Abstract
When the number of projections does not satisfy the Shannon/Nyquist sampling requirement, streaking artifacts are inevitable in x-ray computed tomography (CT) images reconstructed using filtered backprojection algorithms. In this letter, the spatial-temporal correlations in dynamic CT imaging have been exploited to sparsify dynamic CT image sequences and the newly proposed compressed sensing (CS) reconstruction method is applied to reconstruct the target image sequences. A prior image reconstructed from the union of interleaved dynamical data sets is utilized to constrain the CS image reconstruction for the individual time frames. This method is referred to as prior image constrained compressed sensing (PICCS). In vivo experimental animal studies were conducted to validate the PICCS algorithm, and the results indicate that PICCS enables accurate reconstruction of dynamic CT images using about 20 view angles, which corresponds to an undersampling factor of 32. This undersampling factor implies a potential radiation dose reduction by a factor of 32 in myocardial CT perfusion imaging.
Keywords: dynamic CT, image reconstruction, compressed sensing
According to the standard image reconstruction theory in medical imaging, in order to avoid view aliasing artifacts, the sampling rate of the view angles must satisfy the Shannon/Nyquist sampling theorem. The universal applicability of the Shannon/Nyquist sampling theorem lies in the fact that no specific prior information about the image is assumed. However, in practice, some prior information about the image is typically available. When the available prior information is appropriately incorporated into the image reconstruction procedure, an image may be accurately reconstructed even if the Shannon/Nyquist sampling requirement is significantly violated. For example, if one knows a target computed tomography (CT) image is circularly symmetric and spatially uniform, only one view of parallel-beam projections is needed to accurately reconstruct the linear attenuation coefficient of the object. Another example is that if one knows that a target image consists of only a single point, then only two orthogonal projections are sufficient to accurately reconstruct the image point. Along the same line of reasoning, if a target image is known to be a set of sparsely distributed points, one can imagine that the image may be reconstructed without satisfying the Shannon/Nyquist sampling requirements. Of course, it is a highly nontrivial task to formulate a rigorous image reconstruction theory to exploit the sparsity hidden in the above extremal examples. Fortunately, a new image reconstruction theory, compressed sensing (CS), was rigorously formulated to systematically and accurately reconstruct a sparse image from an undersampled data set.1,2 It has been mathematically proven that an N×N image can be accurately reconstructed using on the order of SlnN samples provided that there are only S significant pixels in the image.
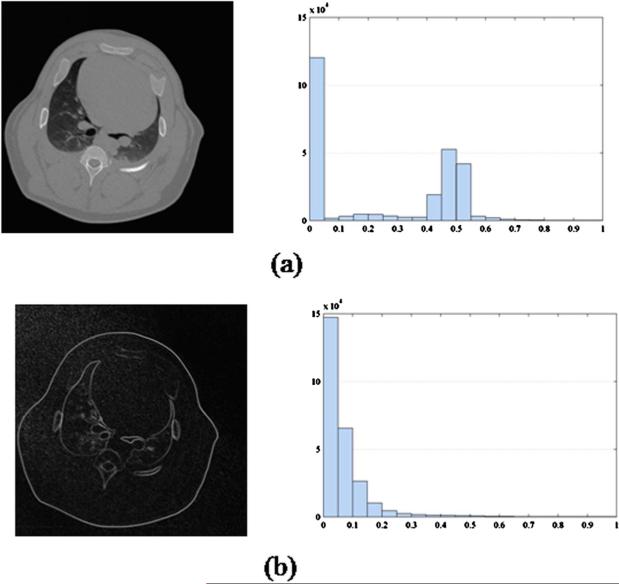
Although the mathematical framework of CS is elegant, the relevance in medical imaging critically relies on the answers to the questions: (1) Are medical images sparse? (2) If a medical image is not sparse, can we use some transforms to make it sparse? In fact, a real medical image is often not sparse in the original pixel representation. For example, in a contrast enhanced CT exam, images before contrast injection or after contrast enhancement are not sparse as shown by the histograms of the pixel values in Fig. 1(a). However, medical imaging physicists and clinicians have known for a long time that a subtraction operation can make the resultant image significantly sparser. In the newly proposed CS reconstruction theory, mathematical transforms have been applied to a single image to sparsify the image. These transforms are referred to as sparsifying transforms. For example, the image in Fig. 1(a) can be sparsified by applying a discrete gradient operation which is defined as
| (1) |
where X(m,n) is the image value at the pixel (m,n) and DxX=X(m+l,n)−X(m,n) and DyX=X(m,n+1)−X(m,n). This image specified by ▽m,nX(m,n) is referred to as the gradient image. As shown in Fig. 1, the discrete gradient image is significantly sparser. To be more quantitative, if we define a significant pixel as the one with more than 10% of the highest pixel value in an image, then we can use a histogram to demonstrate the distribution of the number of significant pixels in the original image and the discrete gradient image (Fig. 1). The discrere gradient image is three times sparser than the original image. This example demonstrates that a medical image can be made sparse even if the original image is not very sparse.
