (Ala 1 in iCF10) sits in a hydrophobic pocket formed by the Cβ of Ser 275, Leu 276, Pro 292 and Thr 296. Replacement with an alanine in iCF10 incompletely fills the cavity so that Pro 292 and Thr 296 are no longer stabilized by hydrophobic interactions. Similarly Val 2 of cCF10 sits in an adjacent very tight hydrophobic pocket formed by Ile 280 and Phe 289. Replacement with an isoleucine in iCF10 produces an unfavourable van der Waals contact with Phe 289. Similarly the larger side-chain of Ile 5 of iCF10 (Val 5 of cCF10) would clash with Tyr 298 in the cCF10 complex but has adequate space next to the flexible aliphatic methylenes of Lys 314. These contacts likely explain the induced conformation of PrgX in the PrgX/cCF10 complex and uninduced conformation of PrgX in the iCF10 complex.
Our results describe, in molecular detail, the functional role for the previously enigmatic pheromone inhibitor peptides. These data also provide insight not only into regulatory function of enterococcal pheromone plasmids, but may also help illuminate the molecular mechanisms used by other peptide signalling systems. These systems include a variety of quorum sensing systems in other Gram-positive pathogens (Lyon and Novick, 2004). By understanding how these systems work at the molecular level, it may be possible to use the information in rational design of chemical inhibitors for therapeutic purposes. For example, in the case of the pCF10 system, a plasmid-encoded virulence factor is induced through the pheromone-sensing system when the host bacterium is grown in the mammalian bloodstream (Chandler et al., 2005b). A synthetic iCF10 mimetic could block expression of virulence in the host without directly killing the bacteria, potentially reducing selective pressure for emergence of drug resistance.
Experimental procedures
Bacterial strains and growth conditions
Enterococcus faecalis strain OG1RF (Dunny et al., 1978) was used for β-galactosidase assays and formaldehyde cross-linking assays. E. faecalis cells were grown in M9-YE (Dunny et al., 1985) broth for all experiments in this study. Antibiotic (all purchased from Sigma) concentrations used for E. faecalis growth and selection were: erythromycin (erm) 10 μg ml−1, tetracycline (tet) 10 μg ml−1, rifampicin (rif) 200 μg ml−1, spectinomycin (spec) 250 μg ml−1, streptomycin (sm) 1000 μg ml−1. Escherichia coli strains BL21 (DE3) and B834 (DE3) (Stratagene) were used for expression and purification of His-PrgX. Kanamycin was used for selection of pET28b-based His-PrgX expression plasmids in E. coli strains at 30 μg ml−1. His-PrgX expression strains of E. coli were grown at 30°C with shaking when used for protein purification.
β-Galactosidase assays
β-Galactosidase assays were performed as described by Kozlowicz et al. (2004), based on Miller (1972) with minor alterations in peptide concentrations used. Briefly, E. faecalis strains were grown overnight in M9-YE with antibiotics and iCF10 at a concentration of 250 ng ml−1 if needed (Fig. 3), and diluted 1:4 with fresh medium containing the same components. After dilution, strains were grown for 1 h at 37°C, static. Cells were induced using cCF10 at 5 ng ml−1 and iCF10 was added immediately after cCF10 addition at the concentrations indicated in figure legends. Strains were grown for an additional 90 min at 37°C, static, after peptide addition. Thereafter the cells were treated according to Miller (1972). Assays with E. faecalis cells were conducted with 100 μl of cells for 7–8 min β-galactosidase measurements reported represent means from duplicate assays, error bars represent one standard deviation of the mean. Assays were repeated a minimum of three times, representative results from one experiment, performed in duplicate, are presented.
Western blotting analysis
Western blotting was performed as described previously (Bae et al., 2004). Briefly, cells at exponential phase were collected and treated with lysozyme. Reducing sample buffer was added to a 1× final concentration, and the samples were boiled and separated using 12% SDS-PAGE. Proteins were transferred to nitrocellulose membrane and probed with anti-His-PrgX (Bae et al., 2000). Horseradish peroxidase-goat anti-rabbit antibody (Zymed) was used as a secondary antibody. Supersignal chemiluminescent substrate (Pierce) was used to detect the signals.
In vivo oligomerization of PrgX in E. faecalis
Oligomerization of PrgX was tested in vivo in E. faecalis using 1% formaldehyde cross-linking and Western blotting, as described above and in Bae and Dunny (2001). Peptides were added at the concentrations indicated in the figure legend.
