Abstract
As previously published, after aerosol infection with Mycobacterium tuberculosis H37Rv, New Zealand white rabbits established infection with active bacillary replication, but later contained disease to a paucibacillary state through an effective adaptive response consistent with latency. Despite the heterogeneity among outbred rabbits, the resistant response was uniform. Immunosuppression resulted in reactivation with increased lung bacillary burden. Using this rabbit model, we isolated bacillary RNA from infected rabbit lungs and assessed transcriptional profiles of bacillary genes using RT-PCR to examine genes differentially regulated during active replication, persistence, steroid-induced reactivation, and post-steroid immune reconstitution. Genes involved in hypoxia response (fdxA), resuscitation promoting factors (rpfB), and DNA repair pathways (Rv2191) may be important in bacillary persistence. Further investigation into these gene pathways is warranted.
Keywords: mycobacterium tuberculosis, gene expression, animal model, rabbit, latency, reactivation
Introduction
Mycobacterium tuberculosis (Mtb) is a successful pathogen because it can survive adverse conditions and persist in low numbers. Host immunosuppression or waning immunity allows the latent bacilli to reactivate to active, transmissible disease. An improved understanding of the genes involved in the maintenance of this low-level dormant infection would be invaluable to our understanding of the molecular pathogenesis of this disease.
The publication of the genome of Mtb has catalyzed a body of work examining genes up-regulated under various conditions hypothesized to exist in vivo in the human granuloma. Because incomplete inhibition of phagosome maturation may lead to acidic pH, the transcriptional response to an acid environment has been studied in vitro1 In addition, because it has been postulated that the central caseous necrosis of the tuberculous granuloma, the pathologic hallmark of human infection, results in limitations in both oxygen and nutrients, there have been several investigations of Mtb gene expression during nutrient starvation and hypoxia. Finally, it has been postulated that Mtb is subjected to both reactive nitrogen (RNI) and oxygen intermediates (ROI) in vivo as an antibacterial host mechanism,2 although transcriptional profiling of the nitrosative and oxidative stress responses in the mouse model of chronic infection have not shown up-regulation of detoxification pathways for RNI and ROI.3
Gene expression profiling in Mtb has been done using multiple models and approaches. In model systems in which larger amounts of RNA could be harvested, microarrays have often been used to analyze the largest pool of possible gene transcripts. For example using infected macrophages, Schnappinger and colleagues noticed up-regulation of a two-component anoxia regulon, DosR-DosS which is induced in response to hypoxia as well as nontoxic nitric oxide concentrations.4 Animal models have proven more challenging, but results from pooled bacillary RNA from large numbers of mice have been published.5, 6
We have developed an aerosol model of latent tuberculosis using New Zealand white rabbits which are resistant to infection with Mtb.7 In this paper, we sought to analyze bacillary RNA harvested from infected lungs at different time points after aerosol infection using RT-PCR to measure the expression of genes that were previously reported to be induced in vitro with nutrient starvation8, hypoxia using the Wayne model 9, acid pH 1, resuscitation promoting factors 10 and 4 iron-regulated genes 11. Genes important for intracellular growth in macrophages 4 and induced in vivo in mice 5 were also selected.
Materials and Methods
Animals and Infection
Forty-two New Zealand white rabbits were aerosol-infected with Mtb H37Rv with an average inhaled dose of 3.57 ± 0.05 logCFU at the United States Army Research Institute for Infectious Diseases (USAMRIID) as previously described.7 Three rabbits were necropsied on the day after aerosol infection, and then 6 rabbits every 5 weeks thereafter until 20 weeks. Intramuscular dexamethasone was administered to 12 rabbits from week 10 to week 15. These rabbits were then allowed to immune reconstitute until 20 weeks as previously reported.