Fig. 1.
Images (left column) and the corresponding histograms (right column) of the image pixel values, (a) Image before sparsifying transform. (b) Discrete gradient image of the image (a).
The basic idea in compressed sensing (CS) image reconstruction theory can be summarized as follows: instead of directly reconstructing a target image, the sparsified version of the image is reconstructed. In the sparsified image, significantly fewer image pixels have significant image values. Thus, it is possible to reconstruct the sparsified image from an undersampled data set without streaking artifacts. After the sparsified image is reconstructed, an “inverse” sparsifying transform is used to transform the sparsified image back to the target image. In practice, there is no need to have an explicit form for the inverse sparsifying transform. In fact, only the sparsifying transform is needed in image reconstruction.
To be more specific, a sparsifying transform is denoted as Ψ. CS image reconstruction is implemented by solving the following constrained minimization problem:
| (2) |
where is the l1 norm of an N-dimensional vector z. The vector X is the vectorized target image. The matrix A is the system matrix to describe the x-ray projection measurements. The vector Y is the line integral values. Namely, the CS image reconstruction selects an image which minimizes the norm of the sparsified image among all images which are consistent with the physical measurements AX=Y. Note that when the projection data are highly undersampled, the available number of equations in Eq. (2) is much less than the number of unknowns in the image vector X. In CS image reconstruction, frequently used sparsifying transforms are discrete gradient transforms3 and wavelet transforms.4 The discrete gradient sparsifying transform has recently been utilized in the CS method to reconstruct CT images.5,6
In this letter, the CS image reconstruction algorithm was generalized to reconstruct dynamic CT image sequences. In dynamic CT imaging, the same image slice or the same image volume is sequentially scanned many times in order to measure the dynamical change in the image object. Dynamic data acquisitions may be prescribed in an interleaved pattern such that the union of the dynamic data sets form a composite data set which satisfies the Shannon/Nyquist sampling requirements. In this letter, a prior image (XP), reconstructed using the standard filtered backprojection (FBP) reconstruction algorithm from the union of the interleaved dynamic projection data sets, was utilized to constrain the CS image reconstruction method. Thus, the proposed method is referred to as prior image constrained compressed sensing (PICCS). In the reconstructed prior image, the dynamical information is lost but the static structures in the image are well reconstructed without undersampling streaking artifacts. The subtraction of the target image at each time frame from this prior image will cancel all of the static structures in the target image. Therefore, the target image can be sparsified by the subtraction operation: X−XP. After the subtraction, the known sparsifying transforms, Ψ1, used in CS are utilized to further sparsify the subtracted image X−XP. Namely, the PICCS image reconstruction algorithm is implemented by solving the following constrained minimization problem:
| (3) |
Here the sparsifying transforms, Ψ1 and Ψ2, can be any transform including those previously used in the CS literature.1,2 In this letter, for simplicity, the discrete gradient in Eq. (1) is selected for both Ψ1 and Ψ2 to demonstrate the performance of the PICCS algorithm. In order to mitigate the effect of potential artifacts in the prior image on the final reconstructed image, the conventional CS objective function has been incorporated into the PICCS algorithm with a relative weight of (1−α). In this letter, the control parameter α was selected to be 0.91. When α is set to be 0, the PICCS algorithm [Eq. (3)] reduces to the conventional CS algorithm [Eq. (2)].
