Summary
Premature anaphase onset is prevented by the mitotic checkpoint through production of a “wait anaphase” inhibitor(s) that blocks recognition of cyclin B and securin by Cdc20-activated APC/C, an E3 ubiquitin ligase which targets them for destruction. Using physiologically-relevant levels of Mad2, Bub3, BubR1, and Cdc20, we demonstrate that unattached kinetochores on purified chromosomes catalytically generate a diffusible Cdc20 inhibitor or inhibit Cdc20 already bound to APC/C. Furthermore, the chromosome-produced inhibitor requires both recruitment of Mad2 by Mad1 that is stably bound at unattached kinetochores and dimerization-competent Mad2. We show that purified chromosomes promote BubR1 binding to APC/C-Cdc20 by acting directly on Mad2, but not BubR1. Our results support a model in which immobilized Mad1/Mad2 at kinetochores provides a template for initial assembly of Mad2 bound to Cdc20 that is then converted to a final mitotic checkpoint inhibitor with Cdc20 bound to BubR1.
Introduction
To ensure accurate segregation, the major cell cycle control mechanism in mitosis, the mitotic checkpoint (or the spindle assembly checkpoint), delays anaphase onset until all chromosomes have properly attached to spindle microtubules. The checkpoint-derived inhibitor(s) blocks premature destruction of key mitotic components. This is achieved by selectively inhibiting Cdc20 stimulated recognition of the mitotic regulators cyclin B and securin by a multisubunit E3 ubiquitin ligase, the Anaphase Promoting Complex/Cyclosome (APC/C). Checkpoint silencing and subsequent deactivation of the checkpoint arrest releases APC/CCdc20 for ubiquitination of cyclin B and securin, with anaphase triggered by their subsequent degradation by the proteosome (reviewed in (Peters, 2006)).
By correlating the timing of anaphase onset with spindle microtubule capture by the last unattached chromosome (Rieder et al., 1994), laser ablation of the last unattached kinetochore (Rieder et al., 1995) and micromanipulation (Li and Nicklas, 1995), unattached kinetochores were first implicated as essential for generation of the wait anaphase inhibitor. Key proteins essential for mitotic checkpoint signaling include Mad1, Mad2, Bub3, CENP-E, Zw10, Rod, and the kinases Mps1, Bub1 and BubR1 (reviewed by (Musacchio and Salmon, 2007)), each of which is at least transiently localized to unattached kinetochores during early mitosis. Fluorescence recovery after photobleaching (FRAP) demonstrated that Mad2, BubR1, and Cdc20 cycle on and off kinetochores rapidly (Howell et al., 2000; Howell et al., 2004; Kallio et al., 2002; Shah et al., 2004). Additionally, several APC/C subunits are at least partially localized onto unattached kinetochores (Acquaviva et al., 2004; Jorgensen et al., 1998), supporting the possibility that one or more of its components are sensitized for checkpoint inhibition there.
Inhibition of Cdc20 activation of APC/C has previously been attributed to Mad2 (Fang et al., 1998a) or BubR1 (Tang et al., 2001), both of which can bind Cdc20 directly and in so doing have been shown to reduce APC/CCdc20 ubiquitination activity accordingly. A complex, named the Mitotic Checkpoint Complex (MCC) and proposed to be comprised of Mad2, BubR1, Bub3 and Cdc20, has been reported to inhibit APC/C much more potently than Mad2 alone (Sudakin et al., 2001). However, the existence of MCC-like complexes has been noted outside of mitosis (Sudakin et al., 2001) or in the absence of a functional kinetochore (Fraschini et al., 2001). The simplest view is that an interphase mechanism independent of kinetochores generates a premade inhibitor(s) of Cdc20 that requires Mad2 and BubR1 and whose half life sets a minimum time before anaphase onset (Meraldi et al., 2004).
A “template” model for kinetochore-dependent activation of Mad2 (De Antoni et al., 2005), a modified version of the “two-state” model (Yu, 2006), has emerged from use of cultured cells and purified components in the absence of chromosomes and the discovery that Mad2 can undergo a large conformational change in which its carboxy terminal “seatbelt” domain encloses either Mad1 or Cdc20, thereby converting the initial Mad2 from an “open” (or “N1”) to a “closed” (or “N2”) conformation (Luo et al., 2000; Luo et al., 2002; Luo et al., 2004; Sironi et al., 2002; Sironi et al., 2001). The closed conformation has been proposed to sequester Cdc20 from binding to and activating APC/C for recognition of cyclin B through direct capture of Cdc20 by Mad2 (Luo et al., 2004). The carboxy terminal domain of Mad1 has been shown to directly bind a molecule of Mad2 in the closed conformation (Luo et al., 2002; Luo et al., 2004; Sironi et al., 2002; Sironi et al., 2001). FRAP has revealed that Mad1 at kinetochores, presumably bound to Mad2, is non-exchangeable, while two equally sized pools of kinetochore-associated Mad2 either cycle on and off rapidly (within a few seconds) or are more stably bound (Shah et al., 2004). Mad2 mutants impaired in dimerization were subsequently shown to be unable to support either mammalian mitotic checkpoint signaling in vivo (De Antoni et al., 2005; Mapelli et al., 2006) or capture of Cdc20 (using a 27 amino acid peptide to mimic Cdc20) by Mad2 in a complex with a 233 amino acid Mad2-binding fragment of Mad1 (De Antoni et al., 2005).
Use of in vitro FRAP demonstrated recruitment of a rapidly exchangeable Mad2 by a second stably associated Mad2 bound in the closed conformation to full length or a fragment of Mad1 (Vink et al., 2006). There is, however, no evidence for facilitating conversion of soluble Mad2 to Mad2-Cdc20 and the rate of Mad2 dissociation from the immobilized Mad1/Mad2 is independent of the Cdc20 peptide (Vink et al., 2006). Moreover, the majority of the cellular Mad2, which is present at approximately equimolar concentrations to Cdc20 during mitosis (Tang et al., 2001), remains in the open conformation that is less capable of interacting with Cdc20 (Luo et al., 2004). Thus, neither the identity(ies) of the checkpoint-derived wait anaphase inhibitor nor how unattached kinetochores participate in its production is established. We have now used purified components to determine that unattached kinetochores catalyze production of a diffusible wait anaphase inhibitor using a Mad2 template to prime Cdc20 for BubR1 binding.
Results
Mad2 recruitment to unattached kinetochores on purified chromosomes
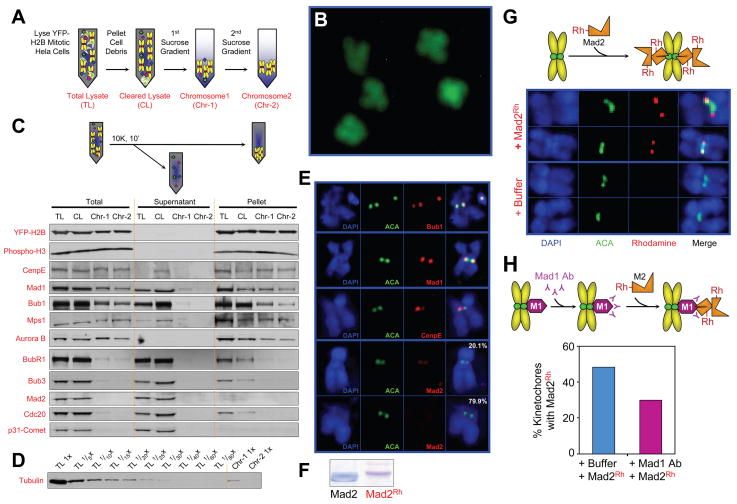
To reconstruct kinetochore-mediated mitotic checkpoint signaling with all purified components, successive sucrose gradients were used to isolate chromosomes with unattached kinetochores from colcemid arrested, mitotic Hela cells stably expressing histone H2B-YFP (Fig. 1A). Morphologically intact, condensed chromosomes were obtained, as observed by YFP fluorescence of unfixed chromosomes (Fig. 1B). Tubulin was reduced to less than 1/40th of its concentration in the initial cell extracts (Fig. 1D), while histones (including H2B-YFP and the mitotic-specific phosphorylated histone H3) and kinetochore-associated kinesin-like motor protein CENP-E were nearly quantitatively retained (Fig. 1C). Components previously reported by FRAP to be stably bound to unattached kinetochores, including Bub1 and Mad1 (Howell et al., 2004; Shah et al., 2004), were also retained, as was Mps1 and a proportion of Aurora B (Fig. 1C).
Fig. 1. Unattached kinetochores on purified chromosomes recruit Mad2.