Total RNA extraction and bacterial gene expression in rabbit lung by Real-Time PCR
For total RNA purification, the tissues snap-frozen in Trizol were homogenized in liquid nitrogen. After bead beating with 0.1mm zirconia/silica beads using a mini bead beater (Biospec, Products), the total RNA was extracted with chloroform and precipitated with isopropanol. Microbial RNA was separated from the total rabbit lung RNA using MICROBEnrich (Ambion, Austin, TX) and then treated with DNaseI (the RNA is not amplified with this kit). Due to the relatively small quantity of bacillary RNA that could be harvested from the lung of latently infected rabbits, the purity and integrity of the RNA samples were analyzed using the microcapillary gel electrophoresis with fluorescent detection (Bioanalyzer, RNA 6000 LabChip kit, Agilent Technologies, Wilmington, DE, USA) using the RNA 6000 Nano Kit. First-strand cDNA synthesis was performed using 1μg of MICROBEnriched mycobacterial RNA as template and random hexamers (Bio-Rad, Hercules, CA) as primers. Expression of Mtb genes was carried out with SYBR Green I assay by real-time PCR using gene-specific primers. (Supplemental Table) For each amplification run (iCycler, Bio-Rad), the calculated threshold cycle (CT) for each gene amplicon was compared to the plasmid standard to calculate the absolute amount of gene transcript. The CT of the 16s rRNA gene amplified from a 1:100 dilution of the same sample was similarly used to calculate the absolute amount of gene transcript because the amount of 16s rRNA was much higher than the transcript amount of the genes of interest. Specificity of the RT-PCR products was confirmed by gel electrophoresis and single products of the desired length with single-product specific melting temperatures were obtained for each primer pair. No primer-dimer formations were generated. Non-RT RNA samples and RNA from uninfected rabbit lung and peripheral blood mononuclear cells were included as negative controls.
All comparisons of non-normally distributed continuous data were analyzed with the Mann-Whitney U test using Prism (GraphPad Software, CA)
Results
Rabbits develop paucibacillary latent disease over the course of 20 weeks, and reactivate infection with immunosuppression
After aerosol infection, all rabbits established infection with evidence of delayed-type hypersensitivity responses to tuberculin in the skin 4 weeks after infection, and with replication of bacilli up through 5 weeks after infection as previously published.7 (Figure 1) With the acquisition of specific immunity, the burden of organisms as well as the number of grossly visible pulmonary lesions decreased over time with features consistent with latency. (Figure 1A & B) To induce reactivation (Figure 1C & D), intramuscular corticosteroids were administered to 12 rabbits from week 10 to week 15. At the conclusion of the treatment (week 15), more rabbits had culturable bacilli in the right upper lung lobe in the dexamethasone treated group than in the untreated group. Subsequently, to examine immune restoration disease, steroids were discontinued at week 15 in the remaining 6 rabbits. At necropsy 5 weeks later (week 20), steroid-induced lymphopenia was recovering. Despite improving lymphopenia, all 6 rabbits were right upper lung lobe homogenate culture-positive. Three (of the 6) rabbits had evidence of an inflammatory syndrome with very large granulomas with multiple caseous centers consistent with an immune reconstitution inflammatory syndrome (IRIS).12
Figure 1.
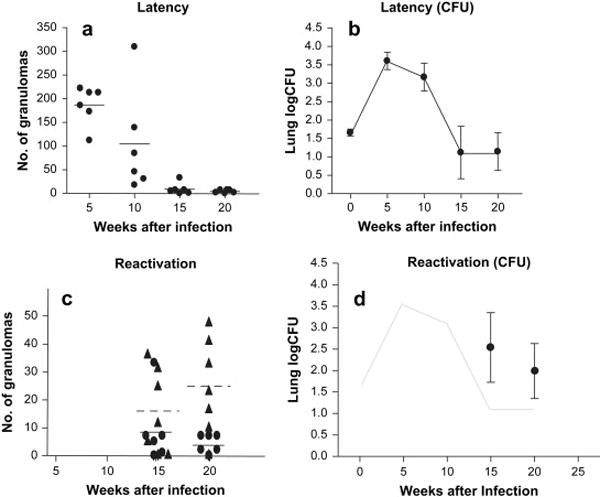