The above constrained minimization was numerically implemented in two separate steps: In the first step, the standard algebraic reconstruction technique (ART)7 was used to reconstruct image X(k) based on equation AX=Y. In the second step, the weighted summation [Eq. (3)] of the total variation (TV), viz., the norm of the discrete gradient image, of the subtracted image X−XP and the TV of the image X is minimized using the standard steepest descent method.8 These two steps were iteratively implemented in an alternating manner. The final image is displayed after the squared difference of reconstructed images at two neighboring ART and TV minimization steps is smaller than a predetermined threshold value. In order to compare the performance of the PICCS and CS algorithms, the control parameter α was set to be zero, yielding the CS algorithm, while the other reconstruction parameters remain the same.
The proposed PICCS image reconstruction algorithm was validated using x-ray projection data acquired from an anesthetized 47 kg male swine. The scanning protocol utilized in this animal study was approved by the Institutional Review Board, and complies with the guidelines for care of animals given by the National Institutes of Health. A state-of-the-art clinical 64-slice scanner (GE Healthcare Lightspeed VCT, Waukesha, Wisconsin) was used to scan the animal. A “cine mode” with a 0.4 s gantry rotation period was employed in data acquisition. The scan was performed with x-ray parameters of 120 kVp and 500 mA. The acquired raw data were preprocessed with the manufacturer's software (GE Healthcare, Waukesha, Wisconsin).
The acquired projection data during the 50 s scan were retrospectively gated using a simultaneously recorded electrocardiogram signal. In this study, a cardiac window with a width of 261 ms centered at 75% of the R-R interval was utilized for gating. This cardiac window corresponds to a short scan angular range which corresponds to 642 view angles. Using the short scan datasets at each heartbeat, the cardiac images were reconstructed using the standard short scan FBP method9 for all heartbeats. One heartbeat corresponds to one image frame in the analysis of contrast dynamics.
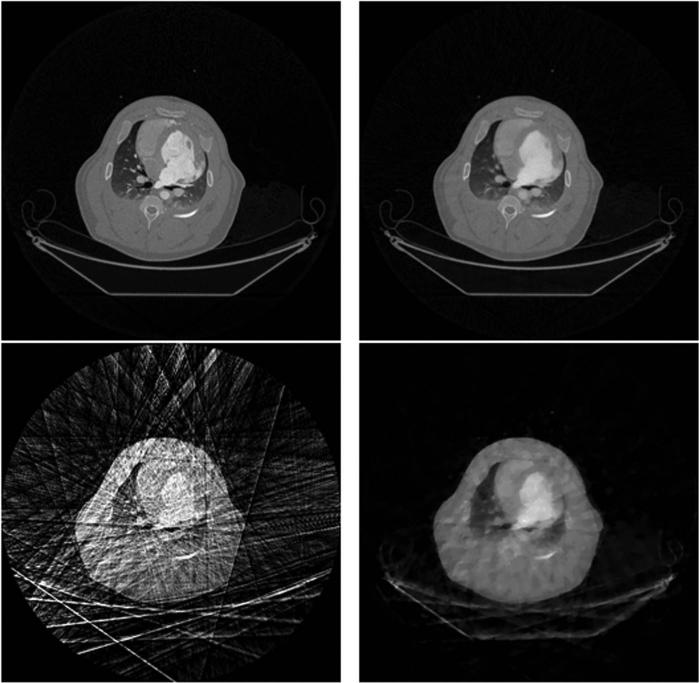
In order to validate the PICCS algorithm, the fully sampled short-scan data set at each heartbeat was down sampled by a factor of 32, which corresponds to 20 views of projection at each heartbeat. The undersampled view angles at each heartbeat have been selected to be approximately uniformly distributed over the short scan angular range. From one heartbeat to another, the undersampled view angles have been selected to be approximately interleaved with each other such that their union provides a fully sampled data set. From this data set, a prior image was reconstructed using the FBP algorithm. The PICCS algorithm was applied to reconstruct image frames at every heartbeat from the undersampled data. A representative image frame was reconstructed using the FBP method over a fully sampled data set (642 views of projections), FBP, CS, and PICCS over the undersampled data set (20 views of projections). As shown in Fig. 2, severe streaking artifacts appear in the undersampled FBP image. These artifacts are so strong that both the major anatomical structures and the patient bed are barely identifiable. Although streaking artifacts are significantly mitigated in the undersampled CS image, the image is significantly blurred and severe patching appears in the reconstructed image. Even the bones cannot be reconstructed using CS algorithm with 20 views of projections. In contrast, the undersampled PICCS image is nearly artifact free. When the PlCCS image is compared with the fully sampled FBP images with 642 views of projections the quality of reconstruction is nearly identical.
