SUMMARY
Electrostatic potentials along the ribosomal exit tunnel are non-uniform and negative. The significance of electrostatics in the tunnel remains relatively uninvestigated, yet is likely to play a role in translation and secondary folding of nascent peptides. To probe the role of nascent peptide charges in ribosome function, we used a molecular tape measure that was engineered to contain different numbers of charged amino acids localized to known regions of the tunnel, and measured chain elongation rates. Positively-charged arginine or lysine sequences produce transient arrest (pausing) before the nascent peptide is fully elongated. The rate of conversion from transiently arrested to full-length nascent peptide is faster for peptides containing neutral or negatively-charged residues than for those containing positively-charged residues. We provide experimental evidence that extra-ribosomal mechanisms do not account for this charge-specific pausing. We conclude that pausing is due to charge-specific interactions between the tunnel and the nascent peptide.
Keywords: electrostatics, translation, nascent peptide, peptide elongation, S4 transmembrane segment, Kv channels
INTRODUCTION
Peptides are made and acquire secondary structure in the ribosomal exit tunnel 1–8. This is not a unilateral act by the peptide, nor is the ribosomal tunnel an innocent bystander. The tunnel collaborates as an active participant in translation 9–13. The precise mechanisms for this teamwork are unknown. Two aspects of this collaboration are implicated by our recent studies: a tunnel-peptide embrace and dynamic electrostatic potentials of a peptide-bearing tunnel 10,14. The electrostatic potential inside the tunnel is negative and varies in magnitude along the length of the tunnel. Moreover, when a charged residue is introduced into the nascent peptide in the tunnel, its charge can be detected as a change in potential at the adjacent position in the peptide chain, but no farther away. These findings suggest a new paradigm: a dynamic wave of electrostatic potential is induced during peptide elongation if the nascent chain contains charged amino acids. For example, as an arginine approaches and then moves away from a given tunnel location during chain elongation, it will transiently decrease the negative potential of the region. This dynamic wave of electrostatic potentials will depend on the primary sequence of the nascent peptide.
An electrostatic potential in the tunnel may have pleiotropic consequences. It can determine the concentration of ions, the induction and stabilization of helical structures, the orientation and pKa of charged side-chains of both nascent peptides and ribosomal proteins that protrude into the tunnel, as well as the organization of polar molecules such as water. These consequences themselves might alter the rates of elongation of nascent peptides. There are a growing number of examples of nascent peptides that pause or arrest in the tunnel 9,11,15–19. Most of these so-called arrested peptides contain charged amino acids. Pausing may play important physiological roles. For example, both slowed and accelerated translation rates affect protein folding. Aberrantly accelerated rates can lead to misfolding (e.g., for chloramphenicol acetyltransferase 20). It has been suggested that such accelerated rates of translation allow a particular part of a polypeptide chain to appear earlier in time than usual, which leads to an interaction of this premature region with the preceding region that has not yet folded properly 20. That is, temporary pausing may permit more efficient domain folding without the complicating influence of the peptide’s C-terminus. The importance of pausing has also been demonstrated for the signal recognition particle receptor α-subunit. In this case, pausing facilitates targeting and binding of the signal recognition particle receptor α-subunit to the endoplasmic reticulum membrane 21. Here, too, correct folding of the subunit may require translational pausing. Further support for the hypothesis that misfolding of proteins could start during protein translation is the phenomenon of cotranslational ubiquitination and degradation of proteins 22–24. The signal recognition particle itself, when it binds to eukaryotic ribosome-nascent chain complexes, reduces elongation rates of nascent chains 25–27, thereby regulating the efficiency of protein translocation into the endoplasmic reticulum and cell growth 28. At the extreme, severely slowed translation, could result in degradation of the mRNA and/or nascent peptide 18,29 and has been associated with translational frameshifting 30 and protein misfolding 31. Even moderately altered rates of translation, due to common versus rare codons, RNA secondary structures, or peptide transit 32–43, might control the amounts of a particular protein and/or regulate its folding.
Here, we test whether, in addition to the aforementioned mechanisms, charged residues alter the rates of translation of a nascent peptide. We use a well-studied molecular tape measure 2 that has been modified to contain the S4 region of a voltage-gated bacterial potassium channel, KvAP 44. S4 contains a series of arginines spaced every third amino acid. We varied the number and location of charged residues, and determined the consequences for nascent chain elongation rates. We find that positively-charged residues slow translation and produce a transiently arrested species. This is not a function of size or shape of the side-chain, but rather of the charge itself. This electrostatic effect is mediated by the ribosomal tunnel and suggests that electrostatic considerations may influence cotranslational events in the biogenesis of voltage-gated potassium channels.
RESULTS
We began our studies with the fourth transmembrane segment, S4, of a voltage-gated potassium (Kv) channel because this segment has up to seven positive charges and has been characterized with respect to its membrane insertion in the endoplasmic reticulum 45,46. We substituted the N-terminal portion of S4 from the bacterial KvAP channel into an all-extended nascent peptide (Figure 1A), a molecular tape measure that spans the length of the tunnel in 33 residues and remains attached to tRNA and the ribosome 2. We chose to study nascent peptides derived from DNA that has been truncated with a restriction enzyme (e.g., BstEII-cut) so that the mRNA lacks a stop codon. The newly synthesized peptide will then remain attached to tRNA and the ribosome, thus avoiding the complicating issue of multiple rounds of translation. The distance of each residue from the peptidyl-transferase center (ΔPTC) in the unsubstituted tape measure is known 2 and approximated by multiplying the ΔPTC for each position times 3-3.4Å /amino acid 47. This S4-containing construct includes the 4 arginines, referred to as R1, R2, R3, R4, that contribute to voltage sensing in voltage-gated potassium (Kv) channels 48,49.
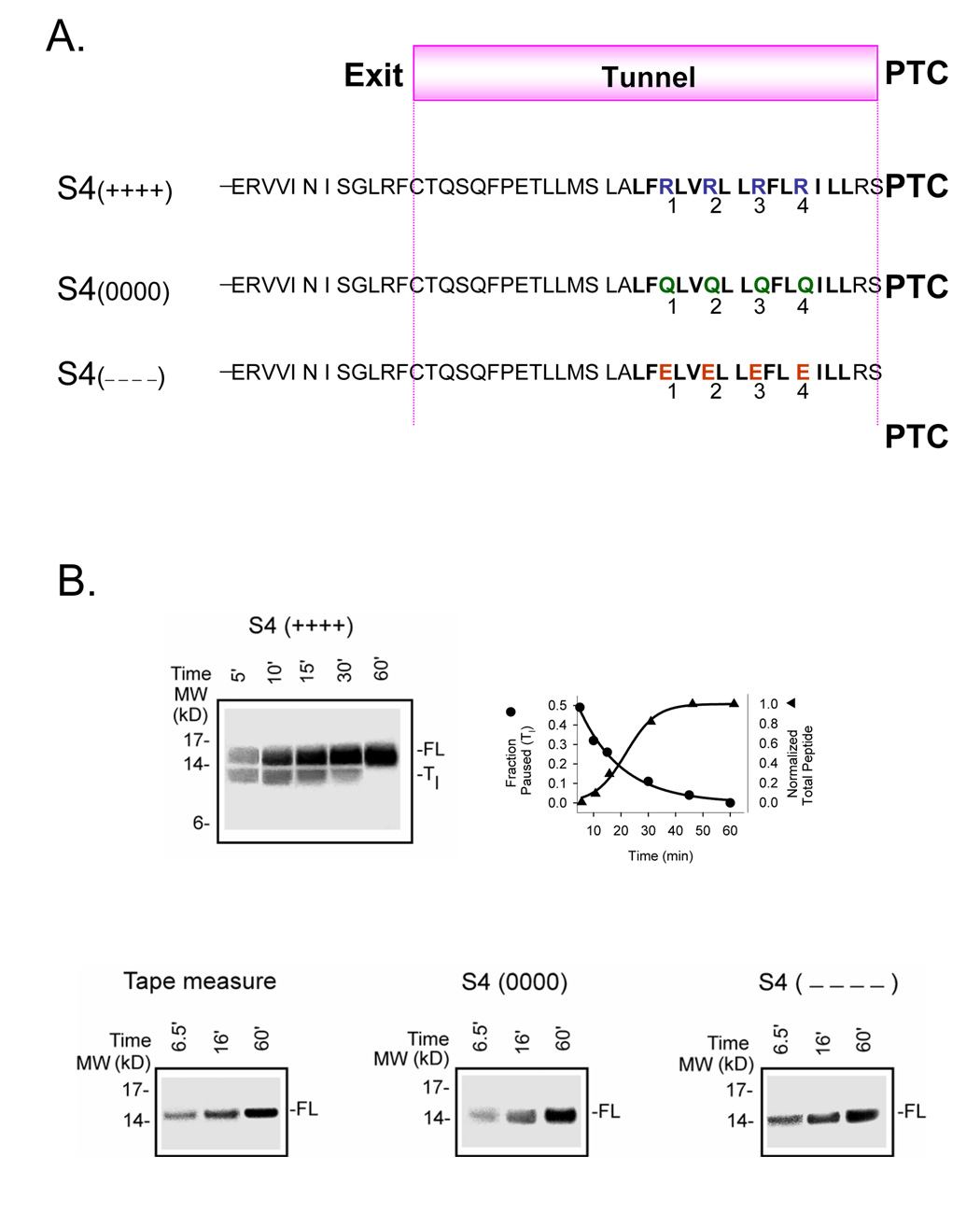
Figure 1. Time course of translation of S4-substituted tape measure transcripts.