(A) Schematic of chromosome purification from mitotic HeLa cells stably expressing YFP-H2B histone. Cells were collected after 16 hours in colcemid, lysed, cell debris removed by pelleting and the chromosome containing supernatant was fractionated on sequential sucrose gradients. (B) Morphology of purified chromosomes detected by fluorescence of YFP-H2B on coverslips without fixation. (C) Protein constituents of purified chromosomes assessed by immunoblotting after pelleting. (D) Tubulin levels remaining in purified chromosomes, along with a dilution series of the initial cellular input. (E) Indirect immunofluorescence for detection of Mad1, Bub1, CENP-E and Mad2 on isolated chromosomes. (Blue) Chromosomes stained with DAPI; (Green) Anticentromere (ACA) antibodies; (right panel) merged image. (F) Purified recombinant Mad2 before and after covalently labeling with rhodamine, assessed by Coomassie staining. (G) Purified chromosomes were incubated with rhodamine-labeled Mad2, fixed, stained for (blue) DAPI and (green) ACA, and imaged by deconvolution microscopy. (H) Chromosomes were incubated for 10 min with anti-Mad1 antibody, then with rhodamine-labeled Mad2, and finally fixed, stained, imaged as in (G), and scored for Mad2 localization.
Using anti-centromere antisera (ACA) and immunofluorescent staining of fixed, DAPI stained chromosomes, Bub1, Mad1 and CENP-E were demonstrated to remain kinetochore-bound (Fig. 1E). Components for which FRAP had revealed rapid cycling, including BubR1, Bub3, and Cdc20, were undetectable by immunoblotting of the purified chromosome fractions (Fig. 1C) (their levels in purified chromosomes were all ≤ 1.1% of the cytosolic levels – Supp. Fig. 1A,C). Although a portion of Mad2 is known to be stably bound at an unattached kinetochore for at least two minutes in vivo (Shah et al, 2004), Mad2 nearly completely dissociated from most kinetochores during the 7 hours required for chromosome isolation (Fig. 1C), with faint levels detectable by immunofluorescence remaining only on a minority (20.1%) of isolated chromosomes (Fig. 1E).
Bacterially produced Mad2 was isolated under conditions promoting primarily the retention of the open monomeric conformation, the conformation thought to be representative of cytosolic Mad2 (Luo et al., 2004; Mapelli and Musacchio, 2007). After covalent ligation of rhodamine (Fig. 1F; see Supp. Fig. 3A for activity assessment) and addition to purified chromosomes, rhodamine-Mad2 bound to unattached kinetochores of the majority of chromosomes (Fig. 1G; see also Supp. Fig. 3C for quantification). At least a portion of this Mad2 binding was mediated through kinetochore associated Mad1, as demonstrated by partial blocking even by brief pre-incubation of the chromosomes with an antibody against Mad1 (Fig. 1H).
Generation by unattached kinetochores of an APC/C-Cdc20 inhibitor
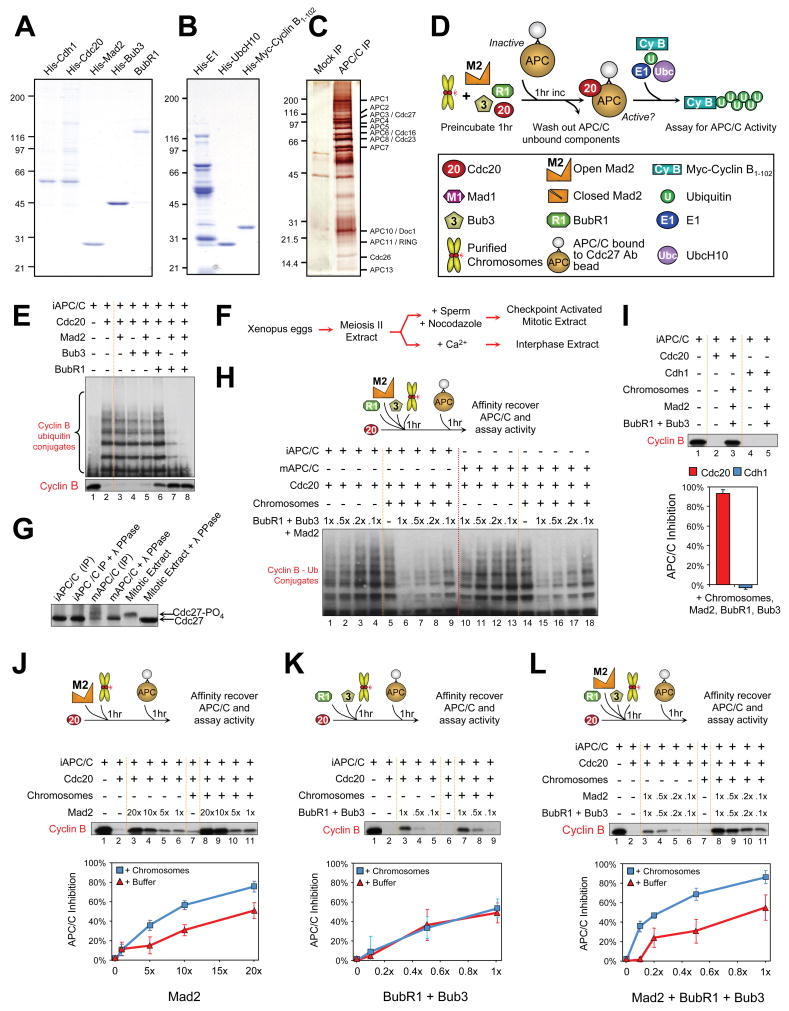
To determine how unattached kinetochores produce a mitotic checkpoint signal, we established an in vitro assay for Cdc20-stimulated ubiquitination by APC/C. Human homologues of Mad2 and BubR1, as well as Bub3, Cdc20, and the APC/C G1-specific activator Cdh1 (Fang et al., 1998b) were purified after production in bacteria or in insect cells using baculovirus (Fig. 2A,B). APC/C was immunoprecipitated (Fig. 2C) from Xenopus interphase or mitotic extracts with an antibody against the Cdc27 subunit. Immunoblotting of mitotic APC/C revealed a slowed mobility of Cdc27 both in the initial mitotic extract and after isolation (Fig. 2G), a shift known to reflect mitotic phosphorylation (Kraft et al., 2003). Addition of Cdc20 to either APC/C equivalently activated the ubiquitination of cyclin B1-102 (Fig. 2E,H), which could be quantified either by the presence of slower mobility cyclin B species (Fig. 2E, top; 2H) or the intensity of the remaining un-ubiquitinated cyclin B (Fig. 2E, bottom).
Fig. 2. Kinetochores amplify production of an APC/CCdc20 inhibitor.
(A) Purified recombinant human Cdh1, Cdc20, Mad2, Bub3 and BubR1, assessed by Coomassie staining. (B) Purified recombinant human E1, UbcH10, and N-terminal of cyclin B, assessed by Coomassie staining. (C) Interphase Xenopus APC/C after immunoprecipitation with immobilized antibodies to Cdc27 and visualized by silver stain. (D) Schematic of APC/C ubiquitination activity assays. (E) Equal molar amounts of Mad2, Bub3, and BubR1 were incubated with Cdcd20 either alone or in various combinations, in the absence of unattached kinetochores. APC/C activity was assessed either as the degree of cyclin B ubiquitination (top panel) or as the depletion of the unubiquitinated pool (bottom panel). (F-H) Kinetochores on purified chromosome amplify inhibition of mitotic APC/CCdc20. (F) Schematic of Xenopus extract preparation for isolation of mitotic APC/C by immunoprecipitation. (G) Immunoprecipitated mitotic APC/C: hyperphosphorylation retards mobility relative to interphase APC/C after immunoblotting for Cdc27. Hyperphosphorylation is lost upon phosphatase treatment. (H) Comparison of mitotic and interphase APC/C activity (quantified by diminished abundance of lower mobility ubiquitin-conjugated cyclin B species) assessed after addition of increasing amounts of BubR1, Bub3, and Mad2 to Cdc20, either in the presence or absence of chromosomes. (I) BubR1, Bub3, Mad2 and chromosomes were incubated with either (red bar) Cdc20 or (blue bar) Cdh1 activators, prior to APC/C activity determination. Incubations with Cdc20 rendered the APC/C almost fully inactive, while Cdh1 incubations had no effect on the activity of APC/C. (J-L) Chromosomes (blue squares) at a final concentration equaling ten unattached kinetochores per cell volume or just buffer (red triangles) were added to increasing concentrations of (J) Mad2, (K) Bub3/BubR1, or (L) both, incubated for 1 hr prior to addition of APC/C, and then assayed for APC/C ubiquitination of myc-cyclin B1-102. APC/C activity was quantified by the intensity of remaining unubiquitinated cyclin B. Mad2 inhibition of APC/C in the presence of chromosomes increased, while BubR1 inhibition remained unchanged. Chromosomes further amplified APC/C inhibition when added to the combination of Mad2, BubR1, and Bub3 at physiological concentrations.