Crystallization, structure determination and refinement
His-PrgX and mutant derivatives were expressed and purified as described by Bae et al. (2002). Crystals were grown by the hanging-drop vapour-diffusion method. Synthetic peptides were purchased from Mimotopes (Raleigh NC); the sequence of iCF10 is Ala-Ile-Thr-Leu-Ile-Phe-Ile. To grow the crystals, 0.5 mg of iCF10 was mixed with 1 ml of 7 mg ml−1 solution of PrgX in 0.005 ppm. Triton X-100, 20 mM HEPES (pH 7.2) and incubated overnight. From this solution, crystals of complex were grown by the hanging-drop vapour diffusion method using a reservoir of 10–28% polyethylene glycol 4000, 50 mM Tris malate, pH 5.6–8.
The crystal data were collected on our in-house R-Axis IV++ detector at 100 K and processed with either HKL2000 (Otwinowski and Minor, 1997) or D*TREK (Pflugrath, 1999) program. Structures were solved by the molecular replacement method using the program EPMR (Kissinger et al., 1999). The PrgX monomer (Shi et al., 2005) was used as the search model and the structural models were subsequently refined using CNS (Brunger et al., 1998). After map averaging and partial refinement, the iCF10 molecules were modelled into the electron density using O (Jones, 1978). The final Rwork/Rfree for ΔXT/iCF10 and Y231C/iCF10 complex are 23.8%/28.5% and 23.8%/27.1% respectively. The co-ordinates have been deposited with RCSB with the accession codes 2GRL (for the ΔCT complex) and 2GRM (for the Y231C complex).
Supplementary Material
Fig. S1. Structure of PrgX in complex with iCF10 (balls).
Fig. S2. C-terminal region of PrgX monomer bound to iCF10.
The following supplementary material is available for this article online
Table S1. Amino acid residues of PrgX comprising α-helical sub-domains.
Acknowledgments
This work was supported by PHS Grants GM49530 (to G.M. D.) and AI57585 (to C.A.E.) and by a grant-in aid to G.M.D. from the University of Minnesota Graduate School. B.K.K. was supported by Grant T32 G08347 and by a Doctoral Dissertation Fellowship from the University of Minnesota Graduate School. K.S. is supported in part by T32 DE07288. Diffraction data were collected using the facilities of the Kahlert Structural Biology Laboratory. Computational facilities were provided by the Basic Sciences Computer Laboratory of the Minnesota Supercomputing Institute.
Footnotes
This material is available as part of the online article from http://www.blackwell-synergy.com
References
- Bae T, Dunny GM. Dominant-negative mutants of prgX: evidence for a role for PrgX dimerization in negative regulation of pheromone-inducible conjugation. Mol Microbiol. 2001;39:1307–1320. doi: 10.1111/j.1365-2958.2001.02319.x. [DOI] [PubMed] [Google Scholar]
- Bae T, Clerc-Bardin S, Dunny GM. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J Mol Biol. 2000;297:861–875. doi: 10.1006/jmbi.2000.3628. [DOI] [PubMed] [Google Scholar]
- Bae T, Kozlowicz B, Dunny GM. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J Mol Biol. 2002;315:995–1007. doi: 10.1006/jmbi.2001.5294. [DOI] [PubMed] [Google Scholar]
- Bae T, Kozlowicz BK, Dunny GM. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol Microbiol. 2004;51:271–281. doi: 10.1046/j.1365-2958.2003.03832.x. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chandler JR, Dunny GM. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides. 2004;25:1377–1388. doi: 10.1016/j.peptides.2003.10.020. [DOI] [PubMed] [Google Scholar]
- Chandler JR, Flynn AR, Bryan EM, Dunny GM. Specific control of endogenous cCF10 pheromone by a conserved domain of the pCF10-encoded regulatory protein PrgY in Enterococcus faecalis. J Bacteriol. 2005a;187:4830–4843. doi: 10.1128/JB.187.14.4830-4843.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler JR, Hirt H, Dunny GM. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci USA. 2005b;102:15617–15622. doi: 10.1073/pnas.0505545102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JW, Dunny GM. Transcriptional analysis of a region of the Enterococcus faecalis plasmid pCF10 involved in positive regulation of conjugative transfer functions. J Bacteriol. 1995;177:2118–2124. doi: 10.1128/jb.177.8.2118-2124.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Leonard BA. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527– 564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- Dunny GM, Brown BL, Clewell DB. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc Natl Acad Sci USA. 1978;75:3479–3483. doi: 10.1073/pnas.75.7.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Zimmerman DL, Tortorello ML. Induction of surface exclusion (entry exclusion) by Streptococcus faecalis sex pheromones: use of monoclonal antibodies to identify an inducible surface antigen involved in the exclusion process. Proc Natl Acad Sci USA. 1985;82:8582– 8586. doi: 10.1073/pnas.82.24.8582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunny GM, Antiporta MH, Hirt H. Peptide pheromone-induced transfer of plasmid pCF10 in Enterococcus faecalis: probing the genetic and molecular basis for specificity of the pheromone response. Peptides. 2001;22:1529–1539. doi: 10.1016/s0196-9781(01)00489-2. [DOI] [PubMed] [Google Scholar]
- Fujimoto S, Clewell DB. Regulation of the pAD1 sex pheromone response of Enterococcus faecalis by direct interaction between the cAD1 peptide mating signal and the negatively regulating, DNA-binding TraA protein. Proc Natl Acad Sci USA. 1998;95:6430–6435. doi: 10.1073/pnas.95.11.6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- Jones TA. A graphics model building and refinement system for macromolecules. J Appl Cryst. 1978;11:268–272. [Google Scholar]
- Kissinger CR, Gehlhaar DK, Fogel DB. Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. 1999;D55:484–491. doi: 10.1107/s0907444998012517. [DOI] [PubMed] [Google Scholar]
- Kozlowicz BK, Bae T, Dunny GM. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol Microbiol. 2004;54:520–532. doi: 10.1111/j.1365-2958.2004.04286.x. [DOI] [PubMed] [Google Scholar]
- Lazazzera BA. The intracellular function of extracellular signaling peptides. Peptides. 2001;22:1519–1527. doi: 10.1016/s0196-9781(01)00488-0. [DOI] [PubMed] [Google Scholar]
- Leonard BA, Podbielski A, Hedberg PJ, Dunny GM. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyon GJ, Novick RP. Peptide signaling in Staphylococcus aureus and other Gram-positive bacteria. Peptides. 2004;9:1389–1403. doi: 10.1016/j.peptides.2003.11.026. [DOI] [PubMed] [Google Scholar]
- Miller JH. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- Mori M, Sakagami Y, Ishii Y, Isogai A, Kitada C, Fujino M, et al. Structure of cCF10, a peptide sex pheromone which induces conjugative transfer of the Streptococcus faecalis tetracycline resistance plasmid, pCF10. J Biol Chem. 1988;263:14574–14578. [PubMed] [Google Scholar]
- Nakayama J, Ruhfel RE, Dunny GM, Isogai A, Suzuki A. The prgQ gene of the Enterococcus faecalis tetracycline resistance plasmid pCF10 encodes a peptide inhibitor, iCF10. J Bacteriol. 1994;176:7405–7408. doi: 10.1128/jb.176.23.7405-7408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Takanami Y, Horii T, Sakuda S, Suzuki A. Molecular mechanism of peptide-specific pheromone signaling in Enterococcus faecalis: functions of pheromone receptor TraA and pheromone-binding protein TraC encoded by plasmid pPD1. J Bacteriol. 1998;180:449–456. doi: 10.1128/jb.180.3.449-456.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J, Cao Y, Horii T, Sakuda S, Akkermans AD, de Vos WM, Nagasawa H. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol Microbiol. 2001;41:145–154. doi: 10.1046/j.1365-2958.2001.02486.x. [DOI] [PubMed] [Google Scholar]
- Otwinowski O, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. 1999;D55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Ruhfel RE, Manias DA, Dunny GM. Cloning and characterization of a region of the Enterococcus faecalis conjugative plasmid, pCF10, encoding a sex pheromone-binding function. J Bacteriol. 1993;175:5253–5259. doi: 10.1128/jb.175.16.5253-5259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi K, Brown CK, Gu Z, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci USA. 2005;102:18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver KE, Clewell DB. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: effects of host strain and traA, traB, and C region mutants on expression of an E region pheromone-inducible lacZ fusion. J Bacteriol. 1990;172:2633–2641. doi: 10.1128/jb.172.5.2633-2641.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Structure of PrgX in complex with iCF10 (balls).
Fig. S2. C-terminal region of PrgX monomer bound to iCF10.
The following supplementary material is available for this article online
Table S1. Amino acid residues of PrgX comprising α-helical sub-domains.