A) The number of granulomas in all lung lobes was highest at 5 weeks after aerosol infection with Mtb and decreased at later time points. B) The bacillary logCFU (±SE) in the lung showing highest bacillary burden at 5 weeks and decreasing thereafter. C) In rabbits treated with dexamethasone from week 10 to 15, the number of granulomas in all lung lobes increased at week 15 after aerosol infection (triangles, dotted line represents the median). The number of granulomas in the lungs of untreated controls in each rabbit necropsied is shown as a filled circle. D) bacillary logCFU in the lungs of 6 rabbits at 15 weeks and 20 weeks after 5 weeks of dexamethasone treatment (data previously published7)
In vivo Mycobacterial Gene Expression Profiles in Rabbit Lungs
Bacillary RNA was successfully isolated from all 18 rabbit lung samples from animals necropsied at 5, 10, and 15 weeks. Purity of the bacillary RNA was checked using the Agilent bioanalyzer which showed that >95% of eukaryotic RNA had been removed using the MICROBEnrich technique and that the RNA had not degraded. This analysis was done on RNA isolated from rabbits necropsied at 5 weeks given the relatively abundant amount of RNA at this time point and significant degradation of RNA was not observed. Accurate and equal amounts of RNA were used for reverse transcription. Concentrations of RNA ranged from 0.92-3.70 μg/μl. At 20 weeks, (steroid) untreated rabbits had bacillary numbers that were uniformly too low to obtain measurable 16s rRNA transcripts. In our previous paper, we showed that only granulomas with caseous centers were culture positive at later time points. Lung parenchyma and non-caseous granulomas were culture-negative. Therefore, we found that we could isolate bacillary RNA from rabbit lungs that had visible caseous granulomas in the left lower lobe from which RNA was harvested (all the rabbits at 15 weeks) By 20 weeks after infection, no detectable 16s rRNA transcripts were found, although these specimens were not bioanalyzed for degradation. In the dexamethasone-treated rabbits, however, bacillary RNA was obtained from the lungs of all 12 rabbits at both 15 and 20 weeks because of increased bacillary burdens.
We measured the expression of each of these transcripts over the course of infection. To obtain the values in Table 1, all specimens were included in the mean and standard deviations shown in the table. Transcripts of sigA were also measured but were not reliably detected in all samples. The specific gene expression at 5 weeks was compared to expression at the other time points.
Table 1.
RT-PCR Ratios
| Gene | Rv | Condition | 5 weeks | 10 weeks | 15 weeks | 15 weeks steroids | 20 weeks steroids |
|---|---|---|---|---|---|---|---|
| Hypothetical Protein (DNA repair) | Rv2191 | Macrophages | 0.063 ± 0.040 (3) | 0.005 ± 0.003 (3) | 1.665 ± 0.996 (6) P=0.05a | 0.959 ± 0.810 (6) P=0.012a | 0.313 ± 0.208 (6) P=0.05a |
| aceAa (isocitrate lyase) | Rv1915 | Macrophages | 0.012 ± 0.006 (4) | 0.009 ± 0.005 (4) | 0.003 ± 0.002 (2) | 0.007 ± 0.006 (3) | 0.005 ± 0.004 (2) |
| fbpA (Antigen 85A) | Rv3804c | Macrophages | - | 0.0006 ± 0.0015 (1) | - | 0.0044 ± 0.0090 (3) | 0.0005 ± 0.0013 (1) |
| Hypothetical Protein | Rv1735c | Macrophages | - | 0.0009 ± 0.0014 (3) | - | - | - |
| Conserved Hypothetical Protein | Rv2050 | Starvation | 0.0002±0.0001 (2) | 0.008 ± 0.006 (2) | 0.010 ± 0.005 (4) | 0.021 ± 0.018 (4) | |
| Conserved Hypothetical Protein | Rv3131 | Starvation, acid | 0.0002 ± 0.0002 (1) | 0.0006 ± 0.0003 (3) | 0.008 ± 0.007 (3) | ||
| lat | Rv3290c | Starvation | 0.0002 ± 0.0002 (1) | 0.004 ± 0.002 (3) | 0.0001 ± 0.0001 (1) | 0.0009 ± 0.0009 (1) | |
| Hypothetical Protein | Rv2660c | Starvation | 13.00 ± 31.84 (1) | 5.54 ± 13.57 (1) | - | - | 0.77 ± 1.87 (1) |
| Hypothetical Protein | Rv0188 | Starvation | 0.0004 ± 0.0009 (1) | 0.0045 ± 0.0109 (1) | - | - | - |