Basal inhibition of APC/CCdc20 ubiquitination activity by our purified checkpoint proteins was initially assayed in the presence of equal molar amounts of Mad2, BubR1, or Bub3 to approximate the relative in vivo stoichiometries (see Supp. Fig. 1). Under these conditions addition of Mad2, Bub3 or BubR1 alone did not significantly inhibit ubiquitination of cyclin B, while the combination of Mad2 and BubR1 did, independent of Bub3 (Fig. 2E, compare lanes 3-6 with lanes 7,8). Inhibition was selective for Cdc20-mediated activation of APC/C, as similar addition to APC/CCdh1 left ubiquitination activity undiminished (Supp. Fig. 2A). BubR1 was a significantly better inhibitor of Cdc20 activation of APC/C: a >10 fold molar excess of Mad2 over Cdc20 was necessary to achieve >50% inhibition, whereas 10 times less of BubR1 with or without Bub3 was required for equivalent APC/CCdc20 inhibition (Fig. 2J,K; Supp. Fig. 2B). Nevertheless, there was synergism between Bub3/BubR1 and Mad2. Inhibition by the combination of Bub3/BubR1 and Mad2 was greater than the additive effect of each alone. Suppression of cyclin B ubiquitination by BubR1 was enhanced by Mad2 such that a quarter of the amount of BubR1 was required for equivalent inhibition of Cdc20 in the presence of Mad2 (Supp. Fig. 2B, lane 8 vs 13), even though the same amount of Mad2 alone produced no inhibition (Supp. Fig. 2B, lane 6 vs 13), similar to an earlier report in which Mad2 and Bub3/BubR1 cooperate to inhibit Cdc20 even in the absence of unattached kinetochores (Fang, 2002).
Addition to APC/CCdc20 of increasing levels of Mad2 alone produced dose dependent inhibition of cyclin B ubiquitination (Fig. 2J, lanes 3-6), but even a 20 fold excess of Mad2 over Cdc20 produced only 50% inhibition. In the absence of chromosomes, BubR1 yielded comparable inhibition of APC/CCdc20 at 10-20 fold lower levels than required for Mad2. Although purified chromosomes alone minimally inhibited APC/CCdc20, addition of them to a concentration approximating ten unattached kinetochores per cell volume amplified the inhibition produced at all concentrations tested of added Mad2 (Fig. 2J, lanes 8-11), but not BubR1 (Fig. 2K). Equivalent inhibition of mitotic and interphase APC/CCdc20 by a combination of Mad2, Bub3 and BubR1 was seen at all concentrations both in the presence and absence of chromosomes (Fig. 2H). For example, although equimolar additions of Mad2, Bub3, and BubR1 produced minimal inhibition of Cdc20-dependent activation of APC/C, unattached kinetochores produced equivalent inhibition at 10 fold lower levels of added Mad2 and BubR1/Bub3 (Fig. 2H, compare lanes 1 versus 9 and 10 versus 18). Moreover, chromosome–dependent inhibitory activity was selective for Cdc20, as APC/CCdh1 ubiquitination of cyclin B was unaffected even at maximal doses of Bub3/BubR1, Mad2 and chromosomes (Fig. 2I). Chromosome amplification of APC/CCdc20 inhibition was greatest at the lowest concentrations of added Mad2 and BubR1/Bub3, with maximal inhibition 35 fold that seen in the absence of chromosomes (Fig. 2L, 0.1x concentration).
Mad2 dimerization is required for kinetochore amplification of Cdc20 inhibition
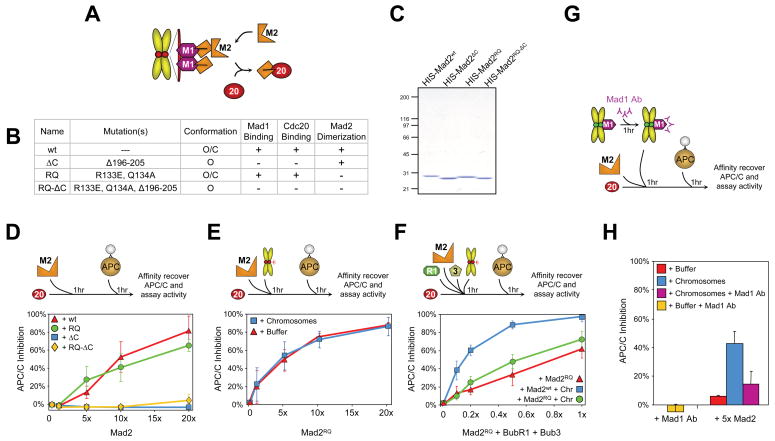
To determine if recruitment of Mad2 to unattached kinetochores via Mad1 (Chen et al., 1998) and Mad2 binding directly to Cdc20 were required for chromosome amplification of APC/CCdc20 inhibition, APC/C activity assays were performed with wild type Mad2 or a Mad2 mutant that can bind to kinetochore-associated Mad1 or Cdc20, but is incompetent for dimerization onto Mad1/Mad2 complexes (Fig. 3A). For this, we chose the Mad2RQ mutant [carrying the two amino acid substitution R133E, Q134A], previously demonstrated as incapable of supporting full mitotic checkpoint function in vivo (De Antoni et al., 2005) (Fig. 3B,C). Incubation of rhodamine-labeled Mad2RQ (Supp. Fig. 3B) with purified chromosomes yielded Mad2 localization to kinetochores with equal frequency as wild type Mad2 (Supp. Fig. 3C), yet with approximately half the intensity (Supp. Fig. 3D). Thus, Mad2RQ bound directly to Mad1 at kinetochores mostly depleted of endogenous Mad2 (Fig. 1C), yet was unable to recruit a second Mad2 molecule to form Mad2 dimers. In the absence of chromosomes, Mad2RQ inhibited Cdc20-stimulated APC/C ubiquitination almost as well as wild type Mad2 did (Fig. 3D; Supp. Fig. 4A). This Cdc20 inhibition required direct binding of Mad2 to Cdc20: Mad2 mutants (Mad2ΔC or Mad2RQ-ΔC; Fig. 3B,C) missing the carboxy-terminal 10 amino acids that are required for Cdc20 binding (Luo et al., 2000) did not inhibit APC/CCdc20 at any concentration (Fig. 3D; Supp. Fig. 4A).
Fig. 3. Mad1-dependent Mad2 conformational change and dimerization are required for kinetochore-mediated amplification of a Cdc20 inhibitor.
(A) Schematic of Mad2 template model (De Antoni et al., 2005). (B) Table of Mad2 mutant properties (De Antoni et al., 2005; Fang et al., 1998a; Luo et al., 2000; Luo et al., 2004; Sironi et al., 2002). (C) Purified recombinant human Mad2ΔC, Mad2RQ, and Mad2RQ-ΔC visualized by Coomassie staining. (D) Increasing quantities of (red triangles) Mad2wt, (blue squares) Mad2ΔC, (green circles) Mad2RQ, or (yellow diamonds) Mad2RQ-ΔC were incubated for 1 hour with Cdc20 prior to APC/C addition and assayed for APC/C ubiquitination of myc-cyclin B1-102. (E) Mad2RQ inhibition of Cdc20 activation of APC/C assessed after incubating increasing quantities with Cdc20 either in the (blue squares) presence or (red triangles) absence of chromosomes before assaying cyclin B ubiquitination. (F) Testing chromosome amplification of a Cdc20 inhibitor after increasing concentrations of (green circles) Mad2RQ or (blue squares) Mad2wt, along with BubR1, Bub3, and Cdc20, either in the presence or (red triangles) absence of chromosomes. (G) Purified chromosomes were incubated with Mad1 antibody for 1 hour prior to addition of a five-fold excess of Mad2 to Cdc20, and finally addition to APC/C ubiquitination assays and (H) inhibition of AP/CCCdc20 was measured.
In contrast to wild type Mad2, the dimerization deficient Mad2RQ was incapable of supporting chromosomal amplification of APC/C inhibition at any added concentration (Fig. 3E; Supp. Fig. 4B). Moreover, when tested for synergy with Bub3/BubR1 and chromosomes in inhibiting APC/CCdc20, Mad2RQ was much less efficient than wild type Mad2 (Fig. 3F; Supp. Fig. 4C), thus supporting action by kinetochores on Mad2 for amplifying generation of a Cdc20 inhibitor.