A. Schematic sequence of S4 (++++), S4(0000), and S4(−−−−) constructs. The substituted tape measure is a portion of the indicated sequence that resides inside the ribosomal exit tunnel. S4(++++) is the designation for a substituted tape measure in which 19 residues have been replaced with a 19-residue sequence from the fourth transmembrane segment (S4) of Kv channels. The bolded sequence is the proximal portion of the S4 from KvAP, a bacterial Kv channel. The four arginines that translocate across the membrane electric field in the function of Kv channels are labeled 1–4. S4(0000) designates an S4-substituted tape measure in which R1–R4 have been mutated to glutamines (Q1–Q4). S4(−−−−) designates an S4-substituted tape measure in which R1–R4 have been mutated to glutamates (E1–E4). All sequences extend an additional 51 residues (not shown) to the N-terminus. B. Translation rates. Translation was carried out by standard means and aliquots quenched at the indicated times (min), treated with RNase, and fractionated on NuPAGE protein gels (Bis-Tris 12%; top gel, lanes 1–5 loaded with 10, 6, 3, 2, and 1.5 µl, respectively; bottom gels, lanes 1–3 loaded with 6, 3, and 1.5 µl, respectively) for the constructs shown in A. The higher molecular weight band in the S4(++++) gel is the full-length nascent peptide (FL). The lower band in this gel is a transient species (Tl). The fraction of Tl peptide, calculated as the intensity of the lower band divided by the sum of the intensities of the lower and higher bands in a given lane, is plotted as a function of translation time (top row, right), along with normalized total peptide, calculated as the sum of both bands divided by the sum at 60 min. The 45-min time point was omitted from the S4(++++) gel display. The data were fit (solid curve) with a single exponential function and a sigmoidal function, respectively. The three gels (bottom row) for the unmodified tape measure, S4(0000), and S4(−−−−) show only one band, the full-length peptide.
A tape measure containing KvAP S4 with four arginines, R1–R4, is denoted as S4 (++++). The time course of its translation is shown in Figure 1B as protein bands on a NuPAGE gel. The protein appears as a full-length peptide (FL, higher band) and a transient shorter peptide (Tl, late transient (defined below), lower band) that decays exponentially (filled circles, right plot, Figure 1B). The transient appearance of lower molecular weight bands indicates slowing of the rate of chain elongation such that the peptide pauses at a shorter length. This is sometimes referred to as “arrest”. A series of discrete bands (peptide intermediates) suggests that the rate of translation of a peptide is not constant, but that the rate varies in a stop-and-go fashion 33,36,37,39,50–52 32,53 with different dwell times for various intermediates. The Tl peptide appears to be ~1.5 kD shorter than the full-length peptide, however, positive charges alter the mobility of the peptide in an electrophoretic gel (Supplementary Figure 1A). Thus, we cannot readily estimate the pause site or relative importance of individual arginines from the gel migration pattern until we calibrate the relative contributions of peptide mass and charge (see below, Figure 5). If these four arginines, R1–R4, are neutralized or made negative by substitution, respectively, with 4 glutamines (denoted S4(0000)) or 4 glutamates(denoted S4(−−−−)), then no pausing is observed (Figure 1B).
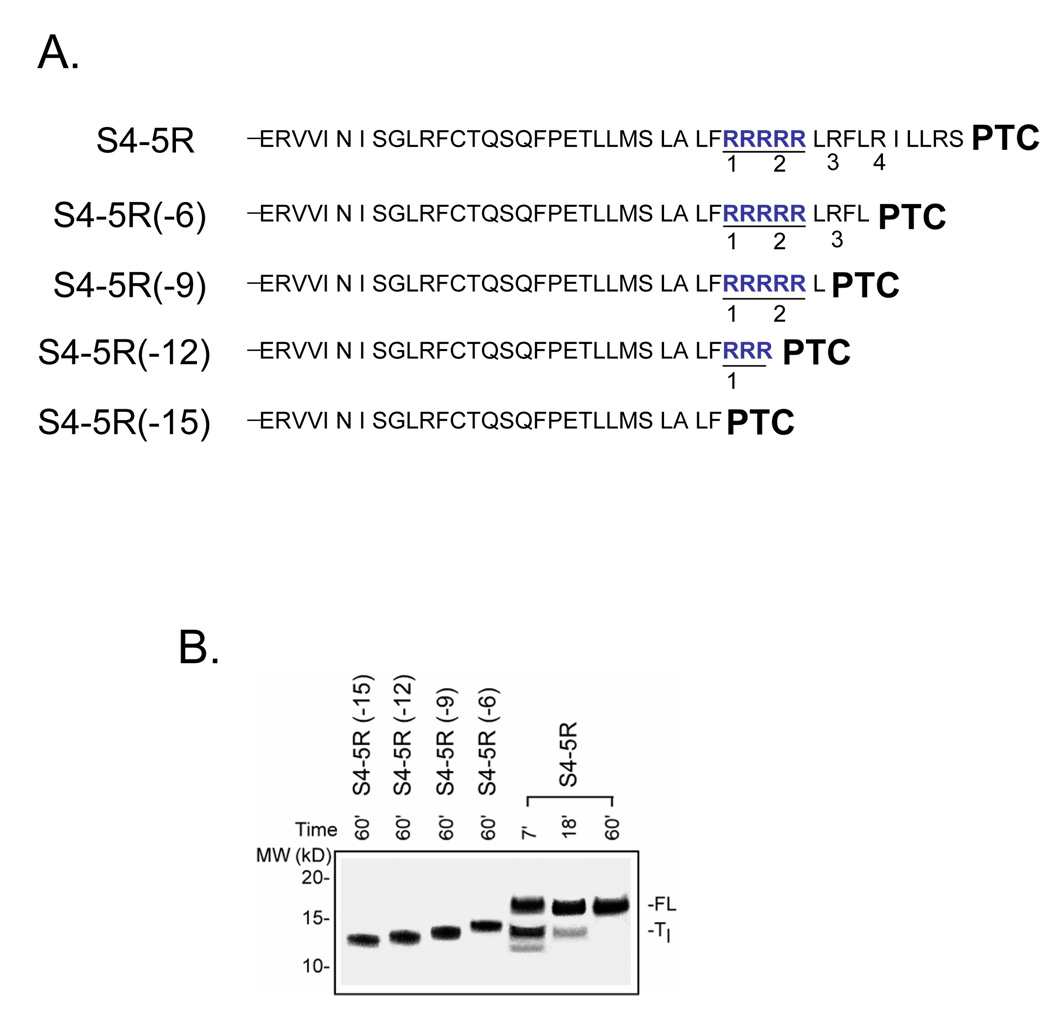
Figure 5. Identity of Tl intermediate.
A. Schematic of truncated peptides used as calibration standards. The number in parenthesis indicates the number of amino acids deleted from the C-terminus of the S4-5R substituted tape measure. B. Migration of standards. The constructs shown in A. were translated for 60 min (produces final full-length peptide) as described in Figure 1 and run on a NuPAGE gel (Bis-Tris 12%; lanes 1–4 loaded with 2, 2, 2, 2 µl, respectively) along with samples from a parallel translation of S4-5R (lanes 5–7 loaded with 20, 10, and 2.5 µl, respectively) using the same batch of reticulocyte lysate and identical reagents and conditions. The gel was run for a longer time to improve resolution of the bands and more accurate assessment of the mass of the paused peptide. The standards (lanes 1–4) exhibit an ascending ladder pattern with increasing mass and charge of the peptide.
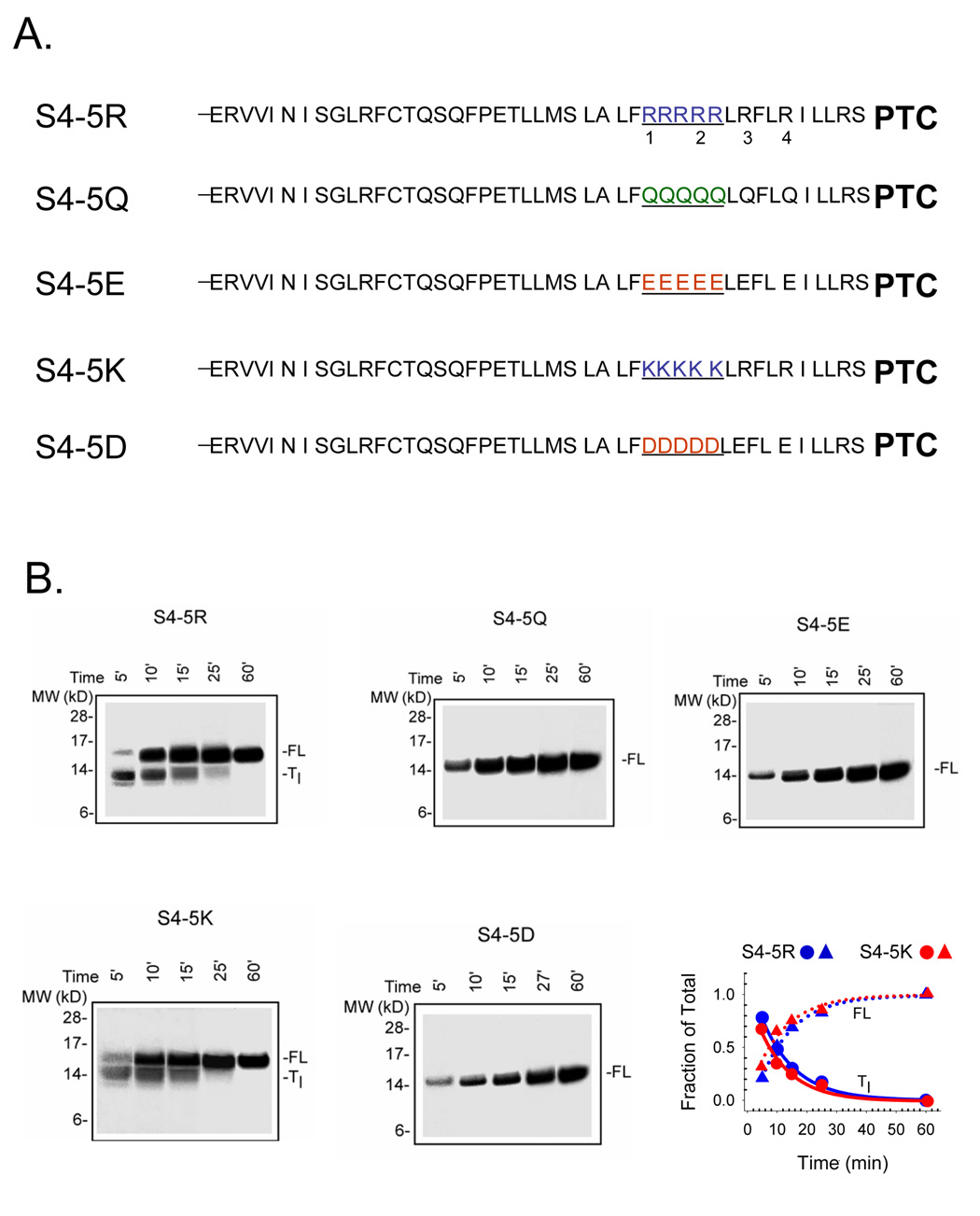
Both the fraction of total peptide paused and the kinetics of disappearance of the paused peptide are dependent on the number of consecutive arginines. Addition of 3 more arginines, two positioned between R1 and R2, and one upstream and adjacent to R2 (Figure 2A), slows translation more than the presence of R1–R4 alone and produces more striking pausing (compare Figure 1B and Figure 2B). Equal amounts of full-length and transient peptide are present at 5 min in S4(++++), but later, at 10.2 ± 1.7 min (n = 7) for S4-5R, as determined from the intersection of FL and Tl curves (e.g., plot in Figure 2B). Is it the charge or the size/shape of the side-chain that influences pausing? To discriminate these possibilities, we substituted lysines for the 5 consecutive arginines and aspartate for the 5 consecutive glutamates as shown in Figure 2A. The additional gels in the bottom row of Figure 2B demonstrate that charge alone is sufficient to cause pausing as the guanidium side-chain (arginine) versus the amine side-chain (lysine) gave similar transient peptide, with similar kinetics of disappearance and formation of full-length peptide, as shown in the plot to the right. The expected delay in the full-length curves cannot be observed with the time resolution and sensitivity used in these experiments (however, see below and Figure 3). The aspartate and glutamate, which differ only in length, gave similar results.