| Conserved Hypothetical Protein | Rv2626c | Hypoxia | 0.085 ± 0.051 (3) | 0.077 ± 0.039 (3) | 0.944 ± 0.663 (2) | 2.304 ± 3.066 (2) | 2.892 ± 2.865 (2) |
| Hypothetical Protein (transcriptional protein) | Rv0792c | Hypoxia | 0.017 ± 0.009 (4) | 0.006 ± 0.003 (2) | 0.001 ± 0.001 (1) | 0.002 ± 0.002 (1) | |
| fdxA | Rv2007c | Hypoxia | - | 0.0006 ± 0.0015 (1) | 0.0044 ± 0.0090 (3) | 0.0005 ± 0.0013 (1) | |
| icl1 (isocitrate lyase) | Rv0467 | Hypoxia | 0.0005 ± 0.0013 (2) | - | - | - | - |
| rpfB (resuscitation promoting factor) | Rv1009 | Resuscitation | 0.018 ± 0.011 (3) | 0.539 ± 0.389 (4) | 0.075 ± 0.048 (2) | 0.268 ± 0.173 (3) | 0.028 ± 0.015 (3) |
| rpfC (resuscitation promoting factor) | Rv1884c | Resuscitation | 0.048 ± 0.025 (4) | 0.139 ± 0.137 (2) | 0.089 ± 0.091 (2) | 0.306 ± 0.202 (4) | |
| Conserved Hypothetical | Rv3402c | Iron-induced | 0.067 ± 0.054 (4) | 0.005 ± 0.002 (3) | 0.009 ± 0.007 (2) | 0.056 ± 0.047 (3) | 0.005 ± 0.003 (3) |
| Conserved Hypothetical | Rv3615c | Acid | 0.0010 ± 0.0024 (1) | 0.0049 ± 0.0078 (3) | - | - | 0.258 ± 0.559 (3) |
| Monoxygenase | Rv3083 | Acid | 0.043 ± 0.106 (1) | - | - | - | - |
| Conserved Hypothetical Protein (DNA repair) | Rv2816c | Other | 0.283 ± 0.243 (4) | 0.226 ± 0.128 (3) | 1.956 ± 0.984 (4) P=0.181 | 1.082 ± 0.717 (4) | 34.769 ± 33.182 (3) |
P-value compared to 5 week value
Numbers denote the number of samples in which a particular gene transcript was detected by RT-PCR is listed in parentheses. 16S rRNA transcripts were detected for all 6 samples at every time point listed in table.
Each outbred rabbit is genetically unique, resulting in heterogeneity in both the lesions and the infection course compared to inbred animals. Therefore, in presenting the data we showed both the relative ratios of normalized transcripts as well as the number of samples at a given time point in which a particular gene transcript was present compared to the total number of samples analyzed at that time point. In addition, most of the transcripts were relatively rare and were only able to be detected within a range of 30-35 cycle times (CT). Table 1 shows all the genes (of 51 genes tested) for which at least 1 specimen had a transcript by RT-PCR. Transcripts from only 11 of the genes listed in Table 1 were detected by RT-PCR in multiple samples at multiple time points. Of the 5 resuscitation promoting factors in Mtb that have been shown to increase the growth of stationary phase cultures,10, 13, 14 we were able to detect transcripts from rpfB at every time point with a trend towards increased expression in reactivation. Icl2 (aceAa), but not icl, as well as fdxA (a gene up-regulated during hypoxia9 were also measured in 3 samples late in infection.
Rv2191, a gene up-regulated during mouse infection,5 was the only gene found in all 6 lung specimens during latency (15 weeks), reactivation (15 week dexamethasone) and immune reconstitution (20 weeks dexamethasone). Rv2191 was significantly up-regulated at both 15 weeks (when tubercle number and lung CFU are significantly lower than at peak infection time 5 weeks after aerosol infection) and 20 weeks during immune reconstitution (when the lung CFU was significantly higher than untreated controls) compared to the 5 week time point (P<0.05) (Table 1, Figure 2). We compared the amount of mRNA in each of the 6 samples found at the latent time point (15 weeks) to the number of tubercles in the left lower lobe from which the RNA was obtained. The absolute amount of Rv2191 mRNA in 1μg of MICROBEnriched RNA was inversely correlated with the number of visible tubercles (R2=0.62). Therefore, fewer tubercles and less culturable CFU correlated with higher expression of Rv2191.