Kinetochore-bound Mad1 is required for chromosome-mediated amplification of a Cdc20 inhibitor
To further test the Mad1 role in the chromosome-dependent amplification of an APC/CCdc20 inhibitor, a Mad1 antibody raised against the region that spans the Mad2 binding domain was added to isolated chromosomes in an effort to inhibit its function at kinetochores (Fig. 3G). This substantially reduced Mad2 recruitment to kinetochores (Fig. 1H), as anticipated. In the absence of chromosomes or Mad2, Mad1 antibody had no effect on APC/CCdc20 ubiquitination (Fig. 3H). In the presence of a five fold excess of Mad2 (the concentration chosen for the greatest enhancement of inhibition upon addition of chromosomes), chromosomes amplified APC/CCdc20 inhibition seven fold, but this amplification was almost eliminated by Mad1 antibody addition (Fig. 3H; Supp. Fig. 4D). Thus, Mad1 at kinetochores is required both for kinetochore recruitment of Mad2 and chromosome-mediated enhancement of inhibition of APC/CCdc20.
Kinetochore-enhanced inhibition of Cdc20-bound to APC/C
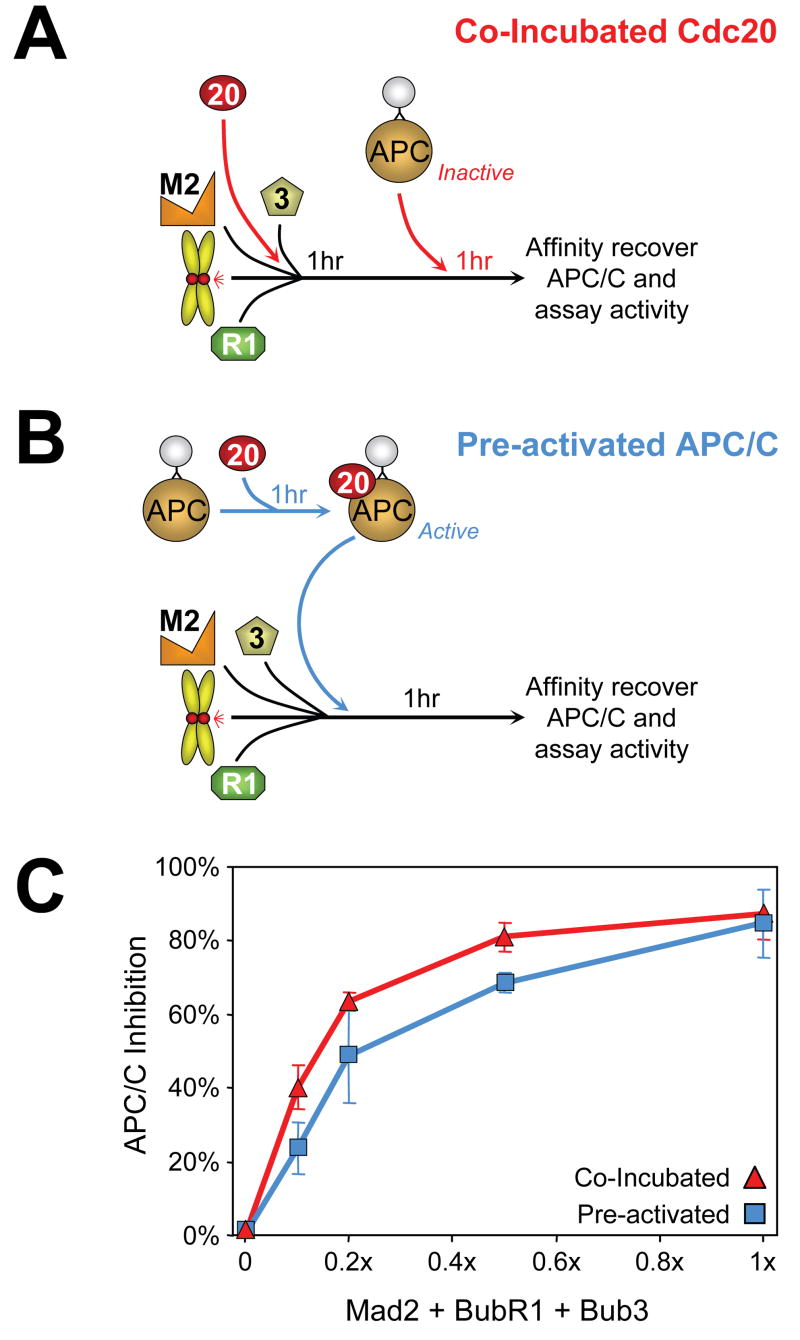
Models of APC/C inhibition in mitosis initially focused upon sequestration of Cdc20 (Fang et al., 1998a; Tang et al., 2001) as the mode of preventing Cdc20 activation of APC/C. An alternative is for inhibition by a checkpoint derived inhibitor binding directly to Cdc20 already bound to APC/C. To distinguish between these models, inhibition of APC/CCdc20 activity was assessed either by pre-incubation of chromosomes, Bub3/BubR1, Mad2, and Cdc20 followed by addition of APC/C (Fig. 4A) or by addition of chromosomes, Bub3/BubR1, and Mad2 to Cdc20 pre-bound to APC/C (Fig. 4B). At all concentrations of checkpoint components, inhibition of APC/CCdc20-mediated ubiquitination of cyclin B1-102 was comparable following co-incubation with Cdc20 or after pre-activation by binding of Cdc20 to APC/C (Fig. 4C; Supp. Fig. 5). Thus, APC/CCdc20 can be directly inhibited by a kinetochore derived inhibitor(s) and a model of simple sequestration of Cdc20 cannot be the sole means by which APC/C is held inactive for cyclin B ubiquitination.
Fig. 4. Inhibition of APC/C activation is not achieved solely by sequestration of Cdc20.
(A) Checkpoint components including Cdc20 and chromosomes were incubated, followed by APC/C addition and assay for its activity. (B) Immunoprecipitated APC/C was first incubated with Cdc20 to form an active complex (“Pre-activated APC/C”). APC/C was affinity recovered and subsequently incubated with chromosomes and increasing amounts of BubR1, Bub3, and Mad2, and APC/C activity was assayed. (C) Quantitation of (red triangles) co-incubated and (blue squares) pre-activated Cdc20-stimulated APC/C activity.
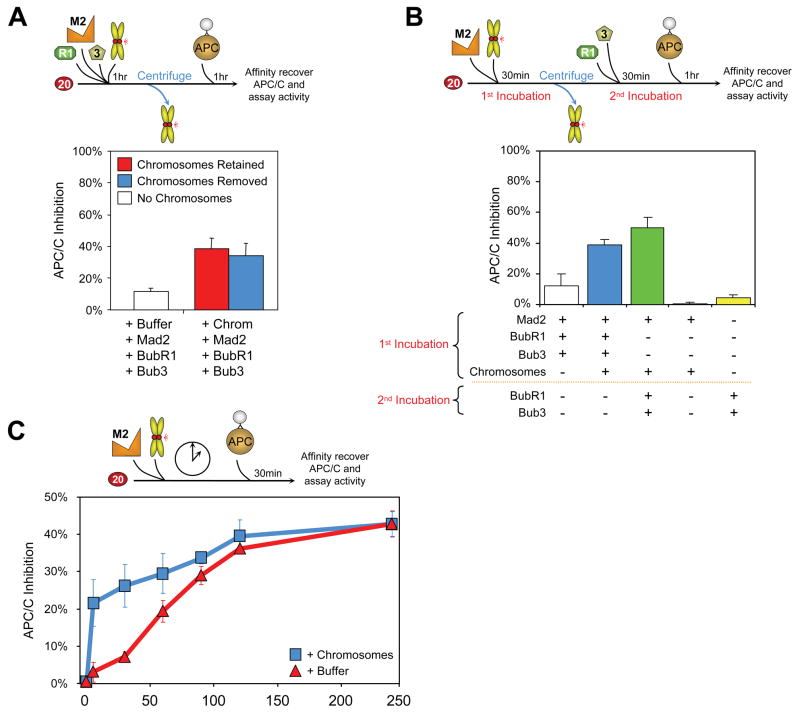
A soluble kinetochore-derived Cdc20-BubR1-Bub3 inhibitor
To test whether a soluble kinetochore derived inhibitor of Cdc20 can be produced, chromosomes with unattached kinetochores were incubated with BubR1, Bub3, Mad2, and Cdc20, but then removed prior to the addition of APC/C (Fig. 5A; Supp. Fig. 6A). In the absence of chromosomes, incubation of Cdc20 and a low level of BubR1, Bub3, Mad2 and Cdc20, followed by subsequent addition of APC/C, produced almost fully active APC/CCdc20. Parallel incubation of the same amounts of BubR1, Bub3, Mad2 and Cdc20 but now in the presence of chromosomes, amplified APC/C inhibition greater than threefold, independently of whether chromosomes were removed prior to APC/C addition. Thus, a soluble inhibitor(s) amplified by unattached kinetochores is produced independently of APC/C.