Figure 2. Effect of additional consecutive charges.
A. Schematic sequences of S4-substituted tape measures. S4-5R designates a construct with 5 consecutive arginines (highlighted in blue), including R1 and R2, as indicated. S4-5Q, S4- 5E, S4-5K, and S4-5D designate constructs with 5 consecutive glutamines (green), glutamates (red), lysines (blue), and aspartates (red), respectively, each including replacement of R1 and R2. B. Translation time course for the constructs shown in A. as described in Figure 1B (Bis-Tris 12%; all gels, lanes 1–5 loaded with 20, 10, 5, 3, and 1.5 µl, respectively). For S4-5R and S4-5K, the fraction of Tl transient peptide, calculated as the intensity of the lower band divided by the sum of the intensities of the lower and higher bands in a given lane, is plotted (lower right) as a function of translation time, along with fraction of full-length peptide (1-fraction Tl). S4-5R (blue) and S4-5K (red) are plotted as circles and triangles, respectively, to give Tc values of 10 and 8 min. S4-5Q, S4-5E, and S4-5D displayed only one band at the full-length molecular weight and therefore were not plotted.
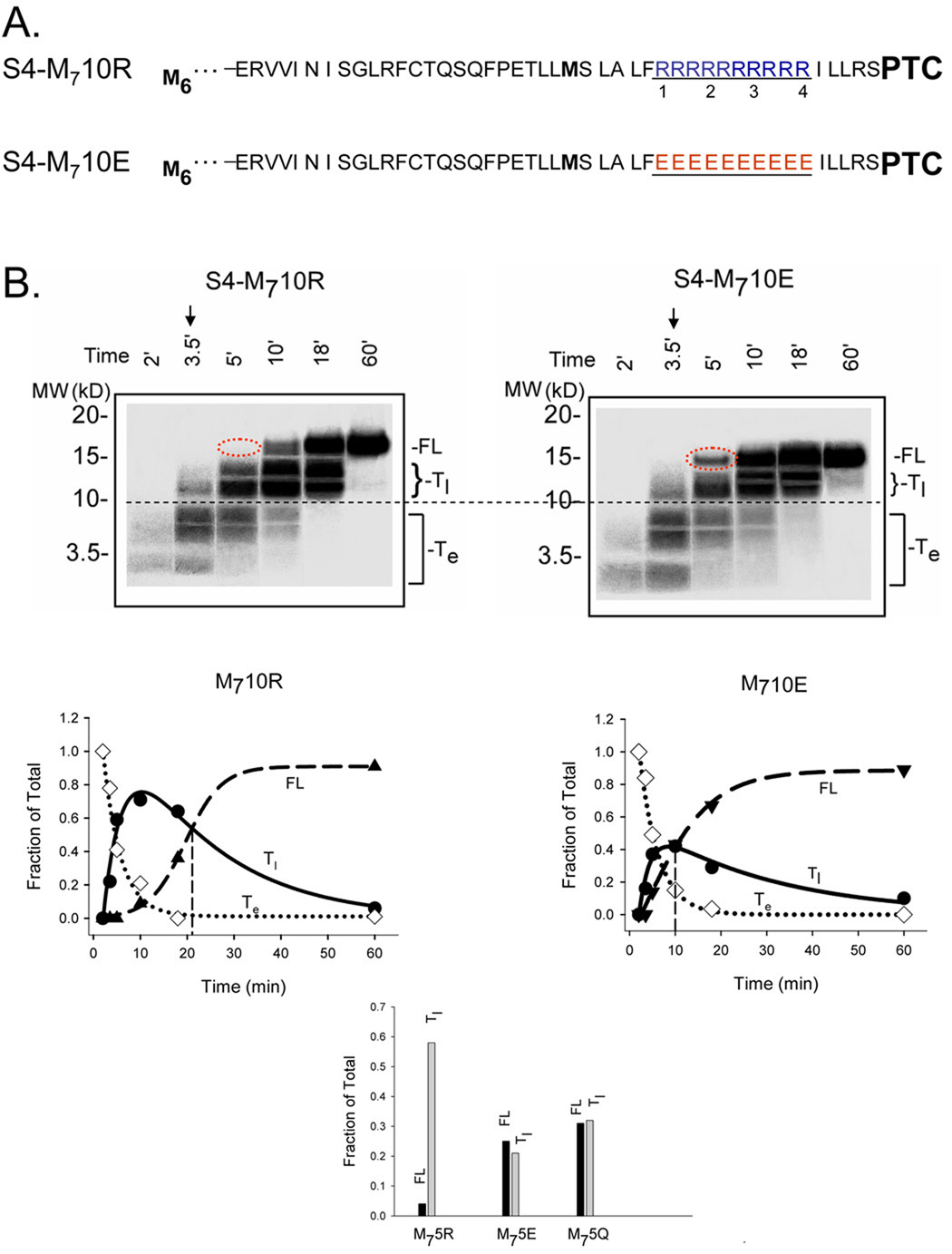
Figure 3. Pulse-chase studies of S4-substituted tape measures.
A. Schematic sequences of methionine-enriched S4-substituted tape measures with 10 consecutive charges. S4-M710R and S4-M710E designate constructs with a total of 7 methionines (bolded) and either 10 consecutive arginines (blue) or 10 consecutive glutamates (red), which replace R1–R4 and the intervening residues in the original S4-substituted tape measure. B. Translation time course for the constructs shown in A. Translation was carried out as described in the Materials and Methods. At the time indicated by the arrow (3.5 min), 1 mM non-radioactive methionine (103-fold dilution of radioactive methionine) was added and additional time points sampled. Samples were fractionated on NuPAGE gels (Bis-Tris 12%, top row; lanes 1–6 loaded with 8, 8, 6, 3, 3, and 3 µl, respectively). The dashed red oval highlights the absence and presence, respectively, of a FL band for S4-M710R and S4-M710E. The data are plotted as fraction of total peptide at the indicated times (below gels). The fraction of a specified population (full-length (FL), early transient (Te), late transient (Tl)) peptide was calculated as the intensity of the specified band divided by the sum of the intensities of all the bands in a given lane. The intensity of Te was multiplied by 1.167 to normalize peptide species containing 6 methionines to the FL containing 7 methionines. The FL data were fit with a sigmoidal function, the Te data with an exponential decay function, and the Tl data with a double exponential (4-parameter) function to give the corresponding solid curves. The vertical line indicates the Tc value, the time at which the fraction of FL equals the fraction of Tl. The bar graph shows results obtained at 5 min from similar experiments using M75R, M75E, and M75Q, which were analyzed as described above.
Two questions arise concerning the Tl band. First, does its disappearance reflect conversion of a transiently paused intermediate or the preferential degradation of an arrested peptide? Second, does its absence in the case of S4-5Q, S4-5E, and S4-5D reflect a lack of paused intermediate or a briefer dwell time in the paused state that is not detectable with our current time resolution and sensitivity? A single experiment addressed both these issues. We monitored conversion of transient bands into full-length in pulse-chase experiments. To optimize these studies we used two strategies. First, we introduced a string of consecutive additional methionines to the N-terminus of the tape measure, as was done previously for a fused polypeptide sequence containing domains from α-globin, luciferase, and arginine attenuator peptide54. We added five N-terminal methionines, thus increasing the total number of methionines in the nascent peptide to seven and consequently increasing the specific radioactivity of the nascent peptide. This enabled us to detect low levels of protein at short translation times (e.g., within 3 minutes). The second strategy entailed using 10 consecutive charges to exaggerate the paused phenotype. We engineered the constructs shown in Figure 3A. Translation was begun and at 3.5 min, non-radioactive methionine (1 mM, 103-fold dilution of radioactive methionine) was added and samples assayed during the next 60 min. Peptide bonds synthesized subsequent to isotopic dilution of methionine will incorporate few radioactive residues. Hence, as the peptide elongates, the radioactivity initially manifest as lower bands will appear as a higher band, i.e., the lower bands will disappear and convert to higher bands. This indeed is the case, as shown in the gels in Figure 3B. If simple degradation of the arrested peptide accounted for the disappearance of the lower bands, then no high molecular weight bands would have appeared. At 5 min (lane 3, dashed red oval), no full-length S4-M710R (left gel) has appeared, yet a significant amount of S4-M710E (right gel) is visible. The rate of formation of full-length S4- M710R is considerably slower than that of full-length S4- M710E. An internal control is present in these constructs because S4- M710R and S4- M710E are identical for the first ~80 residues in the N-terminal portion of the substituted tape measure. When translated, this portion of the substituted tape measure produces shorter, transient peptides, which we designate as transient early (Te) bands (below the horizontal dashed line). The Te bands in the two constructs are virtually identical and serve as internal calibrants common to both S4-M710R and S4-M710E. Quantification of the full-length (FL) and transient species (Te and Tl) are plotted below each gel. Fits of an exponential decay to the disappearance of the Te bands (open diamonds) give time constants of 4.3 and 4.1 min−1, respectively, for S4-M710R and S4-M710E. Although the additional methionines provide the necessary sensitivity, they also appear to prolong the initiation and formation of Te and Tl species (e.g., compare amounts of protein at 5 min to those shown in Figure 2), but do so equally for S4-M710R and S4-M710E as indicated by the similarity of the internal calibration bands. One possible explanation is that the concentration of methionine in the translation mixture, due to the radioactive methionine, is 20 to 40-fold lower than each of the other non-radioactive amino acids. A plot of the fraction of each peptide species present in the total translation mixture indicates a crossover point where full-length (FL) and late transiently paused species (Tl) are equal. We refer to this crossover point as Tc. The average values of Tc for duplicate experiments for S4-M710R and S4-M710E are 19.7 ± 1.7 and 9.5 ± 0.5 min (± average error), respectively. Tc occurs at later times for arginine vis-à-vis glutamate. A comparison of tape measures substituted with 5 arginines, glutamines, or glutamates similarly confirms our results that positively-charged side-chains underlie transient pausing. For S4-M75E and S4-M75Q, the fractions of FL and Tl are approximately equal at 5 min (Tc ~ 5 min), whereas Tc > 5 min for S4-M75R (bar graph, Figure 3B). We conclude that all constructs pause to yield Tl intermediates, but the dwell times in this paused state are prolonged for the positively-charged S4-containing peptides.