Figure 2.
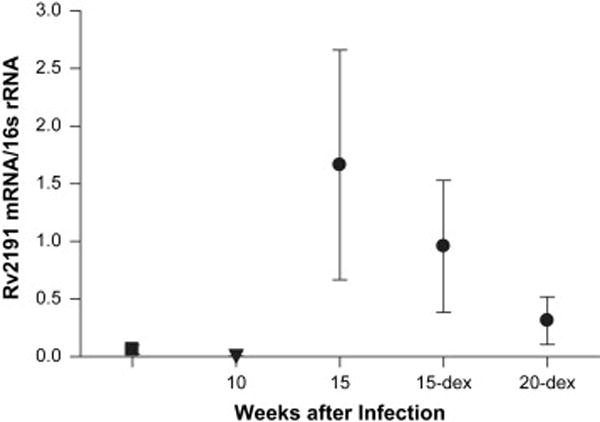
Ratio of Rv2191 to 16S rRNA. The same amount of MICROBEnriched RNA was used from each lung sample, analyzed on the Icycler in triplicate and then averaged across the 6 specimens per time point. All specimens for which a specific transcript could not be detected was included as “0”. All samples had detectable 16S rRNA.
Discussion
In the rabbit aerosol model of tuberculosis, the lung bacillary burden initially increases and then decreases after the acquisition of specific immunity. Some bacilli persist in a proportion of rabbits up to 36 weeks after aerosol infection.7 In this model, most culturable bacilli at late time points appear to be contained in non-clearing caseous granulomas. Since all caseous granulomas found at the latest time points contained culturable bacilli, we surmise that the bacillary RNA analyzed in this study derives from caseous granuloma in the lung lobes that were snap frozen and processed for RNA. One limitation of the study is that we cannot both culture and harvest RNA from the same lung lobe specimen.
Despite relatively low numbers of bacilli, we were able to analyze bacillary gene expression in tuberculous rabbit lungs at all time points except 20 weeks in animals who were not immunosuppressed. Since the assays were insensitive due to relatively low amounts of bacillary mRNA, the transcripts that we detected indicated a relatively high expression of these genes. Four gene transcripts were detected in at least 3 samples at the late, 15 week time point: rpfB, icl2, fdxA, and Rv2191. In human tissues, rpfB was shown to be 3-fold up-regulated in human granuloma compared to in vitro grown bacteria15 and may be important for both bacillary persistence and early reactivation. The 3 lungs in which transcript for rfpB were found were similar in their level of expression and were higher than at the 15 week time point. In a multi-gene deletion mutant analysis, deletion of rpfB had greater attenuating effect for growth in mouse lung than deletion of rpfD.16, 17 Isocitrate lyase, a bacillary enzyme up-regulated in the slow metabolic downshift to anaerobiasis,18 is also important in late-state murine tuberculosis 19, 20 and has been detected in human tuberculous granulomas.21 Although transcripts of fdxA, a DosR-regulated gene, were found at late time points, transcripts from acr were not found in rabbit lungs as has been described in the murine model.4, 20 This difference could be due to inherent pathologic differences between the 2 animal models; rabbit pulmonary granulomas develop caseous necrosis and rabbits contain infection over time with their adaptive response. It is also possible that the infrequent time point sampling led to under-detection of acr transcripts or that Acr protein accumulates over time and does not require high expression of acr at times when the bacillary burden is low.