Fig. 5. Catalytic production by unattached kinetochores of a diffusible Cdc20 inhibitor through sequential involvement of Mad2 and BubR1.
(A) Chromosomes were incubated with BubR1, Bub3, Mad2, and Cdc20 (1:1:1:5); the chromosomes were subsequently removed by centrifugation, and (blue) the supernatant fraction was assayed for activation of APC/C for cyclin B ubiquitination. (Red) Parallel assay was done without chromosome removal or (white) without initial chromosome addition. (B) Chromosomes were initially incubated with Mad2 and Cdc20 (1:5); the chromosomes were subsequently removed, and BubR1/Bub3 added to the supernatant fraction after chromosome removal (green). The resulting ubiquitination activity was compared to (blue) inhibition produced by incubating chromosomes with Mad2, BubR1, and Bub3 prior to chromosome removal or (white) no chromosomes added. (C) Cdc20 was incubated with a 5-fold excess Mad2 and chromosomes. At the indicated time points, (blue squares) a fraction of the incubation was removed and assayed for APC/C activity. Chromosome-mediated catalysis was compared to (red triangles) chromosome-independent inhibitor production over time.
Unattached kinetochores act on Mad2 for amplifying a Cdc20 inhibitor
To test if unattached kinetochores act directly on Mad2, chromosomes and Cdc20 were incubated with Mad2, the chromosomes were then removed, BubR1/Bub3 was added, and finally APC/C was added and assayed for ubiquitination of cyclin B. Comparable to the stimulated inhibition of Cdc20 found when all components were incubated together with chromosomes (Fig. 2L, 5A), a 4 fold stimulation of inhibitory activity was generated when only Mad2 and Cdc20 were co-incubated with chromosomes (Fig. 5B; Supp. Fig. 6B). Thus, amplification of a Cdc20 inhibitor with purified components does not require unattached kinetochores acting directly on either APC/C or BubR1/Bub3, but rather can be produced by direct interaction with Mad2 and/or Cdc20.
Catalytic production by unattached kinetochores of a Mad2 Cdc20 inhibitor
A central unresolved question is whether unattached kinetochores act catalytically in the production of a Cdc20 inhibitor. A central requirement of a catalytic model would be for unattached kinetochores to accelerate the rate of production of a Cdc20 inhibitor, while an extended incubation without kinetochores could ultimately yield comparable inhibition mediated by an uncatalyzed, spontaneous process. To test the catalytic model, Mad2 and Cdc20 were added to a level sufficient to yield 40% inhibition after extended incubation without addition of chromosomes. In the absence of chromosomes, a linear increase in inhibition of Cdc20 was produced over the first 120 minutes, ultimately plateauing at about 40% inhibition by 4 hours (Fig. 5C; Supp. Fig. 6C). As required for chromosome-dependent catalysis, the presence of a concentration of chromosomes corresponding to 10 unattached kinetochores per cell accelerated the initial rate of inhibitor production 8 fold (initial slopes of 2.5 versus 0.3 % inhibition/min in the presence and absence of chromosomes, respectively) (Fig. 5C), with the final level of inhibition similar to that produced spontaneously.
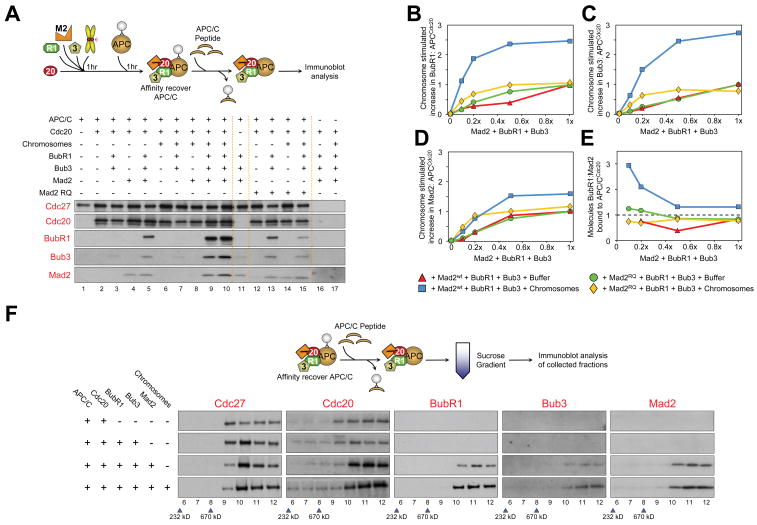
Kinetochores facilitate Bub3/BubR1 binding to APC/CCdc20
To probe whether the unattached kinetochores on purified chromosomes altered the composition of proteins bound to APC/C, a peptide-derived antibody to Cdc27 (Herzog and Peters, 2005) was used to affinity purify APC/C. After preincubation of Cdc20, BubR1, Bub3, Mad2 and chromosomes, bead-bound APC/C was added, the beads were recovered, and APC/C and proteins bound to it were released by addition of a competing Cdc27 peptide. Immunoblotting revealed that similar amounts of Cdc20 co-immunoprecipitated with APC/C regardless of the concentrations of co-incubated checkpoint components (Fig. 6A).
Fig. 6. Unattached kinetochores facilitate BubR1, Bub3, and Mad2 association with APC/CCdc20.
(A)APC/C was incubated with pre-incubated combinations of Cdc20, Mad2, BubR1, Bub3 and chromosomes, recovered, peptide-eluted from Affiprep beads, and analyzed for bound components by immunoblotting. (B-E) APC/C was incubated with Cdc20 and increasing amounts of BubR1, Bub3, and Mad2wt (or Mad2RQ) either in the (blue squares) presence or (red triangles) absence of chromosomes, and treated as in (A). The amounts of eluted (B) BubR1, (C) Bub3, and (D) Mad2wt (or Mad2RQ) were measured against a dilution series of purified protein and quantified relative to the amount of Cdc20 bound to APC/C. Values were plotted as fold change over the initial (1x) non-chromosome incubated eluted amounts. (E) The relative stoichiometry of BubR1 molecules to Mad2 molecules associated with the APC/C complex. BubR1 relative stoichiometry to Mad2 increased above 1:1 when chromosomes were present, but remained approximately 1:1 or below when Mad2wt was replaced with Mad2RQ regardless of the presence of chromosomes. (F) APC/C incubated with combinations of Cdc20, Mad2, BubR1, Bub3 and chromosomes was recovered, eluted from Affiprep beads, fractionated over a sucrose gradient, and analyzed by immunoblotting for bound components.
While a proportion of Mad2 has previously been reported to be associated with APC/C (Fang et al., 1998a; Kallio et al., 1998), Mad2 binding to APC/C was not Cdc20 dependent (lane 11). A small, variable proportion of Mad2 and Mad2RQ bound to APC/C independent of chromosomes, revealing a direct affinity of Mad2 for APC/C that was modestly increased by the presence of chromosomes (Fig. 6A,D,E). Neither BubR1 nor Bub3 bound APC/C in the absence of Mad2. Bub3/BubR1 did, however, bind to APC/C in a Mad2 dependent manner that at all concentrations tested was further enhanced by up to 4 fold by chromosomes (Fig. 6A, compare lanes 4 with 9 and 10; Fig. 6B,C). However, the number of BubR1 molecules bound to APC/CCdc20 after incubation with chromosomes was always greater than the number of bound Mad2 molecules, as seen by a stoichiometry of greater than 1:1 (Fig. 6E), inconsistent with MCC-like complexes which would contain equal stoichiometries of Mad2 and BubR1/Bub3. This chromosome-enhanced BubR1/Bub3 binding was not supported by the dimerization incompetent Mad2RQ (Fig. 6A, lanes 13 versus 15; Fig. 6B,C,E). Sucrose gradient sedimentation of the complexes released from the initial antibody coated beads confirmed a 2-3 fold, chromosome-dependent increase in BubR1/Bub3 bound to APC/C (Fig. 6F). Thus, Bub3/BubR1 binding to APC/C is facilitated by dimerization competent Mad2 and unattached kinetochores through a mechanism that does not affect Cdc20 interaction with APC/C.