What is responsible for transient pausing and the altered rates? Four candidates may be considered. First, a decreased rate of translation could be due to a rate-limiting supply of aminoacylated tRNA. For example, if the level of arginine-tRNA or lysine-tRNA is substantially lower compared with, for example, glutamine-tRNA, this could account for the observed pausing for S4 constructs. Second, ternary complex formation (aa-tRNA/elongation factor/GTP) and/or its binding to the ribosome may be slower and rate-limiting for positively-charged-tRNA compared to neutral or negatively-charged-tRNA. If this occurred, then translation would be slowed. Third, either mRNA secondary structure or rare versus common codon usage might underlie the apparent pausing. For example, if arginine or lysine codons present in the S4 constructs were rare and rate-limiting vis-à-vis those encoding glutamine, glutamate or aspartate, then again the rate of translation would be slowed. Fourth, electrostatic interactions between nascent peptide and tunnel components may be responsible for modulating rates of translation.
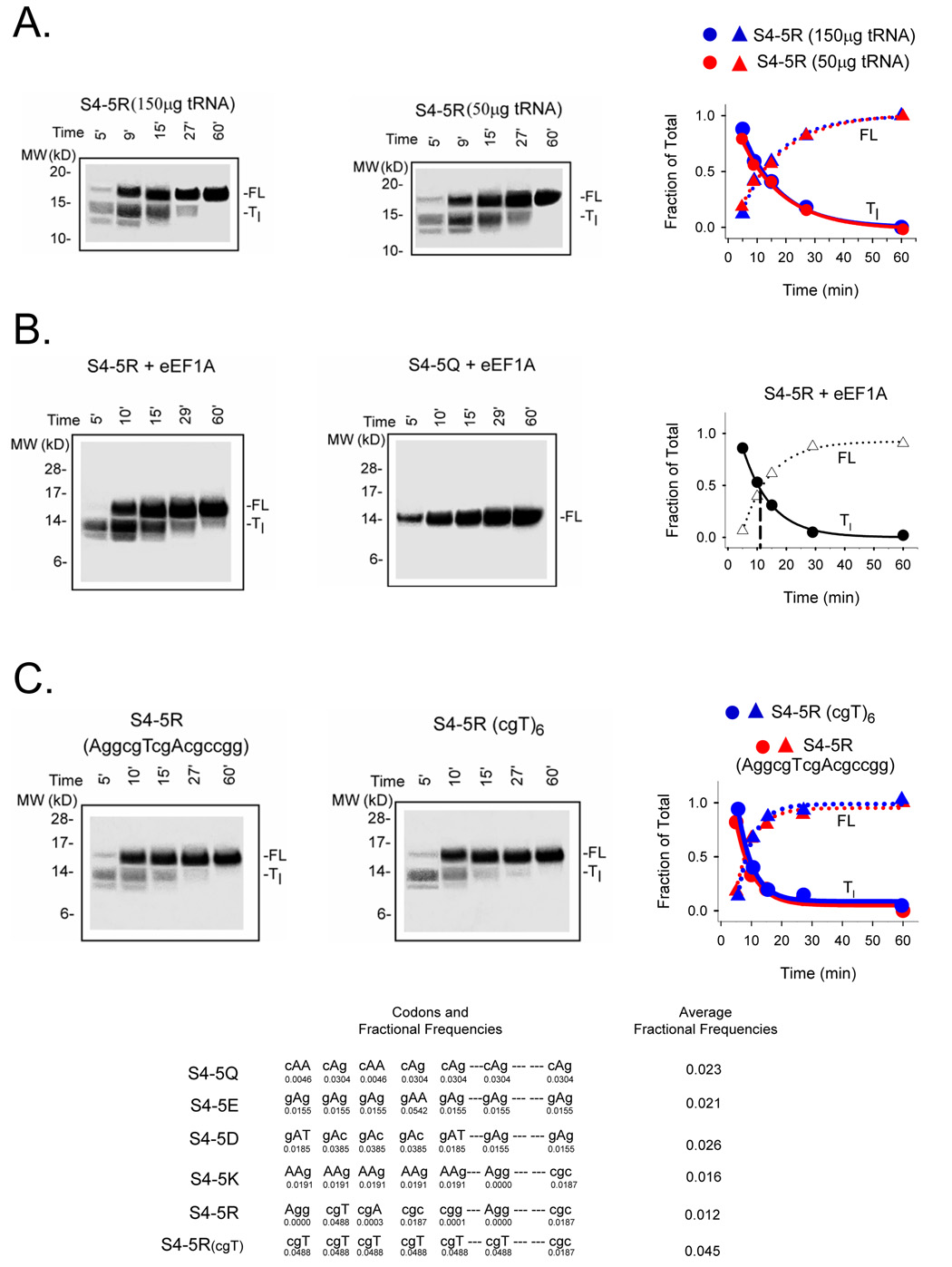
We attempted to examine the first possibility by measuring the relative levels of arginine-tRNA and glutamine-tRNA in our rabbit reticulocyte lysate (RRL). Translation reactions were supplemented with equal molar amounts of either C14-labeled arginine or glutamine followed by fractionation on a 10% TBE-urea gel. Whereas an aminoacylated tRNA band that was sensitive to pre-treatment with RNase (50 µg/ml) was identified for the arginine-tRNA, a similar band could not be detected for glutamine. This was due to specific interference by the superabundant hemoglobin, which precluded a direct comparison of aminoacylated tRNAs. We therefore resorted to an alternative assessment of the aminoacylated tRNA pools. We supplemented the rabbit reticulocyte lysate with additional tRNAs for all 20 amino acids. A mixture of calf liver tRNAs (Novagen), which is identical to the mixture already present at a concentration of 50 µg/ml in the lysate made by Promega, was added at a concentration of 100µg/ml to the lysate to give a total of 150 µg/ml tRNA in the standard translation reaction monitored with S35 methionine. Translation and fractionation of the S4-5R protein products showed a pausing phenotype (Figure 4A) virtually identical to the unsupplemented control S4-5R. Assuming that the rate of aminoacylation of tRNA by the synthetase is fast relative to the translation reaction, a 3-fold higher pool of aminoacylated-tRNAs would be present and in excess throughout the translation reaction. Despite this difference in aminoacylated-tRNA pools, both kinetics and relative amounts of FL and Tl remain the same (right plot). Thus, aminoacylated tRNA is not limiting. Consistent with these observations, we pre-mixed lysate with free amino acids and S35 methioinine for 5 min at 30°C, transferred the suspension to 22°C for 5 additional minutes, and then added S4-5R mRNA to start the translation. Under these conditions, we expect a pre-equilibrated pool of aminoacylated-tRNA. A similar pausing phenotype was still observed (data not shown). We therefore conclude that different levels of aminoacylated-tRNA are not responsible for transient pausing of positively-charged S4 nascent peptides and that the rate-limiting step responsible for pausing is downstream from the formation of aminoacylated tRNA.
Figure 4. Potential factors in charge-dependent pausing.
A. Addition of supplemental tRNAs. Translation reactions were set up exactly as described in the Materials and Methods and Figure 2, but calf liver tRNAs (100 µg/ml; Novagen) was added to give a total of 150 µg/ml tRNAs in the translation reaction. Samples were fractionated on NuPAGE gels (Bis-Tris 12%; lanes 1–5 loaded with 22, 15, 5, 3, and 3 µl, respectively). Data are plotted as fraction of Tl and FL for the indicated times to give a Tc value of ~12 min. B. Addition of supplemental eEF1A. Translations were carried out as described in the Materials and Methods and Figure 2, but in the presence of additional purified eEF1A as described in the text. Samples were fractionated on NuPAGE gels (Bis-Tris 12%; lanes 1–5 loaded with 20, 10, 5, 3, and 1.5 µl, respectively). The data are plotted as fraction of Tl and FL for the indicated times to give a Tc value of ~12 min. C. Rare vs common codons. An S4-5R construct containing 5 consecutive arginines was translated as described in Figure 1. Samples were fractionated on NuPAGE gels (Bis-Tris 12%; lanes 1–5 loaded with 25, 15, 5, 3, and 2.5 µl, respectively). The gel on the left derives from a peptide encoded by the 5 consecutive arginine codons AggcgTcgAcgccgg, some of which are rare codons. The gel on the right derives from a peptide encoded by 5 consecutive cgT codons plus a sixth cgT (R3), which are common. A plot of the fraction of Tl and FL for the indicated times gives Tc values of 10.2 ± 1.8 (n = 2) and 10.4 min, respectively. Below are the average fractional frequencies of occurrence for the 7 codons in each construct (see Figure 2), based on the study of Thanaraj and Argos 37.