DNA excision repair mechanisms, which have previously been demonstrated to have increased in vivo expression in tuberculous mice and in tubercle bacilli within macrophages, was also found to be significantly up-regulated late in rabbit infection. This gene was highly represented in all the specimens tested from late time points. The gene has some homology with the gene encoding the epsilon subunit of DNA polymerase III (dnaQ) and another encoding an exinuclease (uvrC), and may be involved in DNA repair mechanisms. Boshoff and colleagues showed that a deletion mutant of dnaE2 (the α subunit of DNA polymerase III) had impaired in vivo persistence only at 251 days after infection in C57BL/6 mice,22 suggesting that genes involved in DNA repair play an important role in chronic mouse infection when acquired immunity is well established. Rv2191 may be especially important in the persistent granuloma where bacillary DNA damage requiring repair may be more likely. We do not know if the DosR-regulated fdxA was up-regulated in response to hypoxia or to NO stress. It is tempting to speculate that RNI or ROI were involved and, in turn, initiated a cascade of events that may have resulted in DNA damage and may explain the striking presence of Rv2191 transcript in all specimens analyzed at the latest time point. Unfortunately, due to a limited amount of bacillary RNA, we were unable to interrogate other genes in this DNA repair pathway.
Our analysis of bacillary gene expression in the resistant rabbit model of latency and reactivation detected expression of starvation, hypoxia, and acid-induced genes in latent rabbit lungs as well as up-regulation of two of the resuscitation factors during reactivation. At late time points, there was consistently detectable expression of a gene putatively involved in DNA excision repair, Rv2191. Further interrogation of other gene transcripts in this rabbit model may provide novel insights into the metabolic states of Mtb during different bacillary persistence and reactivation and suggest new stage-specific drug targets.
Supplementary Material
Acknowledgments
We would like to acknowledge the editorial advice of Dr. Gyanu Lamichhane, and the helpful discussions with the other members of the Manabe lab, especially Christine Hatem, Alfredo Panebra, and Javier Lopez-Molina. The authors have no conflicting financial interests. This work was supported by funding from the National Institutes of Health (1R01 HL71554).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fisher MA, Plikaytis BB, Shinnick TM. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J Bacteriol. 2002;184:4025–32. doi: 10.1128/JB.184.14.4025-4032.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warner DF, Mizrahi V. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin Microbiol Rev. 2006;19:558–70. doi: 10.1128/CMR.00060-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi L, Sohaskey CD, North RJ, Gennaro ML. Transcriptional characterization of the antioxidant response of Mycobacterium tuberculosis in vivo and during adaptation to hypoxia in vitro. Tuberculosis (Edinb) 2008;88:1–6. doi: 10.1016/j.tube.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schnappinger D, Ehrt S, Voskuil MI, Liu Y, Mangan JA, Monahan IM, Dolganov G, Efron B, Butcher PD, Nathan C, Schoolnik GK. Transcriptional Adaptation of Mycobacterium tuberculosis within Macrophages: Insights into the Phagosomal Environment. J Exp Med. 2003;198:693–704. doi: 10.1084/jem.20030846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talaat AM, Lyons R, Howard ST, Johnston SA. The temporal expression profile of Mycobacterium tuberculosis infection in mice. Proc Natl Acad Sci U S A. 2004;101:4602–7. doi: 10.1073/pnas.0306023101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Talaat AM, Ward SK, Wu CW, Rondon E, Tavano C, Bannantine JP, Lyons R, Johnston SA. Mycobacterial bacilli are metabolically active during chronic tuberculosis in murine lungs: insights from genome-wide transcriptional profiling. J Bacteriol. 2007;189:4265–74. doi: 10.1128/JB.00011-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Manabe YC, Kesavan AK, Lopez-Molina J, Hatem CL, Brooks M, Fujiwara R, Hochstein K, Pitt ML, Tufariello J, Chan J, McMurray DN, Bishai WR, Dannenberg AM, Mendez S. The aerosol rabbit model of TB latency, reactivation and immune reconstitution inflammatory syndrome. Tuberculosis (Edinb) 2007 doi: 10.1016/j.tube.