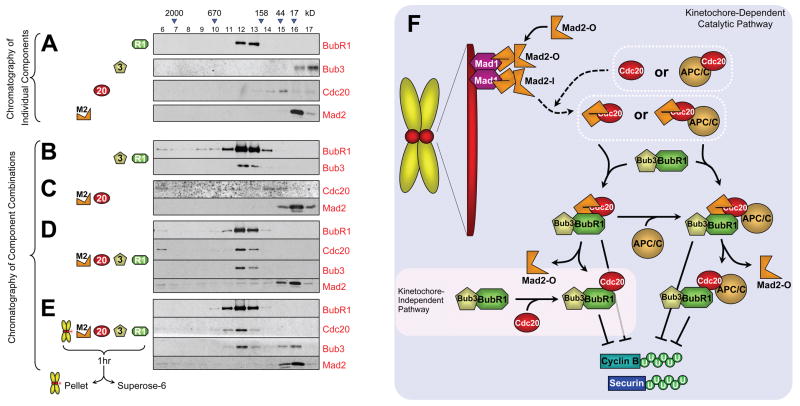
A soluble kinetochore-derived Cdc20-BubR1-Bub3 inhibitor
To determine the composition(s) of complexes produced spontaneously or by action of unattached kinetochores in the absence of APC/C, equal stoichiometries of various combinations of BubR1, Bub3, Mad2, and Cdc20 were incubated with or without purified chromosomes, the chromosomes were removed and the inhibitor-containing supernatant (as in Fig. 4A) was subjected to size exclusion chromatography. As expected, Bub3 shifted into a larger complex, co-eluting with BubR1 under all conditions. Most Cdc20 shifted into a substantially higher molecular weight complex both in the presence (Fig. 7E) and absence (Fig. 7D) of chromosomes, eluting together with BubR1 and Bub3 (centered on fractions 12 and 13). Except for a small proportion of Mad2 bound to Cdc20 when incubated alone with it (Fig. 7C), surprisingly little Mad2 chromatographed with Cdc20 under any condition, eluting instead at a position corresponding to a Mad2 monomeric form, regardless of the presence of chromosomes and/or BubR1/Bub3. Very little Mad2 was found in an MCC-like tetrameric complex with BubR1, Bub3, and Cdc20 in the absence of chromosomes, even when using conditions that produced up to 80% inhibition of Cdc20’s ability to activate APC/C (Fig. 2L). Incubation with chromosomes eliminated even this small amount of MCC-like complex (Fig. 7D,E). On the other hand, incubation with chromosomes produced a proportion of BubR1 and Cdc20 that eluted earlier (e.g., fraction 11), consistent with production of a larger complex or one with a more extended structure so as to produce a higher Stoke’s radius. This complex was comprised of approximately equal molar amounts of BubR1 and Cdc20, but only trace levels of Mad2.
Fig. 7. Kinetochores catalyze the production of a BubR1-Cdc20 inhibitor without stably associated Mad2.
(A) Individual components or (B-E) combinations of Cdc20, Mad2, BubR1, Bub3, and/or chromosomes were incubated for 1hr and subsequently fractionated over a Superose-6 filtration column. (E) Mixtures containing chromosomes were removed by pelleting the chromosomes prior to loading onto the column. Fractions eluted from the column were analyzed for BubR1, Bub3, Cdc20, and Mad2 content by immunoblotting for those components. (F) Model for generation of a “wait anaphase” mitotic checkpoint inhibitor by sequential production of Mad2-Cdc20 and BubR1-Cdc20 inhibitors. Cytosolic Mad2 in an initially open conformation is recruited to unattached kinetochores via an immobilized Mad1:Mad2 heterodimer. This second molecule of Mad2 binds in an activated conformation that is poised for capture of Cdc20 either while kinetochore bound or after release. This transient Mad2-Cdc20 complex promotes handoff of Cdc20 to BubR1, thereby inhibiting ability of that Cdc20 to activate ubiquitination by APC/C of cyclin B, both by sequestering Cdc20 from APC/C and by inhibiting Cdc20 while APC/C bound.
Discussion
Unattached kinetochores catalyze production of a “wait anaphase” inhibitor
While unattached kinetochores have been widely inferred to be the source of a “wait anaphase” mitotic checkpoint inhibitor, we have now demonstrated that kinetochores can, in fact, catalyze production of an initial Mad2-Cdc20 inhibitor, significantly accelerating the initial rate of its production. Unattached kinetochores did not affect inhibition by Bub3/BubR1 in the absence of Mad2. Production of at least two inhibitors can be enhanced by unattached kinetochores: one containing diffusible Cdc20 and another in which Cdc20 is already bound in a megadalton complex to APC/C, consistent with reports that Cdc20 and checkpoint proteins are present in two complexes with differing sizes during mitosis (Braunstein et al., 2007; Morrow et al., 2005; Sudakin et al., 2001; Wang et al., 2006). Both inhibitors prevent recognition by APC/C of cyclin B as an ubiquitination substrate. Disruption of cyclin B ubiquitination by a kinetochore-derived inhibitor even while Cdc20 remains bound to APC/C provides a potential explanation for the differential timing of destruction of cyclins A and B. Instead of simple sequestration of Cdc20, a kinetochore-derived mitotic checkpoint inhibitor bound to APC/CCdc20 may block recognition of cyclin B as an ubiquitination substrate, while permitting APC/CCdc20-mediated ubiquitination and destruction of cyclin A, an event that is known to initiate immediately after mitotic entry (den Elzen and Pines, 2001; Geley et al., 2001).
Despite amplification of Cdc20 inhibition when equal molar levels of BubR1, Mad2 and Cdc20 were added, we found no evidence for assembly of a quaternary MCC-like complex as a bona fide inhibitor produced by unattached kinetochores. Rather, almost all Cdc20 shifted to a complex co-migrating with the majority of BubR1, but containing very little Mad2 (Fig. 7E). Also arguing against a contribution in kinetochore-derived checkpoint signaling, we note that MCC-like complexes in animal cells are present outside of mitosis (Sudakin et al., 2001), and their formation in yeast continues in the absence of a functional centromere/kinetochore (Fraschini et al., 2001). All of this supports an MCC-like, premade Cdc20 inhibitor produced in a kinetochore-independent manner in interphase that restrains APC/C ubiquitination activity for cyclin B just after mitotic entry, which has been referred to as a “timer” (Meraldi et al., 2004).
More importantly, at physiologically relevant concentrations of unattached kinetochores and Mad2, chromosomes catalyzed production of Cdc20 inhibition of cyclin B recognition by APC/C by at least 8 fold relative to inhibitors formed spontaneously in the absence of chromosomes. The actual in vivo effect is likely to be much greater than what we have observed in vitro, since chromosome purification resulted in partial loss of signaling molecules from kinetochores, including a proportion of Mad1 and kinases that include Bub1, BubR1 and Aurora B.
Sequentially produced mitotic checkpoint inhibitors initiated by a Mad1/Mad2 template at kinetochores
Chromosome amplification of Cdc20 inhibition required Mad1 recruitment of Mad2 to kinetochores and dimerization competent Mad2 (Fig. 3E,F), thereby providing a direct demonstration that a Mad1:Mad2 core complex recruits and converts soluble “inactive” Mad2 into a more potent inhibitor of Cdc20. At least part of this is from action of kinetochores on Mad2. Although it has previously been argued that the kinetochore may sensitize the APC/C for checkpoint-mediated inhibition (Acquaviva et al., 2004; Sudakin et al., 2001), direct contact of chromosomes with APC/C was not required to amplify inhibition (Fig. 5A,B). While we by no means exclude a kinetochore-dependent function of BubR1 for roles in microtubule attachment and chromosome alignment (Ditchfield et al., 2003; Elowe et al., 2007; Lampson and Kapoor, 2005) or for further amplification of a kinetochore derived signal (Mao et al., 2003), kinetochore-mediated enhancement of Cdc20 inhibition did not require BubR1 localization to or contact with kinetochores. We conclude that immobilized, kinetochore-bound Mad1/Mad2, but not BubR1, catalyzes conversion at the kinetochore of recruitment of soluble, open Mad2 into a form with its seatbelt domain poised for Cdc20 capture.
Moreover, incubation of physiologically relevant concentrations of each component ultimately produced most Cdc20 bound to BubR1, not Mad2, whether or not chromosomes were present (Fig. 7D,E). In fact, amplification of Cdc20 inhibition by unattached kinetochores was accompanied by a shift to a more rapidly eluting Bub3/BubR1-Cdc20 complex, without a stable pool of Mad2-Cdc20. We propose a model from all of this (Fig. 7F) in which Mad1/Mad2 immobilized at kinetochores templates conversion of an inactive, open Mad2 to one capable of transient capture of Cdc20, followed by relay to BubR1 as sequentially produced mitotic checkpoint inhibitors that may be soluble or APC/C-bound. This evidence supports Mad2-Cdc20, and perhaps an MCC-like complex, as a transient intermediate in kinetochore-mediated checkpoint signaling and one that is a precursor to BubR1-Cdc20. Further, Bub3/BubR1 binds to APC/C but only in a Mad2-dependent manner that is stimulated by unattached kinetochores (Fig. 6A), demonstrating that kinetochores facilitate loading of Bub3/BubR1 onto APC/C. That BubR1-APC/CCdc20 is produced indirectly by unattached kinetochores as the final Cdc20 inhibitor would also support suggestions that BubR1 acts as a non-productive pseudosubstrate of the APC/C (Burton and Solomon, 2007) or mediates Cdc20 proteolytic turnover (King et al., 2007; Pan and Chen, 2004).