The second possibility is that ternary complex formation is limiting in the case of arginine-tRNA. To test for this, we supplemented the RRL translation reaction with elongation factor eEF1A derived from two different sources. In both cases, the eEF1A was first assayed to ensure the eEF1A was fully active 55. As shown in Figure 4B, both unsupplemented and supplemented translation reactions showed similar transient pausing, suggesting that ternary complex formation is not rate-limiting.
A third possible reason for charge-dependent rates of translation could be differences in codon frequency. For example, if the arginine codons present in our constructs are rare, this could introduce a rate-limiting step in the elongation process. The codon sequence for S4 constructs 5Q, 5K, 5R, 5E, and 5D differ with respect to the codons in the seven positions indicated in Figure 4C. We therefore calculated an average fractional frequency of occurrence in each construct based on the study of Thanaraj and Argos 37 who calculated the frequencies of the 61 sense codons as the fractions of the sum of all codons in mRNAs of highly expressed genes from E. coli 56. They use a database of 9,512 codons from 60 mRNA sequences. The average fractional frequency for the 7 codons indicated in Figure 4C (bottom table) in each S4 construct containing 5Q, 5E, 5D, 5K, and 5R is 0.023, 0.021, 0.026, 0.016, and 0.012, respectively. The constructs encoding 5R and 5K have an average fractional frequency that is ~60% of the others. Could this difference account for pausing in the translation of 5R and 5K? Although bacteria have a large variation in codon usage compared to mammals and a higher level of rarely used codons than mammals 57, we nevertheless investigated this possibility by changing codons Agg, cgA, cgc and cgg in the S4-5R construct (Figure 4C) to the commonest arginine codon, cgT, which produced a construct with an average fractional frequency of 0.045, approximately 3 times more than the frequency in either the 5R or the 5K constructs, and twice the average fractional frequency for 5Q, 5E, or 5D. Moreover, this common cgT codon has a translation speed, measured as relative rate of association between the codon and the cognate aminoacyl-tRNA 33, that is faster than the other substituted arginine codons (ranging over an order of magnitude: 1.6 to 17.5-fold for cgc and cgg, respectively). This substitution of 6 cgT codons should accelerate the rate of translation of 5R if the codons themselves contribute to a rate-limiting step. However, as shown in the gels in Figure 4C, pausing of 5R was the same, regardless of the codons, suggesting that altered rate/pausing is due to the peptide side-chains and not to the mRNA codon. Moreover, these results also eliminate the possibility that RNA hairpin formation retards translation. The 5K construct is composed of AAg codons, whereas the 5R construct is composed of cgT codons, yet they produce similar transient pausing, again suggesting that side-chain charge determines pausing rather than RNA codons. Moreover, we calculated the free energy of mRNA folding using the m-fold algorithm (developed by Dr. Michael Zuker http://www.Idtdna.com). The free energies for the strongest potential secondary structure formation in the last 63 bases of the mRNA coding for the C-terminal 21 amino acids attached to the PTC of the S4 constructs 5Q, 5E, 5D, 5K, 5R, and 5R(cgT) are −12.09, −25.06, −19.29, −20.64, −23.3, and −12.79 kcal/mol, respectively. The unsubstituted tape measure mRNA itself has a free energy of −26.05 kcal/mol and therefore has the most driving force to fold, whereas 5Q has the least, similar to 5R(cgT). Thus, hairpin formation of the mRNA coding for positively-charged peptides is not the cause of pausing. We conclude that transient pausing is not due to extra-ribosomal rate-limiting steps in the elongation process. Rather, charge-dependent pausing occurs within the ribosome itself, most likely due to electrostatic peptide-tunnel interactions.
To estimate the ribosomal location of these electrostatic effects, we determined the length of the Tl-paused peptide. Our first task was to evaluate the relative contributions of two factors that alter the migration pattern of the paused S4-5R peptide (Tl). First, the Tl peptide has a smaller mass and will therefore migrate faster. Second, the Tl peptide will contain fewer arginines (because C-terminal codons, which include the 5 arginine codons, are deleted) and will therefore also migrate faster in the applied electric field (negative pole at the top of the gel). We constructed calibration standards of known length by deleting 6, 9, 12, or 15 residues from the C-terminus of the tape measure substituted with S4-5R (Figure 5A) to generate peptides truncated successively up to, and including, R4, R3, R2, or R1, respectively. This yields a family of calibration standards ranging in mass from 12.8 to 14.2 kD that migrate as an ascending ladder with increasing mass and charge (Figure 5B, lanes 1–4). A side-by-side comparison of the calibrants and the S4-5R construct indicates that the Tl peptide (lanes 5–7) migrates to the same position as the S4-5R(−9) calibrant, as if it were ~ 13.5 kD, suggesting that the peptide stalled at R3. This places the remaining arginines (R1, R2 and their immediately adjacent arginines) in the ribosomal tunnel. The precise location depends on the secondary structure of this segment of the peptide. If this segment is extended, then R1 will reside ~18–20Å from the PTC. If it is an α- helix, then R1 will be ~10Å from the PTC. In either case, the charged residues are lodged within the ribosomal exit tunnel.
DISCUSSION
Chain elongation does not proceed at a constant rate, but rather in fits and stops, pausing along the way and giving rise to a series of intermediate peptides 32,33,36,37,39,50–53. This is exhibited most strikingly in the protein gels displaying the reaction products containing 7 methionines, which permits sensitive detection at early times during translation. A constant rate of elongation predicts a more uniform smear of bands rather than discrete bands. The latter are only observed if the relative rate of appearance of a biogenic intermediate compared to its disappearance is significantly greater than one. This stop-and-go traffic is regulated by some or all of the events/participants involved in decoding mRNA, peptide synthesis, structure of the nascent chain, and movement of the elongating nascent chain through the tunnel. The latter two factors will depend on both backbone and side-chain interactions of the nascent peptide with tunnel components and with chaperones.
Tunnel electrostatics influence translation rates
Elongation pausing of nascent peptides containing positively-charged motifs suggests that translation rates depend on electrostatics. What is responsible for this? Two broad categories may be considered. First, there are events outside the ribosome itself, namely synthesis of aminoacylated tRNA (aa-tRNA) and ternary complex formation. Second, there are tunnel-associated events, including events at the PTC (reading of the mRNA codons, A-site entry, accommodation, peptide bond formation, and translocation), compaction of the nascent chain in the tunnel, and/or movement of the peptide along the tunnel. We shall discuss the former category first.
Aminoacylation in extracts, including the RRL cell-free system, is very efficient for all amino acids. Typically, the concentrations of aa-tRNA, including Arg-tRNA, Gln-tRNA, and Glu-tRNA, exceed that of the ribosomes by at least 10-fold 58–60, and the rate of aminoacylation cannot be resolved by manual pipetting because it is too fast (Rodnina, pers. comm.). Moreover, the rates of aminoacyl-tRNA formed by purified synthetases in vitro are similar for methionyl, lysyl, arginyl, leucyl, and isoleucyl-tRNA and ~1.5–2 times faster than glutamyl or glutaminyl-tRNA 61. Thus, aminoacylation should not be rate-limiting for translation of positively-charged S4 segments, consistent with the experimental observations reported in this paper: neither supplementary tRNAs nor pre-equilibration of the rabbit reticulocyte lysate with amino acid substrates abolished S4-5R pausing. We therefore conclude that aminoacylation is not limiting and cannot be the direct cause of pausing.
The aminoacylated tRNA enters the ribosome by first binding elongation factor Tu·GTP (EF-Tu·GTP in prokaryotes and eEF-1 in eukaryotes) to form a ternary complex, which is efficient for all aa-tRNA 62. In prokaryotic systems, each aa-tRNA has evolved its affinity for EF-Tu and the ribosome so that binding of ternary complexes to the ribosome and peptide bond formation seems to be uniformly fast for all natural aa-tRNA 62,63. It is thus unlikely that variations in the rates of translation that we observe are related to poor ternary complex formation. To confirm this conclusion in the RRL system, we supplemented the RRL system with purified eEF1A and compared translation rates for S4-5R and S4-5Q. Results were identical to those obtained in the unsupplemented system, indicating that ternary complex formation is not responsible for the observed pausing.
It is also possible that selection of the correct aa-tRNA is limiting the rate of translation of a particular codon. Translation will typically be slower at the codons that are read by rare aa-tRNA. Another related possibility is that natural mRNAs may have a significant secondary and tertiary structure that the ribosome melts. This may slow translation, particularly at difficult patches, e.g., at RNA hairpins. Such dependence is manifest in prokaryotic ribosomes as pausing during mRNA unwinding 32. Both possibilities, rare codons and mRNA structure, are eliminated by the experiments demonstrating that lysine and arginine each produce similar pausing phenotypes and that substitution of multiple different arginine-encoding codons does not eliminate the pausing. Moreover, a calculation of the free energy of folding for mRNA S4 constructs indicates that secondary RNA structure cannot account for the observed pausing.
We are therefore left with ribosome-associated causes for the dependence of translation rate on electrostatics. These ribosome-associated causes include PTC events, compaction of the nascent chain in the tunnel, and/or movement of the peptide along the tunnel. Let us first consider the role of compaction in pausing. Woolhead et al. 13 propose that in the case of SecM, translation arrest requires both compaction of the nascent SecM sequence and a tunnel-peptide interaction, the former serving to orient critical SecM residues at specific locations in 3- dimensional space for optimal interaction with the tunnel to cause pausing. By analogy, one might speculate that arginine and lysine, with helix propensities of ~ −0.7 kcal/mol compared to glycine 64, compact more readily than glutamine and glutamate with helix propensities of ~−0.3 kcal/mol 64, thus orienting positively-charged side-chains for the requisite pause-inducing interaction with the tunnel. While two lines of evidence, derived from an accessibility assay previously validated to measure compaction in the tunnel 2,10, are consistent with this scenario, a third is not. First, a tape measure substituted with ten consecutive alanines (alanine has the highest helix propensity, −0.77 kcal/mol relative stabilization of α-helical conformation 64) near the PTC forms an α-helix 10 yet has a translation rate that is similar to that of the all-extended unsubstituted tape measure (data not shown). Second, S4-5R and S4-5K compact equivalently to a substituted tape measure containing 5 consecutive alanines (7-Ala5) in a similar tunnel location10, whereas S4-5Q, S4-5E, and the unmodified tape measure are each extended (Supplementary Figure 1B). The polyalanine-containing tape measures compact yet have Tc values less than those for S4-5R and S4-5K and similar to Tc for S4-5Q. A third line of evidence, however, is inconsistent with a scenario in which compaction orients key residues for interaction with the tunnel and consequent pausing: S4(++++) and S4(0000) have similar extended conformations in the tunnel (data not shown), yet S4(++++) pauses while S4(0000) does not (Figure 1). Moreover, two considerations suggest that the tunnel interacts with nascent peptide to influence compaction. S4-5K and S4-5R substituted tape measures differ in overall helix propensity by a factor of two (as determined from an Agadir helix propensity algorithm 65), yet compact similarly and, conversely, S4-5Q and S4-5K tape measures have similar calculated helix propensities, but the former is extended and the latter is compact. We conclude from our studies that pausing is not solely a function of compaction (in fact, compaction appears not to be a prerequisite) but that pausing involves electrostatic interactions in the tunnel.