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–31. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 9.Sherman DR, Voskuil M, Schnappinger D, Liao R, Harrell MI, Schoolnik GK. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha -crystallin. Proc Natl Acad Sci U S A. 2001;98:7534–9. doi: 10.1073/pnas.121172498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tufariello JM, Jacobs WR, Jr, Chan J. Individual Mycobacterium tuberculosis resuscitation-promoting factor homologues are dispensable for growth in vitro and in vivo. Infect Immun. 2004;72:515–26. doi: 10.1128/IAI.72.1.515-526.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manabe YC, Hatem CL, Kesavan AK, Durack J, Murphy JR. Both Corynebacterium diphtheriae DtxR(E175K) and Mycobacterium tuberculosis IdeR(D177K) are dominant positive repressors of IdeR-regulated genes in M. tuberculosis. Infect Immun. 2005;73:5988–94. doi: 10.1128/IAI.73.9.5988-5994.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng VC, Yuen KY, Chan WM, Wong SS, Ma ES, Chan RM. Immunorestitution disease involving the innate and adaptive response. Clin Infect Dis. 2000;30:882–92. doi: 10.1086/313809. [DOI] [PubMed] [Google Scholar]
- 13.Downing KJ, Betts JC, Young DI, McAdam RA, Kelly F, Young M, Mizrahi V. Global expression profiling of strains harbouring null mutations reveals that the five rpf-like genes of Mycobacterium tuberculosis show functional redundancy. Tuberculosis (Edinb) 2004;84:167–79. doi: 10.1016/j.tube.2003.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Mukamolova GV, Turapov OA, Young DI, Kaprelyants AS, Kell DB, Young M. A family of autocrine growth factors in Mycobacterium tuberculosis. Mol Microbiol. 2002;46:623–35. doi: 10.1046/j.1365-2958.2002.03184.x. [DOI] [PubMed] [Google Scholar]
- 15.Rachman H, Strong M, Ulrichs T, Grode L, Schuchhardt J, Mollenkopf H, Kosmiadi GA, Eisenberg D, Kaufmann SH. Unique Transcriptome Signature of Mycobacterium tuberculosis in Pulmonary Tuberculosis. Infect Immun. 2006;74:1233–42. doi: 10.1128/IAI.74.2.1233-1242.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Downing KJ, Mischenko VV, Shleeva MO, Young DI, Young M, Kaprelyants AS, Apt AS, Mizrahi V. Mutants of Mycobacterium tuberculosis lacking three of the five rpf-like genes are defective for growth in vivo and for resuscitation in vitro. Infect Immun. 2005;73:3038–43. doi: 10.1128/IAI.73.5.3038-3043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kana BD, Gordhan BG, Downing KJ, Sung N, Vostroktunova G, Machowski EE, Tsenova L, Young M, Kaprelyants A, Kaplan G, Mizrahi V. The resuscitation-promoting factors of Mycobacterium tuberculosis are required for virulence and resuscitation from dormancy but are collectively dispensable for growth in vitro. Mol Microbiol. 2008;67:672–84. doi: 10.1111/j.1365-2958.2007.06078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wayne LG, Lin KY. Glyoxylate metabolism and adaptation of Mycobacterium tuberculosis to survival under anaerobic conditions. Infect Immun. 1982;37:1042–9. doi: 10.1128/iai.37.3.1042-1049.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McKinney JD, Honer zu Bentrup K, Munoz-Elias EJ, Miczak A, Chen B, Chan WT, Swenson D, Sacchettini JC, Jacobs WR, Jr, Russell DG. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature. 2000;406:735–8. doi: 10.1038/35021074. [DOI] [PubMed] [Google Scholar]
- 20.Gordillo S, Guirado E, Gil O, Diaz J, Amat I, Molinos S, Vilaplana C, Ausina V, Cardona PJ. Usefulness of acr expression for monitoring latent Mycobacterium tuberculosis bacilli in ‘in vitro’ and ‘in vivo’ experimental models. Scand J Immunol. 2006;64:30–9. doi: 10.1111/j.1365-3083.2006.01765.x. [DOI] [PubMed] [Google Scholar]
- 21.Timm J, Post FA, Bekker LG, Walther GB, Wainwright HC, Manganelli R, Chan WT, Tsenova L, Gold B, Smith I, Kaplan G, McKinney JD. Differential expression of iron-, carbon-, and oxygen-responsive mycobacterial genes in the lungs of chronically infected mice and tuberculosis patients. Proc Natl Acad Sci U S A. 2003;100:14321–6. doi: 10.1073/pnas.2436197100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boshoff HI, Reed MB, Barry CE, 3rd, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–93. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