Fig. 2.
Comparison of image quality for FBP method using 642 views of projections (upper left), PICCS (upper right), FBP (lower left) and CS (lower right) using 20 projections. The display window is the same for all images.
In order to further validate the reconstruction accuracy of the PICCS algorithm, the contrast dynamic curves for three regions of interest with 13×13 pixels on the left ventricle, right ventricle, and myocardium were extracted from the 66 reconstructed image frames. The results are presented in Fig. 3. Excellent agreement between the undersampled PICCS results and the fully sampled FBP results has been found.
Fig. 3.
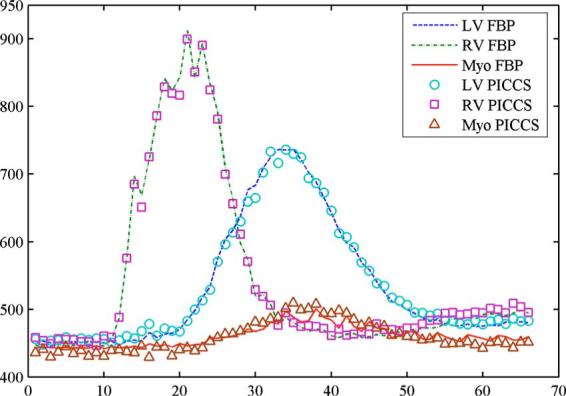
Comparison of the contrast enhancement curves extracted from the fully sampled FBP images (642 views of projections) and undersampled PICCS images (20 views of projections).
In conclusion, the PICCS method has been proposed in this letter to accurately reconstruct dynamic CT images from highly undersampled projection data sets. Preliminary experimental studies have been conducted to validate the proposed PICCS algorithm. The results demonstrated that it is possible to use PICCS to accurately reconstruct dynamic CT images using about 20 views of projections with high reconstruction fidelity for both morphology and contrast dynamics of the image sequence, while other methods such as FBP and CS fail. Relative to the fully sampled data set with about 642 view angles, the undersampling factor of 32 implies a potential 32 times radiation dose reduction in myocardial CT perfusion imaging. Further studies are ongoing to investigate the final radiation dose reduction factor.
References
- 1.Candes EJ, Romberg J, Tao T. Robust uncertainty principles: Exact signal reconstruction from highly incomplete frequency information. IEEE Trans. Inf. Theory. 2006;52:489–509. [Google Scholar]
- 2.Donoho DL. Compressed sensing. IEEE Trans. Inf. Theory. 2006;52:1289–1306. [Google Scholar]
- 3.Block KT, Uecker M, Frahm J. Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Magn. Reson. Med. 2007;57:1086–1098. doi: 10.1002/mrm.21236. [DOI] [PubMed] [Google Scholar]
- 4.Lustig M, Donoho D, Pauly J. Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. 2007;58:1182–1195. doi: 10.1002/mrm.21391. [DOI] [PubMed] [Google Scholar]
- 5.Sidky EY, Kao CM, Pan X. Accurate image reconstruction from few-views and limited-angle data in divergent-beam CT. J. X-Ray Sci. Technol. 2006;14:119–139. [Google Scholar]
- 6.Song J, Liu QH, Johnson GA, Badea CT. Sparseness prior based iterative image reconstruction for retrospectively gated cardiac micro-CT. Med. Phys. 2007;34:4476–4482. doi: 10.1118/1.2795830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Herman G. Image Reconstruction from Projections. Academic; Orlando: 1980. [Google Scholar]
- 8.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical Recipes in C. Cambridge University Press; New York: 1997. [Google Scholar]
- 9.Kak AC, Slaney M. Principles of Computerized Tomographic Imaging. IEEE; New York: 1988. [Google Scholar]