While we cannot entirely eliminate one or the other of the remaining possibilities, PTC events versus movement of the peptide along the tunnel, we nonetheless may conclude the following. The major contribution to pausing occurs in the tunnel, distal to the PTC. This conclusion is based on our demonstration that the arginine-rich region reaches 10–20 Å into the tunnel when the peptide has paused (Tl, Figure 5B). Although there is a minor component (<10 % paused) that stops just before the first arginine enters the tunnel, the major transient intermediate pauses inside the tunnel. We conclude that an important determinant of pausing is electrostatic interactions between the nascent peptide and the tunnel itself.
We can understand the pausing in constructs rich in positively-charged side-chains vis-à-vis neutral or negatively-charged side-chains by considering the free energy landscape that may be encountered along the tunnel during chain elongation. Figure 6 schematically illustrates elongation energies underlying the transition from the Tl intermediate to the full-length peptide in the region 10–20 Å from the PTC, depending on the charges of particular side-chains. The rate-limiting step for elongation at this position of the nascent peptide will be determined by the highest activation free energy barrier, which is side-chain dependent and may be due to any number/combination of factors, including steric clashes, hydrogen bonding, van der Waals forces, hydration energies, and electrostatic interactions. In Figure 6 we represent the hypothetical free energy landscape as two barriers with an energy well between them. We consider peptide elongation as an irreversible movement to the right along the reaction coordinate delineated by the abscissa. Three free energy landscapes are superimposed, black representing the S4-5Q peptide, blue the S4-5R or S4-5K peptides, and red the S4-5E or S4-5D peptides. Two activation free energies are shown, ΔG*1 and ΔG*2. For the neutral side-chains, ΔG*1 > ΔG*2, making the first barrier rate limiting. We show the effect of side-chain charge as an effect on the well depth, i.e., the binding energy for a side-chain interacting with putative negative electrostatic potentials in this region of the tunnel. An acidic side-chain (red) will be destabilized by this negative potential, raising the energy of the well. Notice that the first barrier remains rate limiting in this case. However, an arginine or lysine side-chain (blue) will be stabilized in the well. In the example shown, this will result in pausing, as the second activation energy barrier becomes larger – and rate limiting – during peptide elongation. This energetic representation is greatly simplified, but is intended to show how positive charges of certain side-chains might create a higher energy barrier that would result in pausing, whereas negative charges might have little effect on the rate of peptide extension.
Figure 6. Energy barrier model.
This is a schematic representation of the energetics of transit of S4-substituted tape measures along the region of the tunnel that is 10–20 Å from the PTC. The trajectory for 5R is shown in blue, for 5Q in green, and 5E in red. The height of the first barrier is ΔG*1 and the second is ΔG*2.
Consequences of electrostatic interactions in the tunnel
The introduction of charge into the tunnel will have several immediate consequences. First, as demonstrated previously, the charge will produce a local change in the electrostatic potential of the tunnel 14. Second, the interaction of the charge with the local electrostatic potential may orient the side-chain in the tunnel. Third, the charge may re-orient other dipoles in the tunnel, e.g., ribosomal proteins, rRNA, and/or water and mobile ions. These physico-chemical consequences of charge introduction into the tunnel may, in turn, have biogenic consequences. For example, electrostatic interactions are critical to conformational folding of macromolecules, kinetics of biochemical reactions, and organizing solvent, ions, and molecules with monopole, dipole, and quadrupole moments 66. Given their central role in biology, such electrostatic interactions are likely to be just as important in fundamental translation processes such as peptide elongation, folding, and signaling. Consider two possible effects of electrostatics on secondary folding in the tunnel. First, if some folding takes place in the tunnel, as has been demonstrated 4,10,14, this may be influenced by interactions between charged side-chains and the electrostatic potential in the tunnel, interactions that could induce or stabilize helix formation 14. Moreover, if a folding reaction is accompanied by several charged side-chains moving through a local electrostatic field, this folding reaction will be highly sensitive to the electric field. Second, intramolecular ion-pair formation between oppositely charged side-chains can also stabilize helix formation 3. Whereas a role for electrostatics in signaling in the tunnel has not yet been explored, a role for electrostatics in peptide elongation is compelling.
Other observations also implicate modulation of translation events by electrostatic interactions. For example, several arrest sequences contain charged residues that are critical for pausing events 11,12,15,16,67, and charge changes of amino acids in ribosomal protein L22 that point into the tunnel alter arrest 9,11. These results are consistent with electrostatic interactions modulating translation rates. Moreover, insertion of a polylysine tract into a nascent peptide causes cotranslational arrest and degradation of the peptide 29.
What is the consequence of altered translation rates? Altered rates can lead to misfolding during translation 20,22–24,68 and/or decreased translational fidelity 69. In contrast to these deleterious effects, temporary pausing might lead to efficient domain folding, and/or facilitated targeting and binding to the endoplasmic reticulum membrane 21. Thus, translation rates may need to be regulated carefully within a prescribed range to optimize folding and targeting reactions and avoid the potentially grievous consequences of misfolding. Moreover, optimal translation rates may vary at different positions of the nascent peptide within the ribosomal tunnel. Our results suggest that positively-charged side-chains may be strategically placed along the emerging peptide chain to insure optimal conditions for folding reactions.
MATERIALS AND METHODS
Constructs and in vitro translation
Standard methods of bacterial transformation, plasmid DNA preparation and restriction enzyme analysis were used. The nucleotide sequences of all mutants were confirmed by automated-cycle sequencing performed by the DNA Sequencing Facility at the University of Pennsylvania School of Medicine on an ABI 377 sequencer using Big dye terminator chemistry (ABI). All tape measure DNAs (substituted and unsubstituted) were sequenced throughout the entire coding region.
In all experiments, we used constructs in a molecular tape measure background. The molecular tape measure is the C-terminal 44 amino acids of a 95-amino acid long segment of the N-terminus of Kv1.3 2. Five native cysteines in the tape measure constructs, including Cys71, were replaced with serines to give a cysteine-free background. Two α-helices, α1 (from Leu67 to Leu70) and α2 (from Pro81 to Arg83) in the wild-type T1 domain, were deleted and a new BstEII restriction site was engineered at Arg101 using Stratagene’s QuikChange site-directed mutagenesis kit. This new site inserted a serine between Arg101 and the PTC. When a BstEII-cut tape measure construct (substituted or unsubstituted) is translated, it generates a nascent peptide of 95 amino acids, which migrates at ~15 kD on NuPAGE gels 2. All tape measures (substituted and unsubstituted) were positioned to span the length of the ribosomal exit tunnel using the BstEII restriction enzyme. E64C was made using the QuikChange mutagenesis kit and is positioned at the border of the exit tunnel. A stretch of 15 residues, from R (Δ17) to N (Δ2; 2 residues from the PTC) of the E64C tape measure was replaced with KvAP S4 transmembrane residues LFRLVRLLRFLRILL (C-terminus) using a QuikChange mutagenesis kit. MSLA residues were additional mutations that reinstated the substituted methionine for increased sensitivity on the gel. The underlined arginines are R1, R2, R3, R4, starting at the N-terminus, and were mutated to Q, E, D, or K for various experiments using QuikChange. To further study the effect of the number of charged side-chains, we mutated RLVRL to RRRRR, KKKKK, EEEEE, or DDDDD (5R, 5K, 5E, 5D, respectively). Similarly, we mutated RLVRLLRFLR to RRRRRRRRRR or EEEEEEEEEE (10R and 10E, respectively). To detect low level of signal early in translation during pulse-chase experiments, we added 5 extra methionines to the starting codon of 10R and 10E constructs using QuikChange.
To generate a series of truncated calibration standards for the S4-5R construct, we deleted 6, 9, 12, or 15 residues from the C-terminus using PCR methods. All four constructs share a common 5’ end primer 5’-AgAggATcTggcTAgcgATg-3’, which is situated 250 bp upstream from the starting codon, whereas the different 3’ end primers were designed appropriately upstream from the 3’ end of the construct to yield the desired truncated calibration standards. All PCR fragments were purified and sequenced throughout the entire coding region and used as templates for transcription.
Capped complementary RNA was synthesized in vitro from linearized templates using Sp6 RNA polymerase (Promega). Linearized templates for all biogenic intermediates were generated using BstEII enzyme. Proteins were translated in vitro with [35S]methionine Express (2 µl per 25 µl translation mixture; ~10 µCi µl−1; Amersham) for 1 h at 22 °C in rabbit reticulocyte lysate according to the Promega Protocol and Application Guide. Samples were taken between zero and 60 minutes and quenched with at least 10x volume of ice cold buffer containing 20 mM Hepes (USB, free acid, pH 7.3 at 37°C), 4 mM MgCl2, 100 mM NaCl, and 1 mM DTT, pH 7.4–7.5 at room temperature. The quenched translation product was loaded on a sucrose cushion (120 µl containing 0.5 M sucrose, 100 mM KCl, 5 mM MgCl2, 50 mM Hepes, 1 mM DTT, pH 7.5 at room temperature) and centrifuged using a TLA100.3 Beckman rotor at 70,000 rpm for 20 min at 4°C. The supernatant was removed and the pellet resuspended in 20 µl of buffer (see below).