Combining kinetochore-derived Bub3/BubR1-Cdc20 with evidence for two Cdc20 binding sites on BubR1 (Davenport et al., 2006) further suggests that the spontaneous and kinetochore derived Bub3/BubR1-Cdc20 complexes may represent generation of Cdc20 bound at the two different sites respectively, a point now testable with the appropriate BubR1 mutants.
Experimental Procedures
Protein Purification
Recombinant proteins His-E1, His-UbcH10, His-Mad2wt, His-Mad2RQ, His-Mad2ΔC, His-Mad2RQ-ΔC, and His-Myc-Cyclin B1-102 were purified from bacteria after induction while His-Cdc20, His-Cdh1, His-Bub3 and GST-BubR1 were purified from SF9/Hi5 insect cells post baculovirus infection using a HIS tag purification as previously described (Tang and Yu, 2004). GST-BubR1 purified over glutathione Separose beads was eluted by PreScission protease digestion to cleave the GST tag and separated from oligomeric species by gel filtration. APC/C was immunoprecipitated from Xenopus egg extracts cycled into interphase with calcium addition, or from checkpoint active mitotic extracts post sperm and nocodazole addition.
Chromosome Purification
Hela cells stably expressing YFP-H2B were treated with 50ng/ml colcemid for 16 hrs. Mitotic cells were collected by shake-off and subjected to hypotonic conditions in modified PME buffer (MPME; 5 mM PIPES pH 7.2, 10 mM NaCl, 5 mM MgCl2, 0.5 mM EGTA, and 2 mM EDTA). The swollen cells were resuspended in 10 volumes of MPME buffer supplemented with 10 μg/ml LPC, 0.5 mM spermine, 1 mM Spermidine, 10 mM NaF, 1 mM NaVO4, 1 mM PMSF and 0.1% digitonin. The cells were disrupted using a dounce homogenizer to produce an initial lysate (total lysate). The lysate was centrifuged at 900 x g for 1 min to pellet intact nuclei and cell debris (cleared lysate). The NaCl concentration of the solution was elevated to 100 mM (hsMPME), and the lysate was placed over a sucrose step gradient (30%-40%-50%-60%) prepared with supplemented hsMPME and centrifuged for 15 min at 5000 x g. The flocculent white material at the 40%-50% and 50%-60% interface containing chromosomes was harvested (1st sucrose gradient). The chromosomes were washed in 8 volumes of supplemented hsMPME buffer, sedimented for 15 min at 2900 x g, suspended in supplemented hsMPME, placed over a second sucrose gradient, and reharvested as described above. The chromosomes were washed a second time and resuspended in 10 volumes of chromosome storage buffer (hsMPME containing 50% sucrose, 0.5 mM spermine, 10 mM NaF, 1 mM NaVO4, 10 μg/ml LPC), aliquoted, frozen in liquid nitrogen, and stored at -80°C.
APC/C Ubiquitination / Depletion Assays
APC/C ubiquitination assays were performed as previously described (Tang and Yu, 2004), utilizing different anti-Cdc27 antibodies depending on whether ubiquitination (Fang et al., 1998a) or depletion of cyclin B was measured (Tugendreich et al., 1995). To analyze the depletion assays, film exposures within a nearly linear range based on an internal loading gradient of inactive APC/C were selected. The intensity of bands corresponding to unubiquitinated cyclin B on immunoblots was quantified using NIH Image J software. The intensities of each lane were normalized against inactive APC/C (no Cdc20 added – 0%) and fully active APC/C (with Cdc20, no inhibitors added –100%). Each depletion assay was repeated at least in triplicate and the average represented in graph format, with bars representing standard error of the mean.
Immunofluorescence of Chromosomes
Purified chromosomes were fixed in formaldehyde, placed over a 33% glycerol cushion, and sedimented onto coverslips by centrifugation for 20 min at 5500 x g. The chromosomes were subsequently fixed in ice cold methanol and processed as described (Weaver et al., 2003). The coverslips were stained with the following antibodies: Hpx anti-CENP-E (Brown et al., 1996); sheep SB1 anti-Bub1 (Taylor et al., 2001); BB3-8 anti-Mad1 (De Antoni et al., 2005); rabbit anti-Mad2 (Kops et al., 2004) and ACA sera for identifying centromeres (Antibodies Inc.) Chromosomes were imaged using a DeltaVision deconvolution microscope (Applied Precision). Optical sections were taken at 0.15 intervals and deconvolved using SoftWoRx software (Applied Precision). The images were generated by projecting the sum of the stack of deconvolved images. Images were processed after equivalent scaling.
Labeling of Mad2 and its Localization onto Kinetochores
Mad2 and mutants of it were fluorescently labeled using FluoReporter Rhodamine Red-X Protein Labeling Kit by Molecular Probes. Equivalently labeled batches of Mad2 were selected. Chromosomes were incubated with 480 nM rhodamine-labeled Mad2 fractions for 1hr at room temperature. For Mad1 antibody blocking, the chromosomes were first incubated with the Mad1 antibody for 10 min and subsequently with the labeled Mad2. The chromosomes were treated for imaging as described above. Kinetochore fluorescence was quantified using the average intensity of traced kinetochore shape, as determined by ACA staining, using MetaMorph Imaging software (Molecular Devices).
APC/C Complex Affinity, Elution, and Sucrose Sedimentation
APC/C was immunoprecipitated using a peptide-derived Cdc27 antibody (Herzog and Peters, 2005) conjugated to Affiprep Protein A (BioRad) beads for 2 hrs from Xenopus interphase extracts. The washed APC/C beads were incubated with recombinant checkpoint proteins for 1hr at room temperature. The beads were washed twice with 20 volumes of TBS buffer to remove unbound proteins. The APC/C complex was eluted from the beads by Cdc27 peptide competition as described (Herzog and Peters, 2005) and analyzed by immunoblotting. Sucrose sedimentation of APC/C complexes was performed by scaling up APC/C elution five-fold and placing the eluate over a 5-35% sucrose gradient. The gradients were centrifuged for 4 hr at 50,000 rpm in a Beckman tabletop ultracentrifuge with TLS55 swinging bucket rotor. The fractions were collected from the top and proteins in each fraction precipitated with trichloroacetic acid (TCA) before used in immunoblotting.
Gel Filtration of Complexes of Mad2, BubR1, Bub3 and Cdc20
Equimolar combinations of BubR1, Bub3, Cdc20, and Mad2 proteins were incubated with or without purified mitotic chromosome for 30 min at room temperature and protein complexes generated were resolved by Superose 6 gel filtration. Proteins in each column fraction were concentrated by TCA precipitation and analyzed by immunoblotting.