Gel electrophoresis and fluorography
Pellets for assay were resuspended in buffer containing 50 µg/ml RNase (to remove peptidyl-tRNA to simplify the number of bands on the gel) and incubated at room temperature for 30 minutes, then mixed with NuPAGE loading buffer (1X) and DTT (50 mM final concentration). All final samples were heated at 70 °C for 10 min before loading onto the gel. Electrophoresis was performed using the NuPAGE system and precast Bis-Tris 10% or 12% gels and Mes running buffer. Gels were soaked in Amplify (Amersham) to enhance 35S fluorography, dried and exposed to Kodak X-AR film at −70 °C. Typical exposure times were 16–30 h. Quantification of gels was carried out directly using a Molecular Dynamics PhosphorImager. To compensate for the small amount of protein made at early times after the start of translation, more reaction volume was loaded at earlier time points. The graphs in Figure 1–Figure 4 plot the fraction of total in a given lane, which obviates the need for any normalization procedures.
Supplemental eEF1A and aminoacylated tRNA experiments
Rabbit reticulocyte lysate was warmed to room temperature and pre-mixed with amino acid mixture (minus methionine), 35S-methionine, RNasin ribonuclease inhibitor, and nuclease-free water according to the Promega protocol. Immediately prior to initiating the translation reaction, we supplemented the translation reaction with eEF1A obtained either from the laboratory of W.C. Merrick (lot #3033; 10 µg per 50 µl) or from the laboratory of Tatyana Pestova (originally from Terri Kinzy; 3.5–4 µg per 50 µl). We then added 0.2 µg mRNA per 50 µl translation volume to start the translation. Five samples were taken between zero and 60 minutes and quenched with at least 10x volume of ice cold buffer containing 20 mM Hepes, 4 mM MgCl2, 100 mM NaCl, and 1 mM DTT, pH 7.3. The quenched translation product was loaded on a sucrose cushion (120 µl containing 0.5 M sucrose, 100 mM KCl, 5 mM MgCl2, 50 mM Hepes, 1 mM DTT, pH 7.5 at room temperature) and centrifuged using a TLA100.3 Beckman rotor at 70,000 rpm for 20 min at 4°C. Experiments were carried out to test whether the pool of aminoacylated tRNA was limiting. We supplemented the rabbit reticulocyte lysate with additional tRNA for all 20 amino acids. A mixture of calf liver tRNAs (Novagen), which is identical to the mixture already present at a concentration of 50 µg/ml in the lysate made by Promega, was added at a concentration of 100 µg/ml to the lysate to give a total of 150 µg/ml tRNA in the translation reaction. Translation and isolation of products was carried out as described above for the eEF1A experiments.
Supplementary Material
Supplementary Figure 1. A. Charge-dependent mobility of S4-substituted tape measure. The unsubstituted tape measure, S4(++++), S4(++00), and S4(0000) constructs (lanes 1–4, respectively) were translated as described in the Materials and Methods and Figure 1 for 60 min to give a single, final peptide band that is full-length. The products were run on a Bis-Tris 10% NuPAGE gel. Increasing numbers of arginines cause the peptide to migrate more slowly on the gel. B. Compaction of S4 segments in the tunnel and rate of polyalanine translation. A cysteine accessibility assay previously validated to measure compaction in the tunnel 2,10 was used to measure compaction in S4 constructs. The assay involves modification of a free cysteine with PEG-MAL to produce a gel shift. The fraction of pegylation is quantified (for a given lane on a protein gel) from the ratio of the intensity of the band representing the gel-shifted, higher mass peptide to the sum of the intensities of the bands representing the unmodified parent molecular mass and the gel-shifted higher mass. S4-5R and S4-5K compact equivalently to a molecular tape measure substituted with 5 consecutive alanines (7-Ala5) in a similar tunnel location 10, whereas S4-5Q, S4-5E, and the unsubstituted tape measure are each fully extended.
ACKNOWLEDGEMENTS
We thank Drs. R. Horn, C. Squires, K. Ito, and M.Rodnina for carefully reading of the manuscript, Drs. R. Horn, M. Rodnina, W. Merrick, and O. Uhlenbeck for helpful discussions, and Drs. W. Merrick, T. Pestova, and T. Kinzy for generous donations of purified elongation factor. Supported by NIH GM 52302.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Kosolapov A, Tu L, Wang J, Deutsch C. Structure acquisition of the T1 domain of Kv1.3 during biogenesis. Neuron. 2004;44:295–307. doi: 10.1016/j.neuron.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Deutsch C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry. 2005;44:8230–8243. doi: 10.1021/bi050372q. [DOI] [PubMed] [Google Scholar]
- 3.Tu L, Wang J, Deutsch C. Biogenesis of the T1-S1 linker of voltage-gated K+ channels. Biochemistry. 2007;46:8075–8084. doi: 10.1021/bi700319f. [DOI] [PubMed] [Google Scholar]
- 4.Woolhead CA, McCormick PJ, Johnson AE. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- 5.Mingarro I, Nilsson I, Whitley P, von Heijne G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biology. 2000;1:3. doi: 10.1186/1471-2121-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kowarik M, Kung S, Martoglio B, Helenius A. Protein folding during cotranslational translocation in the endoplasmic reticulum. Molecular Cell. 2002;10:769–778. doi: 10.1016/s1097-2765(02)00685-8. [DOI] [PubMed] [Google Scholar]
- 7.Hardesty B, Kramer G. Folding of a nascent peptide on the ribosome. Progress in Nucleic Acid Research & Molecular Biology. 2001;66:41–66. doi: 10.1016/s0079-6603(00)66026-9. [DOI] [PubMed] [Google Scholar]
- 8.Matlack KE, Walter P. The 70 carboxyl-terminal amino acids of nascent secretory proteins are protected from proteolysis by the ribosome and the protein translocation apparatus of the endoplasmic reticulum membrane. Journal of Biological Chemistry. 1995;270:6170–6180. doi: 10.1074/jbc.270.11.6170. [DOI] [PubMed] [Google Scholar]
- 9.Cruz-Vera LR, Rajagopal S, Squires C, Yanofsky C. Features of ribosome-peptidyl-tRNA interactions essential for tryptophan induction of tna operon expression. Mol. Cell. 2005;19:333–343. doi: 10.1016/j.molcel.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, Deutsch C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005;12:1123–1129. doi: 10.1038/nsmb1021. [DOI] [PubMed] [Google Scholar]
- 11.Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- 12.Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- 13.Woolhead CA, Johnson AE, Bernstein HD. Translation arrest requires two-way communication between a nascent polypeptide and the ribosome. Mol. Cell. 2006;22:587–598. doi: 10.1016/j.molcel.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Kobertz WR, Deutsch C. Mapping the electrostatic potential within the ribosomal exit tunnel. J. Mol. Biol. 2007;371:1378–1391. doi: 10.1016/j.jmb.2007.06.038. [DOI] [PubMed] [Google Scholar]
- 15.Fang P, Wang Z, Sachs MS. Evolutionarily conserved features of the arginine attenuator peptide provide the necessary requirements for its function in translational regulation. J. Biol. Chem. 2000;275:26710–26719. doi: 10.1074/jbc.M003175200. [DOI] [PubMed] [Google Scholar]
- 16.Gong F, Yanofsky C. Reproducing tna operon regulation in vitro in an S-30 system. Tryptophan induction inhibits cleavage of TnaC peptidyl-tRNA. J. Biol. Chem. 2001;276:1974–1983. doi: 10.1074/jbc.M008892200. [DOI] [PubMed] [Google Scholar]
- 17.Gong F, Yanofsky C. Instruction of translating ribosome by nascent peptide. Science. 2002;297:1864–1867. doi: 10.1126/science.1073997. [DOI] [PubMed] [Google Scholar]
- 18.Onouchi H, et al. Nascent peptide-mediated translation elongation arrest coupled with mRNA degradation in the CGS1 gene of Arabidopsis. Genes Dev. 2005;19:1799–1810. doi: 10.1101/gad.1317105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez-Laslop N, Thum C, Mankin AS. Molecular mechanism of drug-dependent ribosome stalling. Mol. Cell. 2008;30:190–202. doi: 10.1016/j.molcel.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 20.Komar AA, Lesnik T, Reiss C. Synonymous codon substitutions affect ribosome traffic and protein folding during in vitro translation. FEBS Lett. 1999;462:387–391. doi: 10.1016/s0014-5793(99)01566-5. [DOI] [PubMed] [Google Scholar]
- 21.Young JC, Andrews DW. The signal recognition particle receptor alpha subunit assembles co-translationally on the endoplasmic reticulum membrane during an mRNA-encoded translation pause in vitro. EMBO J. 1996;15:172–181. [PMC free article] [PubMed] [Google Scholar]
- 22.Lin L, DeMartino GN, Greene WC. Cotranslational biogenesis of NF-kappaB p50 by the 26S proteasome. Cell. 1998;92:819–828. doi: 10.1016/s0092-8674(00)81409-9. [DOI] [PubMed] [Google Scholar]
- 23.Sato S, Ward CL, Kopito RR. Cotranslational ubiquitination of cystic fibrosis transmembrane conductance regulator in vitro. J. Biol. Chem. 1998;273:7189–7192. doi: 10.1074/jbc.273.13.7189. [DOI] [PubMed] [Google Scholar]
- 24.Zhou M, Fisher EA, Ginsberg HN. Regulated Co-translational ubiquitination of apolipoprotein B100. A new paradigm for proteasomal degradation of a secretory protein. J. Biol. Chem. 1998;273:24649–24653. doi: 10.1074/jbc.273.38.24649. [DOI] [PubMed] [Google Scholar]
- 25.Walter P, Blobel G. Translocation of proteins across the endoplasmic reticulum III. Signal recognition protein (SRP) causes signal sequence-dependent and site-specific arrest of chain elongation that is released by microsomal membranes. J. Cell Biol. 1981;91:557–561. doi: 10.1083/jcb.91.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolin SL, Walter P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 1989;109:2617–2622. doi: 10.1083/jcb.109.6.2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mason N, Ciufo LF, Brown JD. Elongation arrest is a physiologically important function of signal recognition particle. EMBO J. 2000;19:4164–4174. doi: 10.1093/emboj/19.15.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakkaraju AK, Mary C, Scherrer A, Johnson AE, Strub K. SRP keeps polypeptides translocation-competent by slowing translation to match limiting ER-targeting sites. Cell. 2008;133:440–451. doi: 10.1016/j.cell.2008.02.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ito-Harashima S, Kuroha K, Tatematsu T, Inada T. Translation of the poly(A)tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 2007;21:519–524. doi: 10.1101/gad.1490207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Farabaugh PJ. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimchi-Sarfaty C, et al. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 32.Wen JD, et al. Following translation by single ribosomes one codon at a time. Nature. 2008;452:598–603. doi: 10.1038/nature06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Curran JF, Yarus M. Rates of aminoacyl-tRNA selection at 29 sense codons in vivo. J. Mol. Biol. 1989;209:65–77. doi: 10.1016/0022-2836(89)90170-8. [DOI] [PubMed] [Google Scholar]
- 34.Guisez Y, Robbens J, Remaut E, Fiers W. Folding of the MS2 coat protein in Escherichia coli is modulated by translational pauses resulting from mRNA secondary structure and codon usage: a hypothesis. J. Theor. Biol. 1993;162:243–252. doi: 10.1006/jtbi.1993.1085. [DOI] [PubMed] [Google Scholar]
- 35.Schmittgen TD, Danenberg KD, Horikoshi T, Lenz HJ, Danenberg PV. Effect of 5-fluoro- and 5-bromouracil substitution on the translation of human thymidylate synthase mRNA. J. Biol. Chem. 1994;269:16269–16275. [PubMed] [Google Scholar]
- 36.Sorensen MA, Pedersen S. Absolute in vivo translation rates of individual codons in Escherichia coli. The two glutamic acid codons GAA and GAG are translated with a threefold difference in rate. J. Mol. Biol. 1991;222:265–280. doi: 10.1016/0022-2836(91)90211-n. [DOI] [PubMed] [Google Scholar]
- 37.Thanaraj TA, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–1612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Varenne S, Buc J, Lloubes R, Lazdunski C. Translation is a non-uniform process. Effect of tRNA availability on the rate of elongation of nascent polypeptide chains. J. Mol. Biol. 1984;180:549–576. doi: 10.1016/0022-2836(84)90027-5. [DOI] [PubMed] [Google Scholar]
- 39.Chaney WG, Morris AJ. Nonuniform size distribution of nascent peptides. The effect of messenger RNA structure upon the rate of translation. Arch. Biochem. Biophys. 1979;194:283–291. doi: 10.1016/0003-9861(79)90620-9. [DOI] [PubMed] [Google Scholar]
- 40.Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO Journal. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim J, Klein PG, Mullet JE. Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J. Biol. Chem. 1991;266:14931–14938. [PubMed] [Google Scholar]
- 42.Kim JK, Hollingsworth MJ. Localization of in vivo ribosome pause sites. Anal. Biochem. 1992;206:183–188. doi: 10.1016/s0003-2697(05)80031-4. [DOI] [PubMed] [Google Scholar]
- 43.Yonath A. Approaching atomic resolution in crystallography of ribosomes. Annu. Rev. Biophys. Biomol. Struct. 1992;21:77–93. doi: 10.1146/annurev.bb.21.060192.000453. [DOI] [PubMed] [Google Scholar]
- 44.Ruta V, Jiang Y, Lee A, Chen J, MacKinnon R. Functional analysis of an archaebacterial voltage-dependent K+ channel. Nature. 2003;422:180–185. doi: 10.1038/nature01473. [DOI] [PubMed] [Google Scholar]
- 45.Hessa T, White SH, von Heijne G. Membrane insertion of a potassium-channel voltage sensor. Science. 2005;307:1427. doi: 10.1126/science.1109176. [DOI] [PubMed] [Google Scholar]
- 46.Tu L, Wang J, Helm A, Skach WR, Deutsch C. Transmembrane biogenesis of Kv1.3. Biochemistry. 2000;39:824–836. doi: 10.1021/bi991740r. [DOI] [PubMed] [Google Scholar]
- 47.Creighton TE. Proteins. New York: W.H. Freeman and Company; 1993. pp. 171–199. [Google Scholar]
- 48.Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. doi: 10.1016/s0896-6273(00)80143-9. [DOI] [PubMed] [Google Scholar]
- 49.Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 50.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J. Mol. Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 51.Makhoul CH, Trifonov EN. Distribution of rare triplets along mRNA and their relation to protein folding. J. Biomol. Struct. Dyn. 2002;20:413–420. doi: 10.1080/07391102.2002.10506859. [DOI] [PubMed] [Google Scholar]
- 52.Ramachandiran V, Kramer G, Horowitz PM, Hardesty B. Single synonymous codon substitution eliminates pausing during chloramphenicol acetyl transferase synthesis on Escherichia coli ribosomes in vitro. FEBS Lett. 2002;512:209–212. doi: 10.1016/s0014-5793(02)02261-5. [DOI] [PubMed] [Google Scholar]
- 53.Jungbauer LM, Bakke CK, Cavagnero S. Experimental and computational analysis of translation products in apomyoglobin expression. J. Mol. Biol. 2006;357:1121–1143. doi: 10.1016/j.jmb.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 54.Fang P, Spevak CC, Wu C, Sachs MS. A nascent polypeptide domain that can regulate translation elongation. Proc. Natl. Acad. Sci. U. S. A. 2004;101:4059–4064. doi: 10.1073/pnas.0400554101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Merrick WC. Assays for eukaryotic protein synthesis. Methods Enzymol. 1979;60:108–123. doi: 10.1016/s0076-6879(79)60011-3. [DOI] [PubMed] [Google Scholar]
- 56.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brunak S, Engelbrecht J. Protein structure and the sequential structure of mRNA: alpha-helix and beta-sheet signals at the nucleotide level. Proteins. 1996;25:237–252. doi: 10.1002/(SICI)1097-0134(199606)25:2<237::AID-PROT9>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 58.Ryazanov AG, Ovchinnikov LP, Spirin AS. Development of structural organization of protein-synthesizing machinery from prokaryotes to eukaryotes. Biosystems. 1987;20:275–288. doi: 10.1016/0303-2647(87)90035-9. [DOI] [PubMed] [Google Scholar]
- 59.Smith DW, McNamara AL. The transfer RNA content of rabbit reticulocytes: enumeration of the individual species per cell. Biochim. Biophys. Acta. 1972;269:67–77. doi: 10.1016/0005-2787(72)90075-5. [DOI] [PubMed] [Google Scholar]
- 60.Smith DW. Reticulocyte transfer RNA and hemoglobin synthesis. Science. 1975;190:529–535. doi: 10.1126/science.1103288. [DOI] [PubMed] [Google Scholar]
- 61.Mirande M, Cirakoglu B, Waller JP. Seven mammalian aminoacyl-tRNA synthetases associated within the same complex are functionally independent. Eur. J. Biochem. 1983;131:163–170. doi: 10.1111/j.1432-1033.1983.tb07244.x. [DOI] [PubMed] [Google Scholar]
- 62.Dale T, Uhlenbeck OC. Amino acid specificity in translation. Trends Biochem. Sci. 2005;30:659–665. doi: 10.1016/j.tibs.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 63.Olejniczak M, Dale T, Fahlman RP, Uhlenbeck OC. Idiosyncratic tuning of tRNAs to achieve uniform ribosome binding. Nat. Struct. Mol. Biol. 2005;12:788–793. doi: 10.1038/nsmb978. [DOI] [PubMed] [Google Scholar]
- 64.O'Neil KT, DeGrado WF. A thermodynamic scale for the helix-forming tendencies of the commonly occurring amino acids. Science. 1990;250:646–651. doi: 10.1126/science.2237415. [DOI] [PubMed] [Google Scholar]
- 65.Munoz V, Serrano L. Development of the multiple sequence approximation within the AGADIR model of alpha-helix formation: comparison with Zimm-Bragg and Lifson-Roig formalisms. Biopolymers. 1997;41:495–509. doi: 10.1002/(SICI)1097-0282(19970415)41:5<495::AID-BIP2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 66.Sharp KA, Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu. Rev. Biophys. Biophys. Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 67.Lovett PS, Rogers EJ. Ribosome regulation by the nascent peptide. Microbiological Reviews. 1996;60:366–385. doi: 10.1128/mr.60.2.366-385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cortazzo P, et al. Silent mutations affect in vivo protein folding in Escherichia coli. Biochem. Biophys. Res. Commun. 2002;293:537–541. doi: 10.1016/S0006-291X(02)00226-7. [DOI] [PubMed] [Google Scholar]
- 69.Dresios J, Derkatch IL, Liebman SW, Synetos D. Yeast ribosomal protein L24 affects the kinetics of protein synthesis and ribosomal protein L39 improves translational accuracy, while mutants lacking both remain viable. Biochemistry. 2000;39:7236–7244. doi: 10.1021/bi9925266. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. A. Charge-dependent mobility of S4-substituted tape measure. The unsubstituted tape measure, S4(++++), S4(++00), and S4(0000) constructs (lanes 1–4, respectively) were translated as described in the Materials and Methods and Figure 1 for 60 min to give a single, final peptide band that is full-length. The products were run on a Bis-Tris 10% NuPAGE gel. Increasing numbers of arginines cause the peptide to migrate more slowly on the gel. B. Compaction of S4 segments in the tunnel and rate of polyalanine translation. A cysteine accessibility assay previously validated to measure compaction in the tunnel 2,10 was used to measure compaction in S4 constructs. The assay involves modification of a free cysteine with PEG-MAL to produce a gel shift. The fraction of pegylation is quantified (for a given lane on a protein gel) from the ratio of the intensity of the band representing the gel-shifted, higher mass peptide to the sum of the intensities of the bands representing the unmodified parent molecular mass and the gel-shifted higher mass. S4-5R and S4-5K compact equivalently to a molecular tape measure substituted with 5 consecutive alanines (7-Ala5) in a similar tunnel location 10, whereas S4-5Q, S4-5E, and the unsubstituted tape measure are each fully extended.