Supplementary Material
Acknowledgments
We thank Andrea Musacchio for suggesting the catalytic experiment in Fig. 5. We would also like to thank Hongtao Yu and Bing Li for their guidance in APC/C ubiquitination assays. We thank Phil Heiter, Andrea Musacchio, Jan Michael Peters, Steven Taylor, Peter Sorger, Harold Fisk, and Hongtao Yu for providing reagents and Zahid Bonday for providing DNA constructs encoding Cdc20, BubR1, and Bub3. J.S.H. has been supported by a postdoctoral fellowship from the Leukemia and Lymphoma Society and from the Korea Research Foundation Grant funded (M01-2004-000-20329-0). This work has been supported by an NIH grant (GM29513) to D.W.C. Salary support for D.W.C. is provided by the Ludwig Institute for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- Acquaviva C, Herzog F, Kraft C, Pines J. The anaphase promoting complex/cyclosome is recruited to centromeres by the spindle assembly checkpoint. Nat Cell Biol. 2004;6:892–898. doi: 10.1038/ncb1167. [DOI] [PubMed] [Google Scholar]
- Braunstein I, Miniowitz S, Moshe Y, Hershko A. Inhibitory factors associated with anaphase-promoting complex/cylosome in mitotic checkpoint. Proc Natl Acad Sci U S A. 2007;104:4870–4875. doi: 10.1073/pnas.0700523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown KD, Wood KW, Cleveland DW. The kinesin-like protein CENP-E is kinetochore-associated throughout poleward chromosome segregation during anaphase-A. J Cell Sci. 1996;109(Pt 5):961–969. doi: 10.1242/jcs.109.5.961. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Mad3p, a pseudosubstrate inhibitor of APCCdc20 in the spindle assembly checkpoint. Genes Dev. 2007;21:655–667. doi: 10.1101/gad.1511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport J, Harris LD, Goorha R. Spindle checkpoint function requires Mad2-dependent Cdc20 binding to the Mad3 homology domain of BubR1. Exp Cell Res. 2006;312:1831–1842. doi: 10.1016/j.yexcr.2006.02.018. [DOI] [PubMed] [Google Scholar]
- De Antoni A, Pearson CG, Cimini D, Canman JC, Sala V, Nezi L, Mapelli M, Sironi L, Faretta M, Salmon ED, et al. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr Biol. 2005;15:214–225. doi: 10.1016/j.cub.2005.01.038. [DOI] [PubMed] [Google Scholar]
- den Elzen N, Pines J. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J Cell Biol. 2001;153:121–136. doi: 10.1083/jcb.153.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S, Hummer S, Uldschmid A, Li X, Nigg EA. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 2007;21:2205–2219. doi: 10.1101/gad.436007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 1998a;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998b;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fraschini R, Beretta A, Sironi L, Musacchio A, Lucchini G, Piatti S. Bub3 interaction with Mad2, Mad3 and Cdc20 is mediated by WD40 repeats and does not require intact kinetochores. Embo J. 2001;20:6648–6659. doi: 10.1093/emboj/20.23.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geley S, Kramer E, Gieffers C, Gannon J, Peters JM, Hunt T. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J Cell Biol. 2001;153:137–148. doi: 10.1083/jcb.153.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog F, Peters JM. Large-scale purification of the vertebrate anaphase-promoting complex/cyclosome. Methods Enzymol. 2005;398:175–195. doi: 10.1016/S0076-6879(05)98016-6. [DOI] [PubMed] [Google Scholar]
- Howell BJ, Hoffman DB, Fang G, Murray AW, Salmon ED. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells. J Cell Biol. 2000;150:1233–1250. doi: 10.1083/jcb.150.6.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell BJ, Moree B, Farrar EM, Stewart S, Fang G, Salmon ED. Spindle checkpoint protein dynamics at kinetochores in living cells. Curr Biol. 2004;14:953–964. doi: 10.1016/j.cub.2004.05.053. [DOI] [PubMed] [Google Scholar]
- Jorgensen PM, Brundell E, Starborg M, Hoog C. A subunit of the anaphase-promoting complex is a centromere-associated protein in mammalian cells. Mol Cell Biol. 1998;18:468–476. doi: 10.1128/mcb.18.1.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio M, Weinstein J, Daum JR, Burke DJ, Gorbsky GJ. Mammalian p55CDC mediates association of the spindle checkpoint protein Mad2 with the cyclosome/anaphase-promoting complex, and is involved in regulating anaphase onset and late mitotic events. J Cell Biol. 1998;141:1393–1406. doi: 10.1083/jcb.141.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallio MJ, Beardmore VA, Weinstein J, Gorbsky GJ. Rapid microtubule-independent dynamics of Cdc20 at kinetochores and centrosomes in mammalian cells. J Cell Biol. 2002;158:841–847. doi: 10.1083/jcb.200201135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King EM, van der Sar SJ, Hardwick KG. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS ONE. 2007;2:e342. doi: 10.1371/journal.pone.0000342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Foltz DR, Cleveland DW. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci U S A. 2004;101:8699–8704. doi: 10.1073/pnas.0401142101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. Embo J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Kapoor TM. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat Cell Biol. 2005;7:93–98. doi: 10.1038/ncb1208. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Luo X, Fang G, Coldiron M, Lin Y, Yu H, Kirschner MW, Wagner G. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat Struct Biol. 2000;7:224–229. doi: 10.1038/73338. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Rizo J, Yu H. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol Cell. 2002;9:59–71. doi: 10.1016/s1097-2765(01)00435-x. [DOI] [PubMed] [Google Scholar]
- Luo X, Tang Z, Xia G, Wassmann K, Matsumoto T, Rizo J, Yu H. The Mad2 spindle checkpoint protein has two distinct natively folded states. Nat Struct Mol Biol. 2004;11:338–345. doi: 10.1038/nsmb748. [DOI] [PubMed] [Google Scholar]
- Mao Y, Abrieu A, Cleveland DW. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 2003;114:87–98. doi: 10.1016/s0092-8674(03)00475-6. [DOI] [PubMed] [Google Scholar]
- Mapelli M, Filipp FV, Rancati G, Massimiliano L, Nezi L, Stier G, Hagan RS, Confalonieri S, Piatti S, Sattler M, et al. Determinants of conformational dimerization of Mad2 and its inhibition by p31comet. Embo J. 2006;25:1273–1284. doi: 10.1038/sj.emboj.7601033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli M, Musacchio A. MAD contortions: conformational dimerization boosts spindle checkpoint signaling. Curr Opin Struct Biol. 2007;17:716–725. doi: 10.1016/j.sbi.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Meraldi P, Draviam VM, Sorger PK. Timing and checkpoints in the regulation of mitotic progression. Dev Cell. 2004;7:45–60. doi: 10.1016/j.devcel.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Morrow CJ, Tighe A, Johnson VL, Scott MI, Ditchfield C, Taylor SS. Bub1 and aurora B cooperate to maintain BubR1-mediated inhibition of APC/CCdc20. J Cell Sci. 2005;118:3639–3652. doi: 10.1242/jcs.02487. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Pan J, Chen RH. Spindle checkpoint regulates Cdc20p stability in Saccharomyces cerevisiae. Genes Dev. 2004;18:1439–1451. doi: 10.1101/gad.1184204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by an inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Schultz A, Cole R, Sluder G. Anaphase onset in vertebrate somatic cells is controlled by a checkpoint that monitors sister kinetochore attachment to the spindle. J Cell Biol. 1994;127:1301–1310. doi: 10.1083/jcb.127.5.1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah JV, Botvinick E, Bonday Z, Furnari F, Berns M, Cleveland DW. Dynamics of centromere and kinetochore proteins; implications for checkpoint signaling and silencing. Curr Biol. 2004;14:942–952. doi: 10.1016/j.cub.2004.05.046. [DOI] [PubMed] [Google Scholar]
- Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a 'safety belt' binding mechanism for the spindle checkpoint. Embo J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L, Melixetian M, Faretta M, Prosperini E, Helin K, Musacchio A. Mad2 binding to Mad1 and Cdc20, rather than oligomerization, is required for the spindle checkpoint. Embo J. 2001;20:6371–6382. doi: 10.1093/emboj/20.22.6371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudakin V, Chan GK, Yen TJ. Checkpoint inhibition of the APC/C in HeLa cells is mediated by a complex of BUBR1, BUB3, CDC20, and MAD2. J Cell Biol. 2001;154:925–936. doi: 10.1083/jcb.200102093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Bharadwaj R, Li B, Yu H. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev Cell. 2001;1:227–237. doi: 10.1016/s1534-5807(01)00019-3. [DOI] [PubMed] [Google Scholar]
- Tang Z, Yu H. Functional analysis of the spindle-checkpoint proteins using an in vitro ubiquitination assay. Methods Mol Biol. 2004;281:227–242. doi: 10.1385/1-59259-811-0:227. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Hussein D, Wang Y, Elderkin S, Morrow CJ. Kinetochore localisation and phosphorylation of the mitotic checkpoint components Bub1 and BubR1 are differentially regulated by spindle events in human cells. J Cell Sci. 2001;114:4385–4395. doi: 10.1242/jcs.114.24.4385. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Vink M, Simonetta M, Transidico P, Ferrari K, Mapelli M, De Antoni A, Massimiliano L, Ciliberto A, Faretta M, Salmon ED, et al. In vitro FRAP identifies the minimal requirements for Mad2 kinetochore dynamics. Curr Biol. 2006;16:755–766. doi: 10.1016/j.cub.2006.03.057. [DOI] [PubMed] [Google Scholar]
- Wang Z, Shah JV, Berns MW, Cleveland DW. In vivo quantitative studies of dynamic intracellular processes using fluorescence correlation spectroscopy. Biophys J. 2006;91:343–351. doi: 10.1529/biophysj.105.077891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Bonday ZQ, Putkey FR, Kops GJ, Silk AD, Cleveland DW. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. Structural activation of Mad2 in the mitotic spindle checkpoint: the two-state Mad2 model versus the Mad2 template model. J Cell Biol. 2006;173:153–157. doi: 10.1083/jcb.200601172. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.